Abstract
Campylobacter jejuni, the leading bacterial cause of human gastroenteritis in the United States, displays significant strain diversity due to horizontal gene transfer. Conjugation is an important horizontal gene transfer mechanism contributing to the evolution of bacterial pathogenesis and antimicrobial resistance. It has been observed that heat shock could increase transformation efficiency in some bacteria. In this study, the effect of heat shock on C. jejuni conjugation efficiency and the underlying mechanisms were examined. With a modified Escherichia coli donor strain, different C. jejuni recipient strains displayed significant variation in conjugation efficiency ranging from 6.2 × 10−8 to 6.0 × 10−3 CFU per recipient cell. Despite reduced viability, heat shock of standard C. jejuni NCTC 11168 and 81-176 strains (e.g., 48 to 54°C for 30 to 60 min) could dramatically enhance C. jejuni conjugation efficiency up to 1,000-fold. The phenotype of the heat shock-enhanced conjugation in C. jejuni recipient cells could be sustained for at least 9 h. Filtered supernatant from the heat shock-treated C. jejuni cells could not enhance conjugation efficiency, which suggests that the enhanced conjugation efficiency is independent of secreted substances. Mutagenesis analysis indicated that the clustered regularly interspaced short palindromic repeats system and the selected restriction-modification systems (Cj0030/Cj0031, Cj0139/Cj0140, Cj0690c, and HsdR) were dispensable for heat shock-enhanced conjugation in C. jejuni. Taking all results together, this study demonstrated a heat shock-enhanced conjugation efficiency in standard C. jejuni strains, leading to an optimized conjugation protocol for molecular manipulation of this organism. The findings from this study also represent a significant step toward elucidation of the molecular mechanism of conjugative gene transfer in C. jejuni.
INTRODUCTION
Campylobacter jejuni is a leading bacterial cause of human gastroenteritis in the United States (1). Typical symptoms of human campylobacteriosis include fever, abdominal cramps, and watery diarrhea. In rare cases, patients may develop severe sequelae such as Guillain-Barré syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death (2). C. jejuni is highly prevalent in poultry, and contaminated poultry meat is a significant source of human campylobacteriosis (3). Despite the significance of C. jejuni in public health and food safety, there is still a lack of effective intervention strategies available to prevent and control Campylobacter infections in humans and animal reservoirs, primarily due to enormous strain diversity, complexity of pathogenesis, and emergence of antimicrobial resistance (3, 4).
It has been widely observed that genomic contents vary significantly in C. jejuni strains, although C. jejuni has a small and compact genome (1.6 to 1.8 Mbp with approximately 1,600 genes) (5). For example, 106 genes differentially existed between C. jejuni NCTC 11168 and CG8486 (5), and the size of the pan-genome (the sum of all genes found in a species) of C. jejuni is estimated to be 2,600 genes (6). It has been proposed that horizontal gene transfer (HGT) plays a critical role in the diversity of bacterial genomes, therefore contributing to bacterial evolution and adaptation in different environmental niches. Of the three general HGT mechanisms in bacteria (transformation, conjugation, and transduction) (7), transformation has been well documented in C. jejuni (3, 8). However, information concerning conjugation, an important mechanism for efficient HGT across species, domains, and even kingdoms, is still very limited in C. jejuni (9).
The plasmid or chromosome DNA from donor cells, which may carry foreign genetic components, could be transferred into recipient cells via a complicated conjugation process and then either exist as self-replicative units in recipient cells or integrate into the chromosomes of recipient cells. Thus, the genes involved in virulence and antibiotic resistance may disseminate from donor to recipient cells using this prevalent HGT mechanism. In Campylobacter, a virulence plasmid, pVir, which was found to contain homologs of type IV secretion systems, could be detected from 17% of 104 C. jejuni clinical isolates and was significantly correlated with the occurrence of bloody diarrhea in humans (10). Antibiotic resistance genes, such as kanamycin resistance markers aphA-3 and aphA-7 and tetracycline resistance gene tet(O), are usually located in transmissible plasmids (11). The kanamycin resistance phenotype could be cotransferred with tetracycline resistance by conjugation between C. jejuni strains (12). Another self-transmissible plasmid, pCC31, which carries the tetracycline resistance gene tet(O) and genes encoding homologs of type IV secretion systems, have been observed to involve conjugative transfer between different C. jejuni species (13). Seventy-three percent open reading frames (ORFs) (45/62) in CJIE3, an integrated element in the chromosome of Campylobacter jejuni RM1221, displayed homology to the ORFs on the megaplasmid from Campylobacter coli RM2228 (14), suggesting conjugation-mediated plasmid integration during Campylobacter evolution. Therefore, examination of the factors influencing conjugative gene transfer would help us better understand the pathogenesis, emergence of antimicrobial resistance, and strain diversity in Campylobacter.
