Abstract
We examined the diversity and community structure of members of the halophilic Archaea (class Halobacteria) in samples from central and southern Tunisian endorheic salt lakes and sebkhet (also known as sebkha) systems using targeted 16S rRNA gene diversity survey and quantitative PCR (qPCR) approaches. Twenty-three different samples from four distinct locations exhibiting a wide range of salinities (2% to 37%) and physical characteristics (water, salt crust, sediment, and biofilm) were examined. A total of 4,759 operational taxonomic units at the 0.03 (species-level) cutoff (OTU0.03s) belonging to 45 currently recognized genera were identified, with 8 to 43 genera (average, 30) identified per sample. In spite of the large number of genera detected per sample, only a limited number (i.e., 2 to 16) usually constituted the majority (≥80%) of encountered sequences. Halobacteria diversity showed a strong negative correlation to salinity (Pearson correlation coefficient = −0.92), and community structure analysis identified salinity, rather than the location or physical characteristics of the sample, as the most important factor shaping the Halobacteria community structure. The relative abundance of genera capable of biosynthesis of the compatible solute(s) trehalose or 2-sulfotrehalose decreased with increasing salinities (Pearson correlation coefficient = −0.80). Indeed, qPCR analysis demonstrated that the Halobacteria otsB (trehalose-6-phosphatase)/16S rRNA gene ratio decreases with increasing salinities (Pearson correlation coefficient = −0.87). The results highlight patterns and determinants of Halobacteria diversity at a previously unexplored ecosystem and indicate that genera lacking trehalose biosynthetic capabilities are more adapted to growth in and colonization of hypersaline (>25% salt) ecosystems than trehalose producers.
INTRODUCTION
The class Halobacteria represents a physiologically and phylogenetically distinct lineage within the archaeal phylum Euryarchaeota. Members of the Halobacteria are encountered in a wide range of environments where their absolute requirement for salt is satisfied. Within various hypersaline (>25% salt), thalassohaline (e.g., crystallizer ponds in solar salterns), and athalassohaline (e.g., the Dead Sea, hypersaline lakes, and soda lakes) water bodies, members of the Halobacteria represent the majority of the cellular biomass (1–6). However, in environments with relatively lower salinity and/or fluctuating salinities, e.g., saline soils (salt plains and alpine salt sediments, soils adjacent to salt-processing plants), traditional Asian salted and fermented seafood products (e.g., jeotgal), and marine sponges, they usually coexist as a smaller fraction of the more diverse prokaryotic community inhabiting these settings (7–13). These habitats with moderate or low salinity and/or fluctuating salinity have been the source of species of many recently described novel Halobacteria taxa (14–18) and are partially responsible for the rapid expansion of recognized Halobacteria spp. during the last decade (19, 20).
Patterns of Halobacteria community structure have mostly been examined in a few model hypersaline habitats with relatively limited Halobacteria diversity. These studies documented the dominance of specific Halobacteria genera in high-pH soda lakes (genera Natronococcus, Natronobacterium, Natronomonas, Natrialba, Natronolimnobius, Natronorubrum, Halorubrum, Halalkalicoccus, and Halobiforma) (21, 22) and in Mg2+-rich water bodies (genera Halosarcina, Natronococcus, Halorhabdus, and Natronomonas) (23, 24), as well as in solar salterns and crystallizer ponds (genera Halorubrum, Haloquadratum, Halonotius, and Haloplanus) (1–3, 25). However, with the exception of these few model ecosystems, patterns and determinants of Halobacteria community structure remain poorly understood.
Given the current recognition of the wide range of Halobacteria phylogenetic diversity (26) and the novel habitats in which Halobacteria spp. are encountered (12, 14, 27–33), extrapolation of diversity and community structure studies to these atypical, nonhypersaline habitats is warranted. Such studies would expand our knowledge regarding overall diversity and ecological distribution within the Halobacteria and aid in deciphering the importance of various factors, e.g., salinity, physical characteristics, and geographical location, on shaping their diversity and community structure patterns. The relatively lower number of Halobacteria cells in such habitats often hinders the use of archaeal domain-wide 16S rRNA gene primers for their targeting, a common procedure in surveying Halobacteria diversity in hypersaline settings. To overcome this problem, we have recently designed, validated, and utilized Halobacteria-specific 16S rRNA gene primers for targeted high-throughput pyrosequencing. Further, we developed a pipeline for accurate phylogenetic assignment of obtained sequences to the genus level by comparison to a curated database of validly described Halobacteria species using BLASTN (12). Within saline and hypersaline ecosystems, the level of and spatiotemporal fluctuations in salinity obviously play an important role in selection of taxa (34), although the impact of other factors, e.g., pH, temperature, physical characteristics, availability of dissolved O2, redox potential, and ionic composition (35), could not be discounted.
To survive in high-salinity environments, cells maintain an intracellular osmotic pressure that is equal to or higher than that of the surrounding environment to prevent osmotically induced cell lysis (36). The most prevalent mechanism for osmoadaptation is “salting in,” where cells accumulate molar concentrations of potassium ions to counter the high extracellular osmotic pressure. This strategy appears to be universally adapted by all members of the Halobacteria (37, 38). In addition to salting in, some members of the Halobacteria maintain high intracellular osmotic pressure by synthesis and/or uptake of highly soluble organic solutes that do not interfere with intracellular enzymatic activities and cellular processes. We have recently demonstrated that multiple genera within the Halobacteria biosynthesize and accumulate molar levels of trehalose (or 2-sulfotrehalose) as an osmoadaptive compatible solute (37). Currently, the impact of the possession (or lack thereof) of such a capability within members of the Halobacteria on their ecological fitness, habitat preferences, and, consequently, the overall Halobacteria community structure within a specific saline or hypersaline habitat is unclear.
Here, we sought to examine the diversity and community structure of members of the class Halobacteria in samples from central and southern Tunisian endorheic salt lakes and sebkhet (also known as sebkha) systems using targeted 16S rRNA gene diversity survey and quantitative PCR (qPCR) approaches. We further investigated whether the possession of trehalose biosynthetic capacity is an ecologically relevant trait that impacts fitness and niche colonization process within the Halobacteria. Our results suggest that the possession of trehalose biosynthetic capacity, or lack thereof, is an ecologically relevant trait, with genera possessing the machinery for trehalose biosynthesis as an osmoadaptive strategy appearing to be less suited for survival and propagation at higher salinities.
MATERIALS AND METHODS
Location and sampling.
A total of 23 samples from 4 different saline systems were obtained (Table 1; see also Fig. S1 in the supplemental material). Briefly, these systems are as follows: (i) Chott El-Djerid (33°56.977′N, 8°25.279′E), a large endorheic salt lake located in southwestern Tunisia (10 samples); (ii) Chott El-Fejej (33°49.96′N, 9°2.025′E), a long narrow inlet to Chott El-Djerid (4 samples); (iii) Sebkhet El-Melah (33°23.655′N, 10°55.745′E), a salt flat southwest of Zarzis (2 samples); and (iv) Sebkhet Douz (33°27.469′N, 9°0.465′E), a salt flat located in south central Tunisia (7 samples). Temperature and pH were recorded on site, and ∼50 g was sampled (using sterile spatulas) into sterile Falcon tubes, placed on ice, and transported to the laboratory, where the samples were kept frozen (−20°C) until DNA extraction. Salinities were measured using a hand-held SW series VistaVision refractometer (VWR, Radnor, PA) as previously described (39) and ranged from very low (2% to 3%, 3 samples from 1 site) to low (5% to 6.6%, 4 samples from 2 sites), medium-low (7.6% to 9.5%, 2 samples from 2 sites), medium-high (10.5% to 12.7%, 3 samples from 3 sites), high (13% to 14%, 4 samples from 2 sites), very high (30%, 2 samples from 1 site), and saturated (37%, 5 samples from 1 site). The physical characteristics of the samples differed between sediment (usually located below a layer of salt crust; n = 15, from 4 sites), saline water (n = 2, from 2 sites), and salt crust (n = 5, from 1 site) samples and 1 biofilm sample.
TABLE 1.
