Abstract
The CRISPR-Cas9 system is a powerful and revolutionary genome-editing tool for eukaryotic genomes, but its use in bacterial genomes is very limited. Here, we investigated the use of the Streptococcus pyogenes CRISPR-Cas9 system in editing the genome of Clostridium cellulolyticum, a model microorganism for bioenergy research. Wild-type Cas9-induced double-strand breaks were lethal to C. cellulolyticum due to the minimal expression of nonhomologous end joining (NHEJ) components in this strain. To circumvent this lethality, Cas9 nickase was applied to develop a single-nick-triggered homologous recombination strategy, which allows precise one-step editing at intended genomic loci by transforming a single vector. This strategy has a high editing efficiency (>95%) even using short homologous arms (0.2 kb), is able to deliver foreign genes into the genome in a single step without a marker, enables precise editing even at two very similar target sites differing by two bases preceding the seed region, and has a very high target site density (median interval distance of 9 bp and 95.7% gene coverage in C. cellulolyticum). Together, these results establish a simple and robust methodology for genome editing in NHEJ-ineffective prokaryotes.
INTRODUCTION
Targeted genome editing is critical for both fundamental molecular biology and applied genetic engineering. Even though current methods (i.e., allele exchange, group II intron retrotransposition, and recombineering) can be used for genome modification in many microbes (1, 2), they have some limitations: (i) traditional stepwise recombination-dependent allele exchange is time-consuming and has low efficiency (3), which can be worse when host transformation efficiency is low and/or usable selection markers are limited; (ii) insertion/deletion-based mutagenesis of large DNA fragments can potentially cause polar effects on downstream genes (4, 5); and (iii) insertion of large DNA fragments, such as metabolic pathway transfer, is difficult with current genome engineering tools, which require existing recombination sites and/or recombinases (1, 6). Thus, a facile and efficient method capable of performing precise, markerless, and versatile genome manipulations is needed to expedite microbial studies.
The clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein (Cas) system is an RNA-guided immune system in many bacteria that is able to recognize and cleave invasive DNAs (7). The type II-A CRISPR-Cas system of Streptococcus pyogenes, which requires a mature CRISPR RNA (crRNA), a trans-activating crRNA (tracrRNA), and DNA endonuclease Cas9, has been harnessed for targeted genome editing in many organisms (8–14). Mechanistically, under the guidance of the tracrRNA-crRNA duplex or latterly engineered single guide RNA (gRNA), S. pyogenes Cas9 or Cas9 nickase (Cas9n) can cut any target DNA having a 5′-N20NGG-3′ region (see Fig. S1A in the supplemental material), where N represents any nucleotide and N20 represents the protospacer appended with a protospacer-adjacent motif (PAM) (NGG) at the 3′ end (8). The cleavage site will then be repaired by nonhomologous end-joining (NHEJ) or homologous recombination (HR) (2, 15). Thus far, Cas9-based tools have shown their versatility for foreign gene knock-in and gene inactivation by DNA deletion or insertion, with attractive features such as ease of use, high efficiency, strong adaptability, and multiplex targeting ability (2, 15). However, reports of their application in bacterial genome editing are quite limited (13, 16–20). By coupling Cas9-mediated cleavage with HR repair, the genomes of Escherichia coli (19), Streptococcus pneumoniae (13), four Streptomyces species (17, 18, 20), and Tatumella citrea (19) were edited at a high efficiency. Cas9-assisted elimination of unmutated cells, after single-stranded DNA recombineering, significantly improved the editing efficacy in E. coli and Lactobacillus reuteri (13, 16). Using the inefficient repair of double-strand breaks (DSBs) in some microbes, reprogrammed Cas9 has been applied as an antimicrobial to selectively kill some strains (21–23). Naturally, the lethal effect of Cas9-induced DSBs does not allow genome editing in repair-defective microbes; however, exploiting a strategy to circumvent this lethality will theoretically allow genome editing in many microbes.
As a model system of a mesophilic cellulolytic bacterium, Clostridium cellulolyticum can directly convert lignocellulosic biomass to valuable end products (i.e., lactate, acetate, ethanol, and hydrogen) (24). It holds promise of producing renewable green chemicals from cellulose to replace petroleum-based products (25). However, genome editing of C. cellulolyticum for metabolic engineering is still challenging due to the lack of efficient editing tools. Despite the predicted presence of the type II-C CRISPR-Cas system in C. cellulolyticum (26), without a basic understanding of this system (e.g., protospacer length, PAM, and gRNA features), we cannot immediately examine its use in genome editing. Here, we tested the use of the single gRNA-directed S. pyogenes Cas9 to edit the C. cellulolyticum genome and found an inefficiency of host NHEJ and HR in repairing Cas9-induced DSBs. Then, we developed a single-nick-assisted HR strategy using a Cas9 nickase and a plasmid-borne donor template to efficiently modify targeted genomic loci by DNA deletion and insertion. This strategy also presented the ability to integrate foreign genes in a single step without a marker, making this a promising step in facilitating genome-level metabolic engineering coupled with synthetic biology in the future.
MATERIALS AND METHODS
Synthetic promoter design.
Promoter sequences in the C. cellulolyticum genome were predicted by PePPER (27). Then, over 100 predicted σA promoters were aligned to create 39-nucleotide (nt)-long DNA logos using WebLogo (28). Based on the alignment result, at each position the nucleotide with the highest usage frequency was selected to build a mini-P4 promoter (5′-TTGACAAATTTATTTTTTAAAGTTAAAATTAAGTTG-3′). To test promoter activity, P4 was used to drive an anaerobic fluorescent protein-encoding gene (afp). Between the P4 promoter and the afp open reading frame is a short sequence containing a ribosome RNA binding site (RBS) (5′-TTAGGAGGTACCCCG-3′).
Plasmid construction.
The P4 promoter generated by annealing extension PCR using P4F and P4R was ligated into the pCR8/GW/TOPO TA vector (Invitrogen). The RBS-containing promoter fragment amplified by using PromF and PromR was assembled with an EcoRI- and BamHI-linearized pLyc017 backbone (29) using a Gibson assembly kit (NEB), generating pP4-AFP.