C. jejuni NCTC 11168, a widely used standard C. jejuni strain and the first C. jejuni isolate for which the whole genome has been sequenced (15), displays high efficiency for natural transformation with donor DNA from the same species (16) but low transformation efficiency for a plasmid derived from different bacterial species (e.g., Escherichia coli) (17). In addition, it has been widely observed that C. jejuni NCTC 11168 displays extremely low conjugation efficiency, posing a challenge for molecular manipulation of this organism. In other bacteria, a temporary heat shock has been observed to enhance the efficiency of transformation (18, 19) and even the efficiency of conjugation (20). These previous findings prompted us to examine the effect of heat shock on C. jejuni conjugation using NCTC 11168 as a representative recipient strain in this study.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The following primary Campylobacter jejuni isolates were used for this study: human isolate NCTC 11168 (15), 81-176 (21), 81116 (22), PG1187-VC96, CG8486, BH-01-0142, CG8421 (generous gift from Patricia Guerry), and chicken isolate RM1221 (14), and 91 (23). The C. jejuni strains were grown routinely in Müller-Hinton (MH) broth or on agar at 42°C in a Heracell 150i Tri-Gas incubator (85% N2, 10% CO2, 5% O2) (Thermo Scientific). Escherichia coli DH5 strain containing RK212.2 helper plasmid (24) was grown routinely in Luria-Bertani (LB) broth with shaking (250 rpm) or on agar at 37°C overnight. When needed, culture media were supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (12.5 μg/ml), and chloramphenicol (6 μg/ml).
Construction of E. coli donor strain.
The triparental E. coli-C. jejuni interbacterial conjugation system, which includes an E. coli donor strain bearing a transmissible plasmid, an E. coli helper strain bearing a helper plasmid, and a C. jejuni recipient strain, has been established in previous studies (25). However, construction of a new E. coli donor strain containing both transmissible plasmid and helper plasmid for biparental conjugation would reduce the confounding factors and simplify the conjugation experiments as described in this study. To achieve this goal, the donor E. coli strain was constructed by transforming the shuttle vector pRY107 (26) into the E. coli DH5 strain containing RK212.2 helper plasmid (24) using a conventional transformation method (27). Transformants were selected on LB plates containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), and tetracycline (12.5 μg/ml). One colony, named JL1116, was selected, confirmed by plasmid extraction, and used for subsequent conjugation experiments.
Heat shock.
To treat cells with heat shock, the microcentrifuge tube containing 0.5 ml of C. jejuni recipient cells was incubated at 44, 46, 48, 50, 52, or 54°C in a water bath for 5, 15, 30, 45, or 60 min. Following the heat shock treatment, the microcentrifuge tubes were immediately transferred to a water bath at room temperature (25°C) and incubated for 2 min to cool down cells; the treated C. jejuni cells were subsequently mixed with E. coli donor cells for biparental conjugation as detailed below. The control cells that were not subjected to heat shock were also in the same type of tube, suspended in the same broth medium, and exposed to the same atmospheric conditions as the heat shock-treated cells. For each specific treatment, two independent experiments were performed with duplicate samples in each independent experiment.
Biparental conjugation.
The biparental conjugation procedure is similar to the triparental conjugation method (28). Briefly, 100 μl of logarithmic-phase C. jejuni culture was spread on MH plates for overnight incubation at 42°C under microaerophilic conditions. In the meantime, the E. coli donor culture was inoculated into LB broth supplemented with ampicillin, kanamycin, and tetracycline for overnight incubation at 37°C in a rotary shaker (250 rpm). Following overnight incubation, 500 μl of the E. coli donor culture was inoculated into 5 ml of LB broth supplemented with the same antibiotics for further incubation until the optical density at 600 nm (OD600) reached 1.2. The overnight recipient C. jejuni cells were harvested from MH plates using MH broth and adjusted to an OD600 of about 1.0 to 1.5. To set up conjugation, 0.5 ml of log-phase E. coli donor cells was pelleted, washed once with antibiotic-free MH broth, and finally suspended in 0.5 ml of MH broth, which was subsequently mixed with 0.5 ml of the harvested C. jejuni recipient cells. These donor and recipient cell mixtures were pelleted down and resuspended in 100 μl of MH broth, which was then spotted onto a MH agar plate until the liquid was completely absorbed. The MH plates containing cell mixtures were incubated for 7 h under microaerophilic conditions. Following incubation, the mixed C. jejuni-E. coli cells were harvested from MH agar plates using 700 μl MH broth. The cell suspension was serially diluted in MH broth, and dilutions were plated onto two different MH plates: one containing Campylobacter-specific selective agents (SR117E; Oxoid) to recover all C. jejuni recipient cells and the other containing the selective antibiotics plus kanamycin (50 μg/ml) to recover the C. jejuni transconjugants that have acquired the pRY107 plasmid from the E. coli donor. The conjugation efficiency was determined on the basis of the number of transconjugant CFU per recipient CFU. Each conjugation experiment was performed in two independent experiments with duplicate measurements in each experiment. Statistical analysis was performed as described in a previous publication (29). Specifically, the differences of conjugation frequencies under different conditions were analyzed using one-way analysis of variance (ANOVA) on arcsine-transformed data, followed by Student's t test performed using SAS (v9.4). A P value of <0.05 was considered significant.
To determine how long the phenotype of the heat shock-enhanced conjugation can persist, the C. jejuni NCTC 11168 recipient cells that were treated with the standard heat shock procedure (50°C for 30 min) were further incubated under normal Campylobacter growth conditions for variable lengths of time (0 to 24 h). Subsequently, the recipient cells were mixed with donor cells to determine conjugation efficiency as described above.
Natural transformation.
