Abstract
Lanthionine-containing peptides (lanthipeptides) are a rapidly growing family of polycyclic peptide natural products belonging to the large class of ribosomally synthesized and posttranslationally modified peptides (RiPPs). Lanthipeptides are widely distributed in taxonomically distant species, and their currently known biosynthetic systems and biological activities are diverse. Building on the recent natural product gene cluster family (GCF) project, we report here large-scale analysis of lanthipeptide-like biosynthetic gene clusters from Actinobacteria. Our analysis suggests that lanthipeptide biosynthetic pathways, and by extrapolation the natural products themselves, are much more diverse than currently appreciated and contain many different posttranslational modifications. Furthermore, lanthionine synthetases are much more diverse in sequence and domain topology than currently characterized systems, and they are used by the biosynthetic machineries for natural products other than lanthipeptides. The gene cluster families described here significantly expand the chemical diversity and biosynthetic repertoire of lanthionine-related natural products. Biosynthesis of these novel natural products likely involves unusual and unprecedented biochemistries, as illustrated by several examples discussed in this study. In addition, class IV lanthipeptide gene clusters are shown not to be silent, setting the stage to investigate their biological activities.
INTRODUCTION
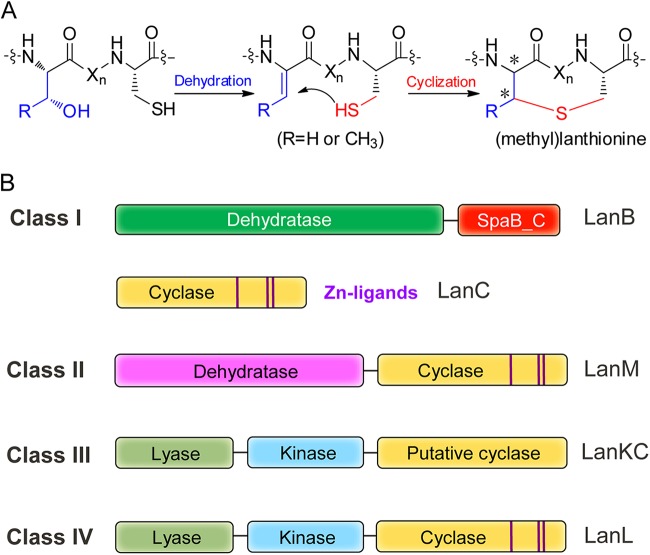
Lanthipeptides are a rapidly growing family of polycyclic peptides characterized by the presence of the thioether cross-linked amino acids lanthionine and methyllanthionine (1–5). These compounds are widely distributed in taxonomically distant species and display very diverse biological activities, ranging from antimicrobial to antiallodynic (5–7). Antibacterial lanthipeptides, such as the commercially used food preservative nisin, are termed lantibiotics (8). Lanthipeptides are generated from a ribosomally synthesized linear precursor peptide (generically termed LanA) and therefore belong to the large class of natural products that are ribosomally synthesized and posttranslationally modified peptides (RiPPs) (9). The precursor peptide LanA consists of a C-terminal core peptide where all posttranslational modifications take place and an N-terminal leader peptide that is important for posttranslational modifications and that is subsequently removed by proteolysis (10, 11). The installation of the (methyl)lanthionine thioether bridges is achieved by the initial dehydration of Ser and Thr residues in the precursor peptides, followed by stereoselective intramolecular Michael-type addition of Cys thiols to the newly formed dehydroamino acids (Fig. 1A).
FIG 1.
Biosynthesis of lanthipeptides. (A) The mechanism of (methyl)lanthionine formation. (B) The four currently known classes of lanthipeptide synthetases. Xn represents a peptide linker. The conserved zinc-binding motifs are highlighted by the purple lines in the cyclase domains. SpaB_C is an elimination domain present as a stand-alone protein in thiopeptide biosynthesis and as the C-terminal domain of LanB enzymes in lanthipeptide biosynthesis.
Four classes of biosynthetic enzymes are known to catalyze lanthionine formation (2, 12) (Fig. 1B). Class I lanthionine synthetases consist of a dehydratase and a cyclase that are generically termed LanB and LanC, respectively (8). Class II enzymes are generically named LanMs, which are single polypeptides containing an N-terminal dehydratase domain that bears no homology to any functionally known enzymes and a C-terminal LanC-like cyclase domain (13, 14). Class III and class IV synthetases are trifunctional enzymes termed LanKC (15) and LanL (16), respectively. These enzymes contain an N-terminal lyase domain and a central kinase domain but differ in their C termini. LanC and the C-terminal domains of LanM and LanL contain a conserved zinc-binding motif (Cys-Cys-His/Cys) (17, 18), whereas the C-terminal cyclase domains of LanKC lack these conserved residues (Fig. 1B). The four distinct classes of biosynthetic machinery reflect the functional importance of lanthionine scaffolds and a convergent evolution process to produce them (12). Notably, products generated by these four classes of lanthionine synthetases are not limited to lanthipeptides. For example, many lanthipeptide-like biosynthetic gene clusters encode precursor peptides that have no Cys residues, and their products are therefore not lanthipeptides (19).
Over the past 5 years, several studies have reported bioinformatic genome mining efforts based on the then-available genomes. These studies have typically focused on an enzyme-specific query, such as class I LanB and LanC enzymes (20), class II LanM proteins (21), or the bifunctional LanT proteins, which transport lanthipeptides and remove their leader sequences (22). Building on the recent natural product gene cluster family (GCF) project (23), here we report large-scale analysis of lanthipeptide-like biosynthetic gene clusters that is driven by similarities in entire clusters rather than specific enzymes. We focused the analysis on lanthipeptides from Actinobacteria, which are relatively understudied compared to family members from Firmicutes. Actinobacteria carry a wealth of natural product biosynthetic gene clusters whose products encompass highly diverse chemical structures. This phylum has been the source and/or inspiration for the majority of pharmaceutically useful natural products (24, 25). Although Actinobacteria constitute only a small proportion of known lanthipeptide producers, they encode a diverse set of posttranslational modifications that have not been observed for lanthipeptides from other phyla (2) (e.g., lysinoalanine formation in cinnamycin [26], tryptophan chlorination and proline dihydroxylation in microbisporicin [27, 28, also called NAI-107 [29], and glycosylation in NAI-112 [30]). The unique modifications in actinobacterial lanthipeptide biosynthesis suggest great potential for exploring unknown lanthipeptide chemical space in this phylum. Our analysis suggests that lanthipeptides are likely much more diverse than is currently appreciated and contain many novel posttranslational modifications. More intriguingly, our analysis suggests that in some cases lanthipeptide synthetases have been repurposed for producing natural products other than lanthipeptides, expanding natural product diversity.
MATERIALS AND METHODS
Chemicals, media, and general methods.
Unless otherwise noted, reagents were purchased from Fisher Scientific. Yeast extract was purchased from Becton Dickinson and N-Z amine type A from Sigma-Aldrich. Acetonitrile for matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) analysis was purchased from Avantor Performance Materials. MALDI–time of flight (MALDI-TOF) MS was carried out on a Bruker UltrafleXtreme TOF/TOF. Reversed-phase liquid chromatography electrospray ionization (ESI) tandem MS was carried out and processed using a Synapt ESI quadrupole TOF mass spectrometry system (Waters).
Bioinformatics.
