Abstract
Association of multiple sclerosis (MS) with the human leukocyte antigen (HLA) class II haplotype DRB1*1501-DQB1*0602 is the most consistently replicated finding of genetic studies of the disease. However, the high level of linkage disequilibrium (LD) in the HLA region has hindered the identification of other loci that single-marker tests for association are unlikely to resolve. In order to address this issue, we generated haplotypes spanning 14.754 Mb (5 cM) across the entire HLA region. The haplotypes, which were inferred by genotyping relatives of 152 patients with MS and 105 unaffected control subjects of Tasmanian ancestry, define a genomic segment from D6S276 to D6S291, including 13 microsatellite markers integrated with allele-typing data for DRB1 and DQB1. Association to the DRB1*1501-DQB1*0602 haplotype was replicated. In addition, we found that the class I/extended class I region, defined by a genomic segment of ∼400 kb between MOGCA and D6S265, harbors genes that independently increase risk of, or provide protection from, MS. Log-linear modeling analysis of constituent haplotypes that represent genomic regions containing class I (MOGCA-D6S265), class III (TNFa-TNFd-D6S273), and class II (DRB1-DQB1) genes indicated that having class I and class II susceptibility variants on the same haplotype provides an additive effect on risk. Moreover, we found no evidence for a disease locus in the class III region defined by a 150-kb genomic segment containing the TNF locus and 14 other genes. A global overview of LD performed using GOLD identified two discrete blocks of LD in the HLA region that correspond well with previous findings. We propose that the analysis of haplotypes, by use of the types of approaches outlined in the present article, should make it possible to more accurately define the contribution of the HLA to MS.
Introduction
Multiple sclerosis (MS [MIM 126200]) is the most common neurological disease of young white adults of northern European ancestry (prevalence 1:1,000). It is a complex disease of the CNS of unknown etiology. Population-based studies indicate that familial aggregation of MS is due to shared genetics (Ebers et al. 1995); however, concordance studies in twins suggest that genetic factors are not solely responsible for disease susceptibility (Ebers et al. 1986; Mumford et al. 1994). Involvement of the HLA complex in susceptibility to MS was identified, through allelic association to HLA-A*03 (telomeric “classical” class I region), almost 30 years ago (Jersild et al. 1972). Subsequently, the initial association with HLA-A*03 was shown to be secondary to HLA-B*07 (centromeric “classical” class I region), which, in turn, was shown to be in linkage disequilibrium (LD) with the class II alleles HLA-DR2 and -DQw6. These serological typings have since been refined to the DR15 and DQ6 subtypes of DR2 and DQw6, respectively, which correspond to phenotypic expression of the DRB1*1501-DRB5*0101 (DR15) and DQA1*0102-DQB1*0602 (DQ6) genotypes (Olerup and Hillert 1991). More recently, the class I region has been reidentified as harboring genes that independently modulate susceptibility to MS (Fogdell-Hahn et al. 2000). Linkage and association studies of the HLA region have consistently supported the involvement of the DRB1*1501-DRB5*0101-DQA1*0102-DQB1*0602 haplotype in susceptibility to MS, even in populations of diverse ethnic origin. Indeed, recent work has indicated that a number of different clinical correlates are associated with the DR2 susceptibility haplotype (McDonnell et al. 1999; Celius et al. 2000; Masterman et al. 2000). Nevertheless, the contribution of HLA genes to MS susceptibility appears to be much less than in type I diabetes; however, this may be difficult to quantify, given the evidence of epistatic interactions between HLA and non-HLA genes (Chataway et al. 1998).
There has been conjecture concerning the involvement of the TNFa gene, which maps in the extended haplotype defined by DRB1*1501-DQB1*0602 (Coraddu et al. 1998). Current theories on the mechanisms of tissue injury in MS hint at a role for the TNFa gene (telomeric class III region) in mediating the destruction of oligodendrocytes by macrophages or microglia. However, conflicting reports continue regarding its involvement in an extended haplotype independent of the HLA class II association (Roth et al. 1994; Allcock et al. 1999; Fernandez-Arquero et al. 1999).
