Abstract
Cyanobacteria are photoautotrophic microorganisms which fix atmospheric carbon dioxide via the Calvin-Benson cycle to produce carbon backbones for primary metabolism. Fixed carbon can also be stored as intracellular glycogen, and in some cyanobacterial species like Synechocystis sp. strain PCC 6803, polyhydroxybutyrate (PHB) accumulates when major nutrients like phosphorus or nitrogen are absent. So far only three enzymes which participate in PHB metabolism have been identified in this organism, namely, PhaA, PhaB, and the heterodimeric PHB synthase PhaEC. In this work, we describe the cyanobacterial PHA surface-coating protein (phasin), which we term PhaP, encoded by ssl2501. Translational fusion of Ssl2501 with enhanced green fluorescent protein (eGFP) showed a clear colocalization to PHB granules. A deletion of ssl2501 reduced the number of PHB granules per cell, whereas the mean PHB granule size increased as expected for a typical phasin. Although deletion of ssl2501 had almost no effect on the amount of PHB, the biosynthetic activity of PHB synthase was negatively affected. Secondary-structure prediction and circular dichroism (CD) spectroscopy of PhaP revealed that the protein consists of two α-helices, both of them associating with PHB granules. Purified PhaP forms oligomeric structures in solution, and both α-helices of PhaP contribute to oligomerization. Together, these results support the idea that Ssl2501 encodes a cyanobacterial phasin, PhaP, which regulates the surface-to-volume ratio of PHB granules.
INTRODUCTION
Cyanobacteria are photosynthetic microorganisms capable of oxygenic photosynthesis. ATP and reduction equivalents derived from photosynthetic electron flow are utilized to fix carbon dioxide and generate 3-phosphoglycerate (1). This metabolite can be utilized for either gluconeogenesis or glycolysis, providing the necessary carbon skeletons for biosynthesis of amino acids and other metabolites required for cell growth (2), when growth conditions are suitable and nutrients are abundant. In fact, carbon flux is greatly affected by the availability of macronutrients like nitrogen (3–5) and phosphorus, which may limit growth (6). Under nutrient-limiting conditions, cyanobacteria undergo a stress adaptation process termed chlorosis (7). This process leads to the degradation of light-harvesting complexes, causing reduced photosynthetic activity and thereby reduced metabolic activity (8). Furthermore, carbon flux is redirected toward glycogen synthesis upon macronutrient starvation (5). In addition, some cyanobacterial strains like Synechocystis sp. strain PCC 6803 (referred to here as Synechocystis) accumulate polyhydroxybutyrate (PHB) as a carbon and redox storage compound (9). PHB is synthesized in three biosynthetic steps, and all three enzymes catalyzing the reactions are known (10, 11). The first step involves a condensation of two acetyl coenzyme A (acetyl-CoA) groups to acetoacetyl-CoA by PhaA (slr1993). In the second step, PhaB (slr1994) reduces acetoacetyl-CoA to hydroxybutyryl-CoA, utilizing NADPH as the electron donor (10). In the last step of biosynthesis, hydroxybutyryl-CoA is polymerized to polyhydroxybutyrate by a class III PHB synthase, which is a heterodimer of PhaE (slr1829) and PhaC (slr1830) that builds the catalytically active enzyme in cyanobacteria (11). The PhaEC heterodimer (12) (or tetramer [13]) synthesizes PHB, thereby building up PHB granules, to which it is attached (4). This marks the boundaries of our understanding of PHB accumulation in cyanobacteria on a molecular level. However, the PHB granule surface is more complex and is coated by various different proteins, as observed with Ralstonia eutropha H16 (currently named Cupriavidus necator; referred to here as R. eutropha) (14). Some of these proteins are transcriptional regulators (PhaR) which modulate the transcriptional response upon biosynthesis of PHB (15); others are responsible for degradation and utilization of the intracellular stored PHB and are termed PHB depolymerases (PhaZ) (16). In addition to these functionally defined PHB-associated proteins, R. eutropha possesses at least 8 additional proteins, termed phasins, covering a large portion of the PHB granules (17, 18). These proteins have a variety of functions. Originally, phasins were identified as regulators of PHB granule size and number within a cell (19). More recently, specific phasins were identified as mediating the attachment of PHB granules to the nucleoid, as in the case of PhaM (20) in R. eutropha or PhaF from Pseudomonas putida KT2442 (21), thereby providing a mechanism for equal distribution of PHA granules to daughter cells. A phasin from Azotobacter sp. strain FA-8 (PhaPAz) was described as promoting stress resistance in Escherichia coli (22), which is most likely associated with its chaperon-like activity (23). Even though many phasin proteins were identified, so far only a few have been studied biochemically in more detail (24). Genes encoding proteins homologous to phasins from R. eutropha or P. putida are absent in the genome of Synechocystis; however, Hein et al. (11) isolated PHB granules from Synechocystis and were able to detect many proteins of different molecular masses by SDS-PAGE. Of these, only one protein could be identified, the product of the open reading frame (ORF) ssl2501. However, subsequent studies found this protein repeatedly associated with thylakoid membranes (25, 26); therefore, the subcellular localization and consequently the biological function of this protein remain unclear. Homologs of Ssl2501 are present in several cyanobacterial strains, and the homologous protein in Arthrospira platensis NIES-39 (NIES39_Q00050) is predicted to have a so-called phasin 2 motif, based on the KEGG sequence similarity database (SSDB) (27). This prompted us to clarify whether the Ssl2501 protein may be a functional phasin in Synechocystis.
MATERIALS AND METHODS
Cultivation conditions.
Standard cloning procedures were done in E. coli XL1 Blue (Stratagene) grown in LB medium at 37°C. Cyanobacterial strains were grown in BG11 medium (28) as described previously, with 40 to 50 μmol photons m−2 s−1 at 27°C and supplemented with 5 mM NaHCO3 (28, 29). Growth was monitored by measuring the optical density at 750 nm (OD750). Nitrogen starvation was induced by washing exponentially growing cells once with BG11 medium lacking sodium nitrate (BG110) and transferring them into BG110 at an OD750 of 0.4 as described previously (29). Antibiotics were added to the growth medium when required.
Strains, oligonucleotides, and plasmids used in this study.
The strains used in this study are listed in Table 1. Oligonucleotides used for molecular biology techniques are listed in Table 2. Plasmids used in the study are listed in Table 3.
TABLE 1.
Strains used in the study
| Strain | Relevant feature | Source or reference |
|---|---|---|
| E. coli XL1-Blue | Cloning strain | Stratagene |
| E. coli RP4 | Conjugation strain | 34 |
| E. coli BL21 | Protein overexpression strain | 57 |
| Synechocystis sp. strain PCC 6803 | Wild-type strain | Pasteur culture collection |
| Δssl2501 mutant | Chromosomal deletion of ssl2501 | This study |
| ssl2501 | Δssl2501 mutant complemented with pVZ322-2501 | This study |
| ssl2501gfp | Δssl2501 mutant complemented with pVZ322-2501gfp | This study |
| wt ssl2501Venus | Wild-type strain transformed with pVZ322-ssl2501Venus | This study |
| wt H1 Venus | Wild-type strain transformed with pVZ322-helix1Venus | This study |
| Δssl2501 H1 Venus | Δssl2501 mutant complemented with pVZ322-helix1Venus | This study |
| wt H2 Venus | Wild-type strain transformed with pVZ322-helix2Venus | This study |
| Δssl2501 H2 Venus | Δssl2501 mutant complemented with pVZ322-helix2Venus | This study |
TABLE 2.