GCF analysis, BLAST searches, and phylogenetic analysis were all performed as described previously (23). All GCF information can be found at http://www.igb.illinois.edu/labs/metcalf/gcf/lant.html. Use of these pages requires a JavaScript-enabled web browser. Mouseover of a gene provides a rough annotation and should be confirmed with additional methods before further use. Mouseover also highlights other genes on the page that are in the same homology group. On click, each gene will open a new tab containing the amino acid sequence and a link to BLAST analysis of the sequence. Strain designations such as Streptomyces species B-3253 refer to strains in the Culture Collection (NRRL) of the Agricultural Research Service (ARS). The genomic information can be found by entering B-3253 into the NCBI search field and then searching in the Assembly or BioSample databases. Designations such as Streptomyces species B-3253_23 refer to cluster 23 in this strain.
Network analysis was performed by BLASTP searches, comparing each sequence against each other sequence. A VBA script was written to remove all duplicate comparisons, and the result was imported into the Cytoscape software package (31). The nodes were arranged by using the yFiles organic layout provided with Cytoscape version 2.8.3. Sequence logos were generated online using Weblogo (32).
Cell cultures, sample preparation, and MS analysis.
Dimethyl sulfoxide (DMSO) stocks of Streptomyces katrae ISP5550 were prepared by mixing 600 μl of an S. katrae ISP5550 liquid culture with an equal volume of sterilized 20% dimethyl sulfoxide (DMSO). The resulting solution was mixed, flash frozen, and stored at −80°C. S. katrae ISP5550 was inoculated from DMSO stocks onto solid ATCC172 medium (20 g soluble starch, 5 g yeast extract, 5 g N-Z amine type A, 1 g calcium carbonate, 15 g agar, and 1 liter of Milli-Q water; after autoclaving the medium, sterilized glucose was added to a 1% final concentration) and incubated at 30°C for 4 to 5 days.
The presence of venezuelins was detected using colony MALDI-TOF MS. A small portion (approximately 1 by 1 mm) of colony was transferred onto a Bruker MTP 384 polished steel target plate. On top of the colony, 2 μl of sinapic acid matrix was overlaid and allowed to air dry. Sinapic acid matrix was prepared by saturating an acetonitrile (MeCN)–Milli-Q water (80:20) solution with sinapic acid (Sigma-Aldrich). MALDI-TOF MS analysis of the sample was performed using a Bruker UltrafleXtreme MALDI-TOFTOF-MS maintained by the UIUC School of Chemical Sciences Mass Spectrometry Laboratory. Data acquisition was completed using Bruker's FlexControl program, while FlexAnalysis was used for data analysis. Prior to data acquisition, the instrument was calibrated using peptide calibration standard II (Bruker Daltonics, Billerica, MA) and the matrix suppression mode set to deflection mode. The data were not subjected to processing.
Upon detection of venezuelins by colony MALDI-TOF MS from solid cultures of S. katrae ISP5550, liquid cultures were analyzed for the production of venezuelins. A single colony was transferred into 50 to 100 ml of liquid ATCC172 medium, prepared in the same way as the solid medium with the omission of agar. The liquid culture was incubated with shaking at 30°C for 12 days. Supernatant from the liquid cultures was obtained by centrifuging at 4,650 × g for 15 min. The hydrophobic components of the supernatant were desalted by ZipTip (EMD Millipore) according to the manufacturer's instructions. The ZipTip contents were directly eluted onto a Bruker MTP 384 polished steel target plate on a spot containing 1 μl of sinapic acid matrix. Analysis by MALDI-TOF MS was performed as described above.
In preparation for ESI-MS, peptide components of the supernatant were precipitated using ammonium sulfate. A 60% final concentration of ammonium sulfate was achieved with the addition of 30 g of ammonium sulfate to 50 ml of supernatant. The solution was allowed to incubate at room temperature for 30 min. The resulting precipitate was pelleted by centrifugation at 22,789 × g for 10 min, and the supernatant was decanted. The pellet was redissolved in Milli-Q water and subjected to ESI-MS analysis using an Acquity ultraperformance liquid chromatography (UPLC) system coupled to a Waters Synapt ESI-QTOF. The UPLC was equipped with an Acquity UPLC BEH C8 1.7-μm, 1.0- by 100-mm column. Sample was introduced into the column via an Acquity autoinjector and subjected to a gradient of 2% B from 0 to 2 min, 2% to 98% B from 2 to 12 min, and 98% B from 12 to 15 min (A was 0.1% formic acid in water and B was 0.1% formic acid in MeCN). The Waters MassLynx V4.1 program was used to tune, calibrate, and acquire data. [Glu1]-Fibrinopeptide B human (Sigma-Aldrich) was used as an external calibrant, and the mass of venezuelin was verified to be within 5 ppm of its calculated mass. Collision-induced dissociation was achieved using a series of energy ramps ranging from 30 to 60 eV.
Isolation of venezuelins.
S. katrae ISP5550 liquid culture supernatant was subjected to pressure-based sample concentration rather than ammonium sulfate precipitation. After centrifugation, 40 ml of culture supernatant was further clarified by filtration through a 0.22-μm nylon membrane and then loaded into an Amicon stirred cell equipped with a 1-kDa nominal-molecular-mass limit regenerated cellulose disc (EMD Millipore). The clarified supernatant was concentrated at room temperature from a volume of 40 to 2 ml over approximately 4 h. The concentrated supernatant was flash frozen and stored at −20°C prior to fractionation by analytical reversed-phase high-performance liquid chromatography (HPLC) using an Agilent 1260 Infinity instrument equipped with a Phenomenex Luna 10-μm, C18(2) 100-Å, 250- by 4.6-mm column. The instrument was managed using Agilent Instrument 1 (online) software. The mobile phase consisted of A (0.1% trifluoroacetic acid [TFA] [Acros Organics]) in water and B (0.1% TFA in 80% MeCN–20% water), with a gradient of 2% B from 0 to 5 min, 2 to 100% B from 5 to 50 min, and 100% B from 50 to 60 min. A 0.5-μl aliquot of each fraction eluting after 20 min was spotted with an equal volume of sinapic acid matrix and analyzed by MALDI-TOF MS for the presence of venezuelin. Venezuelin-containing fractions (typically eluting at 35 to 38 min) were flash frozen and dried in vacuo. Three 1-ml injections of concentrated S. katrae ISP5550 supernatant yielded 100 μg of HPLC-purified, dry material.
Detection of lanthionine.
HPLC-purified venezuelin was analyzed by gas chromatography (GC)-MS as previously described (33). In brief, the sample was redissolved in 3 ml of 6 M HCl and heated at 110°C for 10 to 12 h. The reaction mixture was dried and mixed with a solution of acetyl chloride (Sigma-Aldrich) and methanol. Following reflux at 110°C for 45 min, the reaction mixture was dried and 3 ml of dichloromethane (Sigma-Aldrich) was added. After cooling the solution to 0°C, 1 ml of pentafluoropropionic anhydride (Sigma-Aldrich) was added, and the reaction mixture was refluxed for 15 min at 110°C. The reaction mixture was dried and stored at −80°C, and prior to GC-MS analysis, the sample was redissolved in 100 μl of methanol. Particulates were removed by centrifugation at 23,700 × g for 4 min. An Agilent Hewlett-Packard 5973 mass spectrometer equipped with a CP-Chirasil-L Val 0.12-μm, 25-m by 0.25-mm column (7-in. cage; Agilent Technologies) was used to perform the GC-MS analysis. The sample was introduced to the instrument via a split (1:5) injection of 2 to 5 μl with a 2.0-ml/min helium gas flow rate. The initial inlet temperature was 190°C, and a gradient of 20°C/min from 160°C for 5 min to 180°C for 10 min was used. The following mass spectrometer settings were used: 185°C for the MSD transfer line heater, 150°C for the MS Quad analyzer, and 230°C for the ion source. Data were acquired using scan and selected ion monitoring at 365 and 379 Da. The identity of (methyl)lanthionine residues was confirmed by adding authentic methyl(lanthionine) standards to the sample and analyzing the resulting mixture by GC-MS. The mixtures of sample and standard were prepared such that there was an approximately equal contribution of signal arising from the sample and standard.