LD and variable rates of recombination are both recognized features of the HLA locus (Begovich et al. 1992; Martin et al. 1995; Cullen et al. 1997). It has been postulated that regions of the HLA that experience lower recombination frequencies (stronger LD) may indicate the existence of selective pressure to maintain specific allelic combinations in cis. This could occur if specific alleles at closely linked loci are selected for synergistic interactions. For example, it has been shown previously that haplotypic association of specific DQA1 and DQB1 alleles ensures a functional heterodimer (Kwok et al. 1993). Additional evidence of selection has been provided by high-resolution typing of HLA class I and II genes in CEPH families, showing the existence of particular combinations of class I and class II alleles (Bugawan et al. 2000).
The high level of LD in the HLA region has proven to be a factor confounding to gene localization. Nevertheless, analysis of ancestral haplotypes in the extended class I region of the HLA region was employed to locate the HFE gene, mutant forms of which cause hereditary hemochromatosis in 83% of affected individuals (Feder et al. 1996). Ancestral haplotype analysis has also been employed to localize a susceptibility gene (PSORSI) for psoriasis to a 60-kb region at the centromeric end of the class I region (Jenisch et al. 1998; Nair et al. 2000). In addition, the critical region for Behçet disease has been refined, by association analysis, to a 46-kb interval centromeric of HLA-B (Ota et al. 1999). All of these studies provide valuable information for future high-resolution association studies of marker locations and LD patterns in different haplotypes.
We have recruited patients with MS from Tasmania, an island state of Australia. Tasmania was colonized as a penal settlement by the British in 1803. Free settlers were also encouraged to migrate to Tasmania through the early-to-mid–19th century, and convict transportation ceased in 1853. It is estimated that the subsequent outward migration of large numbers of men—and the relative deficit of women—meant that there were at most 10,000 couples of childbearing age in Tasmania in the mid-1800s. It is believed that these “founding families” are responsible for ∼65% of the current residents of Tasmania. Only modest immigration has occurred in the last 150 years. This is supported by census data from 1996 (Australian Bureau of Statistics), which indicate that >80% of Tasmania’s current 470,000 residents were born in Australia and that almost 90% were born in Australia, New Zealand, Great Britain, and Ireland, suggesting that the population is relatively ethnically homogeneous. Tasmania’s founder effect is documented through the study of a large kindred segregating Huntington disease, which has been traced back nine generations to the father of a female migrant who came to Tasmania in 1842 and gave rise to 765 living descendents (Pridmore 1990). These and many other features of Tasmania make it an attractive place to study the genetics of complex disease.
To elucidate the involvement of the HLA complex in MS, we have performed a comprehensive analysis of haplotypes spanning the HLA region in Tasmanian patients with MS. Exploiting a variety of statistical techniques, including log-linear modeling and two-point and multipoint LD mapping, we have made advances toward the characterization and identification of the genetic determinants of susceptibility and protection that exist for MS in the HLA region.
Material and Methods
Inferring Haplotypes
Several methodologies permit the derivation of haplotypes. We chose to use a constellation of close relatives to infer phase—and hence haplotypes—probabilistically. This method, although expensive in terms of genotypes, has a number of beneficial consequences. First, the genotyping of close relatives of a case or control subject, makes it possible to check Mendelian errors, which can identify pedigree and genotyping errors. Second, genotyping information from a number of close relatives renders the sample sufficiently versatile for use with other genetic mapping techniques. Simulation studies performed prior to recruitment indicated that two scenarios provided excellent haplotype reconstruction (data not shown). Genotyping of samples from both parents was common to both scenarios; however, in one scenario, a grandparent was also genotyped, and, in the other, two siblings were the source of additional phase information. In general, relatives in later generations yielded little phase information. Recruitment strategies were based on the findings of these simulation studies.