Oligonucleotides used for molecular biology techniques

TABLE 3.
Plasmids used in the study
| Plasmid | Relevant feature | Source or reference |
|---|---|---|
| pJet1.2 | Cloning vector | Thermo Fisher |
| pJetΔ2501 | Suicide vector for ssl2501 deletion derived from pJet1.2 | This study |
| pVZ322 | Broad-host-range vector | 32 |
| pVZ322-2501gfp | eGFP fused to the C terminus of Ssl2501 | This study |
| pVZ322-2501 | ssl2501 with native promoter | This study |
| pVZ322-2501Venus | Venus fused to the C terminus of Ssl2501 connected by a 10-amino-acid linker sequence | This study |
| pVZ322-helix1Venus | Venus fused to the C terminus of Ssl2501 helix 1, connected by a 10-amino-acid linker sequence | This study |
| pVZ322-helix2Venus | Venus fused to the C terminus of Ssl2501 helix 2, connected by a 10-amino-acid linker sequence | This study |
| pET15bssl2501 | Vector expressing ssl2501 upon IPTG addition | This study |
| pET15bHelix2 | Vector expressing helix 2 of ssl2501 upon IPTG addition | This study |
Construction of the ssl2501 mutant and other strains.
PCR fragments were generated using a high-fidelity polymerase (Q5; NEB) to amplify genes from genomic DNA or plasmids. Primers used to generate the constructs described in this study are listed in Table 3. A ssl2501 mutant was constructed by amplifying up- and downstream genomic regions by PCR from genomic DNA with primers Ssl2501upfor, Ssl2501uprev, Ssl2501dofor, and Ssl2501dorev. A kanamycin resistance cassette was amplified from pVZ322 using primers Kana-for and Kana-rev. The flanking regions were fused at the 5′ and 3′ ends of a kanamycin resistance cassette by long flanking homology PCR (30) and cloned in pJet1.2 (Fermentas), resulting in pJetΔ2501. Synechocystis was transformed with plasmid pJetΔ2501 using its natural competence. The ORF ssl2501 was amplified by PCR from genomic DNA with primers ssl2501for and ssl2501rev and fused to the coding sequence of enhanced green fluorescent protein (eGFP) derived from plasmid pCESL19 (31) by PCR with primers gfpfor and gfprev. The PCR fragment was inserted in pVZ322 (32) linearized with SalI and PstI, producing plasmid pVZ322-2501gfp. Plasmid pVZ322-2501 was constructed the same way. ORF ssl2502 (CyanoBase) was amplified with ssl2501for and ssl2502rev and fused to eGFP in this plasmid, and it expresses native ssl2501, which is upstream of ssl2502. Venus was amplified with primers Venfor and Venrev and translationally fused to the C terminus of ssl2501 (amplified with promotorfor and revssl2501), or the coding sequences of the first (amplified with promotorfor hel1rev) and second (gBlocks [IDT]) α-helices were assembled (33) in pVZ322 at the XbaI site. A Strep-tag was added to the N terminus of ssl2501 (PCR amplified with Ssl2501for and Ssl2501rev) by inserting it in pASK-IBA5+ at the EheI site using Gibson assembly. The coding region of ssl2501in pASKIBA5+ (including the streptavidin tag) was amplified by PCR (primers pET15bfor and pET15brev) and inserted in pET15b at the unique NcoI site using Gibson assembly. Plasmids were propagated in E. coli XL1-Blue and isolated using the Peqlab miniprep kit. Sequence integrity of the plasmids was verified by sequencing. Plasmids able to replicate autonomously in Synechocystis were transferred by triparental mating as described previously (34).
Microscopy and staining procedures.
Microscopy was performed with a Leica DM5500B fluorescence microscope using the 100×, 1.3 numerical aperture oil objective lens. Fluorescence microscopy was performed with three filter cubes. In order to detect eGFP, a BP470 40-nm excitation filter and a BP525 50-nm emission filter were used; this is referred to as the GFP channel. Venus fluorescence was detected using an ET500/20x excitation filter and an ET535/30m emission filter; this is referred to as the yellow fluorescent protein (YFP) channel. To detect Nile red fluorescence, a filter cube with BP535 50-nm excitation filters and a BP610 75-nm emission filter was used; this is referred to as the Cy3 channel. Image acquisition was done with a Leica DFC360FX black-and-white camera. Bright-field images were exposed for 5 ms and 80 to 200 ms in the fluorescence channels. Images were routinely taken as Z-stacks with 0.25 μm distance between images. Z-stacks were used to perform three-dimensional (3D) deconvolution using the built-in function of the Leica ASF software. Images were recolored by the Leica ASF software based on the filter used, and intensity levels were adjusted using AdobePhotoshopCS5. To visualize PHB granules under nitrogen starvation, 12 μl of cell culture was mixed with 6 μl Nile red solution (1 μg/ml in ethanol) and subsequently analyzed under the microscope.
Purification of recombinant proteins from E. coli BL21.
For purification of recombinant proteins, the corresponding plasmids were transformed in E. coli BL21 and plated on LB agar with appropriate antibiotics. Single clones were picked to start an overnight culture, which was then used to inoculate the production culture. The production culture was grown at 37°C to an OD595 of 0.8, and recombinant-protein production was induced by addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultivation was continued at 20°C overnight. Then, the cells were harvested at 4,000 × g for 10 min, suspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2, 2 mM EDTA, 2 mM dl-dithiothreitol, 1 mM benzamidine, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), and lysed using a Branson Sonifier. The cell lysate was cleared by centrifugation at 50,000 × g for 30 min, and the supernatant was loaded on a Strep-Tactin column using a peristaltic pump. The column was washed with five column volumes of washing buffer (100 mM Tris-HCl [pH 7.8], 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 2 mM dl-dithiothreitol, 1 mM benzamidine, and 0.2 mM PMSF), and proteins were eluted through the addition of desthiobiotin to a final concentration of 2.5 mM to the washing buffer. Desthiobiotin was removed by dialyzing the protein solution against 500 ml storage buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 5 mM MgCl2, 0,5 mM EDTA, 50% [vol/vol] glycerol, 2 mM dl-dithiothreitol, 1 mM benzamidine).
SDS-PAGE and Western blotting.
For electrophoretic separation of proteins, SDS-PAGE was performed with 12% polyacrylamide resolving gels as described previously (35, 36). Twenty micrograms of total protein was used for SDS-PAGE with nitrogen-starved cells, and 40 μg was used for exponentially growing cells. For Western blot analysis, proteins were blotted on a methanol-activated polyvinylidene difluoride (PVDF) membrane as described previously (37). Membranes were blocked with 10% (wt/vol) milk powder in TBS buffer (20 mM Tris-HCl [pH 7.4], 0.5% [wt/vol] NaCl) for 30 min. Afterwards the membrane was transferred to 1% (wt/vol) milk powder in TBS buffer containing 0.25 μg/ml primary antibody (anti-GFP polyclonal rabbit antibody; Santa Cruz Biotechnology, Inc.) and incubated in this solution overnight at 4°C. Nonbound primary antibody was removed by washing and secondary antibody (anti-rabbit polyclonal goat antibody–horseradish peroxidase conjugate; Sigma-Aldrich), diluted 1:10,000 in 1% (wt/vol) milk powder in TBS, was applied. Immunoreactive bands on the membrane were visualized using the LumiLight detection system (Roche Diagnostics) and the Gel Logic 1500 imaging system (Kodak) with the associated software.