The cas9 gene from S. pyogenes SF370 was codon optimized and synthesized with a His tag-encoding sequence at the C terminus (Invitrogen). The adapted cas9 fragment was ligated with the modified pLyc017 (empty vector) to generate an Fd::cas9 cassette in the resultant pCas9. The gRNA scaffoldin was also synthesized (Invitrogen) (see Fig. S1B in the supplemental material). All gRNA cassettes were constructed by splicing the RBS-free P4 promoter and the gRNA fragment using splicing by overlap extension (SOEing). The P4::noncustomized gRNA cassette was generated using primers P4gRF and P4gRR for the promoter and gRCKF and gRNAR for the gRNA region and then assembled with the modified pLyc017, generating pGRNA. To target pyrF, mspI, the β-galactosidase gene (β-gal), 3198D, X21, and X22, one target site in each gene or site was selected (see Table S1 in the supplemental material) and P4gRR and gRCKF were replaced by corresponding primers (see Table S2 in the supplemental material). Customized gRNA cassettes were assembled with linearized pCas9, generating pCas9-pyrF, pCas9-mspI, and pCas9-β-gal. The wild-type (WT) Cas9 endonuclease was mutated to Cas9 nickase (D10A) via site-directed mutagenesis by using mutagenic primers Cas9nF and Cas9nR. The cas9 in the above plasmids was replaced by cas9n, generating pCas9n, pCas9n-pyrF, pCas9n-mspI, and pCas9n-β-gal. gRNA cassettes targeting 3198D, X21, and X22 were assembled with linearized pCas9n, generating pCas9n-3198D, pCas9n-X21 and pCas9n-X22, respectively.
To generate all-in-one vectors, user-defined donor templates were constructed by SOEing and then inserted into coexpression vectors. To construct a 2-kb donor template for a 23-bp deletion in the pyrF gene, 1-kb left (LH) and right (RH) homologous arms were first amplified separately using primer pair 0614LF and 0614LR and primer pair 0614RF and 0614RR, respectively, and then the two fragments were spliced to produce the 2-kb donor for assembly with linearized pCas9n-pyrF, generating pCas9n-pyrF-donor. Similarly, pCas9n-mspI-donor, pCas9n-X21-donor, pCas9n-X22-donor, and pCas9n-β-gal-donor vectors with 1-kb, 0.5-kb, 0.2-kb, and 0.1-kb arm sizes were constructed with designed primers (see Table S2 in the supplemental material). A series of pCas9-pyrF-donors with the 0.71-kb Fd::afp expression cassette, 3-kb λ DNA, and 6-kb λ DNA between 1-kb homologous arms were constructed by three-piece SOEing or sequential cloning using pBR322 (NEB) as intermediate plasmid. The pCas9n-3198D-donor with 1.72-kb promoterless α-acetolactate synthase (alsS) between 1-kb arms was constructed by sequential cloning. The promoterless alsS fragment was amplified from pLyc025. All constructs were verified by DNA sequencing for further studies.
Bacterial strains and culture conditions.
E. coli Top10 (Invitrogen) was used for all cloning. E. coli transformants were grown at 37°C in Luria-Bertani medium with chloramphenicol (15 μg/ml) for the pLyc017-derived series or ampicillin (50 μg/ml) for the pBR322-derived series. C. cellulolyticum H10 (ATCC 35319) and its developed strains were cultured anaerobically at 34°C in VM medium with yeast extract (2.0 g/liter) and cellobiose (5 g/liter). If not otherwise specified, methylated plasmids were used for C. cellulolyticum electroporation (29), and then transformants were normally selected by thiamphenicol (TMP) (15 μg/ml). For ΔpyrF mutant identification, selective medium was additionally supplemented with 5-fluoroorotic acid (5-FOA) (500 μg/ml). Single colonies were anaerobically developed on VM plates at 34°C. Serial transfer was conducted by transferring a cell culture (optical density at 600 nm [OD600] of >0.4) to a new medium (1:10, vol/vol), and TMP was added if required. Cell growth was determined with three replicates by monitoring OD600.
ΔpyrF mutants created by the pCas9n-pyrF-donor were initially screened with 5-FOA and then were identified individually by PCR amplicon sequencing. The ΔpyrF mutant created by group II retrotransposition (29) was used as a positive control for phenotype identification. Similarly, ΔX21 and ΔX22 mutants, created by pCas9n-X21-donor and pCas9n-X22-donor, respectively, were identified by PCR amplicon sequencing. The TMP-resistant population generated from pCas9n-mspI-donor, containing ΔmspI mutants, was serially transferred, and then the population genomic DNA was extracted for PCR identification and sequencing. The Δβ-gal mutant population generated by pCas9n-β-gal-donor was additionally identified with amplicon digestion by EcoRV.
To generate plasmid-cured strains, pure ΔmspI and ΔpyrF/afp+ mutants were serially transferred in TMP-free medium. Then, cells were streaked on TMP-free plates for colony development. Plasmid-cured colonies were screened by both PCR amplification of the plasmid-borne region and TMP selection and then verified by transforming unmethylated pGRNA.
Determination of editing efficiency and cargo capacity.
Transformants of each construct (pCas9n-β-gal-donor with various arm sizes) were generated by electrotransforming 0.25 pmol methylated plasmids with two replicates. Each recovered culture (T0) was equally inoculated into the selection medium (T1). Then, two more serial transfers (T2 and T3) were conducted sequentially when the OD600 was 0.4 to 0.5. At each transfer point, cell culture was collected for genomic DNA extraction. The extracted genomic DNA was used as a PCR template to specifically amplify a 2-kb genomic region covering the entire donor, using primers p3 and p4. A portion (1 μg) of each purified amplicon was digested with 10 U EcoRV in NEBuffer 3.1 at 37°C for 3 h, for the purpose of distinguishing the edited and unedited amplicon by gel electrophoresis. Gel images were subject to densitometry analysis using Thermo MYImage. The editing efficiency (percent) was calculated by dividing the intensity of the 2-kb bands from the selected culture by the initial intensity of the bands from the corresponding T0 control, subtracting the result from 1, and then multiplying by 100.
