Abstract
Clostridium botulinum type E toxin is responsible for extensive mortality of birds and fish in the Great Lakes. The C. botulinum bontE gene that produces the type E toxin was amplified with quantitative PCR from 150 sloughed algal samples (primarily Cladophora species) collected during summer 2012 from 10 Great Lakes beaches in five states; concurrently, 74 sediment and 37 water samples from four sites were also analyzed. The bontE gene concentration in algae was significantly higher than in water and sediment (P < 0.05), suggesting that algal mats provide a better microenvironment for C. botulinum. The bontE gene was detected most frequently in algae at Jeorse Park and Portage Lake Front beaches (Lake Michigan) and Bay City State Recreation Area beach on Saginaw Bay (Lake Huron), where 77, 100, and 83% of these algal samples contained the bontE gene, respectively. The highest concentration of bontE was detected at Bay City (1.98 × 105 gene copies/ml of algae or 5.21 × 106 g [dry weight]). This study revealed that the bontE gene is abundant in the Great Lakes but that it has spatial, temporal, and matrix variability. Further, embayed beaches, low wave height, low wind velocity, and greater average water temperature enhance the bontE occurrence.
INTRODUCTION
Clostridium botulinum produces neurotoxins (BoNT)—the most lethal known—that cause botulism, a neuroparalytic disease. These neurotoxins block the release of acetylcholine at the neuromuscular junction and cause acute flaccid muscle paralysis in humans, domestic animals, and wildlife, especially birds (1); as little as 30 to 100 ng of toxin is potentially fatal to animals as well as to humans (2). C. botulinum is a Gram-positive, anaerobic, spore-forming, saprophytic bacterium that produces eight types of neurotoxin (types A to H) (2–4). Types A and B generally involve in food-borne botulism, types C and E predominantly cause avian botulism (5), while type H is a recently discovered strain associated with infant botulism (4).
Avian botulism outbreaks due to type E have been observed around the Great Lakes and are sporadic temporally and spatially (6–10). Avian botulism outbreaks were reported in the early 1900s, but intensive study did not begin until the 1960s when people died from the consumption of improperly smoked Lake Superior fish (11, 12). In 1963 and 1964, C. botulinum type E killed more than 12,000 birds, mainly gulls and loons, in Lake Michigan (13). Type E botulism outbreaks of various levels of intensity have been recorded for Lakes Huron, Ontario, Erie, and Michigan annually since 2006 (14).
C. botulinum is ubiquitous in the environment and frequently detected in soils, surface water, sediments, Cladophora species, fish, and benthic fauna (6, 12, 15–18), and up to 820,000 bontE gene copies/mg have been detected in Lake Erie chironomids living along the lake bottom (19). The offshore and onshore algae/Cladophora play an important role in occurrence and survival of C. botulinum in the Great Lakes, which produce conditions ideal for the growth and survival of numerous microorganisms, including C. botulinum type E (10, 18, 20–23). C. botulinum is present in Cladophora mats as vegetative cells, not as spores, which indicates that Cladophora mats provide a suitable microenvironment for survival and growth of this organism (10). In 2011, Sleeping Bear Dunes National Lakeshore (SLBE) experienced several large-scale bird mortality events affecting primarily fish-eating birds, which were coincident with deposition of sloughed algae in this area (10, 14).
The environmental and physicochemical conditions that drive avian botulism outbreaks are uncertain: low lake levels, shallow bays, reduced water exchange with open water, and increasing lake temperatures may increase the chance of botulism outbreaks (9, 12, 24). C. botulinum growth and toxin production may depend on several factors. C. botulinum type E favors rich humus soil with high moisture retention capacity, and it has been detected at high concentrations in drainage basins in the Great Lakes (6, 7). High concentrations of C. botulinum type E in lake sediment have been correlated with land drainage, water currents, and sediment deposition (25). Increased water temperatures may enhance the type E detection in sediment, potentially indicating bacterial growth under anoxic conditions (9, 19, 26). In addition, other physicochemical parameters such as redox potential, pH, salinity, available phosphorus, and turbidity may contribute to C. botulinum type E growth (19, 27, 28). Relationships among these parameters are complex; a multivariate regression model developed using various sediment and water variables indicated that type C outbreak risk increased with 7.5 to 9.0 pH, negative redox potential, >20°C water temperature, and <2.0 ppm salinity (29). As far we know, no such published environmental predictive model exists for type E botulism.
Toxin-producing strains of C. botulinum are typically identified by mouse bioassay. However, these assays are expensive and tedious; currently, alternate methods, such as PCR or quantitative PCR (qPCR), are available for determining BoNT serotypes (15). Quantification of bontE in Great Lakes environmental samples has been applied successfully, but the studies were typically limited to only one or a few locations (19, 28). Furthermore, Cladophora mats in multiple beaches in the Lake Michigan were studied using most probable number PCR (MPN-PCR) (10).
The primary goals of this study were to (i) quantify the C. botulinum type E toxin-producing bontE gene using qPCR in algal, sediment, and water samples collected at beaches across the Great Lakes, (ii) assess the spatial, matrix, and temporal variability of occurrence of bontE, and (iii) evaluate possible connections to environmental, physicochemical, and meteorological factors, which may affect the occurrence of C. botulinum in different locations. The results of this study will be useful in developing a predictive model which can be used for important management decisions such as warning the beach visitors if there will be a potential outbreak or preparing for collecting of bird carcasses and planning for proper disposal methods.
MATERIALS AND METHODS
Site description.
Samples were collected from 10 beaches in five states from May to September 2012: Maumee Bay State Park (Maumee Bay) at Lake Erie, Bay City State Recreation Area (Bay City) and South Linwood Beach (Linwood) at Lake Huron, Brimley State Park (Brimley) at Lake Superior, Sleeping Bear Dunes National Lakeshore (SLBE) at Esch Road/Otter Creek, Portage Lakefront Beach (Portage) at Indiana Dunes National Lakeshore, and Jeorse Park Beach (Jeorse), Hammond Beach (Hammond), Waukegan Municipal Beach (Waukegan), and Deland Beach (Deland) at Lake Michigan (Fig. 1). Here, these beaches are referred by the names indicated in parentheses.
FIG 1.
Sampling locations: Maumee Bay, OH (lat 41.686005, long, −83.377197); Bay City, MI (43.672081, −83.906380); Linwood, MI (43.716431, −83.939751); Brimley, MI (46.417309, −84.555489); SLBE, MI (44.766720, −86.074959); Deland, WI (43.762501, −87.696007); Waukegan, IL (42.362358, −87.816010); Hammond, IL (41.699200, −87.513138); Jeorse, IL (41.649502, −87.432861); Portage, IN (41.630630, −87.181633). The map was created using ArcMap 10 (ESRI).
Collection of algal, water, and sediment samples.