Natural transformation was performed by using the standard biphasic method of Wang and Taylor (8). Briefly, 500 μl of C. jejuni NCTC 11168 cells (OD600 of 0.5), which were harvested from MH plates using MH broth, was transferred to a 5.0-ml sterile polystyrene tube (Fisher brand) containing 1 ml of MH agar. Following 3 h of incubation of this biphasic culture under microaerobic conditions at 42°C, 1 μg cfrB::kan isogenic genomic DNA of NCTC 11168 cells (30) was added to a liquid culture, which was further incubated for 5 h under microaerobic conditions. To measure natural transformation efficiency, the cell suspension was serially diluted in MH broth, and dilutions were plated onto two different MH plates: plain MH plates to recover all C. jejuni recipient cells and selective plates containing kanamycin (50 μg/ml) to recover the C. jejuni transformants. The transformation efficiency was determined on the basis of the number of kanamycin-resistant colonies per microgram of DNA per recipient CFU. Three independent experiments were performed.
Preparation of cell-free heat-shocked spent medium.
The microcentrifuge tubes containing 0.5 ml of fresh harvested C. jejuni NCTC 11168 cells (OD600 of 1 to 1.5) were subjected to heat shock at 50°C for 1 h as described above. Following heat shock, the cells were pelleted through centrifugation at 13,000 rpm for 5 min. The supernatant, which potentially contains a secreted substance upon heat shock, was filtered through 0.22-μm sterile syringe filters (Millipore) to prepare sterile cell-free spent medium. Subsequently, the above freshly prepared C. jejuni recipient cells (0.5 ml) were pelleted, immediately suspended in 0.5 ml of the cell-free spent medium, and incubated at 42°C for 30 min under microaerophilic conditions. The spent-medium-treated recipient cells were mixed with E. coli donor cells for a conjugation experiment as described above.
Genomic and genetic analyses of CRISPR.
The clustered regularly interspaced short palindromic repeat (CRISPR) sequence analysis was performed in CRISPRcompar Web service (http://crispr.u-psud.fr/CRISPRcompar/). To construct an isogenic CRISPR mutant, the CRISPR array containing direct repeats and spacers were replaced with tetracycline resistance cassette TetO (31). Briefly, a 1.7-kb fragment was PCR amplified from C. jejuni NCTC 11168 using primer pair CRISPR_F (F stands for forward) and CRISPR_R (R stands for reverse) (Table 1) using PfuUltra II Fusion HS DNA polymerase (Stratagene), which was purified and ligated to the SmaI-digested pUC19. The recombinant plasmids were subsequently digested with SwaI (NEB) to release a 639-bp fragment containing a CRISPR array; the backbone fragment was then ligated with tetO PCR product. The resulting suicide plasmid was introduced into C. jejuni NCTC 11168 by natural transformation (13). Transformants were selected on MH plates containing 5 μg/ml of tetracycline, and the insertional mutation was confirmed by PCR using CRISPR_F/Tet_Seq_R (Tet stands for tetracycline, and Seq stands for sequencing) primer pairs (Table 1).
TABLE 1.
Major primers used in this study
| Primer | DNA sequence (5′–3′)a | Product size (bp) | Target gene or function |
|---|---|---|---|
| Cm_PacI_F | CCCTTAATTAATGCTCGGCGGTGTTCCTTT | 801 | Chloramphenicol resistance cassette |
| Cm_PacI_R | CCCTTAATTAAGCGCCCTTTAGTTCCTAAAG | ||
| CRISPR_F | CCACTTTTTCCACCCTTGAA | 1,718 | CRISPR locus |
| CRISPR_R | AGGGCTAGTGTTGATGATTGTG | ||
| TetOF | TTATTTTTGCATAAACAGATGATTAGTG | 2,308 | TetO from C. jejuni 81-176 |
| TetOR | GCAAGCTGTTAAGCTAACTTGT | ||
| Cj0031_F | ATTGAGGCTAAAAAGCCAAATTC | 1,588 | Cj0031 |
| Cj0031_R | GCTCTATCCAAAGCCTTAGCTTC | ||
| Cj0139_US_F | CTTATCCGTGATGGAGTTGTG | 1,000 | Cj0139 upstream 1 kb |
| Cj0139_US_R | GGCAAATTTCCCGGGTACCAAATCCTTAAATTATGTTATTTTTAA (SmaI) | ||
| Cj0140_DS_F | GATTTGGTACCCGGGAAATTTGCCTATCAAATGCACT (SmaI) | 1,000 | Cj0140 downstream 1 kb |
| Cj0140_DS_R | AAAGAAAAAGAAAAAAGTTCATTTTTAA | ||
| Cj0690c_US_F | TTACAAAAAAAAATAACACCACAAGAA | 1,000 | Cj0690c upstream 1 kb |
| Cj0690c_US_R | TAAATCTTTCATCCCGGGGGCTTTCTTTCTAAAGTATTTTTAAAAA (SmaI) | ||
| Cj0690c_DS_F | GAAAGAAAGCCCCCGGGATGAAAGATTTATATAGTTTTTAAAATTTAATTTT (SmaI) | 1,000 | Cj0690c downstream 1 kb |
| Cj0690c_DS_R | AGGAATTTTAGCAGATTTAAATGCG | ||
| HsdR_F | CAATAGCATGGCGGCTAAAT | 3,999 | HsdR |
| HsdR_R1 | TGGTGCAACTAAAATAGCAGGTT | ||
| Tet_Seq_R | CGGGACTGCTACTTTTTGTTC | Confirming insertional orientation | |
| Cm_Seq_F | TATTATGAGGAGGGCGGAAA | Confirming insertional orientation | |
| Cm_Seq_R | CAGGGCGTATTGCCAAAATA | Confirming insertional orientation |
Restriction sites are shown in boldface type and underlined in the primer sequence, and the restriction sites are identified in parentheses after the sequence.
Inactivation of RM genes.