To examine the genetic cargo capacity, a series of vectors (pCas9-pyrF-donor with 0.71-kb Fd::afp expression cassette, 3-kb λ DNA, and 6-kb λ DNA and pCas9-3198D-donor with 1.72-kb alsS) were transformed. During three serial transfers under only TMP selection, resistant populations were subjected to genomic DNA extraction and then the edited genomes in the population were distinguished from wild type (WT) by PCR amplification and gel electrophoresis.
RNA isolation, RT-PCR, and quantitative real-time PCR.
Total RNA was extracted from cellobiose (5 g/liter)-grown C. cellulolyticum (OD600 of ∼0.45) by TRIzol (Invitrogen) and then reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). The cDNA product was diluted as appropriate and used as a template. gRNA expression was examined by reverse transcription-PCR (RT-PCR) using recA as an internal calibrator (98°C for 30 s and 22 cycles of 98°C for 10 s, 56°C for 10 s, and 72°C for 10 s). Quantitative PCR was performed using iTaq SYBR green Supermix with ROX (Bio-Rad) on a Bio-Rad iQ5 thermal cycler. Gene-specific primers for each transcript are listed in Table S2 in the supplemental material. Thermal cycling conditions were as follows: 95°C for 3 min and 40 cycles each of 95°C for 15 s, 56°C for 15 s, and 72°C for 45 s. The relative expression level of target genes compared to recA was calculated by the Pfaffl method (30).
SDS-PAGE analysis.
To examine the expression of full-length His-tagged Cas9 in C. cellulolyticum, single colonies of pCas9 or CK (empty vector) transformants were cultured. Cells were lysed in the SDS loading buffer, and then supernatant cell lysates were subjected to SDS-PAGE using 9% resolving gels (Bio-Rad). Additionally, His-tagged Cas9 protein in the gel was detected by the Pierce 6×His protein tag stain reagent set (Thermo Scientific).
Fluorescence microscopy.
Fresh cultures of wild-type C. cellulolyticum, P4::afp and Fd::afp strains, and plasmid-cured ΔpyrF/afp+ mutants at mid-log phase were analyzed using an Olympus BX51 fluorescence microscope equipped with optical filter sets with excitation at 490 nm and emission at 525 nm for green fluorescence. The images were collected by an Olympus DP71 digital camera.
Bioinformatic analysis of target sites.
All N20NGG sites in the C. cellulolyticum genome (NC_011898.1) and their locations were extracted from both strands. Then, unique and transcribable target sites were selected by filtering out those with >2 identical sites across the genome, a string of six or more T's in the 23-mer sequence (12) and T3 in the 6-mer region upstream of NGG (31), or an extremely low or high GC content (<25% and >80%, respectively) (32). Usable target sites that had at least two base-pair mismatches with the rest of that region of the genome were used for targeting space analyses, including calculating the distances between all adjacent usable sites and histogram plotting. The number of usable sites in all predicted genes was determined for histogram plotting. Gene coverage percentage was calculated by dividing the number of genes that had at least one usable target site by the total gene number. The genome-wide distribution was drawn by GenomeDiagram (33). Following similar procedures, we analyzed the genomes of Clostridium acetobutylicum ATCC 824 (NC_003030.1), E. coli K-12 (NC_000913.3), Bacillus subtilis 168 (NC_000964.3), and L. reuteri DSM 20016 (NC_009513.1).
RESULTS
Expression of CRISPR-Cas9 system in C. cellulolyticum.
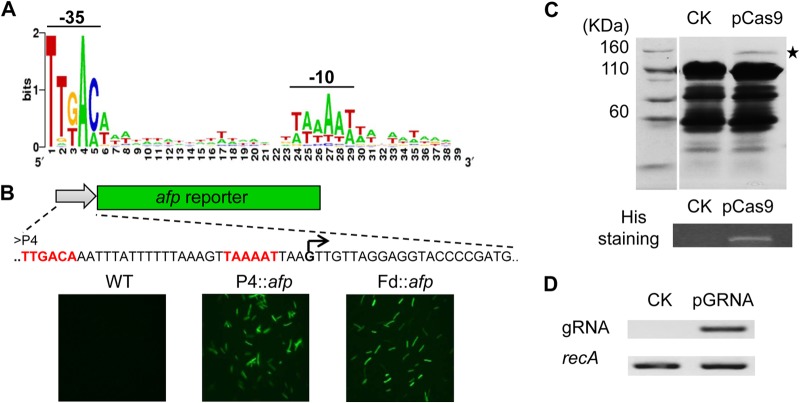
To establish Cas9-based genome editing in C. cellulolyticum, functional promoters are needed to drive the expression of Cas9 and gRNA. To quickly expand the promoter library, synthetic promoter design was applied. Since σA is the primary sigma factor responsible for transcribing most genes in microbial cells (34), in silico analysis of genome-wide σA-dependent promoters was conducted for C. cellulolyticum. An alignment of predicted promoters showed two characteristically conserved regions (−35 and −10) that were separated by a 17-nt T/A-rich spacer (Fig. 1A). A synthetic promoter (P4) comprised of nucleotides with the highest usage frequency at each position was chemically synthesized (length, 36 bp). The activity of P4 was tested in C. cellulolyticum by driving the expression of a reporter gene (afp) encoding the anaerobic fluorescent protein (Fig. 1B). Under fluorescence microscopy, the P4::afp construct presented a fluorescent signal in C. cellulolyticum (Fig. 1B); the fluorescence intensity was comparable to that of the positive control in which a ferredoxin promoter (Fd) from Clostridium pasteurianum was used to control afp expression, generating the Fd::afp construct (29).
FIG 1.