Size exclusion chromatography and cross-linking of proteins.
Prior to size exclusion chromatography or cross-linking, proteins were dialyzed in 100 mM potassium phosphate buffer (pH 7.8) overnight. Ten microliters of dialyzed protein was injected onto a Superdex 200 PC 3.2/30 equilibrated with running buffer (50 mM potassium phosphate buffer [pH 7.8]). Protein elution was detected at 280 nm. Proteins dialyzed in phosphate buffer were cross-linked by addition of 0.1% (wt/vol) glutaraldehyde for 10 min at room temperature. The cross-linking reaction was stopped by addition of Tris-HCl (pH 7.8) to a final concentration of 100 mM. Ten micrograms of protein was used to separate cross-linked proteins on Tricine-PAGE (36).
PHB synthase activity assay.
PHB synthase assays were performed as previously described by Valentin et al. (38), with some modifications. Briefly, 30 ml cells was harvested by centrifugation at 4,000 × g and suspended in lysis buffer (25 mM Tris-HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2, 0,5 mM EDTA, and 1 mM benzamidine). Cells were lysed using FastPrep-24 (MP Biomedical) using 0.1-mm glass beads, and cell debris was removed by centrifugation for 5 s up to 10,000 × g. The resulting cell lysate was separated into a soluble and insoluble fraction by centrifugation at 25,000 × g for 30 min at 4°C. The insoluble material was suspended in lysis buffer, and protein concentrations in the fractions were determined as described by Bradford (39). PHB-biosynthetic activity was monitored by recording the change in absorbance at 412 nm for at last 30 min in the reaction buffer (25 mM Tris-HCl [pH 7.4], 1 mM DTNB [5,5′-dithiobis(2-nitrobenzoic acid)], 20 mM MgCl2, and 100 μM hydroxybutyryl-CoA [Sigma-Aldrich]) after addition of 5 μg protein of the suspended insoluble fraction. The reaction temperature was held constant at 30°C.
PHB quantification.
Intracellular PHB content was measured in principle as described previously (10). Cells were harvested at the dedicated time points by centrifugation (10 min, 4,000 × g, 25°C), washed once with distilled water, and dried for 3 h at 60°C. Dried pellets were boiled for 1 h in 1 ml concentrated H2SO4 diluted with 1 ml 0.014 M H2SO4. Cell debris was removed by centrifugation (10 min at 10,000 × g), and the supernatant was diluted 10-fold in 0.014 M H2SO4. Processed samples were analyzed by high-performance liquid chromatography (HPLC) using a Nucleosil 100 C18 column (125 by 3 mm) and 20 mM phosphate buffer (pH 2.5) as the liquid phase. Crotonic acid was detected at 210 nm, and commercially available PHB processed in parallel was used as a standard.
RESULTS AND DISCUSSION
Ssl2501-eGFP localizes to PHB granules.
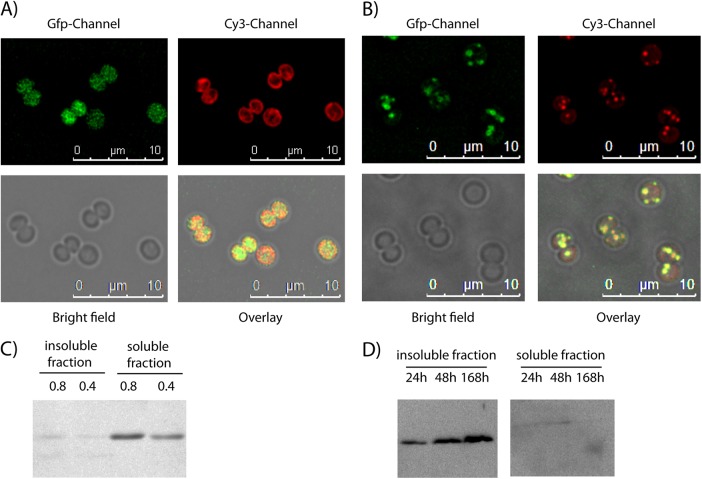
As previous studies identified the Ssl2501 protein at thylakoid membranes and PHB granules, Ssl2501 was translationally fused to eGFP and the recombinant gene was cloned into the Synechocystis shuttle vector pVZ322 in order to study the intracellular localization of this protein in more detail. The resulting plasmid, pVZ322-2501gfp, thus encodes a translational fusion of Ssl2501 with eGFP at its C terminus, which is transcriptionally controlled by the native ssl2501 promoter. By using triparental mating (34), the plasmid was transferred in Synechocystis to study the intracellular localization of Ssl2501. To determine a potential localization of Ssl2501-GFP to thylakoid membranes, cells were grown under photoautotrophic growth conditions in BG11 medium to an OD750 of 0.8 and were then analyzed by fluorescence microscopy (Fig. 1A). After deconvolution of Z-stacked images, no apparent colocalization between thylakoid membranes and Ssl2501-eGFP could be observed. The eGFP signal was distributed in the cytoplasmic space, whereas the fluorescence of thylakoid membranes localized near the cell periphery. To study a possible colocalization with PHB granules, exponentially growing cells were transferred into BG11 medium lacking a nitrogen source (BG110). The nitrogen-free medium induces chlorosis in Synechocystis, leading to the degradation of the photosynthetic apparatus and concomitant accumulation of PHB (9). Cells were nitrogen starved for 5 days before a portion was stained with Nile red to visualize the PHB granules by fluorescence microscopy (Fig. 1B). Under these conditions, the GFP channel showed that Ssl2501-eGFP aggregated in clusters. Pictures from the Cy3 channel visualize Nile red-stained PHB granules. Superimposition of images taken in the GFP channel, the Cy3 channel, and the bright field (to localize the entire cell shape) shows colocalization by yellow color in the overlay. As shown in Fig. 1B, Nile red-stained PHB granules colocalize with Ssl2501-GFP. These data supported the idea that Ssl2501 is a PHB granule-associated protein, as is the case for phasins. To determine whether Ssl2501-GFP remains granule associated in cell lysates, immunoblot analysis was performed with eGFP-specific antibodies. Extracts were prepared from exponentially growing and from nitrogen-starved cells, and the cell lysate was fractionated into soluble and insoluble fractions as described in Materials and Methods. Since PHB synthase activity is located in the insoluble fraction (29), the putative phasin Ssl2501-eGFP should localize in this fraction under conditions of PHB granule formation (nitrogen starvation). In exponentially growing cells devoid of PHB granules, Ssl2501-GFP was primarily localized in the soluble fraction (Fig. 1C), and only traces could be detected in the insoluble fraction, whereas under nitrogen-limiting conditions, the putative phasin was present only in the insoluble fraction, and only trace amounts of Ssl2501-GFP could be detected in the soluble fraction (Fig. 1D). This supports the microscopic data showing that during exponential growth, Ssl2501 is soluble in the cytoplasm, but once PHB permissive conditions appear, the protein is sequestered to PHB granules, rendering it insoluble.