Selected restriction-modification (RM) genes of C. jejuni were inactivated via the allelic exchange using suicide plasmids as described previously (21). Specifically, Cj0031 and hsdR mutants were constructed by insertional mutation with a chloramphenicol resistance cassette (cat). An approximately 1.6-kb fragment of Cj0031 and a 4.0-kb fragment of hsdR were amplified from C. jejuni NCTC 11168 using PfuUltra II Fusion HS DNA polymerase (Stratagene) and primer pair Cj0031_F/Cj0031_R and HsdR_F/HsdR_R1 (Table 1), respectively. These fragments were subsequently cloned into SmaI-digested pUC19. The recombinant plasmids were subsequently digested with SwaI (for Cj0031) and EcoRV (for hsdR) and then ligated with a blunt-ended cat PCR product. The resulting plasmids in which the orientation of cat is the same as that of the specific inserted RM gene were selected as desired suicide plasmids, which were then introduced into C. jejuni NCTC 11168 through natural transformation (8). Transformants were selected on MH plates containing 6 μg/ml of chloramphenicol, and the insertional mutation was confirmed by PCR using primer pairs of Cj0031_F/Cm_Seq_R (Cm stands for chloramphenicol) and Cm_Seq_F/HsdR_R1, respectively (Table 1).
Inactivation of Cj0139-Cj0140 (Cj0139-40) and Cj0690c were achieved by deletional mutation. The 1-kb upstream and 1-kb downstream fragments were amplified from C. jejuni NCTC 11168 using PfuUltra II Fusion HS DNA polymerase (Stratagene) and corresponding primer pairs (Table 1). The upstream and downstream fragments were fused together by overlapping PCR and subsequently cloned into SmaI-digested pUC19. The recombinant plasmids were digested with SmaI and ligated with the blunt-ended cat PCR product (32). The suicide plasmids, in which the orientation of cat is the same as that of inserted RM genes, were introduced into C. jejuni NCTC 11168 by natural transformation (8). Transformants were selected on MH plates containing 6 μg/ml of chloramphenicol, and the insertional mutation was confirmed by PCR using primer pairs Cj0139_US_F/Cm_Seq_R (US stands for upstream) and Cj0690c_US_F/Cm_Seq_R (Table 1).
RESULTS AND DISCUSSION
Conjugation efficiency varied in different C. jejuni strains.
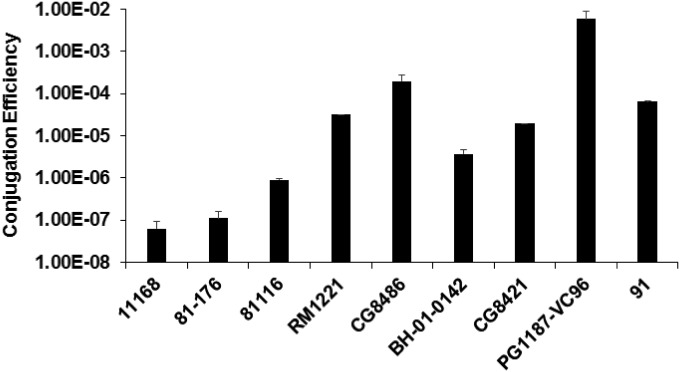
There is variation in conjugation efficiency in C. jejuni strains. The conjugation efficiencies of NCTC 11168 and 81-176, the two widely used standard C. jejuni strains, are (6.3 ± 2.9) × 10−8 and (1.1 ± 0.45) × 10−7CFU/recipient, respectively; the low conjugation efficiency also has been consistently observed in our previous molecular studies (30, 33). In contrast, some strains displayed significantly higher conjugation efficiency than strains NCTC 11168 and 81-176 did (P < 0.05). For example, the conjugation efficiencies of strains RM1221, CG8486, and PG1187-VC96 are (1.8 ± 0.32) × 10−5, (2.2 ± 0.59) × 10−4, and (6.0 ± 2.9) × 10−3 CFU/recipient, respectively.
Heat shock increased conjugation efficiency in C. jejuni NCTC 11168.
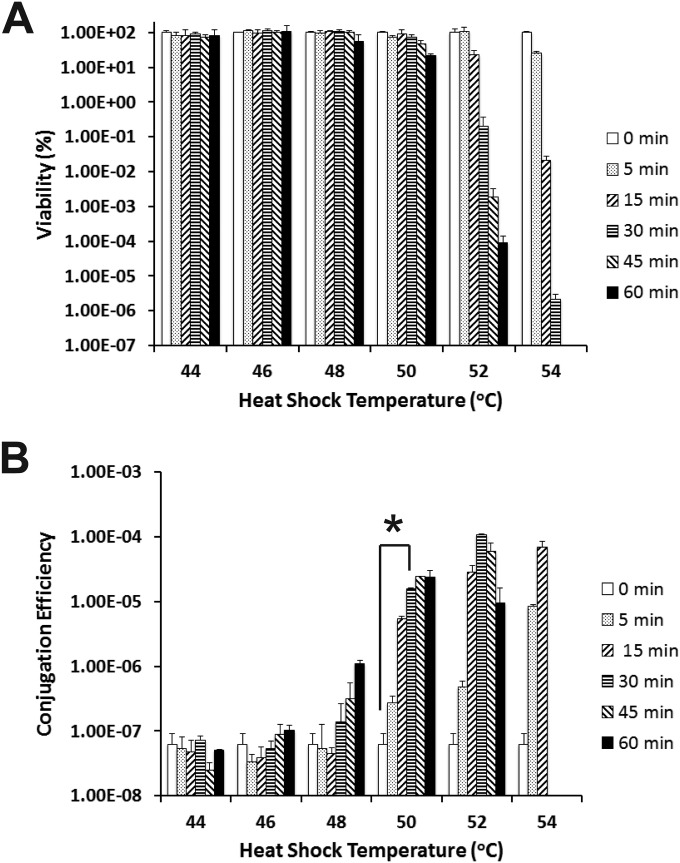
We chose NCTC 11168 as a representative strain to further comprehensively evaluate the effects of different heat shock treatments on conjugation in this study because of its extremely low conjugation efficiency (Fig. 1), its published genome (15), and established genetic tools for this standard strain. In this study, C. jejuni NCTC 11168 recipient cells were subjected to heat shock at different temperatures (44 to 54°C) and different incubation times (5 to 60 min). The corresponding viability and conjugation efficiency of each treatment were determined. As shown in Fig. 2A, treatment of NCTC 11168 cells at a temperature as high as 50°C and with an incubation time as long as 30 min did not greatly affect its viability. The significantly reduced viability (P < 0.05) due to heat shock was observed only for the cells treated for a longer time and/or at a higher temperature, such as 52°C for 30 min (Fig. 2A).