Case Subjects
The Tasmanian MS Research Program was approved by the University of Tasmania’s human research ethics committee, and written consent was obtained from all participants. The program was advertised in local newspapers, radio stations, and in regular newsletters sent to patients with MS by the Multiple Sclerosis Societies of Tasmania and Victoria. Patients with MS were self-referred volunteers, and possession of at least one Tasmanian-born grandparent was a key selection criterion. Information was recorded concerning immediate family history of disease and living close relatives. Recruitment of relatives was prioritized as (in order of preference): parents, grandparents, siblings, spouses, and offspring. We also identified, with the assistance of the MS Society of Victoria, 26 Tasmanian migrants with MS living in Victoria, Australia, who met the recruitment criteria.
Control Subjects
We asked case subjects to nominate control subjects, to maximize matching for age, ethnicity, geographic origin, and socioeconomic background. This approach was taken in order to increase the chances of recruiting close relatives of the control subjects, which was deemed more likely to succeed if the control had a (nongenetic) relationship to the case. Moreover, these control subjects would be more likely to match closely the ancestral background of the patient. When case patients were unable to nominate a person to act as a control, control subjects matched on the basis of age and sex were recruited from the Tasmanian electoral roll.
Clinical Diagnosis
The treating doctors of patients who registered an interest in participating in the present study were asked to complete a confidential medical record summary. They were asked whether they considered the patient to have clinically definite, laboratory-supported definite, clinically probable, or laboratory-supported probable MS or, alternatively, to have no or, at most, only possible MS, according to the diagnostic criteria defined by Poser et al. (1983). Subsequent to this, all respondents were interviewed and examined by one of the six participating neurologists. In addition, when available, magnetic resonance imaging (MRI) results were assessed or, at a minimum, MRI reports were obtained. The Poser criteria of clinically definite or laboratory-supported definite MS were set as the minimum standard by which patients were included in the study, but, in addition, patients were required to have cerebral MRI abnormalities consistent with MS, as defined by the criteria of Paty et al. (1988). All patients with a classification of primary progressive MS had to exhibit progressive neurological disability for at least one year, to have no other better explanation for the clinical features, and to have not only relevant spinal cord abnormalities but also changes on cerebral MRI consistent with demyelination.
DNA Preparation and Genotyping
DNA was extracted from peripheral blood, using a Nucleon extraction kit according to the manufacturer’s instructions. Genomic DNA was resuspended in 10 mM Tris-HCl (pH 8), 1 mM EDTA (pH 8) and was stored at 4°C. PCR was performed, according to published conditions, on 30 ng of genomic DNA by use of AmpliTaq Gold at the Australian Genome Research Facility (AGRF). At the AGRF, genotyping was performed on DNA samples from the case and control subjects and from selected family members, using ABI-Prism 377 sequencing machines. Dye-labeled PCR products were detected using GENESCAN (PE Applied Biosystems) software, and allele calling was performed using GENOTYPER 2.0 (PE Applied Biosystems). Initial error analysis on genotypes was performed as described elsewhere (Ewen et al. 2000).
Simple Sequence-Length Polymorphism Markers
Oligonucleotide primers corresponding to simple sequence-length polymorphisms (SSLPs) for D6S105, MOGCA, D6S265, D6S273, G51152, and RING3CA were designed according to Martin et al. (1998). Primers for D6S276 and D6S291 were obtained from the HD-5 fine-mapping set (PE Applied Biosystems). Primers for D6S1571, D6S1568, D6S439, TNFa, and TNFd were designed according to sequence published by Foissac et al. (2000) and the Genome Database (GDB). Genetic and physical distances between genes and markers were obtained from The National Center for Biotechnology Information, the GDB, and the University of California, Santa Cruz, Human Genome Working Draft. Forward primer sequences of these markers were “pig-tailed” with a heptanucleotide (5′-GTTTCTT-3′) sequence, to facilitate genotyping by minimizing the effects of enzyme-directed, template-independent additions of an “A” nucleotide to PCR products.
HLA Typing
HLA-DRB1 and DQB1 allele typing was performed using methods outlined in the 11th International Histocompatibility Workshop (Kimura and Sasasuki 1991). In brief, exon 2 polymorphisms were determined by use of a series of biotinylated oligonucleotide probes that were hybridized to PCR-amplified DNA immobilized on nylon membranes. Complementary probes were detected using a streptavidin–alkaline phosphatase conjugate and the chemiluminescent substrate CDP-Star (Roche Diagnostics PTY). For each locus, new probes and primers were added to cover sequence polymorphisms not described in the original protocol.