Sloughed algae (typically Cladophora and, rarely, Chara) submerged and floating in water (SUB) and sloughed algae accumulated onshore (AlgOn) were collected weekly (when present) from each of 10 beaches for microbial analysis. “Muck” samples, consisting of decaying algae and other field debris, were taken from the shoreline of Bay City. Both algal and muck samples were scooped with a sterile tongue depressor, placed in 7 oz. Whirl-Pak bags. Water and sediment samples were collected from the Bay City, Jeorse, SLBE, and Portage sites. Water samples were collected in 7-oz. Whirl-Pak bags in waist-deep water approximately halfway between the surface of the water column and the lake bottom. Two sets of routine sediment samples (typically beach sand) were collected at each sampling location: one set in knee-deep water (SED) and one set in moist sand between the water's edge and the high-water mark (sediment onshore [SOS]). Sediment samples under the algal mat (SUM) were collected when algae were present on the beach. Sterile 50-ml tubes were filled about three-quarters full with sediment by pressing the tube into the sand to obtain a core. All samples were collected by using sterile collection techniques, i.e., monitors used latex gloves that were changed between all samples, flame-sterilized forceps, and sampling gear stored in a clean environment. All samples were immediately stored in a cooler on ice and transported to the field office. In the field office, each 100 ml of water was filtered with 0.45-μm-pore-size Isopore filters using sterile Microfil disposable filter cups (Millipore, Billerica, MA), and filters were placed in sterile petri dishes. All samples were stored at −20°C until processed.
Beach monitoring.
Beach transects were located on Federal land or within U.S. Environmental Protection Agency (EPA) areas of concern (30). Each catchment area was identified by using the National Hydrography Dataset (NHD), version 2 (31). If a stream was within 8 km on either side of the sampling location, an upstream area was found by navigating upstream and selecting catchments that corresponded to the upstream area. Variables were then calculated for the catchment and upstream areas, including the percentage of land cover types from the National Land Cover Database 2006 (32). Radar rainfall data (33) were processed using the Environmental Data Discovery and Transformation tool (34), and U.S. Census Bureau population data were obtained by using ESRI's Community Analyst program.
Most physical and meteorological variables were estimated based on standard methods as part of EPA sanitary surveys (36). Wind speed and direction were estimated by using the Beaufort wind scale (www.spc.noaa.gov/faq/tornado/beaufort.html). The sky condition (sunny, mostly sunny, partly sunny, mostly cloudy, or cloudy), the amount of cloud coverage (no clouds, 1/8 to 1/4 coverage, 3/8 to 1/2 coverage, 5/8 to 7/8 coverage, or complete coverage), and rain intensity (misting, light rain, steady rain, heavy rain, or other) were estimated visually.
Wave height was estimated by placing a meter stick just seaward of where the waves were breaking and recording the height of wave crests and troughs. Longshore or littoral current and speed were estimated by noting the time taken for a floating object to move at least 10 m and were expressed as the average of at least three replicates. Undercurrent direction and speed were measured by estimating the direction and intensity of movement (high, medium, or low) of flagging tape attached to a meter stick and submerged in waist-deep water so that the flagging tape moved freely in the water column.
The amount of algal coverage on the beach and in the water was recorded in four categories (none, low [1 to 20%], moderate [21 to 50%], or high [>50%]), and the type (periphyton, globular, free floating, or other), description (attached to rocks, stringy, blobs of floating material, no obvious mass of materials, or other), and color (light green, bright green, dark green, yellow, brown, or other) of the algae were also noted.
AMBLE (avian monitoring for botulism lakeshore events) surveys also prompted the recording of categorical estimates of a variety of beach characteristics (37). Monitors made categorical estimates of precipitation (none, fog, sprinkles, showers, or snow), wind speed (none, felt on skin, small trees swaying, or sand blowing), wind direction, wave height (still, gentle waves, waves 5 to 10 feet up beach, or waves >10 ft up beach), and amount of Cladophora and Chara separately in five categories on the beach (none, couple clumps, isolated spots, scattered continuous, or thick mat) and in the water (none, couple clumps, isolated tufts, scattered continuous on lake bottom, thick, soupy, or solid). They also recorded the number of dead round gobies (none, 1 to 30, or >30) and zebra or quagga mussels on shore (none, couple clumps, isolated spots, scattered continuous, or thick mat). Categorical rankings were expressed as integers (e.g., 1 to 5) for analysis.
Multimeters were used to measure the following water quality variables, as per the manufacturer instructions, at each beach in knee-deep water: conductivity, total dissolved solids (TDS), pH, salinity (Extech ExStik EC500; Test Equipment Depot, Melrose, MA), oxidation reduction potential (Extech ExStik RE300) and dissolved oxygen (Extech ExStik DO600), and water temperature (either EC500 or DO600 Extech meters). Nitrate-nitrogen (NO3-N) and phosphate (PO4) levels were estimated by using a nitrate test kit (NI-11) and an orthophosphate test kit (PO-14; both from Hach, Loveland, CO). Water turbidity was estimated by using custom-made turbidity tubes (38).
Data on beach conditions were also collected from a variety of online resources. Weather data were collected from the weather stations nearest to the beach transects. Weather variables included temperature, dew point, pressure, wind direction, wind speed, wind speed of gusts, humidity, hourly precipitation, daily precipitation, and a description of weather conditions (39). Beach closure, contamination, and Escherichia coli data were harvested from the Wisconsin Department of Natural Resources Wisconsin Beach Health website (http://www.wibeaches.us/) and the BeachGuard websites maintained by the Michigan Department of Environmental Quality (http://www.deq.state.mi.us/beach/), the Ohio Department of Health (http://publicapps.odh.ohio.gov/BeachGuardPublic/), the Indiana Department of Environmental Management (https://extranet.idem.in.gov/beachguard/), and the Illinois Department of Public Health (http://app.idph.state.il.us/envhealth/ilbeaches/public/). See the supplemental material for these data (see Tables S1 to S5 in the supplemental material). AMBLE survey and beach sanitary surveys were conducted at least twice weekly from the end of May to mid-November 2012 and more often in the summer months. Further, monitors measured the extent of nuisance algae (e.g., Cladophora) on the beach and took note of fish, wildlife, and bird activity.
DNA extraction.
Frozen algal and sediment samples were thawed in a refrigerator overnight. About 20 to 25 ml of wet algae was placed in a 50-ml sterile plastic tube and vortexed 15 min at 10,000 × g (Vortex Mixer; Fisher Scientific, Pittsburgh, PA). DNA was extracted from 3 ml of the homogenized algal slurry. Dry weight was determined by using 10% of the remaining algal slurry samples. About 10 g of sediment (SED, SOS, and SUM) was used for DNA extraction, and the sediment dry weight was determined. A PowerMax soil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA) was used for DNA extraction from algae and sediment, while water DNA was extracted from the entire filter representing 100 ml of water by using an UltraClean DNA isolation kit (MO BIO Laboratories), both according to the manufacturer's instructions.
Determination of moisture content of algal slurry and sediment.
Approximately 10% of algal samples were taken to determine the moisture content of algal slurry. The algal slurry was shaken well, and 3 ml of the slurry was pipetted into a preweighed dish. About 2 to 5 g of wet sediment samples were placed on preweighed dish. All samples were kept in the drying oven until the weights were constant and the moisture percentages were calculated.
qPCR standard preparation.
Transformed E. coli competent cells containing the bontE 1467F and 1605R gene fragments (15) were grown in Luria-Bertani (LB) broth (Difco, Sparks, MD) overnight, and plasmid DNA was extracted with a QIAPrep spin miniprep kit (Qiagen, Germantown, MD). Plasmid DNA concentration was measured with a NanoDrop ND-1000 UV spectrophotometer (Thermo Scientific, Wilmington, DE), and the gene copy numbers were calculated.
Reaction conditions for qPCR.