Generation and validation of Cas9 expression system. (A) Alignment of predicted σA-dependent promoters from C. cellulolyticum. Two highly conserved regions (−35 and −10) are separated by a 17-nt T/A-rich spacer. (B) Promoter activity test, in which synthetic promoter P4 drives an anaerobic florescent protein-encoding gene (afp). The right-angled arrow indicates the potential transcriptional start site. The −35 and −10 regions are in red. Fluorescence microscopy of C. cellulolyticum wild type (WT) and transformants carrying P4::afp or Fd::afp constructs. (C) SDS-PAGE analysis of whole-cell proteins from transformants with empty vector (CK) and pCas9. The star denotes the estimated Cas9 band. The full-length His-tagged Cas9 is further verified by His protein staining. (D) RT-PCR analysis of gRNA in both CK and pGRNA strains, using the recA gene as an internal calibrator.
Next, we chose the P4 and Fd promoters to drive gRNA and cas9 gene expression, respectively. The cas9 gene of S. pyogenes was codon adapted to C. cellulolyticum and fused with a His tag at the C terminus. To examine Cas9 expression, we constructed a pCas9 shuttle vector carrying an Fd::cas9 expression cassette. The full-length His-tagged Cas9 protein was successfully expressed as evidenced by SDS-PAGE analysis and His protein staining (Fig. 1C). Additionally, we constructed a pGRNA vector harboring a P4::gRNA expression cassette. This construct was able to generate noncustomized gRNA transcripts as shown by RT-PCR (Fig. 1D). Then, both expression cassettes (Fd::cas9 and P4::gRNA) were combined into a single vector, pCas9-gRNA (see Fig. S1C in the supplemental material). Once the gRNA is customized, the resultant vector is able to coexpress Cas9 and gRNA to edit targeted genomic loci in a single step.
Lethality of Cas9-induced double-strand breaks.
To demonstrate genome editing by gRNA-guided Cas9, a pyrF gene encoding orotidine-5′-phosphate decarboxylase (Ccel_0614) in C. cellulolyticum was chosen as our first target gene since inactivation of this gene would generate uracil auxotrophic and 5-fluoroorotic acid (5-FOA)-resistant phenotypes, which are easily monitored (35). The pCas9-pyrF vector coexpressing Cas9 and the customized gRNA targeting the pyrF gene was electroporated into C. cellulolyticum in parallel with pCas9 and pGRNA-pyrF, both of which served as negative controls expressing only either Cas9 or customized gRNA. Transformation tests revealed that both controls generated antibiotic-resistant transformants but not 5-FOA-resistant transformants (Table 1; see also Fig. S2A and B in the supplemental material); the coexpression vector did not produce cells with both antibiotic and 5-FOA resistance (Table 1; see also Fig. S2A and B). These results suggested that coexpressing Cas9 and gRNA was toxic at least at the selected target site. Then, we tested two more target sites, one in the β-galactosidase gene (β-gal, Ccel_0374) and the other in the mspI endonuclease gene (mspI, Ccel_2866), and determined that coexpression vectors were unable to produce antibiotic-resistant cells (see Fig. S2C). We suspected that the problem might be in the unsuccessful repair of DSBs created by the Cas9-gRNA complex since DSBs can interrupt chromosome replication and cell reproduction. To verify this hypothesis, the wild-type Cas9 was replaced with Cas9n (D10A) (8), generating pCas9n-pyrF. Interestingly, after transformation we observed the propagation of antibiotic-resistant cells, but these cells were not 5-FOA resistant (Table 1; see also Fig. S2A and B), suggesting that the Cas9-induced lethality can be voided by Cas9n and that the single nick created by Cas9n did not enable genome editing via NHEJ. Afterward, we investigated the expression of major NHEJ components (36, 37), including Ku (Ccel_0364), ATP-dependent DNA ligase (Ccel_0365), and DNA polymerase LigD (Ccel_0366). Strikingly, all three genes were expressed at a very low level in comparison to the recA housekeeping gene (4) (see Fig. S3 in the supplemental material). Taken together, these results indicate that C. cellulolyticum NHEJ is inefficient in repairing DSBs, which restricts the use of Cas9 in editing the C. cellulolyticum genome.
TABLE 1.
Use of Cas9 nickase instead of wild-type Cas9 for genome editing in C. cellulolyticum
| Plasmid | Component | Cell growth by resistance typea: |
|
|---|---|---|---|
| TMPr | 5-FOAr | ||
| pCas9 | Cas9 | Y | Ν |
| pGRNA-pyrF | gRNA | Y | Ν |
| pCas9-pyrF | Cas9 + gRNA | Ν | Ν |
| pCas9n-pyrF | Cas9n + gRNA | Y | Ν |
| pCas9-pyrF-donor | Cas9 + gRNA + donor template | Ν | Ν |
| pCas9n-pyrF-donor | Cas9n + gRNA + donor template | Y | Y |
TMPr, thiamphenicol resistant; 5-FOAr, 5-fluoroorotic acid resistant; Y, cell growth; Ν, no cell growth. Growth profiles are shown in Fig. S2 in the supplemental material.
Precise genome editing via an SNHR.