Haplotype Reconstruction
Genotyping errors were detected using PedCheck (O'Connell and Weeks 1998). Inconsistent data were either corrected or deleted. File management of the data, pedigree, and map distance information was performed using an in-house program, LINKPREP, to generate LINKAGE style “.dat” and “.pre” files. GENEHUNTER (Kruglyak et al. 1996) was used to reconstruct inferred haplotypes, which were extracted from the GENEHUNTER output file, haplo.dump, using an in-house PERL script, HAPLO.PL. This program separates case and control haplotypes, and calculates the allele frequencies and counts used in the CLUMP program (Sham and Curtis 1995).
Statistical Analysis
Association tests for single markers were performed using CLUMP (Sham and Curtis 1995) with the T4 statistic being chosen as the test statistic for association. The T4 statistic calculates the maximum χ2 value found by collapsing the contingency table over each allele in turn, forming 2×2 contingency tables and combining alleles whenever allele counts are too small. The distribution of this statistic is not distributed as χ21 variable and its significance must be assessed through the use of Monte Carlo methods, which is how the P values were calculated using CLUMP.
Haplotypes were assessed using odds ratios (ORs) and were calculated using the statistical software R. All haplotypes can be defined as either showing a susceptibility or protective effect. Hence, all ORs were calculated using the appropriate one-sided test for the 2×2 contingency table. The general linear modeling function of Splus5 was used to assess the effects of LD, disease-haplotype interactions and haplotype-haplotype interactions. Multiple testing was corrected for 22 tests, 7 examining the 2×2 tables and 15 examining the T4 statistic from CLUMP. Pairwise LD was assessed for cases and controls using the Global Overview of Linkage Disequilibrium (GOLD) program (Abecasis and Cookson 2000).
Results
Recruitment
A total of 222 patients were assessed, and 181 of these were admitted to the study, after application of the criteria described above. When all relevant data had been compiled and assessed, the final decision concerning inclusion was based on the consensus opinions of two physicians specializing in MS (T.J.K. and H.B.). Of the 41 patients excluded from the study, 17 had normal, nondiagnostic, or atypical MRI results or had not had an MRI performed; 8 had monophasic disease; 7 had other neurological disorders (1 spinocerebellar atrophy, 1 Devic disease, 1 cerebral autosomal dominant arteriopathy with subcortial infarcts and leukoencephalopathy, 1 syringomyelia, and 1 Leber optic atrophy); 2 had connective tissue disorders; 2 had musculoskeletal disorders; 4 had functional disorders; and 1 patient asked to be excluded.
In total, 181 case subjects and 843 of their close relatives (an average of 4.7 controls per case) were recruited. The present article describes data for 152 case subjects only. Sample collection for the remaining 29 case families is in progress. Despite the fact that the average age of case subjects was 45.9 years, most of the relatives recruited were in the “highly informative” category (parents, grandparents, and siblings), rather than spouses and offspring.
Nonbiological relatives (44% spouses and 5% in-laws) accounted for approximately half the control cohort, with friends (18%) and locally selected individuals from the electoral roll (33%) representing the remainder. All control subjects had no family history of MS and had at least one grandparent born in Tasmania. In total, samples for 105 control subjects and 400 of their close relatives were collected.
Assessment of Haplotype Reconstruction
We designed a recruitment strategy for close relatives of case and control subjects that would permit the reconstruction of haplotypes. Haplotypes were inferred from familial genotyping data, using GENEHUNTER (Kruglyak et al. 1996). Genotyping was performed for 152 case and 105 control subjects and their corresponding relatives. Haplotypes were generated across the HLA region for 15 markers, corresponding to 13 SSLP markers and allele typing data for the DRB1 and DQB1 genes (fig. 1). These markers span the HLA region from D6S276 to D6S291, which corresponds to a genetic distance of 5 cM and a physical distance of 14.754 Mb. To gauge the success of the recruitment strategy, we calculated the proportion of haplotypes that would be reconstructed, in theory, if recruitment were “perfect” and compared it with the observed proportion of haplotypes reconstructed. For cases, 292 haplotypes were inferred (96% of the theoretical maximum [304]); and, for controls, 204 haplotypes were inferred (97% of the maximum [210]).