An Applied Biosystems StepOne Plus qPCR (Life Technologies Corp.) was used to amplify a fragment of the bontE gene. All samples were run with six-point standard curves ranging from 10 to 106 gene copies and no-template controls (NTC) in triplicates. Each 25-μl reaction volume contained 5 μl of PCR-grade water (Promega, Madison, WI), 12.5 μl of TaqMan environmental master mixture 2.0 (Applied Biosystems, Carlsbad, CA), 300 nM concentrations of the forward and reverse primers 1467F (5′-AATATTGTTTCTGTAAAAGGCATAAGGAA-3′) and 1605R (5′-AAGTTACTGTATCGTCAATTTCTTTAGGAG-3′) (Sigma-Aldrich, St. Louis, MO), a 200 nM concentration of probe (5′-6FAM-AATGGTGAGTTATTTTTTGTGGCTTCCGAGAAT-TAMRA-3′) (Applied Biosystems) (6FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine), and bovine serum albumin (0.2 μg/μl; Sigma-Aldrich). The qPCR thermocyclic conditions were as follows: 10 min at 95°C for initial denaturing, followed by 45 cycles at 95°C for 15 s for denaturation and 60°C for 60 s for annealing and extension.
qPCR QA/QC.
Several quality assurance and quality control (QA/QC) approaches were performed to validate our qPCR results. (i) DNA was extracted from three algal samples randomly selected to determine matrix inhibition: undiluted, 10×-diluted, and 25×-diluted samples were amplified with a six-point standard curve, and the gene copy numbers were calculated. (ii) The internal control is a quality control parameter that determines whether the assays have amplified appropriately; 100, 1,000, or 10,000 gene copy standards were used for this internal control. (iii) Other quality control parameters for the qPCR standard curve were monitored (see also Table S6 in the supplemental material for additional details).
Bacillus subtilis spore test.
Because of restrictions on C. botulinum growth in regular laboratories in the United States, B. subtilis (NAMSA 6633) spores were used to determine the contribution of spore DNA in final copy number calculations (40). B. subtilis spores (50 μl) were seeded in a 500-ml volumetric flask with sterile water added to a total volume of 500 ml. The seeded cells were mixed well, and four 100-ml aliquots were filtered through a 0.45-μl-pore-size filter. Four blank samples were also filtered separately. Two blanks and two spore-filter-containing tubes were used in a bead beater for 2 min, and two other blanks and spore-containing filters were placed on a vortex mixer at maximum speed for 15 min. All eight samples were used for DNA extraction; in addition, B. subtilis genomic DNA (1 ng/μl) was used as the positive control, and PCR-grade water was used as a no-template control (NTC). A B. subtilis group 16S rRNA gene fragment was amplified with the primers Bsub5F (5′-AAGTCGAGCGGACAGATGG-3′) and Bsub3R (5′-CCAGTTTCCAATGACCCTCCCC-3′) (40) by using Applied Biosystems StepOne Plus qPCR and a comparative standard curve method. Each 25-μl reaction volume contained 5 μl of PCR-grade water (Promega), 12.5 μl of SYBR green master mixture (Applied Biosystems), a 400 nM concentration of forward and reverse primers (Sigma-Aldrich), and bovine serum albumin (0.4 μg/μl; Sigma-Aldrich). The qPCR thermocyclic conditions were as follows: 10 min at 95°C for initial denature, followed by 40 cycles at 95°C for 30 s for denaturation, 65°C for 2 min for annealing, and 72°C for 2 min for extension, and the melting curve was analyzed with a temperature gradient of 0.5°C per min from 60 to 95°C. The cycle thresholds (CT) of the NTC, blanks, and samples were compared with the CT of the positive control.
Handling of triplicates at the lower end of the standard curve.
The limit of quantification (LOQ), i.e., the lowest concentration of a particular genetic marker in the linear range of the qPCR standard curve (41), and the limit of detection (LOD), i.e., the fifth percentile among the observed CT across all blanks and negative-control reactions (42), were calculated. Due to the nature of probability, we cannot expect amplification of all the triplicates at the low end of the standard curve every time. We established a method to classify the triplicates at the lower end of the standard curve. If two or three replicates out of three yielded >3 and <10 gene copies, the samples were categorized as detectable but nonquantifiable samples (DNQ). If samples had two or three replicates that yielded >10 gene copies, those samples were categorized as detectable and quantifiable samples (DQ). If all three replicates or two of three replicates yielded <3 gene copies, the samples were categorized as nondetectable (ND). Throughout this study, “above LOQ” (quantifiable) indicates DQ samples only, and “above LOD” (detectable) indicates both DQ and DNQ samples.
Statistical analysis.
Statistical analyses were conducted for the following categorical and numerical data from all 10 sites by using Systat 13 and SigmaPlot 12.0 (Systat Software, Inc., San Jose, CA): (i) catchment characteristics (the percentage of development, barren lands, forest, agriculture, wetlands, and the population); (ii) water quality (water temperature, conductivity, salinity, pH, dissolved oxygen and turbidity, and NO3-N and PO4); (iii) beach sanitary survey information, i.e., the birds on the beach, bird carcasses, Cladophora accumulation onshore and in the water, Chara (a type of green algae) onshore and water, gobies onshore and mussels onshore); and (iv) hydrological and meteorological data, i.e., air temperature, wave height, precipitation, wind speed and direction, current velocity, and wind velocity (see Table S7 in the supplemental material for these data). Spearman correlations were used to determine the relationship between detection of bontE in all matrices and different meteorological, hydrological, physicochemical, and beach characteristics. Discriminant analysis and factor analysis were used to group the beaches based on their characteristics. Unless specified otherwise, a Student t test was used to identify the differences between the treatments, and a P value of <0.05 was considered statistically significant. A Student t test was performed only for quantifiable bontE gene copies, which were transformed to a log scale.
RESULTS
QA/QC for qPCR assay.
The qPCR standard curve quality control parameters are given in Table S6 in the supplemental material. The assay LOQ was 10 gene copies and the assay LOD was 3 gene copies/5 μl of template DNA. The sample LOD and LOQ for algae and muck were 600 and 2,000 gene copies/ml of slurry; the average dry weight percentage in algae and muck slurry was 3.8%. The sediment LOD and LOQ were 193 and 645 gene copies/g (dry weight); the LOD and LOQ for water were 30 and 100 gene copies/100 ml. Based on the inhibition test, a 10× dilution was selected for all three matrices (data not shown). The results of the spore test revealed spore DNA did not contribute to the qPCR amplification.
Occurrence of bontE at Great Lakes beaches.
The Bay City, Portage, and Jeorse beaches had the highest frequency of detection of bontE in algae, while the Maumee Bay, SLBE, and Brimley sites had intermediate frequency of detection. The Linwood, Hammond, Waukegan, and Deland sites had low detection frequency, and at the latter four sites and at Brimley, all of the results were nonquantifiable (Table 1). The highest quantifiable bontE gene concentration in AlgOn was reported at Bay City, where the average concentration was 99,662 gene copies, and concentrations ranged from 29,336 to 198,559 gene copies/ml of slurry (Table 1). Bay City also had the highest frequency of bontE detection; 90.9 and 63.6% of the AlgOn samples, respectively, were above the LOD and LOQ. Linwood and Bay City are two beaches located only 8 km apart from one another, but at Linwood only 14.3% of SUB and no AlgOn contained bontE above the LOD. Jeorse also showed high bontE detection: 83.3% of AlgOn and 70.0% of SUB samples were above the LOD. Portage also had a high percentage of samples containing bontE above the LOD: 100% of AlgOn, 100% of water, 70% of SED, and 30% of SOS. Jeorse and Hammond are beaches located about 8 km way from each other and, as for Bay City and Linwood, there was a significant variability in the distribution of bontE between these two sites. Algae were not often present at Brimley, but a total of 75% of the Brimley AlgOn samples were above the LOD; only four samples from 3 days were collected, and the samples for all 3 days were positive for bontE. Similarly, at Waukegan four samples for 3 days were available for analysis, and only one of the four samples was above the LOD.