Sample description, numbers of sequences and OTUs, and diversity indices of datasets analyzed
| Sample collection site | Sample |
Data seta |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of sequences | No. of OTUs |
Rarefaction rank |
Good's coverage index |
Simpson index |
Shannon index |
Avg Bray-Curtis index |
||||||||||
| Name | Description | Salinity (%) | 0.03 | 0.06 | 0.03 | 0.06 | 0.03 | 0.06 | 0.03 | 0.06 | 0.03 | 0.06 | 0.03 | 0.06 | ||
| Chott El-Djerid | S1 | Salt crust | 37 | 617 | 53 | 25 | 2 | 1.5 | 0.94 | 0.98 | 0.07 | 0.15 | 3.20 | 2.34 | 0.22 | 0.36 |
| S2 | Salt crust | 37 | 322 | 12 | 8 | 1 | 1.5 | 0.96 | 0.98 | 0.2 | 0.24 | 1.74 | 1.57 | 0.18 | 0.26 | |
| S3 | Salt crust | 37 | 3,956 | 668 | 132 | 6 | 4 | 0.88 | 0.99 | 0.02 | 0.11 | 5.28 | 3.22 | 0.14 | 0.26 | |
| S4 | Sediment | 37 | 455 | 74 | 34 | 7.5 | 7 | 0.88 | 0.96 | 0.06 | 0.09 | 3.52 | 2.77 | 0.19 | 0.30 | |
| S5 | Sediment | 13.1 | 1,845 | 56 | 23 | 13 | 13 | 0.97 | 1.00 | 0.10 | 0.13 | 3.06 | 2.47 | 0.20 | 0.33 | |
| S6 | Sediment | 13.4 | 1,927 | 317 | 139 | 11.5 | 10 | 0.84 | 0.94 | 0.02 | 0.05 | 4.77 | 3.57 | 0.23 | 0.37 | |
| S7 | Sediment | 10.5 | 1,766 | 143 | 39 | 15 | 14 | 0.93 | 0.99 | 0.03 | 0.08 | 3.93 | 2.88 | 0.22 | 0.37 | |
| S10 | Water | 30 | 743 | 324 | 111 | 4 | 6 | 0.66 | 0.92 | 0.01 | 0.04 | 5.21 | 3.76 | 0.18 | 0.33 | |
| S11 | Salt crust | 37 | 1,363 | 297 | 86 | 5 | 4 | 0.85 | 0.98 | 0.02 | 0.06 | 4.93 | 3.41 | 0.25 | 0.38 | |
| S12 | Salt crust | 37 | 6,543 | 414 | 164 | 3 | 4 | 0.92 | 0.97 | 0.06 | 0.13 | 3.98 | 2.72 | 0.15 | 0.22 | |
| Sebkhet Douz | S13 | Water | 9.4 | 8,284 | 515 | 129 | 11.5 | 15 | 0.93 | 0.99 | 0.02 | 0.06 | 4.78 | 3.30 | 0.12 | 0.20 |
| S14 | Sediment | 6.7 | 522 | 62 | 31 | 18.5 | 18.5 | 0.89 | 0.96 | 0.09 | 0.12 | 3.18 | 2.57 | 0.11 | 0.18 | |
| S15 | Sediment | 14.1 | 1,959 | 355 | 91 | 7.5 | 10 | 0.87 | 0.98 | 0.02 | 0.10 | 4.89 | 3.20 | 0.16 | 0.31 | |
| S16 | Biofilm | 2.9 | 108 | 12 | 7 | 22.5 | 23 | 0.91 | 0.99 | 0.21 | 0.26 | 1.84 | 1.51 | 0.08 | 0.10 | |
| S17 | Sediment | 2.7 | 695 | 55 | 24 | 22.5 | 20 | 0.96 | 0.99 | 0.06 | 0.10 | 3.36 | 2.59 | 0.13 | 0.23 | |
| S18 | Sediment | 13.7 | 4,520 | 1,512 | 283 | 9.5 | 10 | 0.76 | 0.97 | 0.01 | 0.11 | 6.33 | 3.44 | 0.14 | 0.27 | |
| S19 | Sediment | 2.2 | 1,532 | 303 | 107 | 20 | 21 | 0.85 | 0.96 | 0.02 | 0.05 | 4.82 | 3.57 | 0.17 | 0.30 | |
| Sebkhet El-Melah | S40 | Sediment | 7.6 | 402 | 132 | 51 | 14 | 17 | 0.75 | 0.94 | 0.04 | 0.10 | 4.10 | 2.97 | 0.06 | 0.1 |
| S42 | Sediment | 10.8 | 5,287 | 485 | 103 | 9.5 | 12 | 0.92 | 0.99 | 0.03 | 0.10 | 4.56 | 2.80 | 0.06 | 0.1 | |
| Chott El-Fejej | S26 | Sediment | 6.6 | 588 | 177 | 66 | 21 | 18.5 | 0.81 | 0.95 | 0.03 | 0.09 | 4.41 | 3.16 | 0.22 | 0.35 |
| S27 | Sediment | 5.5 | 325 | 50 | 27 | 17 | 22 | 0.89 | 0.97 | 0.06 | 0.10 | 3.25 | 2.68 | 0.17 | 0.31 | |
| S28 | Sediment | 6.3 | 243 | 110 | 39 | 18.5 | 16 | 0.66 | 0.94 | 0.02 | 0.08 | 4.31 | 3.00 | 0.19 | 0.32 | |
| S29 | Sediment | 12.7 | 3,133 | 735 | 228 | 16 | 8 | 0.82 | 0.95 | 0.02 | 0.12 | 5.41 | 3.30 | 0.09 | 0.15 | |
Rarefaction curves were used at both 97% (species level [0.03]) and 94% (genus level [0.06]) to rank the diversity of the samples. Samples whose rarefaction curves lie at the top are considered more diverse than samples whose rarefaction curves lie at the bottom. Samples were given diversity rankings ranging from the least diverse (rank 1) to the most diverse (rank 23). Good's coverage index, the Shannon index of sample diversity, and the Simpson index of sample evenness are shown at both the species (0.03) and genus (0.06) levels. The Bray-Curtis index of β diversity between samples was calculated for all possible pairwise comparisons for samples from each site. The average Bray-Curtis index is shown at both the species (0.03) and genus (0.06) levels.
DNA extraction, PCR amplification, sequencing, and analysis.
DNA was extracted using a PowerSoil DNA extraction kit (MoBio, Carlsbad, CA) following the manufacturer's instructions and quantified using a Qubit fluorometer (Life Technologies, Grand Island, NY). For 16S rRNA gene amplification and pyrosequencing, the extracted DNA was used as a template in PCRs that contained the Halobacteria-specific 287F and 589R primers (Table 2). The forward primer was constructed by adding 454 Roche FLX adaptor A (GCCTCCCTCGCGCCATCAG) to the 287F primer as previously described (12). The forward primer also contained a unique bar code (octamer) sequence for multiplexing (12). The reverse primer was constructed by adding 454 Roche FLX adaptor B (GCCTTGCCAGCCCGCTCAGT) to the 589R primer. PCR analysis was performed in 50-μl reaction mixtures that contained 2 μl of the extracted DNA, 1× PCR buffer (Promega, Madison, WI), 2.5 mM MgSO4, a 0.2 mM deoxyribonucleoside triphosphate (dNTP) mixture, 0.5 U of GoTaq flexi DNA polymerase (Promega, Madison, WI), and a 10 μM concentration of each of the forward and reverse primers. PCR was carried out according to the following protocol: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 55°C for 1 min, and elongation at 72°C for 1 min. A final elongation step at 72°C for 10 min was included. All samples were run in at least triplicate, and the resulting PCR products of the expected size were gel purified using a QIAquick gel extraction kit (Qiagen Corp., Valencia, CA) and pooled to give a total of 3 to 5 μg of DNA per sample. Pyrosequencing was performed on a Roche 454-Junior sequencer at the Oklahoma State University Biochemistry and Molecular Biology core facility.
TABLE 2.