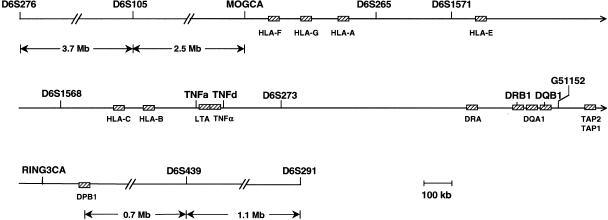
Figure 1.
An integrated physical map of the HLA region, showing the locations of the 13 SSLP markers: pTer-D6S276, D6S105, MOGCA, D6S265, D6S1571, D6S1568, TNFa, TNFd, D6S273, G51152, RINGCA, D6S439, and D6S291-cen. Selected genes (not drawn to scale) involved in functions of the immune system are represented by hatched boxes. The boundary between the “extended” and “classical” class I region lies between MOGCA and HLA-F. The “classical” class I region extends 1.8 Mb centromeric of HLA-F. The telomeric boundary of the class III region lies between HLA-B and the TNF locus. The class III region extends 0.75 Mb to the centromeric boundary with class II, which is located between D6S273 and DRA. The “classical” class II region extends 0.85 Mb, just centromeric of DPB1. This physical map was assembled with information from a number of sources (MHC Consortium 1999; Foissac et al. 2000; National Center for Biotechnology Information).
CLUMP Analysis of Allele Frequencies
Allelic association was performed using the T4 statistic in CLUMP (Sham and Curtis 1995) (table 1). The P value for each marker was evaluated using 100,000 Monte Carlo simulations. Prior to correction for multiple testing (22 tests performed [see tables 1 and 2]), significantly associated markers were found in the extended class I (D6S105 and MOGCA), classical class I (D6S265 and D6S1571), class III (TNFa), class II (DRB1, DQB1, and G51152), and extended class II (D6S439) regions (table 1). After multiple testing was taken into account, only MOGCA (P=2×10-4), TNFa (P=.04), DRB1 (P=2×10-4), DQB1 (P=2×10-4), and G51152 (P=2×10-4) remained significantly associated.
Table 1.
Allele Frequencies in Cases versus Controls[Note]
| Marker | No. ofAlleles | T4a | P | CorrectedPb |
| D6S276 | 14 | 6.5 | .5 | 1.0 |
| D6S105 | 12 | 18.4 | .003 | .07 |
| MOGCA | 13 | 29.2 | 1 × 10−5 | 2 × 10−4 |
| D6S265 | 11 | 14.1 | .009 | .2 |
| D6S1571 | 11 | 13.8 | .01 | .2 |
| D6S1568 | 11 | 5.4 | .7 | 1.0 |
| TNFa | 14 | 20.6 | .002 | .04 |
| TNFd | 8 | 5.3 | .2 | 1.0 |
| D6S273 | 8 | 4.7 | .4 | 1.0 |
| DRB1 | 15 | 32.4 | <1 × 10−5 | <2 × 10−4 |
| DQB1 | 11 | 34.1 | <1 × 10−5 | <2 × 10−4 |
| G51152 | 10 | 35.8 | <1 × 10−5 | <2 × 10−4 |
| RING3CA | 8 | 1.6 | .9 | 1.0 |
| D6S439 | 11 | 14.5 | .009 | .2 |
| D6S291 | 6 | 1.1 | .9 | 1.0 |
Note.— Comparisons were performed using CLUMP (Sham and Curtis 1995).
The T4 measure was determined with 100,000 simulations of the data for all 15 markers, and the corresponding P value was calculated.
The corrected P value was determined by multiplying the uncorrected P value by the number of tests displayed in this table added to those displayed in table 2 (22 tests overall).
Table 2.