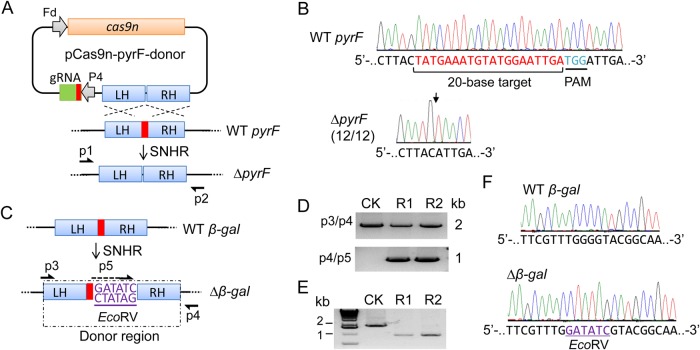
Homology-directed repair is another way to fix DNA lesions when a homologous template is present (38). To mutate the pyrF gene by small DNA deletion, we designed a homologous donor template with a length of 2 kb carrying a 23-bp deletion in the middle and cloned it into pCas9-pyrF and pCas9n-pyrF, generating all-in-one pCas9-pyrF-donor and pCas9n-pyrF-donor plasmids (Fig. 2A; see also Fig. S1C in the supplemental material). In this way, editing templates can be maintained during plasmid replication. Transformation tests showed that even though the editing templates were present, Cas9-induced DSBs did not produce any resistant cells; however, Cas9n-induced single nicks, coupled with HR, produced resistant cells under both antibiotic and 5-FOA selection (Table 1; see also Fig. S2A and B), suggesting that ΔpyrF mutants may be generated. After spread plating, we randomly picked 12 colonies for sequencing and found that all were ΔpyrF mutants containing a precise deletion of the 23-bp target sequence in the gene (Fig. 2B). Using the same strategy, we targeted the mspI gene (see Fig. S4A in the supplemental material), which encodes an endonuclease of the restriction-modification system in C. cellulolyticum (39). After constructing and transforming the pCas9n-mspI-donor carrying a 2-kb donor template with a 23-bp deletion inside, we examined the ΔmspI mutants in the antibiotic-resistant population. PCR amplification revealed that the wild type was specifically detected in the control using an empty vector but was not detected in the resistant population (see Fig. S4B), indicating the deletion of the 23-bp target fragment in that population. Then, DNA sequencing further confirmed a precise deletion in the ΔmspI mutant (see Fig. S4C). After plasmid curing, the ΔmspI mutant was further shown to be transformable with unmethylated plasmids (see Fig. S4D). Taken together, these results demonstrate that this single-nick-triggered HR (SNHR) allows a one-step precise DNA deletion in C. cellulolyticum.
FIG 2.
Precise deletion and insertion of a small fragment. (A) Schematic all-in-one vector for pyrF disruption by a single-nick-triggered homologous recombination (SNHR). The vector consists of an Fd-driven cas9n gene, a P4-driven gRNA-targeting pyrF gene, and a donor template with a 23-bp deletion flanked by 1-kb left homologous (LH) and right homologous (RH) arms. (B) DNA sequence chromatograms showing the deletion of a 23-bp target site in the pyrF gene. The 23-bp region carries a 20-base gRNA sequence and a 3-base protospacer adjacent motif (PAM). Twelve colonies all present precise deletion at the position indicated by a downward black arrow. Amplicon for sequencing was generated using primers p1 and p2, as schematized in panel A. (C) SNHR-mediated insertion of an EcoRV site at a target cut site in the β-gal gene. The donor template shown in the dashed box carries the EcoRV site flanked by 1-kb LH and RH starting from the Cas9n cleavage site. (D) PCR identification of Δgal mutants. The transformant population of empty vector (CK) and pCas9n-β-gal-donor (R1 and R2, two replicates) is identified by two primer pairs as drawn in panel C. (E) EcoRV digestion of p3/p4 PCR products. (F) DNA sequence chromatograms verifying the precise insertion of EcoRV (underlined) in the Δβ-gal mutant.
In genetic engineering, small DNA insertions are useful for integrating short functional elements and introducing frameshift mutations. To test the potential of small DNA insertions, we tried to introduce an EcoRV site (5′-GATATC-3′) into the target site of the β-gal gene (Fig. 2C). A donor template harboring an EcoRV site in the middle flanked by 1-kb homologous arms starting from the cleavage site was constructed and used to generate a pCas9n-β-gal-donor for transformation. EcoRV insertion was initially indicated by differential PCR amplification (Fig. 2D), which generated the intended amplicon only when edited genomes were present. Then, amplicon digestion by EcoRV and amplicon sequencing both confirmed the insertion of EcoRV at the anticipated locus (Fig. 2E and F). Thus, a small insertion is also operable using this strategy.
Assessment of editing efficiency and genetic cargo capacity.
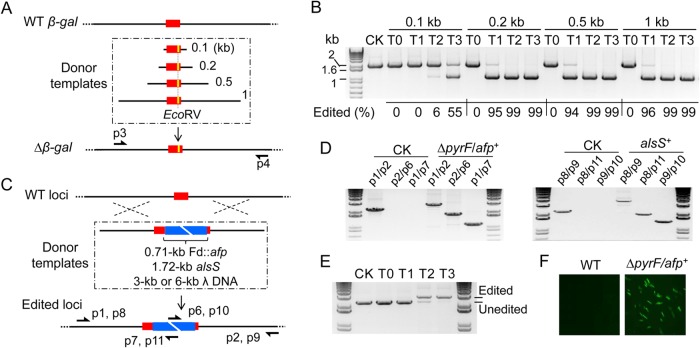
A powerful genome-editing tool should have a high efficiency allowing for marker-independent editing. Here, we evaluated the editing efficacy of this SNHR strategy and the effect of arm size on editing since the length of homologous arms affects recombination frequency (40–42). We constructed a series of donor templates, all of which harbor an EcoRV site in the middle flanked by homologous arms of varied length (0.1, 0.2, 0.5, and 1 kb), and then constructed pCas9n-β-gal-donor vectors (Fig. 3A). Since coexistence of the Cas9n-gRNA complex and the donor template may continuously trigger editing, extending the reaction time and possibly increasing the mutant population abundance, cell cultures from posttransformation recovery (T0) and three serial transfers (T1, T2, and T3) under antibiotic selection were collected for genomic DNA composition analysis. Amplicon digestion by EcoRV reflected the relative abundance of the edited genomes across the whole population (Fig. 3B), demonstrating that (i) the control group using donor-free pCas9n-β-gal never produced any detectable genome editing (unedited, 2 kb; edited, 1 kb); (ii) the 0.1-kb-arm group did not produce edited genomes in T0 or T1, but 6% of the population of T2 carried edited genomes and 55% of the T3 population carried edited genomes; and (iii) in the 0.2-kb, 0.5-kb, and 1-kb groups, editing was not detected in T0 samples but strikingly jumped to over 95% in all T1 samples and then to nearly 100% in T2 and T3. Obviously, the length of the homologous arms exerts an important effect on editing efficiency, and the abundance of edited genomes can be significantly enriched with serial transfers. Once the arm length is greater than 0.2 kb, the editing efficiency of this SNHR strategy was very high (>95%), indicating the ease of markerless genome editing.