Primers used in this study
| Name | Sequence (5′–3′) | Gene amplified | Use(s) | Reference or source |
|---|---|---|---|---|
| 287F | AGGTAGACGGTGGGGTAAC | Halobacteria-specific 16S rRNA gene | Pyrosequencing and qPCR | 12 |
| 589R | RGCTACGRACGCTTTAGGC | |||
| OtsB-F | GAYTTCGACGGBWCCCT | Halobacteria-specific otsB (trehalose-6-phosphatase) gene | qPCR | This study |
| OtsB-R | GGBAAYCACGGNYTSGA |
Sequence quality filtering, OTU identification, and phylogenetic assignments.
Sequence quality control was handled in the program mothur (40) as described previously (12). Briefly, sequences with an average quality score below 25, sequences that did not have the exact primer sequence, sequences that contained an ambiguous base (N), sequences having a homopolymer stretch longer than 8 bases, and sequences shorter than 80 bp were considered of poor quality and removed from the data set. High-quality reads from each sample were aligned against the SILVA alignment database available at the mothur website as a template using a Needleman-Wunsch pairwise alignment algorithm. Filtered alignments were used to generate an uncorrected pairwise distance matrix, followed by binning the sequences into operational taxonomic units (OTUs) at 3% and 6% cutoffs, corresponding to the species and genus levels, respectively. Rarefaction curves were computed in mothur using a resampling-without-replacement approach. Good's coverage indices were also calculated in mothur.
For phylogenetic placement, all sequences were queried using the BLASTN function of the downloaded NCBI standalone BLAST program (version 2.2.26) against a data set of 207 16S rRNA gene sequences representing the validly published species within the class Halobacteria (as of October 2014). A sequence was assigned to a specific genus if it was at least 94.0% similar to the reference 16S rRNA gene sequence belonging to that genus. Sequences with percent similarity of <94% to any known validly described genus were considered novel. Percentages of abundances of genera in each sample were used to construct a heat map for genera representation using the phyloseq package in R (41).
NMDS.
To examine patterns of genus-level co-occurrence and compare community structures and memberships between different data sets, sequences from all samples within a single site (range, 4,289 to 19,537; average, 11,783 ± 7,905) were pooled and binned into OTUs at the putative genus (6%) level. Membership patterns within these OTUs were used to compute pairwise diversity estimates. Bray-Curtis indices (at the putative genus level) were calculated for all possible sample pairs. The indices were then employed in constructing nonmetric multidimensional scaling (NMDS) plots using the command nmds in mothur. The obtained axes for all samples across all sites studied were represented on the same scatter plot, and the proximities of sample points to each other in ordination space were used as an indication of the similarity in community structure.
Deducing the proportion of trehalose-producing genera using 16S rRNA gene pyrosequencing data sets.
Our previous study (37) has provided experimental and genomic evidence that multiple Halobacteria genera are capable of trehalose or 2-sulfotrehalose biosynthesis as an osmoadaptive strategy and that the otsAB operon (encoding trehalose-6-phosphate synthase/trehalose-6-phosphatase) mediates trehalose biosynthesis in the Halobacteria. Further, analysis of the occurrence patterns of otsAB genes demonstrates a distinct phylogenetic pattern, where the otsAB operon was identified in all members of Halobacteria clade I (as defined in references 42 and 43), as well as in the genera Halococcus, Haladaptatus, Halalkalicoccus, and Halosimplex. On the other hand, otsAB was absent in all members of Halobacteria clade II (42, 43) and in all members of Halorhabdus-Halomicrobium-Haloarcula clade III (as defined in reference 44), as well as within the genera Halobacterium and Natronomonas. This distinct pattern allows estimation of the proportion of otsAB-bearing versus otsAB-lacking genera within a specific sample using 16S rRNA genus-level assignments. A list of otsAB-harboring versus otsAB-lacking genera is presented in Table S1 in the supplemental material. Using the genus-level assignments obtained for each sample and the otsAB distribution patterns described above, we calculated the relative abundances of otsAB-harboring genera in all data sets examined.
Nevertheless, the presence or absence of the otsAB operon could not be deduced for 18 genera that were not evaluated previously in prior studies and that do not have representatives with sequenced genomes. For those genera, a phylogenetics prediction system based on 16S rRNA sequence phylogeny was implemented. Genera within this group that phylogenetically belong to clade I, or are closely related to any of the genera Halococcus, Haladaptatus, and Halalkalicoccus, were predicted to harbor the otsAB system. These include the genera Haloarchaeobius, Halorubellus, Halorussus, Natronoarchaeum, Salinarchaeum, and Salinirubrum. Those genera phylogenetically belonging to clade II or to clade III or that are closely related to Halobacterium, or Natronomonas, were predicted to lack the otsAB system. These include the genera Halapricum, Haloarchaeum, Halobellus, Halolamina, Halomarina, Halomicroarcula, Halonotius, Halopelagius, Halopenitus, Halorientalis, Halovenus, and Salarchaeum. These two groups of genera were labeled “predicted otsAB-harboring” genera and “predicted otsAB-lacking” genera, respectively (see Table S1 in the supplemental material). Using this system, all Halobacteria genera were classified into otsAB-harboring genera (17 genera), predicted otsAB-harboring genera (6 genera), otsAB-lacking genera (10 genera), and predicted otsAB-lacking genera (12 genera) (see Table S1).
Quantification of total Halobacteria community and otsAB-harboring community.
The total Halobacteria community within each sample was quantified using a qPCR protocol targeting the Halobacteria 16S rRNA gene. The same primer pair (287F and 589R) used in pyrosequencing-based diversity survey was utilized in qPCR.
The abundance of Halobacteria community capable of synthesizing trehalose was assessed by quantifying otsB gene copy numbers. Specific otsB primers were designed in Primrose (45) on the basis of all available Halobacteria otsB gene sequences (n = 48) (June 2014). The specificity of the primer pair was initially evaluated in silico by comparison to the nr database using BLASTN (46) with the exclusion of class Halobacteria. Specificity of the primer pair was further experimentally verified in two samples (S1 and S19) by PCR amplification, cloning of the PCR product, and sequencing of 12 clones randomly selected from each sample. Primer sequences are shown in Table 2.
qPCR was conducted using a MyIQ thermocycler (Bio-Rad Laboratories, Hercules, CA) and SybrGreenER qPCR mix (Life Technologies, Carlsbad, CA). The same amplification protocol was used to quantify both 16S rRNA and otsB genes. The 25-μl reaction mixtures contained 2 μl of DNA template, 0.5 μM (each) forward and reverse primers, and 10 μl of the qPCR mix. The reactions were heated at 50°C for 2 min, followed by heating at 95°C for 8.5 min. This was followed by 65 cycles, with one cycle consisting of 15 s at 95°C, 60 s at 52°C, and 30 s at 72°C. Haladaptatus paucihalophilus strain DX253T genomic DNA was used as a positive control, as well as to construct a standard curve to deduce the gene copy number/mg DNA. For each sample, the threshold cycle (CT) values obtained for the 16S rRNA gene and the otsB gene were used to calculate the corresponding gene copy number as well as the fraction of the otsAB system-harboring Halobacteria community (calculated as the ratio of the otsB/16S rRNA gene copy numbers).
RESULTS
Sampling results.
Twenty-three distinct samples were analyzed (Table 1; see also Fig. S1 in the supplemental material). These samples belong to 4 different ecosystems in central and southern Tunisia, display different physical characteristics (ranging between salt crystals, hypersaline water, sediments below salt crusts, and biofilm samples), and range in salinities from 2.2% to 37% (Table 1). While some of the samples yielded only a relatively low number of sequences, the calculated genus-level coverage for all samples was always >92% (average, 96.8%) (Table 1).
Halobacteria community and genus-level assignments.