RESULTS
Phylogenetic analysis of LanC-like enzymes.
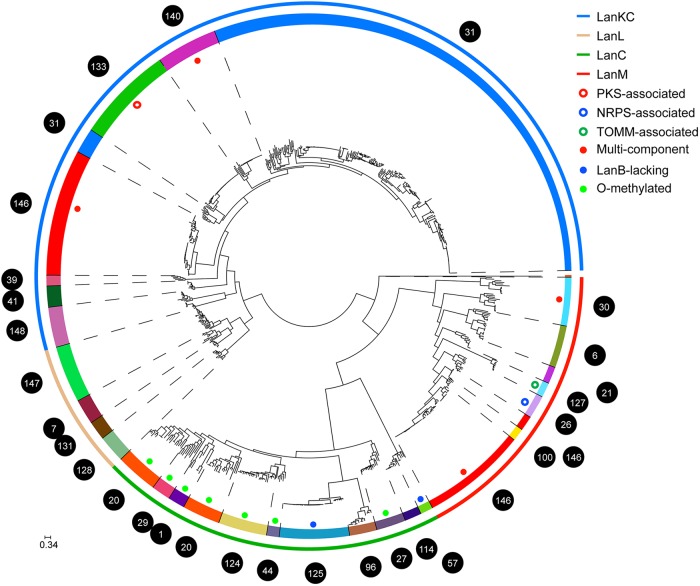
Mining of 830 actinobacterial genomes, including 344 newly sequenced genomes obtained for the GCF project (23), revealed 1,163 lanthipeptide-like gene clusters, which were grouped into more than 100 different families according to their sequence and organizational similarities (see Fig. S1 in the supplemental material) (23). Strikingly, the GCFs that produce the known lanthipeptides produced by Actinobacteria make up a very small fraction, illustrating the tremendous diversity of yet-to-be-investigated GCFs. To first investigate the phylogenetic relationship of LanC-like enzymes from different gene clusters, we constructed a maximum-likelihood (ML) tree using enzymes from 29 large families that each are found in at least five different species (or operational taxonomic units with a clustering threshold of 99% identity in ribosomal proteins [23]). This analysis makes it possible to infer a gene's involvement in a GCF based on conservation over evolutionary time (34). Gene conservation within a GCF relies on the inclusion of multiple species in the analysis, as genome diversity within a single Streptomyces species is much reduced and synteny extends nearly the entire length of the chromosome (35). The tree shows distinct phylogenetic distributions of enzymes from different classes, as LanC and the C-terminal domains of LanM, LanL, and LanKC fall into four different clades (Fig. 2). This observation is consistent with our previous proposal that the four classes of lanthipeptide synthetases have evolved from different ancestors (12, 36). Interestingly, three cases of novel GCFs emerge within an existing GCF: GCF100 branches within the LanM clade of GCF146 (this GCF also contains a LanKC-encoding gene and hence appears twice in Fig. 2), GCF1 and GCF29 branch within GCF20, and GCF133 and GCF140 branch within GCF31. Below we highlight several families of lanthipeptide-like gene clusters that are most interesting in that they deviate from the currently known lanthipeptide biosynthetic systems and likely encode novel and unusual biochemistry in the biosynthesis of lanthionine-related natural products.
FIG 2.
An ML tree of LanCs and LanC-like domains. In the outer ring, the four different classes of enzymes are depicted by the color defined in the legend, whereas the different GCFs are shown in the inner ring with random colors. The GCF numbers are defined in reference 23 and can be found at http://www.igb.illinois.edu/labs/metcalf/gcf/lant.html. PKS, polyketide synthetase; NRPS, nonribosomal peptide synthetase; TOMM, thiazole/oxazole-modified microcins.
Novel multicomponent lanthipeptide systems.
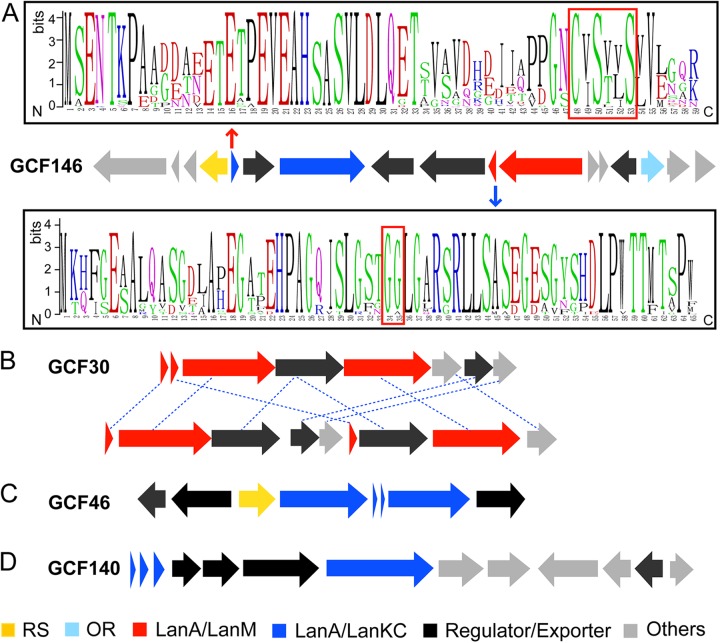
Two-component lanthipeptides are normally potent antimicrobial agents in which the two posttranslationally modified peptides exert strong synergistic biological activity (37). To date, isolated two-component lantibiotics have been confined to Firmicutes, but several families of lanthipeptide gene clusters in the Actinobacteria likely encode two-component lanthipeptides. A very recent study also suggested the presence of two-component lanthipeptides in three Streptomyces strains (22). Among the most interesting class is GCF146 (70 members), which is currently the second-largest lanthipeptide GCF (see Fig. S1 in the supplemental material). Each member encodes a LanM and a LanKC, and the core peptides of the putative LanKC substrates have a strictly conserved CxSxxS motif (Fig. 3A). Previous studies have shown that LanKC proteins can turn SxxSxxxC motifs into a labionin scaffold by initial Michael addition of the C-terminal Cys to a central dehydroalanine (formed from Ser) and subsequent addition of the resulting enolate onto the N-terminal dehydroalanine of the motif (38–42). The CxSxxS motif in GCF146 could be a variation of the labionin-forming sequence. Another interesting finding is that a radical SAM protein containing a canonic CxxxCxxC motif (43, 44) is strictly conserved in these gene clusters, which may catalyze an unprecedented posttranslational modification in lanthipeptide maturation. The putative LanM substrates in GCF146, however, do not contain cysteine residues (Fig. 3A) and are similar to the recently characterized NpnAs (19). Like in the Npn cluster, an oxidoreductase containing the characteristic NAD-binding Rossmann fold is also encoded in the gene cluster. A possible role for this oxidoreductase is to produce d-amino acids by stereoselective reduction of the dehydroamino acids resulting from LanM catalysis (45–47). These findings set the stage for investigation whether the mature products of the LanM and LanKC substrates indeed form a novel functional two-component lanthipeptide system in which one peptide does not contain lanthionine/labionin structures.
FIG 3.