FIG 1.
(A) Localization of Ssl2501-eGFP (GFP channel) in Synechocystis during exponential growth at an OD750 of 0.8 and the autofluorescence of thylakoid membranes (Cy3 channel). (B) Synechocystis cells that had been nitrogen starved for 5 days and expressed Ssl2501-eGFP (GFP channel) were stained with Nile red (Cy3 channel) to visualize PHB. The yellow color in the overlay indicates a colocalization between Ssl2501-GFP and the Nile red-stained PHB granule. (C) Western blot detection of Ssl2501-eGFP during exponential growth at OD750 of 0.8 and 0.4 using an anti-GFP primary antibody. (D) Western blot detection of Ssl2501-eGFP in nitrogen-starved cells using an anti-GFP primary antibody. Cell fracturing and separation in soluble and insoluble fractions were performed as described in Materials and Methods for the PHB synthase assay.
Deletion of ssl2501 alters PHB granule number and diameter.
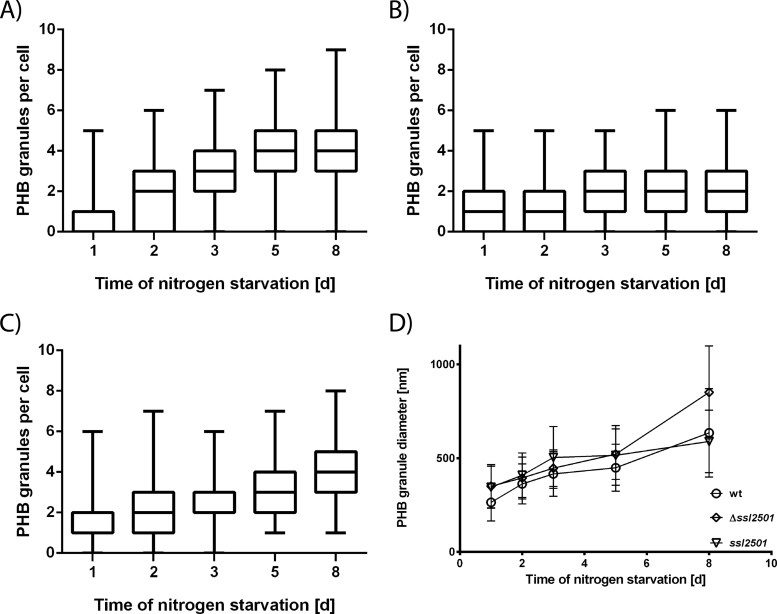
As one function of phasins is thought to be the regulation of PHB granule surface-to-volume ratio and the number of PHB granules within the cell (40), a genomic deletion of ssl2501 could affect the size and amount of PHB granules within a cell. To assess this question, we constructed an ssl2501 mutant in which the ssl2501 open reading frame is replaced with a kanamycin resistance cassette. The complete segregation of the mutant was confirmed by PCR, as shown in Fig. S1 in the supplemental material. Wild-type Synechocystis and the ssl2501 mutant were grown to an OD750 of 0.6 and transferred to nitrogen-depleted medium to induce PHB accumulation. In the course of nitrogen starvation, PHB granules were stained with Nile red, and the number of granules per cell was determined using fluorescence microscopy. Figure 2A and B show the distribution of PHB granules within a cell population at given time points during nitrogen starvation in the wild type and the ssl2501 mutant, respectively. After 24 h of nitrogen starvation, up to five PHB granules could be seen in a wild-type cell, whereas 75% of the cells had no more than two granules. With progression of nitrogen starvation, the number of PHB granules per cell increased and with it the number of cells which had more than two PHB granules. The median increased from one PHB granule per cell at day 1 of nitrogen starvation to four granules after 5 days of nitrogen starvation. The mutant behaved similarly in the beginning of nitrogen starvation, as it induced PHB granule formation, and the distribution of granules in the cell population was similar to that in the wild type. However, prolonged nitrogen starvation revealed a distinct mutant phenotype: The majority of cells accumulated only a few PHB granules. The median increased from one to two PHB granules per cell but not further, in contrast to the wild type. In addition, the heterogeneity in PHB granule numbers was reduced in the mutant, especially at the later time points of nitrogen starvation. Cells lacking ssl2501 had a maximum of five PHB granules, whereas wild-type cells could have up to nine PHB granules. Representative images of the mutant and wild-type strain are shown in Fig. S2 in the supplemental material. This supported the idea that Ssl2501 is a phasin and is involved in PHB granule formation. To exclude the possibility that the observed phenotype is caused by a secondary effect and not the mutation introduced in ssl2501, the mutant was complemented with plasmid pVZ322-2501, which expresses native ssl2501 controlled by the native promoter. The complemented strain was transferred in nitrogen-depleted BG110 and PHB granules were counted during nitrogen starvation (Fig. 2C). The PHB granule distribution in the complemented strain resembled the wild-type situation. The majority of cells had up to two PHB granules after 1 day of nitrogen starvation, and the number of PHB granules within a cell gradually increased throughout nitrogen starvation. This is also evident from the median, which increased gradually from one to four PHB granules per cell, as in the wild type.
FIG 2.
(A to C) Number of PHB granules per cell during nitrogen starvation in the wild type (A), the ssl2501 mutant (B), and the ssl2501 mutant complemented with plasmid pVZ322-2501 (C). Measurements represent the distribution of PHB granules in at least 150 cells at a given point. (D) Mean PHB granule diameter during nitrogen starvation in the wild type (open circles), the ssl2501 mutant (diamonds), and the ssl2501 mutant complemented with plasmid pVZ322-2501(inverted triangles). Difference in PHB granule diameters is statistically significant from day 6 on (P < 0.0001). Each data point represents the mean of at least 170 individual measurements of the PHB granule diameter.
Another possible function of phasins is to regulate the surface-to-volume ratio of a PHB granule. Hence, the mutant might have bigger PHB granules than the wild type. This hypothesis was tested by measuring PHB granule diameter at given time points using fluorescence microscopy (Fig. 2D). Since PHB had to be stained with Nile red, the measurement relies on fluorescence microscopy (with its own limitations) and a single measurement is only a rough estimation of one PHB granule diameter. Nevertheless, this analysis results in a quite accurate determination of average PHB granule diameter due to the relatively large size of Synechocystis cells and acquisition of Z-stack images with in average 25 layers. From each single cell, the different layers of the Z-stack were analyzed to find the largest diameter of an individual granule. In this way, approximately 200 PHB granules were measured (for statistical details, see Table S1 and Fig. S3 in the supplemental material). Similar to other phasin deletion mutants (19, 40), PHB granules in the ssl2501 mutant were in fact apparently larger in diameter than those in the wild type, and the difference in granule size increased with prolonged nitrogen starvation. The mean PHB granule diameter of the wild type after 8 days of nitrogen starvation was estimated to be 640 nm (standard deviation [SD], 240 nm), whereas the mean PHB granule diameter of the mutant was 850 nm (SD, 250 nm). It should be noted that the large standard deviation reflects the natural variation of PHB granule size. The difference in granule size was statistically analyzed by a two-way unpaired t test, and the difference in size was statistically significant from day 6 on, with a P value smaller than 0.0001. This disparity disappeared upon complementation of the mutant strain with pVZ322-2501. The mean PHB granule diameter of the complemented strain was very similar to that of the wild type, and the mean PHB granule diameter after 8 days of nitrogen starvation was estimated to be 620 nm (SD, 190 nm), resembling the value obtained for the wild type. This further supports the hypothesis that ssl2501 encodes a cyanobacterial phasin and that its product is a regulatory protein that modulates PHB granules in vivo. As the observed phenotypes of the ssl2501 deletion and the in vivo colocalization of Ssl2501 with PHB fulfill the criteria for classical phasins (19, 40), the protein is referred to as PhaP. However, it should be noted that in most PHB-producing bacteria, typically more than one type of phasin protein is present (41). It is, therefore, possible that in Synechocystis, further phasin-like proteins might be present, which could influence PHB biogenesis.