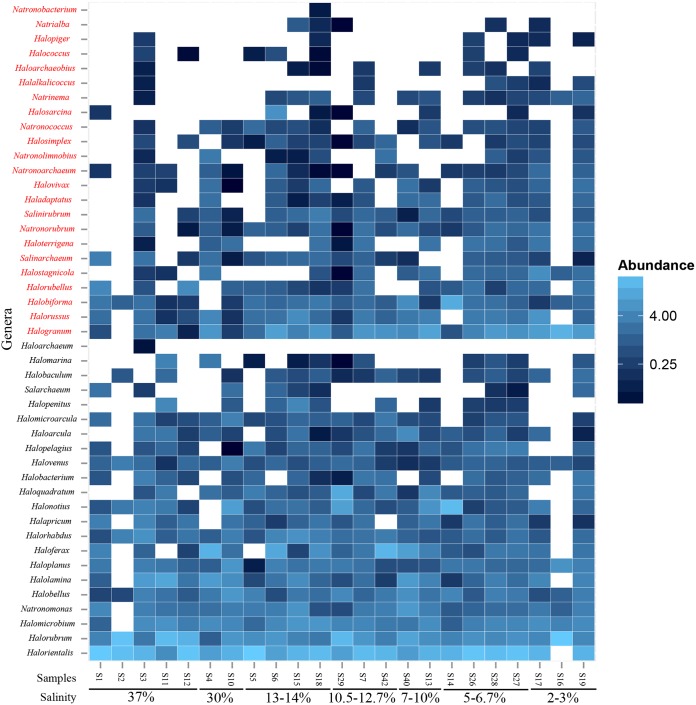
A total of 45 Halobacteria genera were identified in the entire data set, attesting to the indiscriminant performance of the Halobacteria-specific primers used. No non-Halobacteria-affiliated sequences were identified in all data sets. The number of genera within each sample ranged between 8 and 43 (average, 30), indicating the high level of Halobacteria phylogenetic diversity within each sample (Fig. 1; see also Table S2 in the supplemental material). Sequences unaffiliated with currently recognized Halobacteria genera represented only 14% of all sequences. In spite of the large number of genera identified per sample, a general pattern was observed in which a low number of genera always represented the majority of sequences encountered followed by a long tail of less-abundant genera. For example, within each sample, sequences belonging to the three most abundant genera represented 38% to 92% of the community and those belonging to the five most abundant genera represented 51% to 97.8%.
FIG 1.
Heat map of percent abundance of Halobacteria genera in analyzed samples. Salinity is shown for different samples. Genera in red are those known or predicted to harbor the otsAB system for trehalose biosynthesis.
Depending on their occurrence and relative abundance, Halobacteria genera observed in this study could be broadly classified into the following groups (see Table S3 in the supplemental material).
(i) Consistently abundant genera (group 1).
These genera (Halorientalis, Halorubrum, and Halogranum) represented >10% of the community in a few samples (n > 4) and represented >5% of the community in the majority of the samples (see Table S3 in the supplemental material).
(ii) Moderately abundant genera (group 2).
These genera (Haloferax, Halonotius, Halobiforma, Haloquadratum, and Halolamina) represented >10% of the community in a few samples (n < 4) and represented 1% to 5% of the community in the majority of samples (see Table S3 in the supplemental material).
(iii) Genera with occasional moderate abundance (group 3).
These genera (Natronomonas, Halobellus, Halomicrobium, Haloplanus, Halorussus, Halorhabdus, Halostagnicola, and Halorubellus) represented >5% of the community in 1 to 2 samples and represented 1% to 5% of the community in the majority of samples (see Table S3 in the supplemental material).
(iv) Consistently low-abundance genera (group 4).
These genera (Halobacterium, Haloterrigena, Halovenus, Halopelagius, and Halapricum) represented 1% to 5% of the community in the majority of samples (see Table S3 in the supplemental material).
(v) Rare genera with occasional low abundance (group 5).
These genera (Natronorubrum, Salarchaeum, Halovivax, Haladaptatus, Halobaculum, Natronoarchaeum, Natronococcus, Salinarchaeum, Salinirubrum, Natrinema, Natronolimnobius, Halomarina, Halosimplex, and Halomicroarcula) represented 1% to 5% of the community in just a few samples (n = 2 to 8) and represented <1% in the remaining samples (see Table S3 in the supplemental material).
(vi) Consistently rare genera (group 6).
These genera (Natrialba, Halalkalicoccus, Halococcus, Haloarchaeobius, Halopiger, Natronobacterium, and Haloarchaeum) always represented <1% of the total community and were encountered in only a few samples (n = 1 to 8) (see Table S3 in the supplemental material).
Diversity estimates and patterns.
Various diversity estimates (Shannon diversity index and Simpson evenness index for taxonomic alpha diversity; Bray-Curtis and rarefaction curve ranking for beta diversity) were computed (Table 1). Rarefaction curve-based ranking was chosen for comparative diversity purposes since this approach overcomes bias originating from variations in the size of data sets (47). The computed diversity ranks were clearly negatively correlated with salinity in the entire data set at both the genus (Pearson correlation coefficient = −0.92) and species (Pearson correlation coefficient = −0.90) levels (Fig. 2). This was also true when the diversity-salinity relationship was examined for an individual site (Pearson correlation coefficients between −0.55 and −0.97 at the species level and between −0.94 and −0.98 at the genus level; see Fig. S2 in the supplemental material) or for a specific physical condition (e.g., sediment samples; see Fig. S3).
FIG 2.
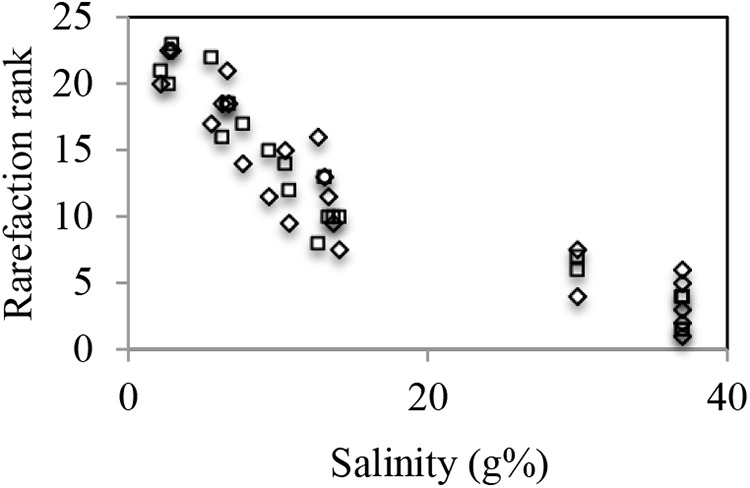
Salinity-diversity relationship. Sample salinity (x axis) was correlated to the diversity ranking of each sample (from 1 [least diverse] to 23 [most diverse]) (y axis), computed at both 97% (□) and 94% (◇) sequence similarity cutoffs.
Community structure analysis.
Nonmetric multidimensional scaling (NMDS) was used to identify community structure and co-occurrence patterns between different samples. The results (Fig. 3; see also Fig. S4 in the supplemental material) suggest that salinity plays an important role in shaping the community structure. Samples with very low salinity (2% to 3%) (n = 3 of 3 in this salinity range) clustered together, as did samples with moderately low salinity (5.5% to 9.4%) (n = 6 of 6 in this salinity range) and samples with moderately high salinity (10.5% to 14%) (n = 6 of 7 in this salinity range), as well as samples with high to saturated salinity (>30%) (two distinct clusters of n = 4 and n = 3). On the other hand, neither the physical characteristics of the sample (see Fig. S3A) nor the geographical location (see Fig. S3B) played a clear role in shaping community structure.
FIG 3.
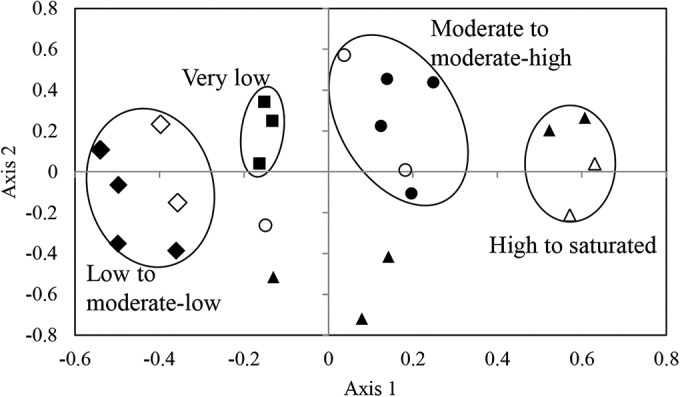
Nonmetric multidimensional scaling based on pairwise Bray-Curtis dissimilarity indices. Each symbol represents one sample, and sample symbols reflect their salinities. ■, very low salinities (2% to 3%); ⬥, low salinities (5.5% to 6.7%); ♢, moderate-low salinities (7.6% to 9.4%); ○, moderate salinities (10.5% to 12.7%); ●, moderate-high salinities (13% to 14%); △, high salinity (30%); ▲, saturated salinity (37% [salt crusts]).