Statistical Analysis of Haplotype Frequencies
| Haplotypea | HLAClass | Effectb | P | Corrected Pc | OR (95% CI)d |
| 3-1-×-×-×-×-× | I | S | 3 × 10−5 | 7 × 10−4 | 3.1 (1.8, ∞) |
| 12-3-×-×-×-×-× | I | S | .09 | 1 | 1.5 (.9, ∞) |
| 1-6-×-×-×-×-× | I | P | .001 | .03 | .3 (.0, .6) |
| 4-5-×-×-×-×-× | I | P | .06 | 1 | .7 (.0, 1.0) |
| ×-×-11-4-5-×-× | III | S | 7 × 10−5 | .002 | 2.8 (1.7, ∞) |
| ×-×-2-5-5-×-× | III | P | .01 | .3 | .4 (.0, .8) |
| ×-×-×-×-×-14-12 | II | S | 5 × 10−9 | 1 × 10−7 | 4.3 (2.7, ∞) |
Haplotypes were stratified for analysis on the basis of results shown in figure 2. Class I, III, and II loci were analyzed independently in cases versus controls, with the presence of any allele at other loci signified by “×.”
S = susceptibility; P = protective.
The corrected P value was determined by multiplying the uncorrected P value by the number of tests displayed in this table added to those displayed in table 1 (22 tests overall). Each corrected P value has been adjusted for 22 tests, as in table 1.
All ORs were calculated using all other haplotypes as the reference group or using one-sided tests, depending on whether the haplotype conferred putative susceptibility or protective effects.
Analysis of Haplotype Frequencies
Since the HLA region is characterized by high levels of LD, we analyzed a subset of those markers showing evidence of association with MS as haplotypes rather than as single markers (fig. 2A–C). To allow a comparison with other studies, the haplotypes studied in more depth corresponded to regions of the HLA complex previously identified as harboring determinants of disease susceptibility. SSLP markers 400 kb apart selected for haplotype analysis of the class I region were MOGCA (upstream of the MOG gene, 100 kb telomeric of HLA-F) and D6S265 (70–100 kb centromeric of HLA-A). For the class III region, the SSLP markers TNFa (3.5 kb telomeric of lymphotoxin, TNFd (8 kb downstream of TNFa), and D6S273 (exon 2 and exon 3 of G6D, 150 kb centromeric of the TNF locus) were chosen (Jongeneel et al. 1991; Udalova et al. 1993; Foissac et al. 2000). Although D6S265, TNFd, and D6S273 were not significantly associated alone, they permit the analysis of genomic segments that harbor a number of genes involved in the function of the immune system. Class II haplotypes were defined by polymorphisms in the DRB1 and DQB1 genes. Allele binning information for markers described in figure 2A–C can be found in the Appendix.
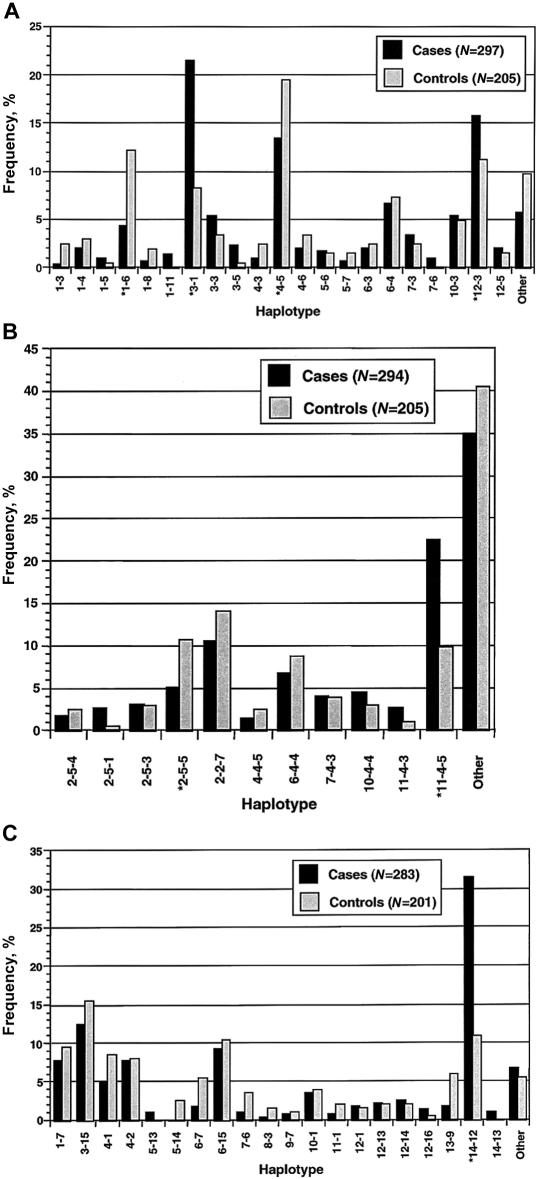
Figure 2.