FIG 3.
Evaluation of editing efficacy and cargo capacity. (A and B) Effect of arm size on editing efficacy. (A) Design of donor templates with various arm sizes (0.1 to 1 kb), in which the target site (red) is modified to contain an EcoRV site (yellow). The all-in-one vectors with these templates introduce EcoRV into the β-gal gene via SNHR. (B) Editing efficacy evaluation by EcoRV digestion of p3/p4 PCR product. The percentage of edited genome in the whole population of control with donor-free vector (CK), recovered cells (T0), and TMP-resistant cells from three serial transfers (T1 to T3) is calculated by densitometry analysis. (C to F) Genetic cargo capacity evaluation by delivering foreign DNA fragments with various sizes into the genome. (C) Design of four donor templates with 0.71-kb Fd::afp, 1.72-kb promoterless alsS, and 3-kb and 6-kb λ DNA (blue) between 1-kb arms. Using SNHR, the alsS fragment and the remainder are inserted into two different sites, 3198D and pyrF, respectively. (D) PCR identification of ΔpyrF/afp+ and alsS+ mutants generated by the insertion of Fd::afp and alsS fragments, using wild type (CK) as a control. Primer pairs are indicated and drawn in panel C. (E) Enrichment of ΔpyrF/afp+ mutant in the population during serial transfer (T0 to T3) using wild type (CK) as control. (F) Fluorescence microscopy of plasmid-cured ΔpyrF/afp+ mutant.
We then examined the genetic cargo capacity of this strategy in delivering foreign DNA into the genome, which is of critical importance for future genome-level metabolic engineering. We constructed a series of all-in-one vectors in which donor templates contained 1-kb homologous arms and foreign fragments of various size (0.71-kb Fd::afp expression cassette, 1.72-kb promoterless α-acetolactate synthase [alsS], and 3-kb and 6-kb λ DNA) (Fig. 3C). After conducting transformation and serial transfer, we successfully integrated the Fd::afp construct and alsS fragment into the targeted loci (Fig. 3D) but not the larger λ DNA fragments. Meanwhile, we examined enrichment during serial transfer for the Fd::afp construct. The edited cells (ΔpyrF/afp+ mutant) quickly accumulated to nearly 100% after three serial transfers (Fig. 3E). The inserted afp gene in the plasmid-cured ΔpyrF/afp+ mutant was well expressed as shown by fluorescence signal (Fig. 3F). Therefore, the SNHR method can efficiently deliver foreign genes in a single step without a marker.
Precise editing at nonspecific target sites.
The specificity of the 23-bp target sites greatly affects the precision of the Cas9-based editing tools; without this specificity, unwanted off-target mutations will occur (43–45). Since the four target sites tested above are highly specific, they are not ideal for examining editing specificity of this SNHR method. Instead, two highly similar target sites, X21 and X22, were selected from a cipC scaffoldin gene (Ccel_0728). These sites differ by only two bases in the 5′ region preceding the same 12-bp seed region (Fig. 4A; also see Fig. S5 in the supplemental material). Loss of specificity in the seed region will dramatically decrease editing precision such that off-target mutations would occur (43, 44). For each target site, a corresponding donor template was constructed to introduce a deletion of a 12-bp DNA fragment spanning the protospacer adjacent motif. After transformation and plating, we picked individual colonies for site-specific amplification and sequencing. Results (Fig. 4B and C) showed that (i) the editing system targeting X21 exhibited a 100% on-target editing ratio (12/12) for introducing a deletion there, and no off-target mutations (0/12) were detected at X22, and (ii) the editing system targeting X22 also presented a 100% on-target editing ratio (10/10), and no off-target mutations (0/10) occurred at X21. Obviously, this method presented an extraordinary editing precision at nonspecific target sites. This feature does not need the high-specificity target sites for precise genome editing required by other Cas9-based methods (43–45).
FIG 4.
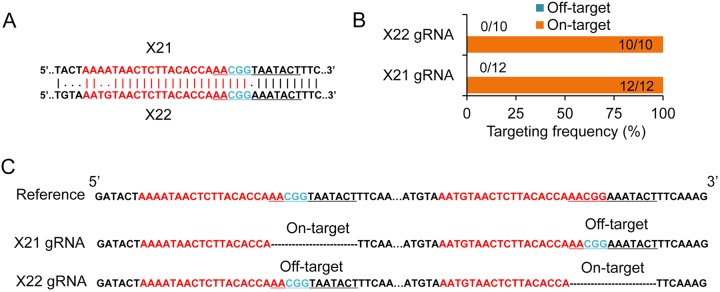
Targeting specificity test. (A) Pairwise alignment of two target sites, X21 and X22 (colored region). The 12-bp-deletion regions are underlined. (B) On-target and off-target frequency in mutants generated by X21 and X22 gRNAs. There are 12 and 10 individual colonies analyzed for X21 and X22, respectively. (C) Results of amplicon sequencing at both sites in each mutant.