GCFs that likely produce two-component lanthipeptides. (A) A representative gene cluster of GCF146 and the sequence logos of the precursor peptide substrates. The predicted LanKC substrate/enzyme pair is shown in blue, and the predicted LanM substrate/enzyme pair is shown in red. The strictly conserved CxSxxS motif of the putative LanKC substrate and the GG/A leader peptide cleavage site of the putative LanM substrate are highlighted by red boxes. (B) A representative gene cluster of GCF30 that likely encodes a haloduracin-like system. The gene clusters appear in two different types, and homologous enzymes are linked by blue dashed lines. (C) A representative gene cluster of GCF46 that may encode class III two-component lanthipeptides. (D) A representative gene cluster of GCF140 that may encode three precursor peptides and a LanKC. The putative functions of the gene products are shown by colors. The logos of the precursor peptides in GCF30, GCF46, and GCF140 are shown in Fig. S2 to S4, respectively, in the supplemental material. The boundaries of each gene cluster are predicted based on the conserved genes within each GCF and are not clearly defined. RS, radical SAM; OR, oxidoreductase.
GCF30 (12 members) consists of gene clusters appearing in two different types of organization, both of which encode two LanMs and two LanA substrates (Fig. 3B). Both LanAs have a Gly-Gly/Ala motif characteristic for leader peptide removal and multiple Cys and Ser/Thr residues (see Fig. S2 in the supplemental material). Therefore, the products of these gene clusters are likely similar to well-known two-component lantibiotics such as haloduracin and lichenicidin produced by bacilli (20, 37, 48–51). One GCF30 member from Streptomyces viridochromogenes was also noted in a recent genome mining study focusing on LanT analogs (22).
GCF46 (6 members) consists of gene clusters encoding two LanAs and two LanKC enzymes (Fig. 3C). The two LanKCs share high sequence similarities (about 40% identity) with each other, and the two LanAs are similar but not identical (see Fig. S3 in the supplemental material). Both LanAs have highly conserved CxxSxxS motifs, suggesting a potential first class III two-component lanthipeptide. A radical SAM protein is also encoded in the gene cluster, whose function remains to be determined. Notably, the two LanAs in the gene clusters from GCF46 are significantly shorter than other LanAs and span only about 30 amino acids (see Fig. S3 in the supplemental material). These short precursor peptides are possibly truncated LanAs, which have recently been found in several lanthipeptide biosynthetic systems and were shown to be genuine substrates (19).
Another intriguing lanthipeptide biosynthetic system was found in GCF140 (29 members), whose gene clusters each encode three putative precursor peptides, a LanKC, and a set of hypothetical enzymes (Fig. 3D). Notably, all three precursor peptides lack a Cys residue in the core peptide region, whereas one peptide has a Cys residue in the leader peptide (see Fig. S4 in the supplemental material). It remains to be determined whether these putative precursor peptides are substrates of the corresponding LanKC enzyme.
Lanthionine synthetases associated with polyketide and nonribosomal peptide biosynthesis.
The third-largest lanthipeptide family in the investigated genome collection is GCF133 (48 members) (see Fig. S1 in the supplemental material), which represents a very unusual biosynthetic cluster. Each member of this family encodes a LanKC and a series of polyketide synthetase (PKS)-like enzymes, including an acyl carrier protein (ACP), two 3-oxoacyl-ACP synthases (KS), a malonyltransferase (MT), a short-chain dehydrogenase/reductase (SDR) likely to be a ketoreductase (KR), and a putative enoyl coenzyme A (enoyl-CoA) hydratase (Fig. 4A). These enzymes are reminiscent of a type II PKS system, which, unlike modular PKSs, are complexes of monofunctional enzymes (52). A lanthipeptide precursor peptide could not be found in the cluster, but the LanKC is likely to be involved in the biosynthesis of the product of GCF133, as its encoding gene is embedded in the middle of the cluster (Fig. 4A). A P450 enzyme is also encoded in the gene cluster, which shares significant similarity (37% identity and 53% similarity) with EryF, which is involved in erythromycin biosynthesis (53). In addition, the GCF encodes an enzyme sharing significant similarities to nonheme dinuclear Fe or Fe/Mn enzymes related to the small subunit of ribonucleotide reductase (RNR). It is currently too early to propose potential products of GCF133. Regardless, the biosynthesis almost certainly involves unprecedented and intriguing biochemistry.
FIG 4.
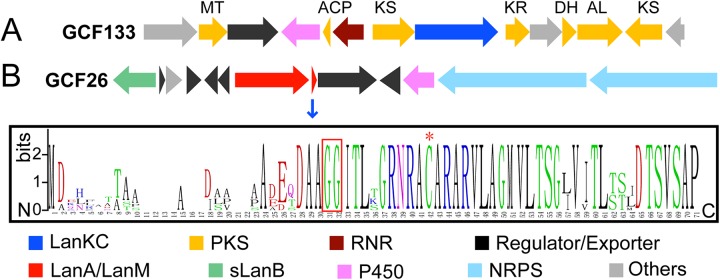
Association of genes encoding lanthipeptide synthetases with PKS and NRPS genes. (A) A representative gene cluster of GCF133 that likely encodes lanthipeptide synthetase-PKS hybrid systems. (B) A representative gene cluster of GCF26 that likely encodes lanthipeptide synthetase-NRPS hybrid systems, and a logo depicting the precursor peptide sequences. The empty sites in the leader peptide region are a consequence of high sequence divergence (i.e., no conserved residues). The sequence alignment of GCF26 precursor peptides is shown in Fig. S5 in the supplemental material. The GG leader peptide cleavage site and the conserved Cys residue in the putative core region are highlighted by a red box and a red asterisk, respectively. The putative functions of gene products are shown by colors. MT, malonyl transferase; ACP, acyl carrier protein; KS, ketosynthase; KR, ketoreductase; DH, dehydratase/enoyl-CoA hydratase; AL, ATP-dependent ligase; RNR, ribonucleotide reductase-like (these RNR-like proteins appear to contain the ligands for a dinuclear metal cluster but not a tyrosine for radical formation); sLanB, small LanB. The boundaries of each gene cluster are predicted based on the conserved genes within each GCF and are not clearly defined.
We have recently shown that the nisin dehydratase NisB consists of an N-terminal domain that utilizes glutamyl-tRNA to glutamylate the side chains of Ser and Thr residues that are targeted for dehydration and a small C-terminal domain (SpaB_C domain) that is required for glutamate elimination to form the dehydroamino acids (54). Notably, a set of small LanBs (sLanBs) lacking the elimination domain are apparently associated with nonribosomal peptide synthetases (NRPSs), possibly adding amino acids to the growing peptide in a tRNA-dependent manner (54). While the majority of these sLanBs reside on their own polypeptide chain, the NRPS GCF348 (23) contains a gene cluster wherein the sLanB resides in a module of an NRPS along with an adenylation domain and a peptidyl carrier protein, providing further support for integration of sLanB into the assembly line of certain NRPS biosynthetic pathways.
We also noted a unique family, GCF26 (8 members), that encodes an sLanB, a LanM, and an NRPS (Fig. 4B). A putative LanA substrate is also encoded in these clusters, which has a conserved Gly-Gly proteolytic leader peptide removal site and multiple Ser/Thr residues but only one Cys in the predicted core peptide (Fig. 4B; see Fig. S5 in the supplemental material). These observations suggest the possibility of cross talk between ribosomal and nonribosomal peptide natural products, which has recently also been reported in another context for pheganomycin biosynthesis (55). Interestingly, the LanM enzymes encoded in GCF26 all lack the normally highly conserved zinc ligands that are important for cyclization (see Fig. S6 and Table S1 in the supplemental material). Of the 211 LanM enzymes encoded in the genomes investigated here, 196 have the canonical conserved CC(H/C) zinc-binding ligand set. Of the 15 that do not, 12 are encoded in clusters with NRPS genes (GCF26 and GCF98) (see Table S1 in the supplemental material). These findings suggest that these LanM enzymes are used only for dehydration processes and not for cyclization.