FIG 1.
Conjugation efficiency among C. jejuni strains. The conjugation efficiency was determined on the basis of the number of transconjugant CFU per recipient. Each bar represents the mean from two independent experiments with duplicate measurements for each experiment. The error bars indicate the standard deviations. The procedure for conjugation is detailed in Materials and Methods.
FIG 2.
Effects of heat shock on the viability and conjugation efficiency of C. jejuni NCTC 11168. (A) Viability of C. jejuni NCTC 11168 in response to different heat shock temperatures for different lengths of time (0 to 60 min). (B) Conjugation efficiency of C. jejuni NCTC 11168 in response to different heat shock temperatures for different lengths of time (0 to 60 min). NCTC 11168 recipient cells were not subjected to heat shock treatment (0 min) as a control. Each bar represents the mean from two independent experiments with duplicate measurements for each experiment. The error bars indicate the standard deviations. The value that was significantly different (P < 0.05) from the value for the untreated cells is indicated by an asterisk, and this heat shock treatment (50°C for 30 min) was used in subsequent experiments in this study.
Notably, although heat shock at a relatively narrow temperature range had little effect on cell viability (Fig. 2A), the conjugation efficiencies of treated cells have already been observed to increase significantly (Fig. 2B). For example, heat shock at 48°C for 60 min slightly decreased viability but significantly increased conjugation efficiency (more than 10-fold) (P < 0.05) (Fig. 2B). Similarly, heat shock at 50°C for 30 min dramatically increased conjugation efficiency approximately 100-fold (P < 0.05) (Fig. 2B). Heat shock at a higher temperature (52°C and 54°C) greatly reduced cell viability; however, conjugation efficiencies still increased dramatically. For example, the conjugation efficiency increased >1,000-fold increase after heat shock at 52°C for 30 min (P < 0.05) (Fig. 2). Since the heat shock at 50°C for 30 min had minimal effect on cell viability but led to significantly enhanced conjugation efficiency, this heat shock condition was chosen as the standard heat shock procedure for other experiments in this study.
Effect of heat shock on conjugation efficiency of other C. jejuni strains.
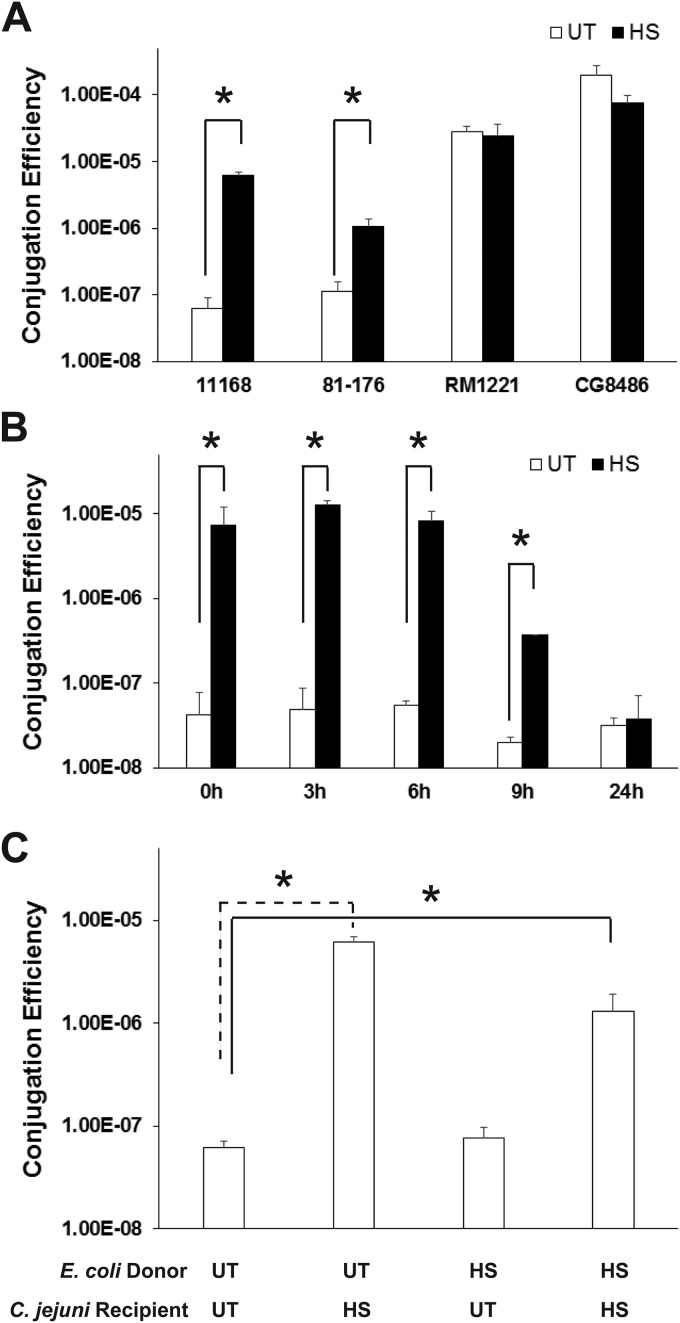
We then determined the effect of heat shock on conjugation efficiency of other C. jejuni strains. As shown in Fig. 3A, as in C. jejuni NCTC 11168, heat shock could significantly increase the conjugation efficiency of C. jejuni 81-176, another commonly used laboratory human isolate, by approximately 10-fold (P < 0.05). Thus, the findings from this study led to an optimized conjugation protocol for successful plasmid transfer in standard C. jejuni strains, enabling genetic manipulation of this bacterium, which is valuable to the Campylobacter research community.