TABLE 1.
Frequency of bontE detection or quantification and average concentration at each sampling site and matrix
| Sampling site | Matrix | Total no. of samples | % of samples |
Avg no. of gene copies/mla (SD) | |
|---|---|---|---|---|---|
| Above LOQ | Above LOD | ||||
| Bay City | AlgOn | 11 | 63.6 | 90.9 | 99,662 (66,838) |
| Muck | 12 | 41.7 | 66.7 | 39,787 (16,738) | |
| SUB | 1 | 0 | 0 | Below LOQ | |
| SED | 9 | 22.2 | 22.2 | 8,335 (8) | |
| SOS | 8 | 12.5 | 37.5 | 12,227 (NA) | |
| SUM | 3 | 0 | 66.7 | Below LOQ | |
| Water | 8 | 100 | 100 | 1,511 (169) | |
| Portage | AlgOn | 7 | 42.9 | 100 | 35,487 (8,124) |
| SED | 10 | 30 | 70 | 19,260 (6,266) | |
| SOS | 10 | 10 | 30 | 14,897 (NA) | |
| Water | 10 | 80 | 100 | 1,900 (4,64) | |
| Jeorse | AlgOn | 12 | 66.7 | 83.3 | 71,941 (45,154) |
| SUB | 10 | 40 | 70 | 104,925 (53,158) | |
| Maumee | AlgOn | 19 | 10.5 | 52.6 | 25,602 (906) |
| SED | 12 | 41.7 | 41.7 | 21,861 (9,262) | |
| SOS | 7 | 28.6 | 28.6 | 21,346 (7,167) | |
| Water | 12 | 16.7 | 50 | 2,981 (1,250) | |
| SLBE | AlgOn | 10 | 10 | 40 | 4,725 (NA) |
| SED | 14 | 7.1 | 14.3 | 34,656 (NA) | |
| Water | 7 | 42.9 | 85.7 | 1,690 (150) | |
| Linwood | AlgOn | 11 | 0 | 0 | Below LOQ |
| SUB | 7 | 0 | 14.3 | Below LOQ | |
| Brimley | AlgOn | 4 | 0 | 75 | Below LOQ |
| Hammond | AlgOn | 12 | 0 | 16.7 | Below LOQ |
| SUB | 8 | 0 | 25 | Below LOQ | |
| Waukegan | AlgOn | 3 | 0 | 33.3 | Below LOQ |
| SUB | 1 | 0 | 0 | Below LOQ | |
| Deland | AlgOn | 12 | 0 | 0 | Below LOQ |
| SUB | 10 | 0 | 10 | Below LOQ | |
| Total | AlgOn | 101 | 20.8 | 46.5 | 68,359 (54,368) |
| SUB | 37 | 10.8 | 32.4 | 104,925 (53,137) | |
| Muck | 12 | 41.7 | 66.7 | 39,787 (16,738) | |
| SED | 45 | 24.4 | 35.5 | 19,855 (9,629) | |
| SOS | 25 | 16 | 32 | 17,454 (6,205) | |
| SUM | 4 | 0 | 50 | 0 | |
| Water | 37 | 56.8 | 81.1 | 1,825 (572) | |
Algae (AlgOn, muck, and SUB), gene copies/ml; sediment (SED, SOS, and SUM), gene copies/g (dry weight); water, gene copies/100 ml. NA, not applicable.
SED from SLBE had the lowest percentage of samples containing bontE above the LOQ (7%), and Maumee Bay SED had the highest percentage above the LOQ (41.7%). Portage SED had the highest percentage of bontE above the LOD (70%).
Bay City and Portage had high frequencies of bontE detection in water; 100% of the samples were above the LOD, while 100% of the Bay City and 80% of the Portage samples were quantifiable. Maumee Bay had the lowest percentage of water samples with bontE above the LOQ and LOD (17 and 50%, respectively) while those for SLBE were 43 and 86%, respectively.
bontE concentration in different matrices.
A total 261 samples were analyzed for bontE gene: onshore algae (n = 101), submerged algae (n = 37), muck (n = 12), sediment (SED, SOS, and SUM [n = 74]), and 37 water samples. The bontE gene concentration was highest in AlgOn and SUB, followed by muck, sediment, and water; all differences between major matrices (water, muck, algae, and sediment) were statistically significant (P < 0.05; Fig. 2). The muck and algal matrices were largely wet; the average dry weight percentage of algae and muck was 3.8% and that of sediment was 84.8%. All of the matrices were compared on a wet volume basis (1 ml). The distribution of bontE in different matrices at different sites is shown in Fig. S1 in the supplemental material.
FIG 2.
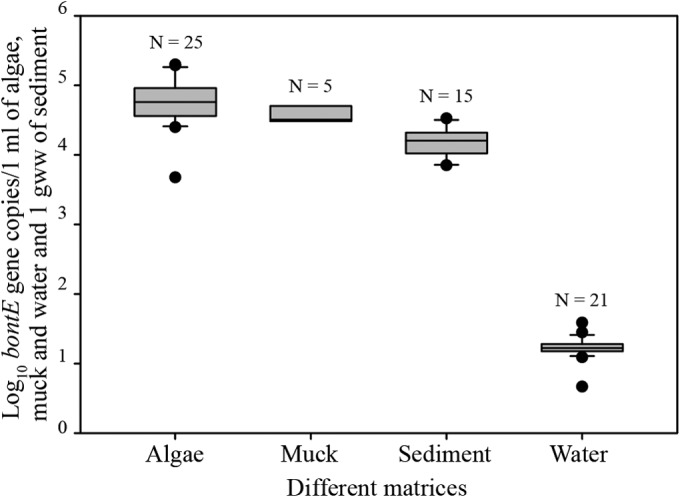
Distribution of bontE in different matrices: algae (SUB and AlgOn), muck from Bay City, sediment (SED and SOS), and water. The horizontal line in the gray bars shows the median value, the whiskers show the 10th and 90th percentiles, and the black dots show the outliers. The number above each bar indicates the number of samples that were above the LOQ. gww, gram wet weight.
Water had the highest detection frequency but the lowest concentration compared to the other matrices. bontE was detected above the LOQ in 56.8% and above the LOD in 81.1% of the water samples. The quantifiable bontE concentration in water ranged from 1,222 gene copies/100 ml at Bay City to 3,865 gene copies/100 ml at Maumee Bay. In all cases, the bontE concentrations in water were significantly lower than in algal, muck, and sediment matrices (P < 0.0001).
Of 101 AlgOn samples, 20.8% were above the LOQ, while 46.5% were above the LOD. The quantifiable bontE concentration varied from 4,725 gene copies/ml of algal slurry at SLBE to 198,559 gene copies/ml at Bay City. At Bay City, where muck was tested, there was a high percentage of bontE detection: 41.7% of samples were above the LOQ, 66.7% of samples were above the LOD, and the bontE concentration in muck ranged from 29,336 to 69,417 gene copies/ml. Among algal samples, SUB had the lowest percentage containing bontE above the LOQ (10.8%) or LOD (32.4%). Only the Jeorse SUB samples had bontE concentrations in the quantifiable range: 34,544 to 158,268 gene copies/ml of slurry.