Clusters containing LanC but not LanB enzymes.
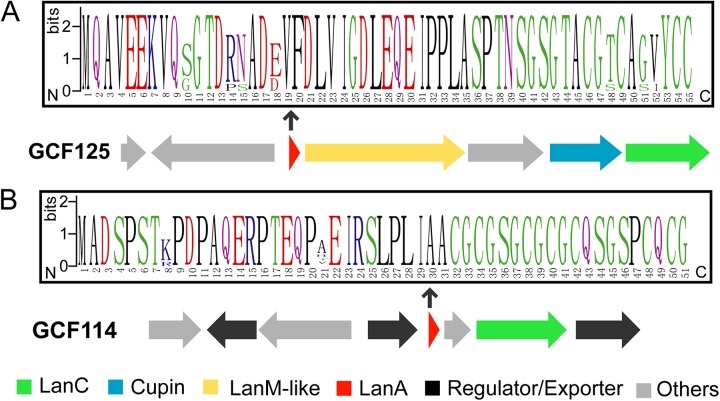
Dehydration of Ser/Thr residues catalyzed by LanBs is a prerequisite for LanC-catalyzed cyclizations (Fig. 1A). However, LanC-like enzymes are widespread in eukaryotes, whereas the corresponding LanB homologs are not present (12, 36). The molecular details of the functions of eukaryotic LanC-like enzymes remain largely elusive (56–61). In prokaryotes, orphan LanCs have also been observed sporadically, which could possibly be evolutionary vestiges (12, 21). In our analysis, we noted that GCF125, the fourth-largest lanthipeptide family in the investigated Actinobacteria genome collection, with 36 members (see Fig. S1 in the supplemental material), encodes LanCs but not LanBs (Fig. 5A). Putative LanA substrates are also encoded in the gene cluster, which are highly conserved in sequence and contain multiple Cys and Ser/Thr residues (Fig. 5A). An enzyme from the cupin family, a large family of enzymes with highly diverse functions (62), is also present in these gene clusters. Interestingly, a hypothetical protein sharing weak similarity with LanM enzymes (BLAST E value of ∼10−6 with the TIGR03897 family consisting of LanMs) is also encoded (Fig. 5A), suggesting a novel lanthipeptide biosynthetic system. This LanM-like stand-alone enzyme is likely a dehydratase and possibly a surrogate for the canonical LanB enzymes usually found in conjunction with LanC proteins. A stand-alone dehydratase domain related to LanM proteins is also found in PoyE, which is involved in polytheonamide biosynthesis (63), although the LanM-like proteins in GCF125 are more divergent from canonical LanM proteins than PoyE. The very high density of Ser, Cys, and Gly residues in the substrate peptides (Fig. 5A) is reminiscent of thiazole/oxazole-modified microcins (TOMMs) (64), but no other RiPP biosynthetic genes were found in the gene clusters.
FIG 5.
Putative LanC-containing lanthipeptide systems without LanB enzymes. (A) A representative gene cluster and the precursor peptide sequence logo of GCF125. (B) A representative gene cluster and the precursor peptide sequence logo of GCF114. The putative functions of gene products are shown by colors. The boundaries of each gene cluster are predicted based on the conserved genes within each GCF and are not clearly defined.
Another notable family whose members encode LanCs without the corresponding LanBs is GCF114 (9 members) (Fig. 5B). No other lanthipeptide synthetase-like enzymes are found in this family. Once again, the putative LanAs encoded in these gene clusters contain multiple Gly, Ser, and Cys residues (Fig. 5B), presenting a potentially very interesting natural product biosynthesis system for future investigations.
Orchestration of lanthipeptide and TOMM biosynthesis.
Thiopeptides are a class of RiPPs characterized by a six-member nitrogen-containing heterocycle that is believed to be formed from cycloaddition of dehydroalanine residues (65). In addition, thiopeptides typically contain azole heterocycles. The dehydroalanines in thiopeptides are formed by a LanB-like enzyme and the azoles by a cyclodehydratase/dehydrogenase pair (66, 67). GCF127 (8 members; currently limited to Kitasatospora) is a unique family that also combines lanthipeptide and TOMM biosynthetic machinery but in a manner different from that for the thiopeptides. This family encodes a LanM and a pair of TOMM biosynthetic enzymes, including a cyclodehydratase and a dehydrogenase. Notably, based on sequence, the LanM has a functional dehydratase domain but the C-terminal domain is not homologous to the canonical LanC-like cyclization domain. The putative cyclodehydratase is a fusion protein containing an N-terminal docking domain and a C-terminal YcaO domain (68), an organization similar to that of the cyclodehydratases involved in thiopeptide and cyanobactin biosynthesis (Fig. 6) (64, 66–71). The dehydrogenase is flavin mononucleotide dependent, and related enzymes catalyze the conversion of azolines to azoles (64, 72). The two TOMM enzymes and LanM in GCF127 are encoded together with a P450 enzyme, a putative precursor substrate, and three transporter proteins.
FIG 6.
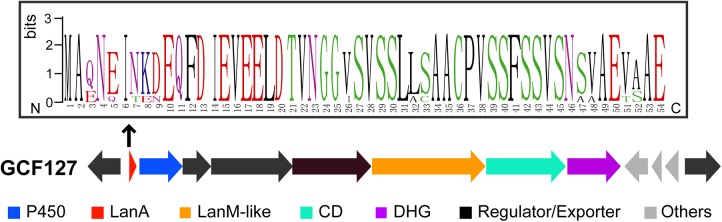
A representative gene cluster of GCF127 that likely encodes lanthipeptide-TOMM hybrid systems. The putative functions of gene products are shown by colors. The boundaries of each gene cluster are predicted based on the conserved genes within each GCF and are not clearly defined. CD, cyclodehydratase; DHG, dehydrogenase.
Possible O-methylation in class I lanthipeptide biosynthesis.
A reoccurring observation in our analysis, one that was also made previously with a smaller genome set (21), is that many class I lanthipeptide biosynthetic gene clusters have a protein homologous to l-isoaspartate O-methyltransferase. These gene cluster families include GCF1, GCF20, GCF27, GCF29, GCF44, and GCF124 (Fig. 7A; see Fig. S7 in the supplemental material). The LanA precursor peptides from each family are distinct from others, but they all contain at least one Asp residue in the core region that is conserved in each GCF (see Fig. S7 in the supplemental material). Whether these Asp residues are related to methyl transfer remains to be investigated. Although methyltransferases in different gene cluster families share significant similarities, they mostly separate into different clusters (Fig. 7B), suggesting that each family of enzymes may have evolved separately. Interestingly, the phylogeny of LanC is different from the clustering of the methyltransferases. For example, the LanCs from GCF1, GCF29, and GCF124 are more closely related to each other and are on a separate branch from GCF27 (Fig. 2), but the methyltransferases from GCF27 and GCF124 are very closely related and separate only with very stringent cutoff (Fig. 7B), suggesting that the LanC and methyltransferase enzymes may not have coevolved. This finding is in contrast to the observation that, except for rare cases, LanC and LanB enzymes appear to have coevolved (12). A notable finding for GCF20 and GCF29 is that a stand-alone SpaB_C protein, similar to the elimination protein in thiopeptide biosynthesis (54), is encoded in the gene cluster (Fig. 7A); this SpaB_C protein likely involves novel chemistry.