PHB synthase activity but not PHB quantity is affected by ssl2501 deletion.
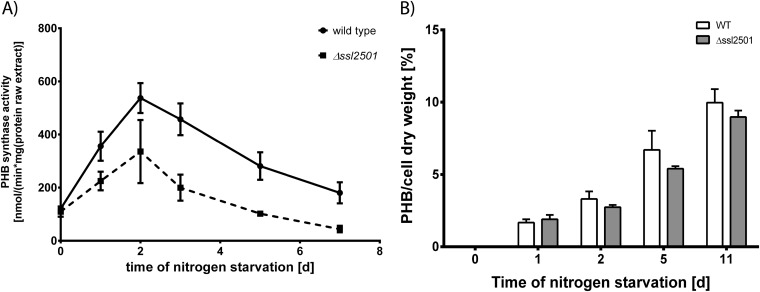
A recent study showed that the phasin PhaM from R. eutropha drastically reduces the lag time of PHB synthase in vitro (42). Therefore, deletion of the phasin ssl2501 might change the biosynthetic activity or amount of PHB that accumulates in cells. Thus, both parameters were determined during nitrogen starvation, as shown in Fig. 3. PHB-biosynthetic assays were performed as described previously (29, 38), by lysing the cells and separating the raw cell extract into soluble and insoluble fractions, followed by measuring the biosynthetic activity in the insoluble fraction, where PHB granules accumulate due to their high density. PHB synthase activity increased during nitrogen starvation, with peak activity at the second day of nitrogen starvation, and then gradually decreased with prolonged nitrogen starvation, as seen in Fig. 3A. This tendency toward an activity peak at day 2 of nitrogen starvation was also seen in the ssl2501 mutant; however, the absolute activity of PHB synthase was lower throughout nitrogen starvation than in the wild type. A typical reaction of a PHB synthase assay is shown in Fig. S4 in the supplemental material. To examine whether the reduced enzyme activity in the ssl2501 mutant was due to decreased levels of PHB synthase, we compared the levels of one subunit of PHB synthase, PhaE, in wild-type and mutant cells by Western blotting (see Fig. S5 in the supplemental material). The levels of PhaE protein in both strains were similar, and no major differences could be observed. Therefore, the effect of decreased PHB synthase activity is not caused by altered enzyme levels but by the absence of PhaP on the PHB granule surface, which might act as a regulator of PHB synthase activity in vivo. The phasin GA24 of Chromatium vinosum was shown to alter the average polymer length synthesized by PHB synthase in in vitro assays, concluding that phasins alter the processivity of PHB synthase (43). The influence of phasins on PHB properties was also observed in the heterologous host E. coli (44). This work demonstrated that phasins regulate the PHB synthase activity in vitro, but the outcome of regulation depends on the individual PHB synthase. While PhaC proteins from Ralstonia eutropha and Delftia acidovorans were inhibited by the presence of phasins, PhaC from Aeromonas caviae was activated by the same phasins (44). The impact of phasins on PHB synthase activity highlights a possible biological function of phasin proteins: protecting the active conformation of PHB synthase and maybe other proteins, according to the recently reported chaperon activity of phasins (23). This function might be important, as PHB is synthesized during periods of imbalanced metabolism or stress, when protein stability is of great importance as protein turnover is reduced. The reduced activity of PHB synthase in the Synechocystis ssl2501 mutant had only a minor influence on the total amount of intracellular PHB, as seen in Fig. 3B. PHB accumulated gradually during nitrogen starvation in both the wild type and the ssl2501 mutant, with only minor differences in PHB quantity being detectable. Since PHB synthase activity in the PhaP mutant was significantly lower than that in the wild type, this result was unexpected. We conclude that in Synechocystis, the quantity of produced PHB is limited not by the activity of the PHB synthase but presumably by the supply of its substrate 3-hydroxybutyrate, which is provided by primary metabolism. This result sets a starting point for further metabolic engineering approaches to divert more carbon from primary metabolism toward this pathway for increased synthesis of PHB, 3-hydroxybutyrate, or 1-butanol as a precursor for fine chemicals (45–47).
FIG 3.
(A) PHB synthase activity of the wild type and mutant strain during nitrogen starvation; (B) intracellular accumulation of PHB in both strains. Measurements are means for three biological replicates. The PHB synthase activity in the ssl2501 deletion strain is lower than in the wild type; nevertheless, the difference of accumulated PHB in both strains is minor and slightly lower in the ssl2501mutant.
Secondary-structure prediction of PhaP.
Next, we tried complementation of the ssl2501 mutant with PhaP-GFP. After induction of PHB synthesis by nitrogen starvation, PhaP-GFP associated with PHB granules and localized to the insoluble fraction. However, PhaP-GFP was not able to complement the mutant phenotype, namely, reduce granule diameter and increase PHB granule number per cell, as shown in Fig. S6 and S7 in the supplemental material. This implied that the large GFP might present a steric hindrance that affects the biological function of PhaP. To test this hypothesis, Ssl2501 was linked to Venus (a YFP variant with improved photochemical properties) (48) by a 10-amino-acid Gly-Ser flexible linker and inserted in pVZ322 yielding pVZ322-2501Venus. After transformation into the ssl2501 mutant, association of PhaP with PHB granules as well as PHB granule size was determined microscopically (see Fig. S8 in the supplemental material). Importantly, the mutant phenotype could now be complemented, supporting the suggestion that the bulky GFP sterically impaired PhaP function, which can be overcome by a flexible linker. Whole-genome yeast two-hybrid assays suggested that PhaP interacts with itself, suggesting oligomerization (49). Oligomerization of other phasins has been described previously (24, 50–52), and these proteins can form dimers (PhaP5 from R. eutropha), trimers (PhaP1 from R. eutropha), tetramers (PhaPAz from Azotobacter sp. strain FA8 and PhaF from Pseudomonas putida KT2440), or dodecamers (PhaM from R. eutropha). Even though the purpose of oligomerization is not understood, it seems to be a conserved feature among phasin proteins and might stabilize the interaction between the proteins and the PHB surface. In order to identify possible structural features responsible for PHB association and self oligomerization, the secondary structure of PhaP was predicted using the PSIPRED server (53, 54). According to this prediction, the protein folds into two α-helices (see Fig. S9 in the supplemental material) and contains no coiled-coil regions as seen in other phasins (24, 51). The first helix starts at Thr3 and ends at Glu26, and the second α-helix starts at Asp39 and is predicted to end at Gln83. Both helices are connected by an unstructured linker. In order to identify possible amphipathic regions responsible for binding to the PHB granule surface, a helix wheel projection was applied to both α-helices. Only the first α-helix is predicted to have amphipathic character (see Fig. S10 in the supplemental material), and six amino acids (Phe5, Phe6, Tyr9, Leu13, Trp16, and Phe20) make up the highly hydrophobic interface of the helix. The second α-helix does not have a distinct hydrophobic region but possesses a highly charged interface, which might be involved in oligomerization or protein-protein interactions (see Fig. S10 in the supplemental material).