The bontE concentration in SED varied from 8,330 gene copies/g (dry weight) at Bay City to 37,141 gene copies/g (dry weight) at Maumee Bay, while 24.4% of the samples had bontE concentrations above the LOQ and 35.5% above the LOD. The SOS concentration varied from 12,227 gene copies/g (dry weight) in Bay City to 26,414 gene copies/g (dry weight) in Maumee Bay. There was no significant difference between the bontE concentration distributions in SED and SOS (P > 0.05). Generally, sediment samples (SED, SUM, and SOS) displayed lower detection frequencies and concentrations (P < 0.001) than algae.
Temporal variability of occurrence of bontE at various sites and matrices.
Because so many samples were below the LOQ, analysis of the temporal variability of C. botulinum in all sites and matrices was not possible; however, samples collected from Bay City and Jeorse had sufficient data to explore the temporal patterns of bontE in AlgOn and water. Figure 3 shows the temporal variability of the bontE found at Bay City. The bontE concentration found in water remained almost constant over time (ranging from 1,222 to 1,765 bontE gene copies/100 ml). AlgOn samples showed a temporal pattern (see Table S8 in the supplemental material), where a high bontE concentration was reported in mid-June (193,377 gene copies/ml), but for the next two consecutive weeks these bontE concentrations were below the LOD. The second week of July we found the highest reported bontE concentration in algae in this study (198,559 gene copies/ml). This changing pattern is not consistent over time, and the bontE concentrations found during 3 months, June, July, and August, were compared. A Student t test showed that the bontE distribution during different months was statistically not significant (P > 0.5). This is a single-summer study; we feel that studying at least three consecutive years is necessary to get more insight into temporal patterns.
FIG 3.
Temporal variability of C. botulinum bontE found at Bay City Recreational Beach. Closed circles represent AlgOn, and open circles represent water samples. Three AlgOn samples detectable but not quantifiable were assigned a value of log10 = 1.
Spearman correlation analysis of all sites showed that bontE found in AlgOn was positively correlated with water temperature (r = 0.3; see Table S9 in the supplemental material). There were only two quantifiable SED (8,330 and 8,341 gene copies/ml in August and September) and one SOS (12,227 gene copies/ml in June) samples found in Bay City. Most sediment samples collected during the middle of the summer contained bontE concentrations below the LOD.
Jeorse was unique in that two storm events were targeted for sampling that occurred in the beginning and middle of August. Most of these event samples contained higher bontE concentrations than the regular samples (see Fig. S2 in the supplemental material). A Student t test showed that the bontE concentrations during the two storm events were significantly higher than during the regular sampling period (P < 0.05). Figure S2 and Table S10 in the supplemental material show a trend for the bontE concentration increasing during storm events, but we suggest that more sampling is required to confirm this hypothesis.
Correlation between bontE detection and meteorological and physicochemical factors.
Several significant correlations were observed between log10 bontE detection in different sites and matrices with different meteorological, hydrological, physicochemical, and beach characteristics (see Table S8 in the supplemental material). A positive correlation was found between the bontE gene concentration in AlgOn and the bontE concentration in SUB (r = 0.47). bontE in SUB was negatively correlated with wind speed, wave height, and mussels (r = −0.52, −0.35, and −0.42, respectively) and positively correlated with pH (r = 0.4). Further, bontE in water was also negatively correlated with wind speed, NO3-N, and turbidity (r = −0.43, −0.37, and −0.49, respectively) and positively correlated with conductivity and salinity (r = 0.41 and 0.48), but bontE found in SOS showed a positive correlation with wave height (r = 0.5). bontE found in Bay City muck, which had a limited number of samples, showed positive correlations with PO4, TDS, and bontE found in water (r = 0.81, 0.60, and 0.95).
Grouping of beaches based on bontE gene concentration.
Beaches were grouped into those having (Bay City, Jeorse, SLBE, Maumee Bay, and Portage) or not having (Deland, Waukegan, Hammond, Brimley, and Linwood) quantifiable results in AlgOn, and initial discriminant analysis conducted by using all the variables in Tables S1 to S5 in the supplemental material did not discriminate between these two groups (Wilk's lambda = 0.2160, P = 0.972); however, many of the variables were correlated (e.g., land cover percentages). Subsequently, six variables that varied substantially between the two groups were selected to represent land cover (barren lands), water quality (mean NO3-N), weather (average water temperature and average wind speed), and hydrometeorology (wave height and the magnitude of depth-averaged water velocity) (Table 2). Discriminant analysis using these six variables distinguished these two groups (Wilk's lambda = 0.037; P = 0.03; Table 2). Beaches with quantifiable bontE in algae had different catchment land uses (more barren lands, less developed area, more forest and shrubs, and more agriculture) and a greater average water temperature, but lower concentrations of NO3-N, wave heights, and water velocity than beaches without detections (Table 2).
TABLE 2.
Group means for environmental characteristics of beaches having or not having quantifiable bontE concentrations in algae
| Parametera | Group mean |
|
|---|---|---|
| Beaches with quantifiable bontE in algae | Beaches without quantifiable bontE in algae | |
| Catchment area (ha) | 19.7 | 5.9 |
| Developed area (%) | 25.8 | 48.1 |
| Barren lands (%)* | 13.1 | 9.60 |
| Forest and shrubs (%) | 25.9 | 12.6 |
| Agriculture (%) | 11.9 | 1.70 |
| Grasslands (%) | 0.70 | 0.30 |
| Wetlands (%) | 18.8 | 21.1 |
| Catchment population (log10) | 2.76 | 3.1 |
| E. coli (CFU/100 ml [geometric mean]) | 113 | 55 |
| Birds (seasonal avg) | 43.6 | 41.0 |
| Cladophora onshore (1 to 5 scale) | 2.03 | 1.82 |
| Mussels on shore (1 to 5 scale) | 1.23 | 1.32 |
| Mean water temp (°C)* | 23.1 | 21.7 |
| Mean conductivity (μS) | 391 | 265 |
| Mean total dissolved solids (ppm) | 273 | 187 |
| Mean pH | 8.73 | 8.53 |
| Mean dissolved oxygen (ppm) | 7.67 | 7.63 |
| Mean NO3-N (mg/liter)* | 0.35 | 0.58 |
| Wave ht (m)* | 0.28 | 0.37 |
| Magnitude of water velocity (depth averaged) (m/s)* | 0.02 | 0.06 |
| Wind avg (mph)* | 6.05 | 5.57 |
| Precipitation daily avg (in.) | 0.10 | 0.13 |
*, six variables were used for discriminant analysis.
Among all 10 sampling sites, Bay City had the highest bontE concentration and frequency of detection. Therefore, the possible pathways of C. botulinum to this site are drainage from the Saginaw River, storm drains, and Tobico Marsh (see Fig. S4 in the supplemental material); further, both northeast and southwest winds (northwest and southeast winds were rare) may play a role in high bontE detection at Bay City (see Fig. S3 in the supplemental material).
DISCUSSION
Variability of bontE distribution by locations.