FIG 7.
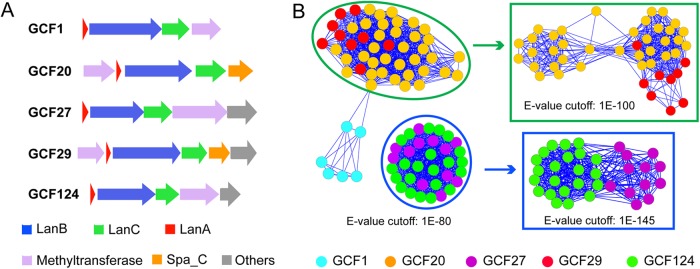
O-Methyltransferases in class I lanthipeptide biosynthesis. (A) Representative gene clusters of GCFs that encode class I lanthipeptide systems with a putative O-methyltransferase. (B) Sequence similarity network analysis of the putative O-methyltransferases in panel A.
Venezuelins produced by Actinobacteria.
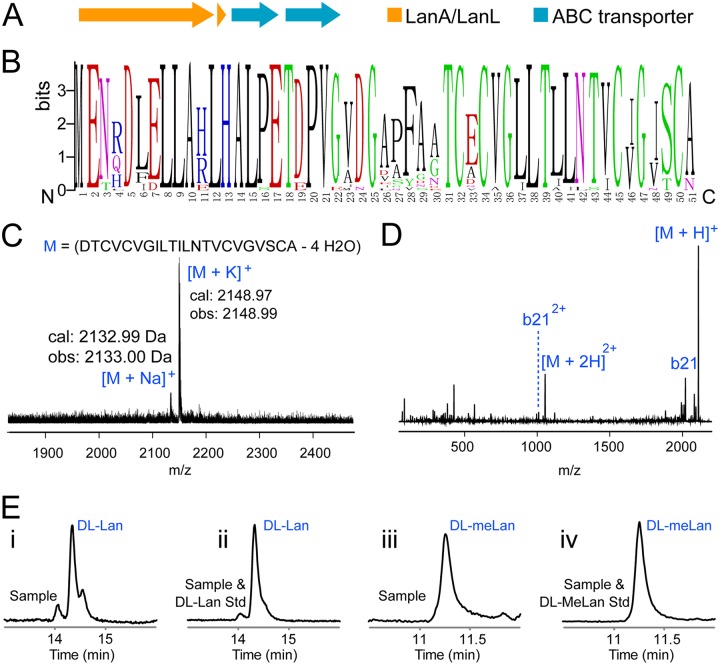
Although they share similar enzyme structure and organization, class IV LanL enzymes are significantly outnumbered by the class III LanKCs in Actinobacteria (Fig. 2). Venezuelin, previously obtained only via in vitro modification of the precursor peptide VenA by VenL followed by proteolytic removal of the leader peptide (16), represents the only class IV lanthipeptide characterized so far. However, production of venezuelin was detected neither in the wild-type strain Streptomyces venezuelae nor after attempted expression of the gene cluster in Streptomyces lividans, raising the question whether venezuelin is a genuine natural lanthipeptide. In our current analysis, we noted that GCF147 (33 members) (see Fig. S1 in the supplemental material) consists mostly of venezuelin-like gene clusters (Fig. 8A), and the putative precursor peptide sequences are highly conserved (Fig. 8B), suggesting the wide occurrence of venezuelin-like lanthipeptide gene clusters (although it was first discovered in S. venezuelae, here we collectively designate all these similar putative lanthipeptides venezuelins). The conserved genes in GCF147 include a LanA gene, a LanL gene, and two ABC transporter genes (Fig. 8A), indicating that (methyl)lanthionine formation is likely the only posttranslational modification on LanA core peptides in this family. We screened for venezuelin production in strains from GCF147 using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Three Streptomyces strains (S. katrae ISP5550, S. lavendulae subsp. lavendulae NRRL B-2508, and Streptomyces sp. strain NRRL B-2375) produced venezuelins in detectable amounts (Fig. 8C; see Fig. S8 in the supplemental material). Venezuelins of different lengths were detected, similar to the observations with class III lanthipeptides (38, 39, 42, 73). The variation in lengths of the observed venezuelins corresponded to the presence or absence of Phe28 and Ala29 (numbering based on VenA sequence) (Fig. 8B; see Fig. S8 in the supplemental material), presumably arising from stepwise removal of (part of) the leader peptides. The venezuelin from S. katrae ISP5550, which contained Ala29, was further analyzed by electrospray ionization (ESI)-MS-MS analysis (Fig. 8D). The lack of any fragmentation showed that the peptide likely consists of overlapping (methyl)lanthionine rings, consistent with the previously proposed ring topology of venezuelins based on extensive mutagenesis studies (16). Additional verification that venezuelin from S. katrae ISP5550 is indeed a lanthipeptide was provided by hydrolysis of the peptide and derivatization of the constituent amino acids as methyl esters and pentafluoropropionamides. Chiral gas chromatography (GC)-MS analysis of the derivatized venezuelin residues revealed the presence of (methyl)lanthionine, supporting the designation of venezuelin as a lanthipeptide (Fig. 8E). The (methyl)lanthionine derivatives all had the dl configuration. Our analysis validates that venezuelins are genuine natural products, and based on the frequency of their biosynthetic genes (see Fig. S1 in the supplemental material), they may be produced by many Actinobacteria. This observation also further supports the previous suggestion that identification of the products of silent clusters may be accomplished by screening several strains that contain the GCF (23).
FIG 8.
Production of venezuelins in Actinobacteria. (A) Representative gene clusters of GCF147. (B) A logo depicting the GCF147 precursor peptide sequences. (C) MALDI-TOF MS analysis of venezuelin produced by Streptomyces katrae ISP5550. MALDI-TOF MS analysis of venezuelins produced by Streptomyces lavendulae subsp. lavendulae NRRL B-2508 and Streptomyces sp. NRRL B-2375 are shown in Fig. S8 in the supplemental material. (D) ESI-MS-MS analysis of venezuelin produced by S. katrae ISP5550. The lack of fragmentation is consistent with a globular, overlapping ring topology. (E) Chiral GC-MS analysis of hydrolyzed and derivatized venezuelin residues, which revealed the presence of dl-lanthionine and dl-methyllanthionine. Trace i, sample showing the presence of dl-lanthionine; trace ii, sample spiked with dl-lanthionine standard; trace iii, sample showing the presence of dl-methyllanthionine; trace iv, sample spiked with dl-methyllanthionine standard.
DISCUSSION
Lanthipeptides are an intriguing class of natural products not only because of their potential medicinal values but also because of their wide occurrence and suitability for genome mining and heterologous expression studies. An extensive analysis of Actinobacteria genomes building on the recent gene cluster family project shows that the chemical space of lanthipeptide-related natural products is far beyond what the research community has tapped thus far. Analysis of just 17 selected GCFs that are among the most abundant GCFs already revealed novel posttranslational modifying enzymes that appear to have been recruited for lanthipeptide biosynthesis, as well as several intriguing systems that illustrate even more cross talk between different RiPP systems than recognized at present (9). The analysis also shows that the classification system currently in use may need to be updated, since several of the GCFs do not fall clearly in the four classes as they are currently defined. For instance, GCF146 appears to be a hybrid of class II and III lanthipeptide gene clusters, and GCFs26 and 125 contain genes encoding enzymes that would currently define them as class I or II. In addition, the analyses suggest cross talk between lanthipeptide biosynthetic enzymes and PKS and NRPS systems.