Salinity-abundance correlation for individual Halobacteria genera.
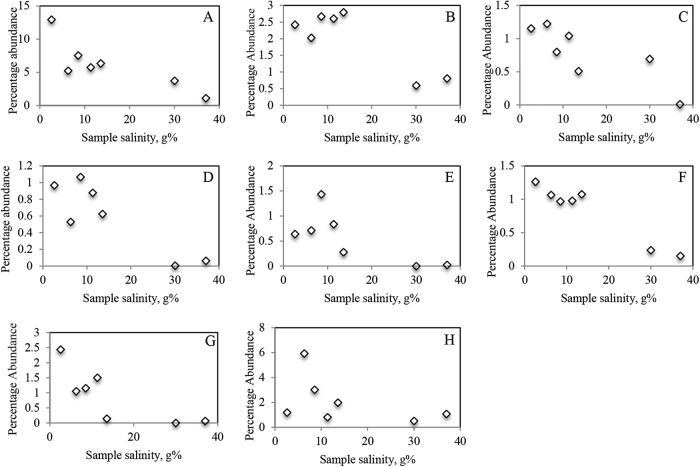
The results described above clearly suggest that salinity plays an important role in shaping the microbial community structure of the Halobacteria. To zoom in on genus-level specific preferences and determine the genera that appear most sensitive and responsible for the observed community structure shifts (Fig. 3), we examined salinity-relative abundance correlations for genera present above an empirical occurrence cutoff defined as a relative abundance of 1% or more in at least 5 samples (n = 28 genera). In general, three distinct patterns were observed (Fig. 4; see also Table S4 in the supplemental material).
FIG 4.
Percentages of abundance of individual Halobacteria genera capable of trehalose biosynthesis as a function of sample salinity. Values shown are averages for samples within the same salinity range as follows: very low (2% to 3%, 3 samples from 1 site), low (5% to 6.6%, 4 samples from 2 sites), medium-low (7.6% to 9.5%, 2 samples from 2 sites), medium-high (10.5% to 12.7%, 3 samples from 3 sites), high (13% to 14%, 4 samples from 2 sites), very high (30%, 2 samples from 1 site), and saturated (37%, 5 samples from 1 site). Results are shown only for otsAB-harboring genera present with >1% abundance in at least 5 samples. (A) Halogranum. (B) Halobiforma. (C) Halorussus. (D) Halostagnicola. (E) Haloterrigena. (F) Halovivax. (G) Haladaptatus. (H) Natronorubrum.
(i) Genera whose percentages of abundance increased with the increase in sample salinity (Pearson correlation coefficient, 0.62 to 0.9).
This group included the genera Haloferax, Halobellus, Halorhabdus, Halapricum, Halovenus, and Halomicroarcula.
(ii) Genera whose percentages of abundance decreased with the increase in sample salinity (Pearson correlation coefficient, −0.65 to −0.97).
This group included the genera Halogranum, Halobiforma, Halorussus, Halostagnicola, Haloterrigena, Halovivax, Haladaptatus, and Natronorubrum.
(iii) Genera with no clear effect of salinity fluctuations on their relative abundance.
This group included the genera Halorientalis, Halorubrum, Halonotius, Haloquadratum, Halolamina, Halomicrobium, Natronomonas, Haloplanus, Haloarcula, Halobacterium, Halorubellus, Salinarchaeum, Salinirubrum, and Halopelagius.
The ecological significance of trehalose biosynthesis capacity.
A close examination of salinity-abundance profiles of various genera (Fig. 4; see also Table S4 in the supplemental material) indicated that all genera whose percentages of abundance increase with salinity (n = 6) lack the otsAB operon and are hence incapable of utilizing trehalose biosynthesis as an osmoadaptive strategy. On the other hand, all genera whose percentages of abundance decrease with salinity (n = 8) belong to genera known for their capability to produce trehalose as a compatible solute (see Table S1 for a list of otsAB distributions within the Halobacteria).
To further examine this hypothesis, we identified in each data set the overall percentage of sequences belonging to genera shown to possess the otsAB operon. The relative proportions of such genera (listed in Table S1 in the supplemental material) ranged between 1.1% and 38.97% (Fig. 5; see also Fig. S5 in the supplemental material) and showed a strong negative correlation with salinity (Pearson correlation coefficient = −0.83).
FIG 5.
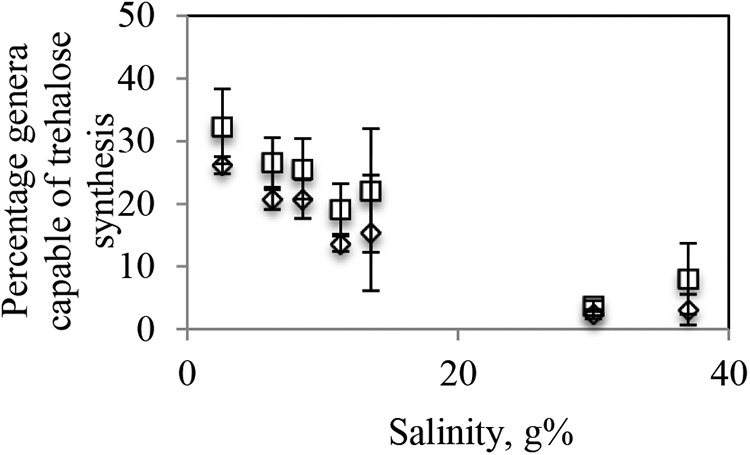
Effect of sample salinity on the total abundance of trehalose-producing genera. The sum of percent abundances of genera possessing an otsAB system determined on the basis of either experimental or genomic evidence (○) or predicted on the basis of phylogenetic affiliations (◇) is plotted on the y axis versus salinity on the x axis. Values shown are averages ± standard deviations for samples within the same salinity range as follows: very low (2% to 3%, 3 samples from 1 site), low (5% to 6.6%, 4 samples from 2 sites), medium-low (7.6% to 9.5%, 2 samples from 2 sites), medium-high (10.5% to 12.7%, 3 samples from 3 sites), high (13% to 14%, 4 samples from 2 sites), very high (30%, 2 samples from 1 site), and saturated (37%, 5 samples from 1 site). Individual sample data are shown in Fig. S3 in the supplemental material.
Finally, we used qPCR to quantify the otsB gene copy number per sample and the normalized Halobacteria otsB/16S rRNA gene ratio. The latter was used as a quantitative index of the relative abundance of trehalose-producing Halobacteria as a fraction of the overall community within a specific sample. A progressive decrease in this ratio was observed in samples with higher salinity (Fig. 6). Therefore, multiple lines of evidence (salinity-relative abundance curves of individual genera, proportions of sequences belonging to otsAB-harboring genera, and qPCR-based quantifications of Halobacteria otsB/16S rRNA ratios in various samples) strongly indicate that genera lacking the otsAB operon are more adapted to growth in and colonization of hypersaline (>25%) environments than trehalose producers.
FIG 6.