We found that C. botulinum type E is ubiquitous, but its occurrence was consistent across neither locations nor matrices. Only a few studies have been conducted throughout the Great Lakes to compare data on spatial variability of the distribution of bontE, and most of these studies concentrated on local areas (10, 19, 28). A study conducted in 1968 at several locations over the Great Lakes also found an uneven distribution of C. botulinum type E in the Great Lakes, and it was more prevalent in Green Bay, followed by the Bay City and the Maumee Bay areas (7). Chun's group also identified spatial variability of C. botulinum at beaches in the Great Lakes (10); they had conducted intensive studies in SLBE in 2011 and found that 73% of the Cladophora samples collected from seven beaches contained C. botulinum type E and the highest concentration detected was 105 MPN/g (dry weight) (10). Our study in 2012 (samples were analyzed from only one beach at SLBE) found a lower frequency of detection, where only 40% AlgOn samples contained bontE; only one sample was quantifiable at 1.24 × 105 gene copies/g (dry weight) (4,725 gene copies/ml of algal slurry). In our study, we found lake hydrodynamics, natural wetlands, storm drains, river outfalls, and barren lands influenced the occurrence of C. botulinum at the Great Lakes beaches.
The overall effect of lake hydrodynamics on C. botulinum occurrence in the Great Lakes beaches was clearly observed in this study. Wind speed, depth-averaged water velocity, and wave height may have a negative effect on the occurrence of bacteria in algae. Deland, Brimley, and Hammond beaches had above average wave heights and wind speeds; Linwood, Deland, Hammond, and Waukegan have higher depth-averaged water velocity than other sites. All samples from these sites were nonquantifiable, and only a low percentage of samples were above the LOD. In contrast, Maumee Bay, SLBE, and Bay City all had low wave heights and greater bontE detection frequency.
The effect of wind direction, lake currents, and beach orientation could explain the drastic difference in bontE detection at close-by locations. Linwood and Bay City are two beaches located only 8 km apart from one another. Linwood has a sheer bluff shoreline with a higher wind speed and depth-averaged current velocity, while Bay City has low waves, low current velocity, and an embayed shoreline. This embayed beach orientation and the lake hydrodynamics at Bay City may cause bontE to accumulate in algae, muck, and sediment and result in higher bontE concentrations in Bay City than in Linwood. The same conditions were observed at the Hammond and Jeorse beaches, which may explain the higher concentration of bontE at Jeorse than Hammond. In addition, we found that wind plays a part in the distribution of bontE at Bay City. Winds from both the northeast and southwest, typical at Bay City, were associated with quantifiable results at Bay City. The northeast wind could bring algae with bontE toward the beach, and a southwest wind could cause rainfall runoff or river effluent to bring C. botulinum to this area. Further intensive study of the hydrodynamic processes of the Great Lakes is essential for understanding the pathways that add bontE to the Great Lakes.
Natural wetlands may enhance the occurrence of C. botulinum. Tobico Marsh is a bird sanctuary that ultimately drains over the beach at Bay City (43). A study in Spain found that feces of water birds contained C. botulinum types C and D, and wetlands provide a good environment for type C and D proliferation (44). Bird fecal matter of Tobico Marsh may contain C. botulinum type E, and it is vital to conduct further studies to detect bontE in fecal matter and potential delivery to Saginaw Bay. In this study, we found that development within the beach catchment was greatest at beaches with less quantifiable bontE detection in algae. Hammond is a well-developed area with about 60% developed lands and only 2% wetlands. The catchments surrounding Deland, Portage, and Waukegan exhibit a similar pattern. Thus, undeveloped lands and natural wetland may enrich C. botulinum proliferation, while well-developed areas may not support the natural process of C. botulinum prevalence and survival.
Further, storm drains, river outfalls, and barren lands also may enrich C. botulinum occurrence. The Saginaw River (carrying about 80% of the basin nutrient discharge) (45) and storm drains, which have outfalls in the Bay City beach area, may also play an important role in adding C. botulinum into this bay. In addition, both the Jeorse and Portage beaches have river outfalls. Our hypothesis is further supported by a study in Spain, which found that wastewater treatment plant effluent was a good source for types C and D and other avian pathogens (44). Other than Bay City, Jeorse also has very high C. botulinum detections and concentrations. About 63% of this beach catchment area contains barren lands. When beaches have a high percentage of nearby barren lands, there is no barrier to prevent the entry of microorganisms into beach water from runoff.
Variability of bontE detection in different matrices.
In this study, we observed a high frequency of detection of bontE in AlgOn. Chun's group in 2011 found comparable results: 74% (n = 53) of the Cladophora samples were positive for bontE (10). Several other studies have also found that that algae support the growth and survival of C. botulinum; algae may provide protection from UV light and attachment surfaces. Algal exudates and thalli may also be good sources of nutrients for bacterial growth and survival (10, 23, 46). Further, decaying algae consume oxygen and provide ideal anoxic conditions for this organism to grow (46), since C. botulinum type E can grow in algal mats at 25°C (23).
Among the three matrices, the lowest concentration of bontE was found in water, but the frequency of detection is high, which suggests that bontE is well distributed in water. The bontE gene could be delivered to water by river or overland runoff, which may carry C. botulinum cells. Earlier studies have reported C. botulinum type E association with drainage and drainage basins (17, 25). The findings from Jeorse support our hypothesis that stream and storm water may carry C. botulinum cells, where bontE concentration in AlgOn was high during rain events and decreased following rain events; at other sites, no storm event samples were collected to identify such variability with rainfall.
Another possible way of adding bontE into water is that when there are strong waves, submerged algal mats may break apart, and C. botulinum attached to the mat may wash away and thus be added to the water (21). A negative correlation found between bontE in submerged algae and wave height supports this hypothesis. Beaches with quantifiable bontE in algae had a lower average wave height and lower current velocities than beaches without quantifiable bontE. In addition, strong waves may stir up sediment that contains C. botulinum and add those cells to water. However, bacteria typically prefer to attach to sediment or plants that may provide a suitable microenvironment for their growth and survival (47). Therefore, these organisms may eventually settle under calm conditions to sediment or attach to algae and result in a low concentration of bontE in water.
Detection of bontE in sediment showed site specificity. The beaches with low wave heights and wind velocities showed high levels of bontE detection in sediment. The wave heights at Maumee Bay were below average, and we found a higher frequency of detection, as well as higher concentrations of bontE in sediment (SED and SOS), at Maumee Bay than at other sites. Despite the high concentrations of bontE in sediments at Maumee Bay, we found that water at this beach had the lowest detection frequency compared to other sites. When the lake is calm, these cells could settle into the sediment as at Maumee Bay. bontE detected in SOS showed a positive correlation with wind speed, suggesting that waves with strong wind bring the C. botulinum to shore or cause it to release from algae to sediment.
Temporal variability of distribution of bontE.
Studying the temporal variability of C. botulinum will provide essential information about the organism's population dynamic, which will be useful for interpreting bird mortality trends. Various environmental or physicochemical conditions may govern the temporal variability at these study sites. The bontE concentration in water did not show considerable seasonal variability. At Bay City, high bontE concentrations were found in early summer and late summer. Overall, the highest bontE concentrations were documented in shoreline environments during July; Chun's study in 2011 also found high bontE concentrations in July (10). At Jeorse, high bontE concentrations were associated with rainfall events, which suggests that runoff water may carry bontE to the beaches or that hydrological variables such as currents or waves associated with storms may influence bontE delivery to the beach. A study with C. botulinum type C found that high pH (7.5 to 9.0), low redox potential (−300 to −200), and warmer temperature (<30°C) increase the risk of having type C outbreaks (29). In our study, we also found that bontE in AlgOn has a positive correlation with water temperature, salinity, and pH. A study conducted at Lake Erie found that a higher temperature, anoxic conditions near sediments, and a low redox potential favor C. botulinum type E growth (19, 28).
Bay City, a case study.