Helix 1 and helix 2 of PhaP localize to PHB granules in vivo.
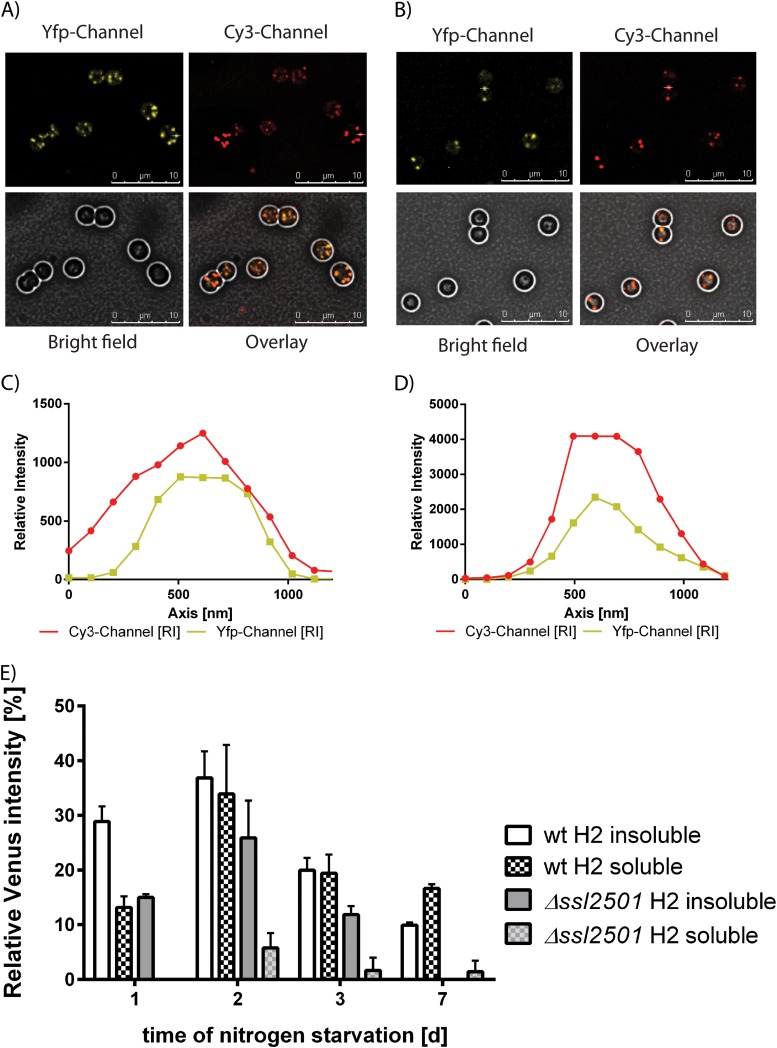
To test whether α-helix 1 or 2 of PhaP is able to localize to PHB granules, both helices were translationally fused to Venus with a 10-amino-acid flexible linker, and the plasmids were transformed into the ssl2501 mutant and wild-type backgrounds. PHB synthesis was induced by nitrogen starvation, and after 3 days, PHB granules in the cells were analyzed by fluorescence microscopy (Fig. 4A and B). The first α-helix is able to bind to PHB granules independently of the genetic background, as indicated by the orange color in the overlay of the Cy3 channel (Nile red) and the YFP channel (Venus). This is also shown in Fig. 4C and D, where an intensity profile of the Cy3 channel and YFP channel of a single PHB granule is plotted. The peak intensities of both channels overlap at the same position, meaning that the first α-helix of PhaP is able to localize to the PHB granule. However, not every PHB granule stained by Nile red has a corresponding signal in the YFP channel. Furthermore, the signal in the YFP channel appears blurred, suggesting a lower affinity of helix 1 for PHB than for the full-length phasin. To independently confirm this observation, extracts were prepared from nitrogen-starved PHB-producing wild-type cells expressing full-length PhaP-Venus and from wild-type and ssl2501 mutant cells carrying helix 1-Venus. The presence of Venus in soluble and insoluble fractions was determined by Western blotting. Band intensities were determined densitometrically and normalized to the Venus intensity in the insoluble fraction of the wild-type strain expressing PhaP-Venus after 1 day of nitrogen starvation (Fig. 4E). Surprisingly, we were not able to detect helix 1-Venus in the insoluble fraction (independent of the genotype) as we would have expected based on microscopic images. Helix 1-Venus was predominantly found in the soluble fraction with decreasing signal intensities as nitrogen starvation proceeded, suggesting that the affinity of the first α-helix for PHB is very low. The same types of experiments were then performed with the wild type and the ssl2501 mutant expressing helix 2-Venus (Fig. 5A and B). The second α-helix of PhaP is able to bind to PHB granules, as can be seen by the orange color of the PHB granules in the overlay image. The association of the helix to PHB is independent of the genetic background, as confirmed by intensity plots of the Cy3 and YFP channel (Fig. 5C and D). The intensities simultaneously rise and fall, thereby confirming PHB granule localization of helix 2. As we have seen with helix 1, localization in vivo does not necessarily correlate with localization in vitro. Hence the localization of helix 2-Venus was tested by Western blotting as described above. The α-helix was found to localize in the soluble and insoluble fraction in the wild type, whereas it localized primarily in the insoluble fraction in the ssl2501 mutant. This suggests that helix 2 competes with native PhaP for occupation of the PHB surface. PHB association of helix 2 seems stable, as it resists cell lysis. Taken together, these data indicate that helix 2 has a higher affinity for PHB than helix 1. Furthermore, the amount of helix 2-Venus in the wild-type background seemed to be higher than in the ssl2501 mutant, suggesting that the presence of full-length PhaP protects helix 2 from degradation, in agreement with the suggested chaperon function of phasins (see above). Taken together, these observations demonstrate that the information to associate with PHB granules is present in both the first and second α-helix of PhaP, as both peptides associate with PHB granules in vivo, but helix 2 seems to be the primary anchor for PHB association.
FIG 4.
(A and B) Localization of PhaP helix 1-Venus in the wild type (A) and ssl2501 mutant (B). (C and D) Signal intensities in the YFP and Cy3 channels of PhaP helix 1-Venus in the wild type (C) and ssl2501 mutant (D). PhaP helix 1-Venus localization was quantified by Western blotting against GFP in the wild type and ssl2501 mutant (E).
FIG 5.