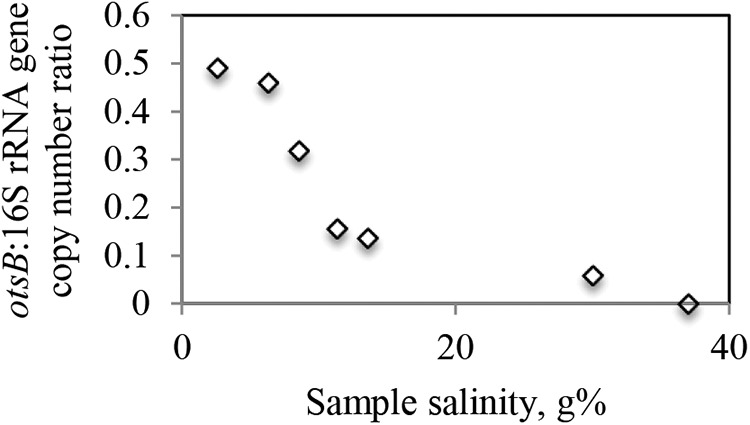
Quantification of otsB-harboring cells as a fraction of the total Halobacteria community. The ratio of the number of copies of the otsB gene to the Halobacteria 16S rRNA gene copy number is plotted as a function of salinity. Values shown are averages ± standard deviations for samples within the same salinity range as follows: very low (2% to 3%, 3 samples from 1 site), low (5% to 6.6%, 4 samples from 2 sites), medium-low (7.6% to 9.5%, 2 samples from 2 sites), medium-high (10.5% to 12.7%, 3 samples from 3 sites), high (13% to 14%, 4 samples from 2 sites), very high (30%, 2 samples from 1 site), and saturated (37%, 5 samples from 1 site).
DISCUSSION
In this study, we examined the diversity and community structure of the halophilic Archaea (class Halobacteria) in samples from central and southern Tunisian endorheic salt lakes and sebkhet systems. Our results suggest that (i) a high level of genus-level phylogenetic diversity exists within samples, with distinct genera consistently representing the majority of sequences in all data sets (Fig. 1; see also Table S2 in the supplemental material); (ii) Halobacteria diversity estimates within samples exhibited strong negative correlation with salinity (Fig. 2; see also Fig. S2 and S3); (iii) salinity was the most important factor shaping the observed microbial community structure, rather than geographical location or sample physical characteristics (Fig. 3; see also Fig. S4); and (iv) genera possessing the machinery for trehalose biosynthesis as an osmoadaptive strategy appear to be less suited for survival and propagation at higher salinities (Fig. 5 and 6; see also Fig. S5).
In general, extremely high genus-level diversity was observed in all samples (8 to 43 genera; average, 30), with sequences affiliated with all 45 currently described Halobacteria genera detected in the entire data set. The highest levels of diversity were observed in sediment and water samples of relatively lower salinity, providing additional evidence for the emerging view that habitats of lower and fluctuating salinity are reservoirs for novel Halobacteria diversity (7–18). The use of 16S rRNA gene primers targeting members of the Halobacteria class for diversity and quantification studies is crucial for the targeted exploration of Halobacteria diversity in such habitats, since the prevailing levels of salinity and putative frequent salinity fluctuation are conducive to the coexistence of additional halotolerant and nonhalophilic microorganisms.
Within all samples, a distinct community structure pattern was observed in which a few genera (groups 1 and 2 in Table S3 in the supplemental material) were present in relatively high abundance, followed by a longer tail of less-abundant and rare genera. Halorubrum, Halogranum, and Halorientalis genera were the three most abundant and consistently encountered genera within all data sets. Halorubrum is a well-described and ubiquitous genus within the Halobacteria, members of which have been isolated and detected (using 16S rRNA diversity surveys) in a wide range of saline environments (2, 9, 25, 48–50). Halorientalis and Halogranum, on the other hand, are two recently identified genera, with few cultured representatives. Halogranum species were first isolated from marine solar salterns in eastern China (51, 52). Recently, Halogranum species were also isolated from evaporitic salt crystals collected along the seashore of Namhae, South Korea (53), as well as from Zodletone Spring in southwestern Oklahoma (unpublished data). Physiological studies and sample origins suggest the genus capability to survive in environments with various salinities. Similarly, Halorientalis species were first isolated from marine solar salterns in eastern China (54) and then recently from a salt lake in Iran (55). All Halorientalis species isolated so far seem to require at least 2.5 M (14.6%) NaCl. The current study demonstrated the dominance of this genus not only in hypersaline environments but also in environments with fluctuating and low salinities. Interestingly, detection of Halogranum species and Halorientalis species in culture-independent studies is currently curtailed by a methodological oddity. Most curated 16S rRNA gene databases (RPD, Greengenes, and GenBank) do not acknowledge these two validly published names (as well as many other additional recently described genera) as part of their Halobacteria taxonomic outline. Therefore, we suspect that, in many cases, sequences affiliated with these two genera are usually deemed “unclassified” in culture-independent diversity surveys. Hence, our current approach for Halobacteria identification, which depends on using 16S rRNA sequences retrieved from all described species within the class Halobacteria as a BLAST database, circumvents this problem.
Interestingly, although some genera appeared to be predominant in all samples regardless of salinities, physical conditions, or locations (see groups 1 and 2 in Table S3 in the supplemental material), we observed distinct shifts in community structure between samples (Fig. 3). We argue that this is a reflection of the fact that, beyond the few highly abundant genera that seem to be salinity indifferent, a highly dynamic community of salinity-sensitive genera with moderate to low abundance (see groups 3, 4, and 5 in Table S3) exists and is responsible for the observed differences in community structure between samples. Indeed, in classifying genera belonging to each of the groups in Table S3 according to their response to salinity (salinity indifferent versus salinity sensitive), we saw that 67% of the group showing consistently high abundance (group 1 in Table S3) and 60% of the group showing moderately high abundance (group 2 in Table S3) are salinity-indifferent genera. Similarly, 67% of the group showing consistently low abundance (group 4 in Table S3) and 60% of the group showing occasionally low abundance (group 5 in Table S3) are salinity-sensitive genera.
Perhaps the most interesting observation in this study is the identified strong negative correlation between salinity and possession of genes for trehalose biosynthesis. otsAB-harboring genera showed a distinct negative correlation between abundance and salinity in genus-level salinity-abundance correlations (Fig. 5 and 6; see also Fig. S5 and Table S4 in the supplemental material). Further, while the combined percentages of abundance of all sequences affiliated with otsAB-harboring genera never exceeded 40% in any data set, a strong negative correlation to salinity was shown (Fig. 5). Finally, quantification of Halobacteria otsB/16S rRNA gene ratio (Fig. 6) confirmed the notion that the percentages of abundance of genera capable of trehalose biosynthesis and accumulation decrease with increases in salt concentrations.
These results demonstrate that the recently recognized divergence between trehalose producers and nonproducers is ecologically relevant. The most prevalent mechanism for osmoadaptation in the Halobacteria is salting in, where cells accumulate molar concentrations of potassium ions intracellularly to counter the high extracellular osmotic pressure. The salting-in strategy has long been demonstrated in model Halobacteria isolates such as Halobacterium salinarum, Haloarcula marismortui, Haloferax volcanii, Haloferax mediterranei, Haloferax gibbonsii, Halorubrum saccharovorum, and Halorubrum trapanicum (56–62) and, more recently, in a large number (n = 18) of Halobacteria taxa (37). Further, a pathway for the dependence on the H+/K+ symporter of the Trk family for potassium uptake has recently been proposed on the basis of an extensive genomic survey of 80 different Halobacteria genomes (63). In addition to the salting-in strategy, recent studies have led to an increasing appreciation of the role played by another osmoadaptive mechanism, compatible solute accumulation, as a supplemental strategy in species of numerous taxa within the Halobacteria (37, 64–66). We have recently demonstrated that the biosynthesis and accumulation of molar levels of trehalose (or 2-sulfotrehalose) occur in multiple genera within the Halobacteria and that the genes mediating the process (the otsAB operon) are present in 61/80 examined genomes (37). Interestingly, trehalose biosynthesis capability within the Halobacteria appears to follow a phylogenetic pattern, where all genera within a major clade either possess or lack trehalose biosynthetic capability. We argue that this pattern is a reflection of the benefits/costs of utilizing this system for osmoadaptation under different environmental conditions. In habitats of low and fluctuating salinity, a compatible solute strategy provides much-needed flexibility in responding to the salinity fluctuations frequently encountered, thus justifying the energetic costs associated with the production of molar quantities of this divalent sugar. In permanently hypersaline habitats, the energetic cost associated with this process leads to a growth rate lower than that seen with non-trehalose producers, decreased ecological fitness, and eventual out-competition from such ecosystems. It is telling that otsAB-harboring genera are rarely identified in typical hypersaline water bodies (1, 2, 25, 67, 68), attesting to the ecological advantages imparted by the loss of this gene system (or of the compatible solute osmoadaptive strategy as a whole) to allow niche colonization and dominance in hypersaline habitats.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation Microbial Observatories Program (grant EF0801858), by a Fulbright Scholarship to A.N., and by the BIODESERT Project (European Community's Seventh Framework Program CSA, EU FP7-CSA-SA REGPOT-2008-2; grant 245746).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01097-15.