The highest concentration of bontE was detected at Bay City, especially in algae. Interestingly, the Saginaw Bay area experiences fewer bird deaths than SLBE despite high bontE detection, suggesting that bird mortality may not be related to toxin production that occurs on or near shore, that target bird groups are usually not present in this area, and/or that toxin transfer mechanisms (food webs) differ between these sites or that C. botulinum concentrations are not high enough to produce toxin. Kimura and others determined the lowest concentration of C. botulinum required for effective toxin production using a combination of mouse bioassays and qPCR and found that a minimum of 1.6 × 106 CFU/g corresponded to effective type E toxin production (48). In our study, only a few samples from Bay City AlgOn contained sufficient concentrations of bontE to theoretically indicate toxin production.
bontE detection in muck at Bay City had high correlations with several essential growth factors such as phosphorus and total dissolved solids (TDS). Muck is porous and may trap phosphorus, TDS, and other elements, which are needed for bacterial growth and survival (48). A study conducted at Bay City identified the possibility of regrowth of E. coli, Enterococcus, C. perfringens, and coliphages in muck (45). Similarly, E. coli and enterococci have been shown to grow in Cladophora extract or washings (20) and rehydrated algal mats (21) under laboratory conditions. Therefore, muck in Bay City may enhance the growth of C. botulinum, but further studies are needed to prove this hypothesis.
Summary.
We found that C. botulinum type E is ubiquitous, but it has temporal, spatial, and matrix variability. Wind speed, magnitude of depth-averaged water velocity, wave height, and beach characteristics played an important role in the distribution variability of C. botulinum at different sites and matrices in the Great Lakes. Algae provide better microenvironmental conditions, such as nutrients, anoxic conditions, and protection from UV light, for the prevalence of C. botulinum than do sediment and water. Further, terrestrial sources may significantly contribute C. botulinum type E population to the Great Lakes beaches. The results of this 1-year study provide insight for the future direction of C. botulinum studies. It is vital to conduct continuous studies for at least for 3 years in order to assess botulinum variability under different climatic and environmental conditions, the terrestrial input of C. botulinum, and its association with storm runoff, onshore and offshore linkage, and lake hydrodynamics. Finally, C. botulinum has a complex distribution pattern, and various physical, chemical, and environmental conditions influence its prevalence. Further, greater awareness of C. botulinum survival, sporulation, and vegetative cell production in different environmental matrices will elucidate this organism's complex distribution pattern in the environment.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Great Lakes Science Center beach monitors for field sample collection and to Lori Fuller of the U.S. Geological Survey (USGS) for assistance with downloading and processing of environmental data. We especially thank Brenda Lafrancois of the National Park Service and Muruleedhara Byappanahalli of the USGS for reviewing the manuscript and for their insightful comments. We thank the two anonymous reviewers for their comments, which helped to improve the quality of the manuscript.
This study was funded by the Great Lakes Restoration Initiative.
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00098-15.
REFERENCES
- 1.Shukula HD, Sharma SK. 2005. Clostridium botulinum: a bug with beauty and weapon. Crit Rev Microbiol 31:11–18. doi: 10.1080/10408410590912952. [DOI] [PubMed] [Google Scholar]
- 2.Peck MC. 2009. Biology and genomic analysis of Clostridium botulinum. Adv Microb Physiol 55:183–265. doi: 10.1016/S0065-2911(09)05503-9. [DOI] [PubMed] [Google Scholar]
- 3.Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. 1994. Bergey's manual of determinative bacteriology, 9th ed Springer, New York, NY. [Google Scholar]
- 4.Barash JR, Aron SS. 2014. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J Infect Dis 209:192–202. doi: 10.1093/infdis/jit450. [DOI] [PubMed] [Google Scholar]
- 5.Peck MC, Stringer SC, Carter AT. 2011. Clostridium botulinum in the post-genomic era. Food Microbiol 28:183–191. doi: 10.1016/j.fm.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Bott TL, Deffner JS, McCoy E, Foster EM. 1966. Clostridium botulinum type E in fish from Great Lakes. J Bacteriol 91:919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bott TL, Johnson J, Foster EM, Sugiyama H. 1968. Possible origin of high incidences of Clostridium botulinum type E in an inland bay (Green Bay of Lake Michigan). J Bacteriol 95:1542–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strom SM, Courtney JKBC, Meyer W, Langenberg JA. Surveillance of botulism E related mortality in watersheds of the Wisconsin Great Lakes. FWS agreement number 30181-9-G027 U.S. Fish and Wildlife Service, Washington, DC: http://www.fws.gov/midwest/fisheries/GLFWRA/30181-9-G027.pdf. [Google Scholar]
- 9.Lafrancois BM, Riley SC, Blehert DS, Ballmann AE. 2011. Links between type E botulism outbreaks, lake levels, and surface water temperatures in Lake Michigan, 1963–2008. J Great Lakes Res 37:86–91. doi: 10.1016/j.jglr.2010.10.003. [DOI] [Google Scholar]
- 10.Chun CL, Ochsner U, Byappanahalli MN, Whitman RL, Tepp WH, Lin GY, Johnson EA, Peller J, Sadowsky MJ. 2013. Association of toxin-producing Clostridium botulinum with the macroalga Cladophora in the Great Lakes. Environ Sci Technol 47:2587–2594. doi: 10.1021/es304743m. [DOI] [PubMed] [Google Scholar]
- 11.Kautter DA. 1964. Clostridium botulinum type E in smoked fish. J Food Sci 29:843–849. doi: 10.1111/j.1365-2621.1964.tb00458.x. [DOI] [Google Scholar]
- 12.Graikoski JT, Bowman EW, Robohm RA, Koch RA. 1970. Distribution of Clostridium botulinum in the ecosystem of the Great Lakes. U.S. Bureau of Commercial Fisheries, Ann Arbor, MI. [Google Scholar]
- 13.Fay LD, Kaufman OW, Ryel IA. 1965. Field observations and laboratory investigations concerning recent Lake Michigan bird mortalities. Report 25. Michigan Department of Conservation and Resource Development, Lansing, MI. [Google Scholar]
- 14.U.S. Geological Survey National Wildlife Health Center. 2014. New and ongoing mortality events nationwide. U.S. Geological Survey, Kearneysville, WV: http://www.nwhc.usgs.gov/mortality_events/ongoing.jsp. [Google Scholar]
- 15.Getchell RG, Culligan WJ, Kirchgessner M, Sutton CA, Casey RN, Bowser PR. 2006. Quantitative PCR assay used to measure the prevalence of Clostridium botulinum type E in fish in the lower Great Lakes. J Aquat Anim Health 18:39–50. doi: 10.1577/H05-013.1. [DOI] [Google Scholar]
- 16.Nol P, Rocke TE, Gross K, Yuill TM. 2004. Prevalence of neurotoxic Clostridium botulinum type C in the gastrointestinal tracts of tilapia (Oreochromis mosambicus) in the Salton Sea. J Wildl Dis 40:414–419. doi: 10.7589/0090-3558-40.3.414. [DOI] [PubMed] [Google Scholar]