(A and B) Localization of PhaP helix 2-Venus in the wild type (A) and ssl2501 mutant (B). (C and D) Signal intensities in the YFP and Cy3 channels of PhaP helix 2-Venus in the wild type (C) and ssl2501 mutant (D). PhaP helix 2-Venus localization was quantified by Western blotting against GFP in the wild type and ssl2501 mutant (E).
Oligomerization of PhaP and helix 2 of PhaP.
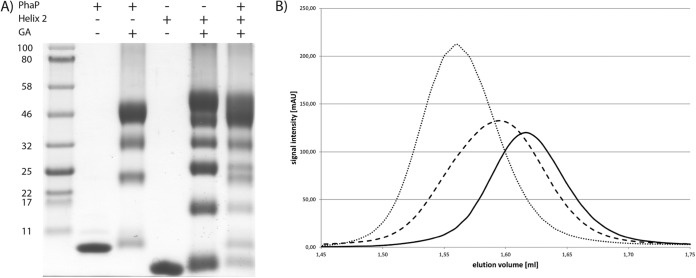
The localization experiments disproved the assumption that the first α-helix would be responsible for PHB binding and the second α-helix for oligomerization. Therefore, we next addressed the question of PhaP oligomerization by in vitro studies using recombinant proteins with an N-terminal Strep-tag II for purification. The full-length phasin and helix 2 of PhaP could be successfully purified to electrophoretic homogeneity, whereas helix 1 seemed to be unstable in E. coli. To confirm the structural prediction of PhaP, CD spectra of PhaP and helix 2 were taken. As shown in Fig. S11 in the supplemental material, the spectra of both proteins revealed characteristic minima at 208 and 222 nm, typical of α-helices. To test whether PhaP is able to interact with itself, as has been proposed (49), the protein was cross-linked using glutaraldehyde and analyzed using Tricine-SDS-PAGE (Fig. 6A). Four distinct bands were visible at the apparent molecular weights of a monomer, a dimer, a trimer and a tetramer, having calculated masses of 12 kDa, 24 kDa, 34 kDa, and 48 kDa, respectively. This result implies that PhaP forms a tetramer in solution. The same experiment was performed with helix 2 with a monomer size of 8.7 kDa. Surprisingly, six distinct bands with all intermediate oligomeric states were visible, suggesting the formation of a hexamer. As the in silico results suggested that helix 2 mediates PhaP oligomerization, we mixed equal amounts of both proteins and cross-linked them with glutaraldehyde. Following electrophoretic separation, mixed hetero-oligomers could be detected. Monomers and dimers of both proteins could be clearly identified as well as the trimeric form of helix 2; in addition, a third band with an apparent molecular mass of 32 kDa was visible, which could correspond to either the trimer of PhaP or a tetramer of the second α-helix, as these are indistinguishable on the gel. A top band was also present which was spread roughly between 44 and 54 kDa and could consist of several hetero-oligomers. Oligomerization was also investigated using size exclusion chromatography, allowing us to observe the interaction in a more dynamic mode (Fig. 6B). PhaP eluted as a single peak with an elution volume of 1.62 ml, corresponding to a molecular mass of approximately 44 kDa. As the calculated mass of a PhaP monomer is 12.3 kDa, a 44-kDa complex fits best with a tetramer, in agreement with the cross-linking experiments. The second α-helix eluted at 1.56 ml, corresponding to a size of approximately 58 kDa. As the monomer of α-helix 2 has a theoretical molecular mass of 8.7 kDa, this corresponds to a hexa- or heptamer forming a complex. In a 1:1 mixture of both proteins, the mixture eluted as a single peak at 1.60 ml, corresponding to a molecular mass of 48 kDa. In contrast to the single proteins, the elution peak of the protein mixture was slightly broader, suggesting that it consists of several mixed oligomers. If oligomerization of PhaP specifically occurred via α-helix 2, we would have expected constant tetramer formation also for isolated helix 2 and for the mixture. However, the preferential formation of hetero-oligomeric forms indicates that this corresponds to the most favorable thermodynamic state of complex formation. The fact that in the absence of helix 1 oligomers of different stoichiometry are formed indicates that helix 1 is involved in oligomerization of PhaP. This highlights the fact that the mode of oligomerization of PhaP is different than that described for other phasins, such as PhaF and PhaPAz, which are thought to oligomerize through coiled-coil regions (24, 51). As PhaP requires both α-helices for correct oligomerization, the tertiary structure most likely resembles a doughnut formed by the eight α-helices. For thermodynamic reasons, partially hydrophobic surfaces are often involved in the formation of tertiary structures, aiding oligomerization (55, 56). As a consequence several modes of interaction of tetrameric PhaP with the PHB granule are conceivable. The exposed coiled region connecting the aligned helices might bind to the PHB granule surface; alternatively, the tertiary structure of PhaP could open its conformation to expose its hydrophobic residues for interaction with the PHB surface, or the hydrophobic core of the PhaP tetramer could accommodate a surface-exposed PHB strand.
FIG 6.
(A) Tricine-PAGE of PhaP, helix 2 of PhaP, and a mixture of both proteins cross-linked by glutaraldehyde. (B) Size exclusion chromatography of PhaP (solid line), helix 2 of PhaP (dotted line), and a mixture of both proteins (dashed line). PhaP elutes at 1.62 ml (44 kDa), helix 2 of PhaP at 1.56 ml (58 kDa), and the mixture at 1.6 ml (48 kDa).
Conclusion.
This work demonstrates that the ORF ssl2501 of Synechocystis encodes a cyanobacterial phasin, PhaP. PhaP attaches to the PHB granule surface and regulates the number and size of PHB granules within a cell. In addition, PhaP acts as a regulator affecting the biosynthetic activity of PHB synthase in vivo. It is predicted to fold into two α-helices, which contribute to the binding of the phasin to PHB. The second α-helix retains the ability to oligomerize but is not solely responsible for oligomerization.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the DFG-funded GRK 1708 “Molecular principles of bacterial survival strategies.”
We thank Marcus Hartmann and Albrecht Reinhard for help with CD spectroscopy and Johanna Weirich and Jan Lennings for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00604-15.