REFERENCES
- 1.Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E. 2010. Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern. Res Microbiol 161:573–582. doi: 10.1016/j.resmic.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. 2004. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol 70:5258–5265. doi: 10.1128/AEM.70.9.5258-5265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maturrano L, Santos F, Rosselló-Mora R, Antón J. 2006. Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol 72:3887–3895. doi: 10.1128/AEM.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oren A. 2010. The dying Dead Sea: the microbiology of an increasingly extreme environment. Lakes Reser Res Manag 15:215–222. doi: 10.1111/j.1440-1770.2010.00435.x. [DOI] [Google Scholar]
- 5.Williams TJ, Allen MA, DeMaere MZ, Kyrpides NC, Tringe SG, Woyke T, Cavicchioli R. 2014. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant Haloarchaea. ISME J 8:1645–1658. doi: 10.1038/ismej.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorokin D, Berben T, Melton E, Overmars L, Vavourakis C, Muyzer G. 2014. Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 18:791–809. doi: 10.1007/s00792-014-0670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caton TM, Caton IR, Witte LR, Schneegurt MA. 2009. Archaeal diversity at the Great Salt Plains of Oklahoma described by cultivation and molecular analyses. Microb Ecol 58:519–528. doi: 10.1007/s00248-009-9507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, Qian P-Y. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J 5:650–664. doi: 10.1038/ismej.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochsenreiter T, Pfeifer F, Schleper C. 2002. Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6:267–274. doi: 10.1007/s00792-001-0253-4. [DOI] [PubMed] [Google Scholar]
- 10.Radax C, Gruber C, Stan-Lotter H. 2001. Novel haloarchaeal 16S rRNA gene sequences from Alpine Permo-Triassic rock salt. Extremophiles 5:221–228. doi: 10.1007/s007920100192. [DOI] [PubMed] [Google Scholar]
- 11.Roh SW, Kim K-H, Nam Y-D, Chang H-W, Park E-J, Bae J-W. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J 4:1–16. doi: 10.1038/ismej.2009.83. [DOI] [PubMed] [Google Scholar]
- 12.Youssef NH, Ashlock-Savage KN, Elshahed MS. 2012. Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl Environ Microbiol 78:1332–1344. doi: 10.1128/AEM.07420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sørensen KB, Canfield DE, Teske AP, Oren A. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol 71:7352–7365. doi: 10.1128/AEM.71.11.7352-7365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savage KN, Krumholz LR, Oren A, Elshahed MS. 2008. Halosarcina pallida gen. nov., sp. nov., a halophilic archaeon from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 58(Pt 4):856–860. doi: 10.1099/ijs.0.65398-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Ren M, Zhang L-L. 2015. Natribaculum breve gen. nov., sp. nov. and Natribaculum longum sp. nov., halophilic archaea isolated from saline soil. Int J Syst Evol Microbiol 65(Pt 2):604–608. doi: 10.1099/ijs.0.060541-0. [DOI] [PubMed] [Google Scholar]
- 16.Cui H-L, Yang X, Mou Y-Z. 2011. Salinarchaeum laminariae gen. nov., sp. nov.: a new member of the family Halobacteriaceae isolated from salted brown alga Laminaria. Extremophiles 15:625–631. doi: 10.1007/s00792-011-0393-0. [DOI] [PubMed] [Google Scholar]
- 17.Inoue K, Itoh T, Ohkuma M, Kogure K. 2011. Halomarina oriensis gen. nov., sp. nov., a halophilic archaeon isolated from a seawater aquarium. Int J Syst Evol Microbiol 61(Pt 4):942–946. doi: 10.1099/ijs.0.020677-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W-Y, Huo Y-Y, Zhang X-Q, Zhu X-F, Wu M. 2013. Halolamina salifodinae sp. nov. and Halolamina salina sp. nov., two extremely halophilic archaea isolated from a salt mine. Int J Syst Evol Microbiol 63(Pt 12):4380–4385. doi: 10.1099/ijs.0.050864-0. [DOI] [PubMed] [Google Scholar]
- 19.Gupta RS, Naushad S, Baker S. 2014. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol 65(Pt 3):1050–1069. doi: 10.1099/ijs.0.070136-0. [DOI] [PubMed] [Google Scholar]
- 20.Oren A. 2012. Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271. doi: 10.1099/ijs.0.038653-0. [DOI] [PubMed] [Google Scholar]
- 21.Grant WD, Sorokin DY. 2011. Distribution and diversity of soda lake alkaliphiles, p 27–54. In Horikoshi K. (ed), Extremophiles handbook, vol 1 Springer, Tokyo, Japan. [Google Scholar]
- 22.Jones BE, Grant WD, Duckworth AW, Owenson GG. 1998. Microbial diversity of soda lakes. Extremophiles 2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- 23.Oren A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol Ecol 39:1–7. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes ME, Oren A, House CH. 2012. Dynamics and persistence of Dead Sea microbial populations as shown by high-throughput sequencing of rRNA. Appl Environ Microbiol 78:2489–2492. doi: 10.1128/AEM.06393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh D, Porter K, Russ B, Burns D, Dyall-Smith M. 2010. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14:161–169. doi: 10.1007/s00792-009-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oren A. 2008. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukushima T, Usami R, Kamekura M. 2007. A traditional Japanese-style salt field is a niche for haloarchaeal strains that can survive in 0.5% salt solution. Saline Syst 3:2. doi: 10.1186/1746-1448-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purdy KJ, Cresswell-Maynard TD, Nedwell DB, McGenity TJ, Grant WD, Timmis KN, Embley TM. 2004. Isolation of haloarchaea that grow at low salinities. Environ Microbiol 6:591–595. doi: 10.1111/j.1462-2920.2004.00592.x. [DOI] [PubMed] [Google Scholar]
- 29.Savage KN, Krumholz LR, Oren A, Elshahed MS. 2007. Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 57(Pt 1):19–24. doi: 10.1099/ijs.0.64464-0. [DOI] [PubMed] [Google Scholar]
- 30.Leuko S, Legat A, Fendrihan S, Wieland H, Radax C, Gruber C, Pfaffenhuemer M, Weidler G, Stan-Lotter H. 2005. Isolation of viable Haloarchaea from ancient salt deposits and application of fluorescent stains for in situ detection of halophiles in hypersaline environmental samples and model fluid inclusions, p 91–104. In Gunde-Cimerman N, Oren A, Plemenitaš A (ed), Adaptation to life at high salt concentrations in Archaea, Bacteria, and Eukarya, vol 9 Springer, Dordrecht, Netherlands. [Google Scholar]
- 31.Jaakkola ST, Zerulla K, Guo Q, Liu Y, Ma H, Yang C, Bamford DH, Chen X, Soppa J, Oksanen HM. 2014. Halophilic Archaea cultivated from surface sterilized middle-late Eocene rock salt are polyploid. PLoS One 9:e110533. doi: 10.1371/journal.pone.0110533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani K, Salgaonkar B, Braganca J. 2012. Culturable halophilic archaea at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquat Biosyst 8:15. doi: 10.1186/2046-9063-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braganca JM, Furtado I. 2009. Isolation and characterization of Haloarchaea from low-salinity coastal sediments and waters of Goa. Curr Sci 96:1182–1184. [Google Scholar]
- 34.Baricz A, Coman C, Andrei AS, Muntean V, Keresztes ZG, Păuşan M, Alexe M, Banciu HL. 2014. Spatial and temporal distribution of archaeal diversity in meromictic, hypersaline Ocnei Lake (Transylvanian Basin, Romania). Extremophiles 18:399–413. doi: 10.1007/s00792-013-0625-6. [DOI] [PubMed] [Google Scholar]