- 17.Huss HH. 1980. Distribution of Clostridium botulinum. Appl Environ Microbiol 39:764–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannett GE, Stone WB, Davis SW, Wroblewski D. 2011. Biodiversity of Clostridium botulinum type E associated with a large outbreak of botulism in wildlife from Lake Erie and Lake Ontario. Appl Environ Microbiol 77:1061–1068. doi: 10.1128/AEM.01578-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Fuentetaja A, Clapsadl MD, Getchell RG, Bowser PR, Lee WT. 2011. Clostridium botulinum type E in Lake Erie: inter-annual differences and role of benthic invertebrates. J Great Lakes Res 37:238–244. doi: 10.1016/j.jglr.2011.03.013. [DOI] [Google Scholar]
- 20.Byappanahalli MN, Shively DA, Nevers MB, Sadowsky MJ, Whitman RL. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol Ecol 46:203–211. doi: 10.1016/S0168-6496(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 21.Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl Environ Microbiol 69:4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii S, Yan T, Shively DA, Byappanahalli MN, Whitman RL, Sadowsky MJ. 2006. Cladophora (Chlorophyta) spp. harbor human bacterial pathogens in nearshore water of Lake Michigan. Appl Environ Microbiol 72:4545–4553. doi: 10.1128/AEM.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byappanahalli MN, Whitman RL. 2009. Clostridium botulinum type E occurs and grows in the alga Cladophora glomerata. Can J Fish Aquat Sci 66:879–882. doi: 10.1139/F09-052. [DOI] [Google Scholar]
- 24.Laycock RA, Longard AA. 1972. Clostridium botulinum in sediments from Canadian Atlantic Seaboard. J Fish Res Board Can 29:443–446. doi: 10.1139/f72-073. [DOI] [Google Scholar]
- 25.Laycock RA, Loring DH. 1972. Distribution of Clostridium botulinum type E in gulf of St. Lawrence in relation to physical environment. Can J Microbiol 18:763–769. [DOI] [PubMed] [Google Scholar]
- 26.Higgins SN, Malkin SY, Howell ET, Guildford SJ, Campbell L, Hiriart-Baer V, Hecky RE. 2008. An ecological review of Cladophora glomerata (Chlorophyta) in the Laurentian Great Lakes. J Phycol 44:839–854. doi: 10.1111/j.1529-8817.2008.00538.x. [DOI] [PubMed] [Google Scholar]
- 27.Ando Y, Iida H. 1970. Factors affecting the germination of spores of C. botulinum type E. Japan J Microbiol 14:361–370. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Fuentetaja A, Clapsadl MD, Einhouse D, Bowser PR, Getchell RG, Lee WT. 2006. Influence of limnological conditions on Clostridium botulinum type E presence in eastern Lake Erie sediments (Great Lakes, USA). Hydrobiologia 563:189–200. doi: 10.1007/s10750-005-0011-1. [DOI] [Google Scholar]
- 29.Rocke TE, Euliss NH, Samuel MD. 1999. Environmental characteristics associated with the occurrence of avian botulism in wetlands of a northern California refuge. J Wildl Manage 63:358–368. doi: 10.2307/3802520. [DOI] [Google Scholar]
- 30.Hartig JH, Thomas RL. 1988. Development of plans to restore degraded areas in the Great Lakes. Environ Manage 3:327–347. [Google Scholar]
- 31.USEPA/USGS. 2012. National Hydrography Dataset Plus, NHDPlus version 2.1. U.S. Environmental Protection Agency/U.S. Geological Survey, Washington, DC: http://www.horizon-systems.com/NHDPlus/NHDPlusV2_home.php. [Google Scholar]
- 32.Fry J, Xian G, Jin S, Dewitz J, Homer C, Yang L, Barnes C, Herold N, Wickham J. 2011. Completion of the 2006 National Land Cover Database for the conterminous United States. Photogrammetric Eng Remote Sensing 77:858–864. http://digital.ipcprintservices.com/publication/?i=78634. [Google Scholar]
- 33.McDonald BE, Graziano TM. 2000. The National Precipitation Verification Unit (NPVU): operational implementation. National Weather Service, Weather Prediction Center, College Park, MD: http://www.wpc.ncep.noaa.gov/. [Google Scholar]
- 34.Environmental Data Discovery and Transformation. 2011. Access and integrate environmental observations for coastal decision support. EnDDaT/U.S. Geological Survey, Kearneysville, WV: http://cida.usgs.gov/enddat/. [Google Scholar]
- 35.Reference deleted.
- 36.USEPA. 2008. Great Lakes beach sanitary survey user manual. EPA-823-B-06-001 U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 37.U.S. Geological Survey, Lake Michigan Volunteer AMBLE (Avian Monitoring for Botulism Lakeshore Events) Team. 2012. Information for volunteers, beach monitoring protocol, and carcass collecting protocol. U.S. Geological Survey, Madison, WI: https://www.nwhc.usgs.gov/amble/files/AMBLE%20-%20Protocols.pdf. [Google Scholar]
- 38.Myre E, Shaw R. 2006. The turbidity tube: simple and accurate measurement of turbidity in the field. Michigan Technological University, Department of Civil and Environmental Engineering, Houghton, MI: http://www.cas.umn.edu/assets/pdf/Turbidity%20Tube.pdf. [Google Scholar]
- 39.NOAA/GLERL. 2015. Great Lakes Coastal Forecasting System (GLCFS). National Oceanic and Atmospheric Administration, Great Lakes Environmental Research Laboratory, Ann Arbor, MI: http://www.glerl.noaa.gov/res/glcfs/. [Google Scholar]
- 40.Wattiau P, Renard ME, Ledent P, Debois V, Blackman G, Agathos SN. 2001. A PCR test to identify Bacillus subtilis and closely related species and its application to the monitoring of wastewater biotreatment. Appl Microbiol Biotechnol 56:816–819. doi: 10.1007/s002530100691. [DOI] [PubMed] [Google Scholar]
- 41.Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 7:249–259. doi: 10.1111/j.1462-2920.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- 42.Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl Environ Microbiol 76:3255–3262. doi: 10.1128/AEM.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrouder KS. 1997. Tobico Marsh. Status of the Fishery Resource Report 97-1, 1997 Michigan Department of Natural Resources, Lansing, MI: http://www.michigan.gov/documents/dnr/97-1_463567_7.pdf. [Google Scholar]
- 44.Anza I, Vidal D, Laguna C, Díaz-Sánchez S, Sánchez S, Chicote A, Florín M, Mateo R. 2014. Eutrophication and bacterial pathogens as risk factor for avian botulism outbreaks in wetlands receiving effluents from urban wastewater treatment plants. Appl Environ Microbiol 80:4251–4259. doi: 10.1128/AEM.00949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saginaw Bay Coastal Initiative. Potential public health risks associated with pathogens in detritus material (“muck”) in Saginaw Bay. Saginaw Bay Coastal Initiative, Bay City, MI: http://www.deq.state.mi.us/documents/deq-ogl-sbci-SBReport-Final-HumanHealthRisks.pdf. [Google Scholar]
- 46.Hecky RE, Smith REH, Barton DR, Guildford SJ, Taylor WD, Charlton MN, Howell T. 2004. The near shore phosphorus shunt: a consequence of ecosystem engineering by dreissenids in the Laurentian Great Lakes. Can J Fish Aquat Sci 61:1285–1293. doi: 10.1139/f04-065. [DOI] [Google Scholar]
- 47.Utsumi M, Nojiri Y, Nozom YTW, Seki H. 1998. Dynamics of attached bacteria at the water-sediment interface in a mesotrophic swampy bog of Japan. J Oceanogr 54:179–184. doi: 10.1007/BF02751693. [DOI] [Google Scholar]
- 48.Kimura B, Kawasaki S, Nakano H, Fujii T. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl Environ Microbiol 67:206–216. doi: 10.1128/AEM.67.1.206-216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