FIG 3.
Characterization of heat shock-enhanced conjugation in C. jejuni. (A) Effect of heat shock (HS) on the conjugation of different C. jejuni isolates. (B) Persistence of the heat shock-enhanced conjugation phenotype in C. jejuni NCTC 11168. After being treated with heat shock (50°C for 30 min), the recipient NCTC 11168 cells were incubated at 42°C in the Tri-Gas incubator for different lengths of time prior to the conjugation experiment. The cells without heat shock treatment (untreated) were also subjected to the same treatment and used as a control. (C) The heat shock-enhanced conjugation is mediated through C. jejuni NCTC 11168 recipient cells only. For all experiments, each bar represents the mean from two independent experiments with duplicate measurements for each experiment. The error bars indicate the standard deviations. Values that are significantly different (P < 0.05) are indicated by a bar and asterisk. Abbreviations: UT, untreated control; HS, heat shocked at 50°C for 30 min prior to conjugation.
However, for the strains with high conjugation frequencies (RM1221 and CG8486) (Fig. 1), heat shock treatment did not lead to further enhanced conjugation efficiency (Fig. 3A). This strain-specific effect is likely due to the lack of a conjugation-limiting component in the C. jejuni strain displaying a high basal conjugation frequency; such a conjugation-limiting component, which is present in the strain with a low conjugation frequency (e.g., NCTC 11168) should be sensitive to heat treatment. This speculation needs to be examined in further research.
Persistence of the heat shock-enhanced conjugation efficiency.
As shown in Fig. 3B, the enhanced conjugation efficiency (approximately 100-fold; P < 0.05) consistently persisted after 6-h incubation under normal culture conditions. Despite the reduced conjugation efficiency by 9 h, the conjugation efficiency of the heat shock-treated cells was still 18-fold higher than that of the control cells (P < 0.05) (Fig. 3B). After 24 h of incubation, the conjugation efficiency of the heat shock-treated cells was comparable to that of the control cells (Fig. 3B).
Heat shock of donor E. coli cells did not enhance conjugation efficiency.
In the specific environment where conjugation occurs, both donor and recipient cells are very likely stressed by the same thermal temperature; this is different from the laboratory conjugation procedure described above in which E. coli donor cells were not subjected to heat shock. Therefore, we further investigated whether heat shock could also affect the E. coli donor cells, subsequently affecting conjugative DNA transfer to C. jejuni recipient cells. As shown in Fig. 3C, heat shock of the E. coli donor cells alone did not affect conjugation efficiency. Similar to the heat shock of C. jejuni recipient cells alone, heat shock of both donor and recipient cells also dramatically enhances the conjugation efficiency (more than 10-fold) (P < 0.05). This result clearly demonstrated that heat shock exerts its effect only on recipient cells for enhanced conjugation efficiency.
Heat shock reduced natural transformation efficiency.
It has been reported that heat shock could enhance transformation efficiency in some bacteria (18, 19, 27). Therefore, it is possible that heat shock may enhance the natural transformation efficiency of the C. jejuni recipient cells and then affect conjugation indirectly. In this study, we also determined the effect of heat shock on the natural transformation efficiency of C. jejuni NCTC 11168. Regardless of heat shock, the NCTC 11168 strain failed to take up the intact pRY107 plasmid extracted from E. coli (data not shown). Surprisingly, when C. jejuni genomic DNA was used as the donor DNA, heat shock treatment consistently led to significantly reduced (P < 0.05) natural transformation efficiency [(1.4 ± 0.54) × 10−5 CFU/μg DNA/recipient] compared to the control cells that were not heat shocked [(7.4 ± 0.94) × 10−5 CFU/μg DNA/recipient]. Thus, the natural transformation process did not contribute to the heat shock-enhanced conjugation efficiency observed in this study.
Heat shock-enhanced conjugation is independent of the CRISPR system.