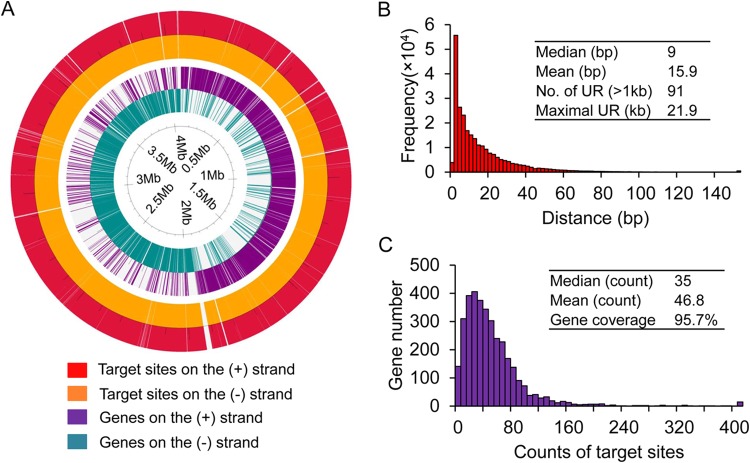
To further assess the potential use of this method for genome editing, we analyzed the targeting space in the genome of C. cellulolyticum. After screening for usable target sites, i.e., those N20NGG sites (N is any base) that are unique, are able to be transcribed, and have a certain degree of specificity, 75% of all extracted N20NGG sites met these criteria (see Table S3 in the supplemental material). The sites were spread across the genome, but there are 91 regions (>1 kb) without any usable target sites with a maximal nontargetable region length of 21.9 kb (Fig. 5A, in the outer two tracks of the map). Further statistical analysis indicated that the median interval distance between target sites was 9 bp (Fig. 5B) and that almost all genes (95.7%) had at least one usable site and the median number of usable sites per gene was 35, without considering fragment length (Fig. 5C). This high targeting coverage was also observed in other bacteria, including E. coli K-12, B. subtilis 168, C. acetobutylicum ATCC 824, and L. reuteri DSM 20016 (see Table S3). Thus, this repurposed CRISPR-Cas9 tool is applicable for editing nearly all encoding genes despite some inaccessible noncoding genomic regions.
FIG 5.
Bioinformatic analysis of targeting space in C. cellulolyticum. (A) Genome-wide distribution of genes and target sites on both DNA strands. White areas in each track indicate gaps between adjacent genes or target sites. The color code is given below the map. (B) Histogram of distance between adjacent usable target sites. Values of mean and median, the number of untouchable regions (UR) with lengths of >1 kb, and the length of the maximal UR are inset within the plot. (C) Histogram of the number of usable target sites in genes. The values of mean, median, and gene coverage are inset.
DISCUSSION
We have developed a highly efficient strategy for genome editing in C. cellulolyticum using Cas9n-mediated single-nick generation and HR. This SNHR strategy is capable of circumventing the DSB lethality to allow versatile editing in hosts with inefficient DSB repair systems. Although NHEJ and HR assist Cas9-mediated genome editing in diverse eukaryotes (2), our study demonstrated that the NHEJ components of C. cellulolyticum were minimally expressed, which resulted in ineffective rejoining of DSBs created by Cas9. Since key components of the NHEJ system, specifically the signature protein Ku, are present in only 27.5% of sequenced microbes (see Fig. S6 in the supplemental material) (37), and even those genomes harboring these genes may not encode functional proteins, as is the case for C. cellulolyticum, the Cas9-/double-nicking-triggered NHEJ system will not work in a majority of prokaryotes. The alternative to NHEJ is template-directed HR, which is a ubiquitous housekeeping process involved in the maintenance of chromosome integrity and the generation of genetic variability, although the exact mechanism of HR may vary (38, 46). Our plasmid-borne homologous donor successfully triggered HR at the nick created by Cas9n but not at the break induced by Cas9. Recent studies showed that single-nick-triggered HR may undergo a distinct mechanism without proceeding through a DSB intermediate of DSB-induced HR (47, 48). It is also possible that DSBs created by Cas9 are more toxic than the single-strand nicks or nick-induced single-end DSBs occurring during DNA replication and may be beyond the host's ability to repair (38). Although little is known about the molecular basis of the C. cellulolyticum type II-C CRISPR-Cas system, our study suggests that the native system did not affect the S. pyogenes type II-A system and might use separate mechanisms (e.g., different PAM and gRNA structure as well as protospacer length) since the gRNA-expressing strain was not able to direct the native Cas9 to accomplish targeted editing. The Cas9 orthogonality demonstrated in E. coli and human cells also supports this point (49).
The SNHR strategy presents unmatched advantages over mainstream bacterial genome-editing tools. Compared with the widely used double-crossover recombination method, it is much faster, more efficient, and more versatile. As we demonstrated, the SNHR strategy allows a one-step generation of an edited genome using a single vector. The high efficiency of this strategy enables markerless editing so that difficulties associated with low transformation efficiency, tedious stepwise screening, and the need for multiple positive-/negative-selection markers can be avoided, unlike in the double-crossover recombination method (1–3). Studies have shown that a low spontaneous recombination frequency in bacteria, which is the basis of double-crossover recombination, can decrease exponentially when reducing the size of homologous arms or increasing the nonhomologous insert between the flanking homologous arms because these changes can affect the efficiency of recombination pathways and RecA binding (40–42, 50). While both SNHR and double-crossover recombination can generate defined mutations (deletion, insertion, and replacement) via HR, the SNHR strategy exhibited a strong ability to use homologous arms as short as 0.2 kb to trigger recombination and deliver DNA fragments within a single step, and so the SNHR method is a more robust method for small gene insertion within a short time frame. However, the genetic cargo capacity is relatively low and needs to be improved in order to integrate the large DNA fragments required for massive metabolic engineering. Group II intron retrotransposition is also widely used for gene disruption in many bacteria (1), yet this method has some targeting limitations, including an obvious bias for intron insertion near the replication origin (51), a relatively sparse targeting space (every few hundred bases on average), and no guarantee of efficiency depending on the insertion site and species (52). In contrast, the SNHR strategy has a very wide targeting space with a median interval distance of 6 to 14 bp in the multiple bacterial genomes analyzed in this study. It also allows editing of over 95% of genes in multiple genomes, demonstrating the great versatility of this editing system. In addition, the customizability of the SNHR strategy, which enables the generation of precise microdeletion, microinsertion, or codon change to inactivate gene function, can minimize the polar effect on downstream genes that can be exerted by intron insertion or insertion/deletion of other large DNA fragments (4, 5). With these demonstrated strengths, the SNHR strategy can overcome the limitations of currently available genetic approaches to engineering bacterial genomes. This new Cas9 technology can be used for in vivo and in situ characterization and alteration of biological functions of interest (e.g., DNA sequence motif, gene, protein domain, and protein localization), in addition to genetic engineering of Clostridia and other industrial microorganisms for metabolic and physiologic improvement.