We emphasize that in addition to these interesting GCFs that contain at least 5 members, a very large number of smaller GCFs as well as singleton clusters were identified in Actinobacteria (see Fig. S1 in the supplemental material) (23), which suggests an even larger diversity than that discussed here. In fact, with the exception of the class III lanthipeptides such as the morphogen SapB and various peptins (both encoded within GCF31, the largest GCF), the known lanthipeptides from Actinobacteria, such as microbisporicin and actagardine, are found in such small GCFs or singleton clusters (see Fig. S1 in the supplemental material), further highlighting that a large amount of structural and functional diversity remains to be discovered. Based on precedent with the small number of previously characterized lanthipeptides from Actinobacteria, we anticipate that a large subset of the lanthipeptide-related natural products uncovered in this work are likely antibiotics, especially those made by machinery that is homologous to class I and II biosynthetic enzymes. Conversely, many others may have other biological activities that remain to be revealed by future studies. Indeed, most of the class III lanthipeptides reported thus far from Actinobacteria have no or weak antimicrobial activities, and quite a few have anti-allodynic/antinociceptive activity (38, 74) or morphogenetic activities (75). Since the venezuelins did not display antimicrobial activity in a previous study (16), class IV lanthipeptides may also have activities that would have eluded antimicrobial screens. The unusual biosynthetic systems highlighted here will help guide future efforts to discover novel biosynthetic mechanisms. More importantly, these studies set the stage for discovery of natural products with new scaffolds that potentially have interesting biological activities.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (GM PO1 GM077596 to W.A.V.D.D. and GM R01 058822 to W.A.V.D.D.). The Bruker UltrafleXtreme MALDI-TOF/TOF mass spectrometer was purchased in part with a grant from the National Institutes of Health (S10 RR027109). J.R.D. was funded through an Institute for Genomic Biology fellowship, and M.C.W. is supported by 1 F32 GM 0112284-01 (NIH).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00635-15.
REFERENCES
- 1.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 2.Knerr PJ, van der Donk WA. 2012. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem 81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 3.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 4.Ross AC, Vederas JC. 2011. Fundamental functionality: recent developments in understanding the structure-activity relationships of lantibiotic peptides. J Antibiot 64:27–34. doi: 10.1038/ja.2010.136. [DOI] [PubMed] [Google Scholar]
- 5.Piper C, Cotter PD, Ross RP, Hill C. 2009. Discovery of medically significant lantibiotics. Curr Drug Discov Technol 6:1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- 6.Dischinger J, Basi Chipalu S, Bierbaum G. 2014. Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 304:51–62. doi: 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Cox CR, Coburn PS, Gilmore MS. 2005. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci 6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 8.Schnell N, Entian KD, Schneider U, Götz F, Zahner H, Kellner R, Jung G. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 9.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oman TJ, van der Donk WA. 2010. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol 6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plat A, Kuipers A, Rink R, Moll GN. 2013. Mechanistic aspects of lanthipeptide leaders. Curr Protein Pept Sci 14:85–96. doi: 10.2174/1389203711314020001. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Yu Y, Velásquez JE, van der Donk WA. 2012. Evolution of lanthipeptide synthetases. Proc Natl Acad Sci U S A 109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siezen RJ, Kuipers OP, de Vos WM. 1996. Comparison of lantibiotic gene clusters and encoded proteins. Antonie Van Leeuwenhoek 69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 14.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. 2004. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science 303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 15.Müller WM, Schmiederer T, Ensle P, Süssmuth RD. 2010. In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew Chem Int Ed 49:2436–2440. doi: 10.1002/anie.200905909. [DOI] [PubMed] [Google Scholar]
- 16.Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. 2010. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol 8:e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. 2006. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 18.Tang W, van der Donk WA. 2012. Structural characterization of four prochlorosins: a novel class of lantipeptides produced by planktonic marine cyanobacteria. Biochemistry 51:4271–4279. doi: 10.1021/bi300255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Yang X, Wang H, van der Donk WA. 2014. High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem Biol 9:2686–2694. doi: 10.1021/cb500622c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Begley M, Cotter PD, Hill C, Ross RP. 2009. Rational genome mining for LanM proteins leads to the identification of a novel two-peptide lantibiotic, lichenicidin. Appl Environ Microbiol 75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh AJ, O'Sullivan O, Ross RP, Cotter PD, Hill C. 2010. In silico analysis highlights the frequency and diversity of type 1 lantibiotic gene clusters in genome sequenced bacteria. BMC Genomics 11:679. doi: 10.1186/1471-2164-11-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M, Sareen D. 2014. Novel LanT associated lantibiotic clusters identified by genome database mining. PLoS One 9:e91352. doi: 10.1371/journal.pone.0091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. 2014. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berdy J. 2005. Bioactive microbial metabolites. J Antibiot 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 25.Monciardini P, Iorio M, Maffioli S, Sosio M, Donadio S. 2014. Discovering new bioactive molecules from microbial sources. Microb Biotechnol 7:209–220. doi: 10.1111/1751-7915.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ökesli A, Cooper LE, Fogle EJ, van der Donk WA. 2011. Nine post-translational modifications during the biosynthesis of cinnamycin. J Am Chem Soc 133:13753–13760. doi: 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foulston LC, Bibb MJ. 2010. Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc Natl Acad Sci U S A 107:13461–13466. doi: 10.1073/pnas.1008285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, Parenti F. 2008. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15:22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Maffioli SI, Iorio M, Sosio M, Monciardini P, Gaspari E, Donadio S. 2014. Characterization of the congeners in the lantibiotic NAI-107 complex. J Nat Prod 77:79–84. doi: 10.1021/np400702t. [DOI] [PubMed] [Google Scholar]
- 30.Iorio M, Sasso O, Maffioli SI, Bertorelli R, Monciardini P, Sosio M, Bonezzi F, Summa M, Brunati C, Bordoni R, Corti G, Tarozzo G, Piomelli D, Reggiani A, Donadio S. 2014. A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol 9:398–404. doi: 10.1021/cb400692w. [DOI] [PubMed] [Google Scholar]
- 31.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross AC, Liu H, Pattabiraman VR, Vederas JC. 2010. Synthesis of the lantibiotic lactocin S using peptide cyclizations on solid phase. J Am Chem Soc 132:462–463. doi: 10.1021/ja9095945. [DOI] [PubMed] [Google Scholar]
- 34.Doroghazi JR, Metcalf WW. 2013. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doroghazi JR, Buckley DH. 2014. Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genomics 15:970. doi: 10.1186/1471-2164-15-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Zhang Q, van der Donk WA. 2013. Insights into the evolution of lanthipeptide biosynthesis. Protein Sci 22:1478–1489. doi: 10.1002/pro.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawton EM, Ross RP, Hill C, Cotter PD. 2007. Two-peptide lantibiotics: a medical perspective. Mini Rev Med Chem 7:1236–1247. doi: 10.2174/138955707782795638. [DOI] [PubMed] [Google Scholar]