The heat shock-enhanced conjugation in C. jejuni reported in this study provides a unique opportunity to further examine the molecular basis of conjugation from the side of the recipient. The first system we examined is the CRISPR system, a widespread bacterial adaptive defense mechanism against phage and plasmid invasion (34). Genomic analysis showed that C. jejuni NCTC 11168 contains a type II CRISPR locus, with three CRISPR-associated proteins, Cj1521c, Cj1522c, and Cj1523c, and a CRISPR array containing five direct repeat sequences (each 36 bp in length with consensus sequence GTTTTAGTCCCTTTTTAAATTTCTTTATGGTAAAAT) interspaced with four spacers (each 30 bp in length). Because C. jejuni 81-176, a strain also displaying heat shock-enhanced conjugation efficiency (Fig. 3A), does not contain a CRISPR based on genomic analysis (data not shown), the CRISPR may be dispensable for the heat shock-enhanced conjugation in C. jejuni. To further examine the role of CRISPRs in conjugation, an isogenic CRISPR mutant of strain NCTC 11168 was constructed, and it was observed to display a conjugation efficiency similar to that of the wild type regardless of heat shock treatment (data not shown). This genetic finding indicates that CRISPR is dispensable for the heat shock-enhanced conjugation in C. jejuni.
The heat shock-enhanced conjugation efficiency is independent of potential extracellular components.
It has been reported that pheromone, a secreted peptide, could regulate conjugative plasmid transfer through intercellular signaling system (35), consequently affecting conjugation efficiency. Thus, we also determined whether the heat shock-treated C. jejuni recipient cells could generate certain extracellular components that enhance conjugation efficiency. To test this, the cell-free spent medium from heat shock-treated recipient cells was prepared and used for conjugation experiments. However, treatment of recipient cells with the cell-free spent medium did not lead to enhanced conjugation efficiency (data not shown), suggesting that the pheromone-like system observed in other bacteria does not play a role in the heat shock-enhanced conjugation in C. jejuni.
Inactivation of selected RM genes did not affect heat shock-enhanced conjugation efficiency.
The RM systems are classical components controlling HGT in bacteria. Therefore, it is reasonable to speculate that heat shock might inactivate the restriction component of RM systems in recipient cells, which leads to more-intact DNA after the conjugative transfer process and consequently, an increased number of transconjugants. In Corynebacterium glutamicum ATCC 13032, a stress-sensitive type II restriction system has been identified to be responsible for the heat shock-enhanced conjugation (20). C. jejuni NCTC 11168 possesses up to seven sets of RM genes (17). Since both C. jejuni NCTC 11168 and 81-176 displayed heat shock-enhanced conjugation (Fig. 3A), the RM systems commonly existed in these two strains, including Cj0031 (type IIG), Cj0139-40 (type IV), Cj0690c (type IIG), and HsdR (type I), were chosen as priority targets for genetic analysis in this study.
However, inactivation of the selected individual RM gene target in this study had little effect on conjugation efficiency whether heat shock-treated cells or untreated cells were used (data not shown). This finding suggests that the heat shock-enhanced conjugation might be caused by other C. jejuni RM systems or the synergistic effect of the four tested RM systems. A previous study has showed that Cj1051c is a major restriction barrier for C. jejuni NCTC 11168 to acquire pMW10-derived plasmid through electroporation (17). However, Cj1051c is absent in the C. jejuni 81-176 genome, which suggests that Cj1051c may not be critical for heat shock-enhanced conjugation. Therefore, to determine the specific roles of C. jejuni RM systems in heat shock-enhanced conjugation, further mutagenesis work on the RM systems in C. jejuni (either single or multiple mutations within a single strain) is needed.
This study may also have important implications in agricultural practices for improving food safety and public health. It has been widely accepted that chicken is a major animal reservoir for C. jejuni. During the scalding and plucking practices in poultry processing, slaughtered chickens are immersed into 53°C water baths for 3 min (36). Based on the findings of this study, a temporary heat shock during processing may significantly increase conjugation efficiency in residual C. jejuni and lead to emergence of undesired resistant C. jejuni. Composting is another agricultural practice that could generate high temperatures; composting is widely used to treat livestock manures, including poultry litter. The internal temperature of the compost could reach 50°C within 18 h (37). The temperature in the core part of a cattle manure composting system can reach above 55°C for more than 35 days, and the temperature in the periphery manure layer can be above 40°C for 4 months. Despite the high temperature generated as a result of the composting process, viable C. jejuni could be detected even after 112 days in the periphery manure layer (38). Therefore, based on the findings of this study and the potential high load of C. jejuni in poultry litter (>109 CFU/g feces), C. jejuni may display enhanced ability for HGT during composting. Notably, animal litter could contain various antibiotic-resistant bacteria, either commensals or pathogens, such as Campylobacter, pathogenic E. coli, Salmonella, and Clostridium perfringens (39). Although composting may reduce the load of certain pathogens in litter, this practice may promote the emergence of an antibiotic-resistant pathogen, such as C. jejuni, due to the heat shock-enhanced conjugation observed in this study. This speculation needs to be examined in further epidemiological studies.
ACKNOWLEDGMENTS
Special thanks to Patricia Guerry for providing pRY107 and RK212.2 plasmids and C. jejuni isolates PG1187-VC96, CG8486, BH-01-0142, and CG8421. We are also grateful to Samantha Brown and Barbara Gillespie for providing technical support.