Comparison of haplotype frequencies between case and control subjects. For the class I region (A), MOGCA-D6S265 haplotypes were analyzed; for the class III region (B), TNFa-TNFd-D6S273 haplotypes were analyzed and for the class II region (C), DRB1-DQB1 haplotypes were analyzed. Haplotypes with a frequency of ⩾1% in cases and controls were plotted for MOGCA-D6S265 and DRB1-DQB1, but only haplotypes with a frequency ⩾2% were plotted for TNFa-TNFd-D6S273. Coding of haplotypes is describe in the Appendix. Asterisks (*) indicate haplotypes selected for association testing.
Forty-two haplotypes were counted in cases and controls for the class I (MOGCA-D6S265) region. For ease of presentation—and because we did not have sufficient power to evaluate differences in frequency between rare haplotypes—only those with a frequency ⩾1% were compared (fig. 2A). A susceptibility haplotype, 3-1, was observed with a frequency of 21.5% in cases and 8.2% in controls. Because the HLA-A gene is flanked distally by MOGCA and proximally by D6S265, we tested whether case and control samples carrying the 3-1 susceptibility haplotype were also carrying the HLA-A*03 allele. Twenty-one cases carrying the 3-1 haplotype were tested for the presence of an HLA-A*03 allele (data not shown). All samples tested positive for HLA-A*03. Since the A*0301 allele comprises 99% of all A*03s, which have a population frequency of ∼20%, there is strong evidence to suggest that the 3-1 haplotype in this sample corresponds to the same ancestral susceptibility haplotype described recently in Scandinavian patients with MS (Fogdell-Hahn et al. 2000). Haplotypes in which discrepancies in frequency between cases and controls provided evidence for additional effects on susceptibility (12-3) and protection (1-6 and 4-5) were selected for statistical analysis.
Overall, 97 different three-marker haplotypes were recorded for the class III (TNFa-TNFd-D6S273) region. The larger number of haplotypes observed for class III than for the class I region is a result of the additional marker used to define a three- instead of a two-marker haplotype. The majority (86) of these haplotypes had a frequency <2%. Again, only the more common haplotypes (frequency ⩾2%) were compared between cases and controls (fig. 2B). The 11 haplotypes analyzed individually represent 65.0% and 59.5% of case and control haplotypes, respectively. A putative susceptibility haplotype, 11-4-5, with a frequency of 22.3% in cases and 9.9% controls, was observed; and a putative protective haplotype, 2-5-5, with a frequency of 5.1% in cases and 10.7% in controls, was also a notable finding at this locus.
HLA class II (DRB1-DQB1) haplotypes with a frequency ⩾1% were analyzed in cases and controls (fig. 2C). Only the 14-12 haplotype (DRB1*1501-DQB1*0602) differed markedly in frequency between cases (31.5%) and controls (11.0%). The phenotypic frequency of the DRB1*1501-DQB1*0602 haplotype among cases (57.7%, n=142) and controls (22%, n=95) was similar to other populations of northern European ancestry (Francis et al. 1991; Hillert and Olerup 1993).