In addition, compared with reported Cas9-based strategies (i.e., Cas9-NHEJ/HR, double nicking-NHEJ/HR) (9, 11, 45), this strategy can enable precise editing at target sites with low specificity. For instance, Cas9n guided by X21 gRNA probably induces at least two nicks in the C. cellulolyticum genome, including at the on-target X21 and the off-target X22, but the donor template of X21 will specifically choose the nick in X21 to repair through HR and then other nicks will be faithfully religated without introducing any unwanted mutations, as usually occurs during NHEJ-dependent DSB repair. That means that the SNHR strategy not only improves editing accuracy but also expands our editing target space. However, strategies still need to be developed to target those genomic regions lacking targeting sites and to increase targeting resolution across genomes, which is problematic for all Cas9-based methods, including SNHR. Considering that different Cas9s have varied PAM preferences (e.g., NGG in S. pyogenes, NNNNGANN in Neisseria meningitidis, and NAAAAN in Treponema denticola) (49), exploiting or engineering Cas9 to have an expanded ability to use multiple short protospacer adjacent motifs, and to decrease the length requirement of protospacers without sacrificing targeting specificity, may offer solutions for allowing accurate editing anywhere.
In conclusion, the single-nick-triggered HR strategy described here allows for markerless gene delivery and versatile editing in a single step with a high editing efficiency and precision. This method provides an exemplary strategy for precise genome editing in prokaryotes that are sensitive to DSB toxicity. This approach will facilitate microbial genome editing for fundamental and applied research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NSF EPSCoR award EPS 0814361, by the Office of the Vice President for Research at the University of Oklahoma, and by the Collaborative Innovation Center for Regional Environmental Quality.
We thank L. A. Marraffini and W. Jiang (Rockefeller University) for kind suggestions. We thank members of the Zhou laboratory for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00873-15.
REFERENCES
- 1.Esvelt KM, Wang HH. 2013. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol 9:641. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu T, Li Y, Van Nostrand JD, He Z, Zhou J. 2014. Cas9-based tools for targeted genome editing and transcriptional control. Appl Environ Microbiol 80:1544–1552. doi: 10.1128/AEM.03786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heap JT, Ehsaan M, Cooksley CM, Ng YK, Cartman ST, Winzer K, Minton NP. 2012. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res 40(8):e59. doi: 10.1093/nar/gkr1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu T, Li Y, He Z, Zhou J. 2014. Dockerin-containing protease inhibitor protects key cellulosomal cellulases from proteolysis in Clostridium cellulolyticum. Mol Microbiol 91:694–705. doi: 10.1111/mmi.12488. [DOI] [PubMed] [Google Scholar]
- 5.Maamar H, Valette O, Fierobe HP, Belaich A, Belaich JP, Tardif C. 2004. Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol Microbiol 51:589–598. doi: 10.1046/j.1365-2958.2003.03859.x. [DOI] [PubMed] [Google Scholar]
- 6.Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, Whitt JT, Keatinge-Clay AT, Lambowitz AM, Ellington AD. 2013. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol 9:685. doi: 10.1038/msb.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. 2013. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selle K, Barrangou R. 2015. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Oh JH, van Pijkeren JP. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobb RE, Wang Y, Zhao H. 8 December 2014. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Zheng G, Jiang W, Hu H, Lu Y. 2015. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. 7 April 2015. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 21.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. 2014. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5(1):e00928-13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desvaux M. 2005. Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev 29:741–764. doi: 10.1016/j.femsre.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Lan EI, Liao JC. 2013. Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour Technol 135:339–349. doi: 10.1016/j.biortech.2012.09.104. [DOI] [PubMed] [Google Scholar]
- 26.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J. 2012. PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics 13:299. doi: 10.1186/1471-2164-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Xu T, Tschaplinski TJ, Engle NL, Yang Y, Graham DE, He Z, Zhou J. 2014. Improvement of cellulose catabolism in Clostridium cellulolyticum by sporulation abolishment and carbon alleviation. Biotechnol Biofuels 7:25. doi: 10.1186/1754-6834-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA. 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard L, White JA, Birch PR, Toth IK. 2006. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics 22:616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- 34.Osterberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu Rev Microbiol 65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 35.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, Hogsett DA, Caiazza NC. 2010. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol 76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitcher RS, Brissett NC, Doherty AJ. 2007. Nonhomologous end-joining in bacteria: a microbial perspective. Annu Rev Microbiol 61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 37.Bowater R, Doherty AJ. 2006. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet 2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui GZ, Hong W, Zhang J, Li WL, Feng YG, Liu YJ, Cui Q. 2012. Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J Microbiol Methods 89:201–208. doi: 10.1016/j.mimet.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Kung SH, Retchless AC, Kwan JY, Almeida RP. 2013. Effects of DNA size on transformation and recombination efficiencies in Xylella fastidiosa. Appl Environ Microbiol 79:1712–1717. doi: 10.1128/AEM.03525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khasanov FK, Zvingila DJ, Zainullin AA, Prozorov AA, Bashkirov VI. 1992. Homologous recombination between plasmid and chromosomal DNA in Bacillus subtilis requires approximately 70 bp of homology. Mol Gen Genet 234:494–497. doi: 10.1007/BF00538711. [DOI] [PubMed] [Google Scholar]
- 42.Bertolla F, Van Gijsegem F, Nesme X, Simonet P. 1997. Conditions for natural transformation of Ralstonia solanacearum. Appl Environ Microbiol 63:4965–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin YN, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. 2014. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocha EP, Cornet E, Michel B. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet 1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD. 2011. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res 39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis L, Maizels N. 2011. DNA nicks promote efficient and safe targeted gene correction. PLoS One 6:e23981. doi: 10.1371/journal.pone.0023981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen P, Huang HV. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong J, Karberg M, Lambowitz AM. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res 31:1656–1664. doi: 10.1093/nar/gkg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perutka J, Wang WJ, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.