This work was supported by Agriculture and Food Research Initiative (AFRI) competitive grant 2012-68003-19679.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nachamkin I, Allos BM, Ho T. 1998. Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev 11:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young KT, Davis LM, DiRita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 4.Lin J. 2009. Novel approaches for Campylobacter control in poultry. Foodborne Pathog Dis 6:755–765. doi: 10.1089/fpd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poly F, Read T, Tribble DR, Baqar S, Lorenzo M, Guerry P. 2007. Genome sequence of a clinical isolate of Campylobacter jejuni from Thailand. Infect Immun 75:3425–3433. doi: 10.1128/IAI.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebure T, Bitar PD, Suzuki H, Stanhope MJ. 2010. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol 2:646–655. doi: 10.1093/gbe/evq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith HO, Danner DB, Deich RA. 1981. Genetic transformation. Annu Rev Biochem 50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J Bacteriol 172:949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyarzabal OA, Rad R, Backert S. 2007. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol 45:402–408. doi: 10.1128/JCM.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tracz DM, Keelan M, Ahmed-Bentley J, Gibreel A, Kowalewska-Grochowska K, Taylor DE. 2005. pVir and bloody diarrhea in Campylobacter jejuni enteritis. Emerg Infect Dis 11:838–843. doi: 10.3201/eid1106.041052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfredson DA, Korolik V. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett 277:123–132. doi: 10.1111/j.1574-6968.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- 12.Gibreel A, Skold O, Taylor DE. 2004. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb Drug Resist 10:98–105. doi: 10.1089/1076629041310127. [DOI] [PubMed] [Google Scholar]
- 13.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507–3517. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 14.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol 3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 16.Vegge CS, Brondsted L, Ligowska-Marzeta M, Ingmer H. 2012. Natural transformation of Campylobacter jejuni occurs beyond limits of growth. PLoS One 7:e45467. doi: 10.1371/journal.pone.0045467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt JP, Grant AJ, Coward C, Maskell DJ, Quinlan JJ. 2012. Identification of Cj1051c as a major determinant for the restriction barrier of Campylobacter jejuni strain NCTC11168. Appl Environ Microbiol 78:7841–7848. doi: 10.1128/AEM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawoud TM, Jiang T, Mandal RK, Ricke SC, Kwon YM. 2014. Improving the efficiency of transposon mutagenesis in Salmonella enteritidis by overcoming host-restriction barriers. Mol Biotechnol 56:1004–1010. doi: 10.1007/s12033-014-9779-4. [DOI] [PubMed] [Google Scholar]
- 19.Edwards RA, Helm RA, Maloy SR. 1999. Increasing DNA transfer efficiency by temporary inactivation of host restriction. Biotechniques 26:892–894. [DOI] [PubMed] [Google Scholar]
- 20.Schafer A, Kalinowski J, Puhler A. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl Environ Microbiol 60:756–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, Galan JE. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun 74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. 2007. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J Bacteriol 189:8402–8403. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang KV, Stern NJ, Saxton AM, Xu F, Zeng X, Lin J. 2011. Prevalence, development, and molecular mechanisms of bacteriocin resistance in Campylobacter. Appl Environ Microbiol 77:2309–2316. doi: 10.1128/AEM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller WG, Bates AH, Horn ST, Brandl MT, Wachtel MR, Mandrell RE. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl Environ Microbiol 66:5426–5436. doi: 10.1128/AEM.66.12.5426-5436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 28.Davis L, Young K, DiRita V. 2008. Genetic manipulation of Campylobacter jejuni. Curr Protoc Microbiol Chapter 8:Unit 8A.2.1–8A.2.17. doi: 10.1002/9780471729259.mc08a02s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beuls E, Modrie P, Deserranno C, Mahillon J. 2012. High-salt stress conditions increase the pAW63 transfer frequency in Bacillus thuringiensis. Appl Environ Microbiol 78:7128–7131. doi: 10.1128/AEM.01105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu F, Zeng X, Haigh RD, Ketley JM, Lin J. 2010. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J Bacteriol 192:4425–4435. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. 2011. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother 66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Xu F, Lin J. 2013. Specific TonB-ExbB-ExbD energy transduction systems required for ferric enterobactin acquisition in Campylobacter. FEMS Microbiol Lett 347:83–91. doi: 10.1111/1574-6968.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng X, Xu F, Lin J. 2009. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect Immun 77:5437–5448. doi: 10.1128/IAI.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 36.Lehner Y, Reich F, Klein G. 2014. Influence of process parameter on Campylobacter spp. counts on poultry meat in a slaughterhouse environment. Curr Microbiol 69:240–244. doi: 10.1007/s00284-014-0575-y. [DOI] [PubMed] [Google Scholar]
- 37.Macklin KS, Hess JB, Bilgili SF, Norton RA. 2006. Effects of in-house composting of litter on bacterial levels. J Appl Poultry Res 15:531–537. doi: 10.1093/japr/15.4.531. [DOI] [Google Scholar]
- 38.Xu W, Reuter T, Inglis GD, Larney FJ, Alexander TW, Guan J, Stanford K, Xu Y, McAllister TA. 2009. A biosecure composting system for disposal of cattle carcasses and manure following infectious disease outbreak. J Environ Qual 38:437–450. doi: 10.2134/jeq2008.0168. [DOI] [PubMed] [Google Scholar]
- 39.Dhanarani TS, Shankar C, Park J, Dexilin M, Kumar RR, Thamaraiselvi K. 2009. Study on acquisition of bacterial antibiotic resistance determinants in poultry litter. Poult Sci 88:1381–1387. doi: 10.3382/ps.2008-00327. [DOI] [PubMed] [Google Scholar]