REFERENCES
- 1.Stanier RY, Cohen-Bazire G. 1977. Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol 31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 2.Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R. 2013. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol 9:e1003081. doi: 10.1371/journal.pcbi.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osanai T, Oikawa A, Shirai T, Kuwahara A, Iijima H, Tanaka K, Ikeuchi M, Kondo A, Saito K, Hirai MY. 2013. Capillary electrophoresis-mass spectrometry reveals the distribution of carbon metabolites during nitrogen starvation in Synechocystis sp. PCC 6803. Environ Microbiol 16:512–524. doi: 10.1111/1462-2920.12170. [DOI] [PubMed] [Google Scholar]
- 4.Hauf W, Schlebusch M, Hüge J, Kopka J, Hagemann M, Forchhammer K. 2013. Metabolic changes in Synechocystis sp. PCC6803 upon nitrogen-starvation: excess NADPH sustains polyhydroxybutyrate accumulation. Metabolites 3:101–118. doi: 10.3390/metabo3010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasunuma T, Kikuyama F, Matsuda M, Aikawa S, Izumi Y, Kondo A. 2013. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J Exp Bot 64:2943–2954. doi: 10.1093/jxb/ert134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markou G, Chatzipavlidis I, Georgakakis D. 2012. Effects of phosphorus concentration and light intensity on the biomass composition of Arthrospira (Spirulina) platensis. World J Microbiol Biotechnol 28:2661–2670. doi: 10.1007/s11274-012-1076-4. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz R, Forchhammer K. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151:2503–2514. doi: 10.1099/mic.0.27883-0. [DOI] [PubMed] [Google Scholar]
- 8.Görl M, Sauer J, Baier T, Forchhammer K. 1998. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology 144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- 9.Panda B, Jain P, Sharma L, Mallick N. 2006. Optimization of cultural and nutritional conditions for accumulation of poly-beta-hydroxybutyrate in Synechocystis sp. PCC 6803. Bioresour Technol 97:1296–1301. doi: 10.1016/j.biortech.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Taroncher-Oldenburg G, Nishina K, Stephanopoulos G. 2000. Identification and analysis of the polyhydroxyalkanoate-specific beta-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803. Appl Environ Microbiol 66:4440–4448. doi: 10.1128/AEM.66.10.4440-4448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hein S, Tran H, Steinbüchel A. 1998. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch Microbiol 170:162–170. [DOI] [PubMed] [Google Scholar]
- 12.Numata K, Motoda Y, Watanabe S, Osanai T, Kigawa T. 2015. Co-expression of two polyhydroxyalkanoate synthase subunits from Synechocystis sp. PCC 6803 by cell-free synthesis and their specific activity for polymerization of 3-hydroxybutyryl-coenzyme A. Biochemistry 54:1401–1407. doi: 10.1021/bi501560b. [DOI] [PubMed] [Google Scholar]
- 13.Müh U, Sinskey AJ, Kirby DP, Lane WS, Stubbe J. 1999. PHA synthase from Chromatium vinosum: cysteine 149 is involved in covalent catalysis. Biochemistry 38:826–837. doi: 10.1021/bi9818319. [DOI] [PubMed] [Google Scholar]
- 14.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handrick R, Reinhardt S, Jendrossek D. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J Bacteriol 182:5916–5918. doi: 10.1128/JB.182.20.5916-5918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- 18.Sznajder A, Pfeiffer D, Jendrossek D. 2015. Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81:1847–1858. doi: 10.1128/AEM.03791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol 177:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 21.Galán B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, García JL, Prieto MA. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol 79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- 22.de Almeida A, Catone MV, Rhodius VA, Gross CA, Pettinari MJ. 2011. Unexpected stress-reducing effect of PhaP, a poly(3-hydroxybutyrate) granule-associated protein, in Escherichia coli. Appl Environ Microbiol 77:6622–6629. doi: 10.1128/AEM.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mezzina MP, Wetzler DE, de Almeida A, Dinjaski N, Prieto MA, Pettinari MJ. 8 October 2014. A phasin with extra talents: a polyhydroxyalkanoate granule-associated protein has chaperone activity. Environ Microbiol. doi: 10.1111/1462-2920.12636. [DOI] [PubMed] [Google Scholar]
- 24.Mezzina MP, Wetzler DE, Catone MV, Bucci H, Di Paola M, Pettinari MJ. 2014. A phasin with many faces: structural insights on PhaP from Azotobacter sp. FA8. PLoS One 9:e103012. doi: 10.1371/journal.pone.0103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava R, Pisareva T, Norling B. 2005. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5:4905–4916. doi: 10.1002/pmic.200500111. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Sun J, Chitnis PR. 2000. Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC 6803. Electrophoresis 21:1746–1754. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer D, Jendrossek D. 2012. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J Bacteriol 194:5909–5921. doi: 10.1128/JB.00779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 29.Schlebusch M, Forchhammer K. 2010. Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 76:6101–6107. doi: 10.1128/AEM.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Muro-Pastor AM, Olmedo-Verd E, Flores E. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp strain PCC 7120. FEMS Microbiol Lett 256:171–177. doi: 10.1111/j.1574-6968.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 32.Grigorieva G, Shestakov S. 1982. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol Lett 13:367–370. doi: 10.1111/j.1574-6968.1982.tb08289.x. [DOI] [Google Scholar]
- 33.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 34.Wolk CP, Vonshak A, Kehoe P, Elhai J. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci U S A 81:1561–1565. doi: 10.1073/pnas.81.5.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Schagger H. 2006. Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentin HE, Lee EY, Choi CY, Steinbüchel A. 1994. Identification of 4-hydroxyhexanoic acid as a new constituent of biosynthetic polyhydroxyalcanoic acids from bacteria. Appl Microbiol Biotechnol 40:710–716. doi: 10.1007/BF00173333. [DOI] [Google Scholar]
- 39.Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 40.Pieper-Fürst U, Madkour MH, Mayer F, Steinbüchel A. 1994. Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol 176:4328–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 42.Pfeiffer D, Jendrossek D. 2014. PhaM is the physiological activator of PHB synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol 80:555–563. doi: 10.1128/AEM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jossek R, Reichelt R, Steinbüchel A. 1998. In vitro biosynthesis of poly(3-hydroxybutyric acid) by using purified poly(hydroxyalkanoic acid) synthase of Chromatium vinosum. Appl Microbiol Biotechnol 49:258–266. doi: 10.1007/s002530051166. [DOI] [PubMed] [Google Scholar]
- 44.Ushimaru K, Motoda Y, Numata K, Tsuge T. 2014. Phasin proteins activate Aeromonas caviae polyhydroxyalkanoate (PHA) synthase but not Ralstonia eutropha PHA synthase. Appl Environ Microbiol 80:2867–2873. doi: 10.1128/AEM.04179-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B, Pugh S, Nielsen DR, Zhang W, Meldrum DR. 2013. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab Eng 16:68–77. doi: 10.1016/j.ymben.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Lan EI, Liao JC. 2012. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc Natl Acad Sci U S A 109:6018–6023. doi: 10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osanai T, Numata K, Oikawa A, Kuwahara A, Iijima H, Doi Y, Tanaka K, Saito K, Hirai MY. 2013. Increased bioplastic production with an RNA polymerase sigma factor SigE during nitrogen starvation in Synechocystis sp. PCC 6803. DNA Res 20:525–535. doi: 10.1093/dnares/dst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 49.Sato S, Shimoda Y, Muraki A, Kohara M, Nakamura Y, Tabata S. 2007. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803 DNA Res 14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeiffer D, Jendrossek D. 2013. Development of a transferable bimolecular fluorescence complementation system for the investigation of interactions between poly(3-hydroxybutyrate) granule-associated proteins in Gram-negative bacteria. Appl Environ Microbiol 79:2989–2999. doi: 10.1128/AEM.03965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maestro B, Galan B, Alfonso C, Rivas G, Prieto MA, Sanz JM. 2013. A new family of intrinsically disordered proteins: structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. PLoS One 8:e56904. doi: 10.1371/journal.pone.0056904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann L, Spinozzi F, Sinibaldi R, Rustichelli F, Potter M, Steinbüchel A. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J Bacteriol 190:2911–2919. doi: 10.1128/JB.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 55.Tsai C-J, Lin SL, Wolfson HJ, Nussinov R. 1997. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci 6:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali MH, Imperiali B. 2005. Protein oligomerization: how and why. Bioorg Med Chem 13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 57.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.