- 35.Podell S, Emerson JB, Jones CM, Ugalde JA, Welch S, Heidelberg KB, Banfield JF, Allen EE. 2014. Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME J 8:979–990. doi: 10.1038/ismej.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oren A. 2013. Life at high salt concentrations, p 421–440. In Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Heidelberg, Germany. doi: 10.1007/978-3-642-30123-0_57. [DOI] [Google Scholar]
- 37.Youssef NH, Savage-Ashlock KN, McCully AL, Luedtke B, Shaw EI, Hoff WD, Elshahed MS. 2014. Trehalose/2-sulfotrehalose biosynthesis and glycine-betaine uptake are widely spread mechanisms for osmoadaptation in the Halobacteriales. ISME J 8:636–649. doi: 10.1038/ismej.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch EA, Langille MGI, Darling A, Wilbanks EG, Haltiner C, Shao KSY, Starr MO, Teiling C, Harkins TT, Edwards RA, Eisen JA, Facciotti MT. 2012. Sequencing of seven haloarchaeal genomes reveals patterns of genomic flux. PLoS One 7:e41389. doi: 10.1371/journal.pone.0041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elshahed MS, Najar FZ, Roe BA, Oren A, Dewers TA, Krumholz LR. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl Environ Microbiol 70:2230–2239. doi: 10.1128/AEM.70.4.2230-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh DA, Bapteste E, Kamekura M, Doolittle WF. 2004. Evolution of the RNA polymerase B′ subunit gene (rpoB′) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol 21:2340–2351. doi: 10.1093/molbev/msh248. [DOI] [PubMed] [Google Scholar]
- 43.Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T. 2010. Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B' (rpoB') gene. Int J Syst Evol Microbiol 60:2398–2408. doi: 10.1099/ijs.0.017160-0. [DOI] [PubMed] [Google Scholar]
- 44.Andam CP, Harlow TJ, Papke RT, Gogarten JP. 2012. Ancient origin of the divergent forms of leucyl-tRNA synthetases in the Halobacteriales. BMC Evol Biol 12:85. doi: 10.1186/1471-2148-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashelford KE, Weightman AJ, Fry JC. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res 30:3481–3489. doi: 10.1093/nar/gkf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Youssef NH, Elshahed MS. 2009. Diversity rankings among bacterial lineages in soil. ISME J 3:305–313. doi: 10.1038/ismej.2008.106. [DOI] [PubMed] [Google Scholar]
- 48.Benlloch S, Acinas SG, Anton J, Lopez-Lopez A, Luz SP, Rodriguez-Valera F. 2001. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb Ecol 41:12–19. [DOI] [PubMed] [Google Scholar]
- 49.Fan H, Xue Y, Ma Y, Ventosa A, Grant WD. 2004. Halorubrum tibetense sp. nov., a novel haloalkaliphilic archaeon from Lake Zabuye in Tibet, China. Int J Syst Evol Microbiol 54:1213–1216. doi: 10.1099/ijs.0.03032-0. [DOI] [PubMed] [Google Scholar]
- 50.Hu L, Pan H, Xue Y, Ventosa A, Cowan DA, Jones BE, Grant WD, Ma Y. 2008. Halorubrum luteum sp. nov., isolated from Lake Chagannor, Inner Mongolia, China. Int J Syst Evol Microbiol 58:1705–1708. doi: 10.1099/ijs.0.65700-0. [DOI] [PubMed] [Google Scholar]
- 51.Cui HL, Gao X, Sun FF, Dong Y, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ. 2010. Halogranum rubrum gen. nov., sp. nov., a halophilic archaeon isolated from a marine solar saltern. Int J Syst Evol Microbiol 60(Pt 6):1366–1371. doi: 10.1099/ijs.0.014928-0. [DOI] [PubMed] [Google Scholar]
- 52.Cui HL, Yang X, Gao X, Xu XW. 2011. Halogranum gelatinilyticum sp. nov. and Halogranum amylolyticum sp. nov., isolated from a marine solar saltern, and emended description of the genus Halogranum. Int J Syst Evol Microbiol 61(Pt 4):911–915. doi: 10.1099/ijs.0.024976-0. [DOI] [PubMed] [Google Scholar]
- 53.Kim KK, Lee KC, Lee JS. 2011. Halogranum salarium sp. nov., a halophilic archaeon isolated from sea salt. Syst Appl Microbiol 34:576–580. doi: 10.1016/j.syapm.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Cui HL, Yang X, Gao X, Xu XW. 2011. Halobellus clavatus gen. nov., sp. nov. and Halorientalis regularis gen. nov., sp. nov., two new members of the family Halobacteriaceae. Int J Syst Evol Microbiol 61(Pt 11):2682–2689. doi: 10.1099/ijs.0.025841-0. [DOI] [PubMed] [Google Scholar]
- 55.Amoozegar MA, Makhdoumi-Kakhki A, Mehrshad M, Fazeli SA, Sproer C, Ventosa A. 2014. Halorientalis persicus sp. nov., an extremely halophilic archaeon isolated from a salt lake and emended description of the genus Halorientalis. Int J Syst Evol Microbiol 64(Pt 3):940–944. doi: 10.1099/ijs.0.058164-0. [DOI] [PubMed] [Google Scholar]
- 56.Christian JH, Waltho JA. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim Biophys Acta 65:506–508. doi: 10.1016/0006-3002(62)90453-5. [DOI] [PubMed] [Google Scholar]
- 57.Ginzburg M, Sachs L, Ginzburg BZ. 1970. Ion metabolism in a Halobacterium. I. Influence of age of culture on intracellular concentrations. J Gen Physiol 55:187–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanyi JK, Silverman MP. 1972. The state of binding of intracellular K+ in Halobacterium cutirubrum. Can J Microbiol 18:993–995. doi: 10.1139/m72-154. [DOI] [PubMed] [Google Scholar]
- 59.Matheson AT, Sprott GD, McDonald IJ, Tessier H. 1976. Some properties of an unidentified halophile: growth characteristics, internal salt concentration, and morphology. Can J Microbiol 22:780–786. doi: 10.1139/m76-114. [DOI] [PubMed] [Google Scholar]
- 60.Mojica FJ, Cisneros E, Ferrer C, Rodriguez-Valera F, Juez G. 1997. Osmotically induced response in representatives of halophilic prokaryotes: the bacterium Halomonas elongata and the archaeon Haloferax volcanii. J Bacteriol 179:5471–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oren A, Heldal M, Norland S, Galinski EA. 2002. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6:491–498. doi: 10.1007/s00792-002-0286-3. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Fillol M, Rodríguez-Valera F. 1986. Potassium ion accumulation in cells of different halobacteria. Microbiologia 2:73–80. [PubMed] [Google Scholar]
- 63.Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE, Facciotti MT. 2014. Phylogenetically driven sequencing of extremely halophilic Archaea reveals strategies for static and dynamic osmo-response. PLoS Genet 10:e1004784. doi: 10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desmarais D, Jablonski PE, Fedarko NS, Roberts MF. 1997. 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic Archaea. J Bacteriol 179:3146–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goh F, Jeon YJ, Barrow K, Neilan BA, Burns BP. 2011. Osmoadaptive strategies of the archaeon Halococcus hamelinensis isolated from a hypersaline stromatolite environment. Astrobiology 11:529–536. doi: 10.1089/ast.2010.0591. [DOI] [PubMed] [Google Scholar]
- 66.Kokoeva MV, Storch KF, Klein C, Oesterhelt D. 2002. A novel mode of sensory transduction in Archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J 21:2312–2322. doi: 10.1093/emboj/21.10.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bidle K, Amadio W, Oliveira P, Paulish T, Hicks S, Earnest C. 2005. A phylogenetic analysis of Haloarchaea found in a solar saltern. BIOS 76:89–96. doi: 10.1893/0005-3155(2005)076[0089:RAAPAO]2.0.CO;2. [DOI] [Google Scholar]
- 68.Podell S, Ugalde JA, Narasingarao P, Banfield JF, Heidelberg KB, Allen EE. 2013. Assembly-driven community genomics of a hypersaline microbial ecosystem. PLoS One 8:e61692. doi: 10.1371/journal.pone.0061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.