- 38.Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Guhring H, Vertesy L, Wink J, Hoffmann H, Bronstrup M, Sheldrick GM, Süssmuth RD. 2010. Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew Chem Int Ed Engl 49:1151–1154. doi: 10.1002/anie.200905773. [DOI] [PubMed] [Google Scholar]
- 39.Völler GH, Krawczyk JM, Pesic A, Krawczyk B, Nachtigall J, Süssmuth RD. 2012. Characterization of new class III lantibiotics—erythreapeptin, avermipeptin and griseopeptin from Saccharopolyspora erythraea, Streptomyces avermitilis and Streptomyces griseus demonstrates stepwise N-terminal leader processing. Chembiochem 13:1174–1183. doi: 10.1002/cbic.201200118. [DOI] [PubMed] [Google Scholar]
- 40.Oldach F, Al Toma R, Kuthning A, Caetano T, Mendo S, Budisa N, Süssmuth RD. 2012. Congeneric lantibiotics from ribosomal in vivo peptide synthesis with noncanonical amino acids. Angew Chem Int Ed Engl 51:415–418. doi: 10.1002/anie.201106154. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, van der Donk WA. 2012. Biosynthesis of the class III lantipeptide catenulipeptin. ACS Chem Biol 7:1529–1535. doi: 10.1021/cb3002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krawczyk JM, Völler GH, Krawczyk B, Kretz J, Brönstrup M, Süssmuth RD. 2013. Heterologous expression and engineering studies of labyrinthopeptins, class III lantibiotics from Actinomadura namibiensis. Chem Biol 20:111–122. doi: 10.1016/j.chembiol.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 43.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frey PA, Hegeman AD, Ruzicka FJ. 2008. The radical SAM superfamily. Crit Rev Biochem Mol Biol 43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 45.Cotter PD, O'Connor PM, Draper LA, Lawton EM, Deegan LH, Hill C, Ross RP. 2005. Posttranslational conversion of l-serines to d-alanines is vital for optimal production and activity of the lantibiotic lacticin 3147. Proc Natl Acad Sci U S A 102:18584–18589. doi: 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suda S, Lawton EM, Wistuba D, Cotter PD, Hill C, Ross RP. 2012. Homologues and bioengineered derivatives of LtnJ vary in ability to form d-alanine in the lantibiotic lacticin 3147. J Bacteriol 194:708–714. doi: 10.1128/JB.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohans CT, Li J L, Vederas JC. 2014. Structure and biosynthesis of carnolysin, a homologue of enterococcal cytolysin with d-amino acids. J Am Chem Soc 136:13150–13153. doi: 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 48.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. 2006. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci U S A 103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dischinger J, Josten M, Szekat C, Sahl HG, Bierbaum G. 2009. Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. PLoS One 4:e6788. doi: 10.1371/journal.pone.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shenkarev ZO, Finkina EI, Nurmukhamedova EK, Balandin SV, Mineev KS, Nadezhdin KD, Yakimenko ZA, Tagaev AA, Temirov YV, Arseniev AS, Ovchinnikova TV. 2010. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry 49:6462–6472. doi: 10.1021/bi100871b. [DOI] [PubMed] [Google Scholar]
- 51.Caetano T, Krawczyk JM, Mosker E, Süssmuth RD, Mendo S. 2011. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol 18:90–100. doi: 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. 2007. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep 24:162–190. doi: 10.1039/B507395M. [DOI] [PubMed] [Google Scholar]
- 53.Cupp-Vickery JR, Poulos TL. 1995. Structure of cytochrome P450eryF involved in erythromycin biosynthesis. Nat Struct Biol 2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 54.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noike M, Matsui T, Ooya K, Sasaki I, Ohtaki S, Hamano Y, Maruyama C, Ishikawa J, Satoh Y, Ito H, Morita H, Dairi T. 2015. A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin. Nat Chem Biol 11:71–76. doi: 10.1038/nchembio.1697. [DOI] [PubMed] [Google Scholar]
- 56.Chung CHY, Kurien BT, Mehta P, Mhatre M, Mou S, Pye Q N, Stewart C, West M, Williamson KS, Post J, Liu L, Wang R, Hensley K. 2007. Identification of lanthionine synthase C-like protein-1 as a prominent glutathione binding protein expressed in the mammalian central nervous system. Biochemistry 46:3262–3269. doi: 10.1021/bi061888s. [DOI] [PubMed] [Google Scholar]
- 57.Hensley K, Venkova K, Christov A. 2010. Emerging biological importance of central nervous system lanthionines. Molecules 15:5581–5594. doi: 10.3390/molecules15085581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong WX, Wang YB, Peng L, Ge XZ, Zhang J, Liu SS, Zhang XN, Xu ZH, Chen Z, Luo JH. 2012. Lanthionine synthetase C-like protein 1 interacts with and inhibits cystathionine beta-synthase: a target for neuronal antioxidant defense. J Biol Chem 287:34189–34201. doi: 10.1074/jbc.M112.383646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W, Wang L, Liu Y, Xu J, Zhu G, Cang H, Li X, Bartlam M, Hensley K, Li G, Rao Z, Zhang XC. 2009. Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione. Genes Dev 23:1387–1392. doi: 10.1101/gad.1789209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang C, Chen M, Pang D, Bi D, Zou Y, Xia X, Yang W, Luo L, Deng R, Tan H, Zhou L, Yu S, Guo L, Du X, Cui Y, Hu J, Mao Q, Worley PF, Xiao B. 2014. Developmental and activity-dependent expression of LanCL1 confers antioxidant activity required for neuronal survival. Dev Cell 30:479–487. doi: 10.1016/j.devcel.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng M, van der Donk WA, Chen J. 2014. Lanthionine synthetase C-like protein 2 (LanCL2) is a novel regulator of Akt. Mol Biol Cell 25:3954–3961. doi: 10.1091/mbc.E14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunwell JM, Purvis A, Khuri S. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. 2012. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 64.Melby JO, Nard NJ, Mitchell DA. 2011. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr Opin Chem Biol 15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bagley MC, Dale JW, Merritt EA, Xiong X. 2005. Thiopeptide antibiotics. Chem Rev 105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 66.Li C, Kelly WL. 2010. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep 27:153–164. doi: 10.1039/B922434C. [DOI] [PubMed] [Google Scholar]
- 67.Walsh CT, Malcolmson SJ, Young TS. 2012. Three ring posttranslational circuses: insertion of oxazoles, thiazoles, and pyridines into protein-derived frameworks. ACS Chem Biol 7:429–442. doi: 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunbar KL, Chekan JR, Cox CL, Burkhart BJ, Nair SK, Mitchell DA. 2014. Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. Nat Chem Biol 10:823–829. doi: 10.1038/nchembio.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A 102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sivonen K, Leikoski N, Fewer DP, Jokela J. 2010. Cyanobactins—ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol 86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Q, Liu W. 2013. Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep 30:218–226. doi: 10.1039/C2NP20107K. [DOI] [PubMed] [Google Scholar]
- 72.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. 1996. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 73.Krawczyk B, Völler GH, Völler J, Ensle P, Süssmuth RD. 2012. Curvopeptin: a new lanthionine-containing class III lantibiotic and its co-substrate promiscuous synthetase. Chembiochem 13:2065–2071. doi: 10.1002/cbic.201200417. [DOI] [PubMed] [Google Scholar]
- 74.Iorio M, Sasso O, Maffioli SI, Bertorelli R, Monciardini P, Sosio M, Bonezzi F, Summa M, Brunati C, Bordoni R, Corti G, Tarozzo G, Piomelli D, Reggiani A, Donadio S. 2014. A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol 9:398. doi: 10.1021/cb400692w. [DOI] [PubMed] [Google Scholar]
- 75.Willey JM, Gaskell AA. 2011. Morphogenetic signaling molecules of the streptomycetes. Chem Rev 111:174–187. doi: 10.1021/cr1000404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.