Statistical Analysis of Haplotype Frequencies
For the loci described above, a subset of seven haplotypes were selected for further analysis. Previous studies guided us in our choice of haplotypes that were relatively common and that showed the greatest disparity between cases and controls. Therefore, we believe that those haplotypes that account for the greatest disease risk have been selected for further analysis. Nevertheless, although it is unlikely that we have missed high-risk haplotypes, the possibility remains.
For the class I region, four haplotypes with putative susceptibility (3-1 and 12-3) or protective (1-6 and 4-5) effects were chosen. Two haplotypes were analyzed further for the class III region (11-4-5 and 2-5-5), and only the 14-12 (DRB1*1501-DQB1*0602) haplotype was selected from class II haplotypes (table 2). Given that the true MS susceptibility loci and/or the main susceptibility haplotype(s) were unknown, there was no natural reference point to which other putative risk haplotypes could be compared. Hence, ORs and Fisher’s exact test for association were calculated from the number of haplotypes for the seven “sub-haplotypes” that were preselected, and ORs were calculated relative to all other haplotypes. For the class I region, the 3-1 haplotype was found to be significantly associated with disease susceptibility (P=7×10-4), whereas the 1-6 haplotype was borderline protective P=.03. The 11-4-5 haplotype from the class III region (P=.002) and 14-12 from the class II region (DRB1*1501-DQB1*0602, P=1×10-7) were significantly associated with disease susceptibility after correction for multiple testing (table 2).
We recognize that preselecting the most-discordant haplotypes visually (fig. 2A–C) represents, in itself, a form of statistical testing and, as such, these tests should be corrected for. However, many of the tests are highly correlated, and it is unclear what multiple-test correction is appropriate. Bonferroni correction represents the strongest form of statistical correction, since all tests are assumed to be independent; however, no impact on the major significant results in tables 1 and 2 occurred when a correction for more tests was performed (data not shown).
Log-Linear Modeling of Haplotype Interaction
The haplotype most associated with MS from each locus was selected for further analysis to determine their mode of interaction. The model included terms to describe association with disease, and LD between haplotypes. The data for cases and controls were stratified according to the most significantly associated haplotypes, 3-1 (class I), 11-4-5 (class III), and 14-12 (class II), producing a 2×2×2×2 (four-way) contingency table (tables 3 and 4) with three factors (the haplotypes) and one response variable (affected or unaffected). Each factor had two levels: having the haplotype or not having the haplotype. Haplotypes with missing data at one or more loci were excluded from the analysis, reducing the data set to 276 case haplotypes and 190 control haplotypes. The log-linear model was fitted using iterative proportional fitting from several different models, such as a saturated model (by deleting terms) and a main-effects–only model (by adding terms). The same final fitted model was always reached, regardless of the starting-point model: μ+dis+h1412+h1145+h31+dis•h31+dis•h1412+h31•h1145+h1145•h1412
Table 3.
Log-Linear Modeling Analysis of Haplotypes[Note]
|
No. of HaplotypesObserved (Expected) for |
||
| Subject, Haplotype,and ConstituentHaplotype | 14-12 | Not 14-12 |
| Case, 11-4-5: | ||
| 3-1 | 26 (27.8) | 7 (6.4) |
| Not 3-1 | 25 (22.6) | 4 (5.5) |
| Case, not 11-4-5: | ||
| 3-1 | 6 (5.4) | 22 (21.4) |
| Not 3-1 | 33 (34.2) | 153 (152.7) |
| Control, 11-4-5: | ||
| 3-1 | 5 (3.2) | 2 (2.6) |
| Not 3-1 | 4 (6.4) | 7 (5.5) |
| Control, not 11-4-5: | ||
| 3-1 | 0 (.6) | 8 (8.6) |
| Not 3-1 | 11 (9.8) | 153 (153.3) |
Note.— The table combines the four-way contingency tables that were constructed to explore the relationship between the constituent haplotypes 3-1 (MOGCA-D6S265: class I), 11-4-5 (TNFa-TNFd-D6S273: class III) and 14-12 (DRB1-DQB1: class II). Only complete haplotypes (276 case and 190 control) with no missing data at any of the loci tested were included in the analysis.
Table 4.
Summary of Class I (3-1) and Class II (14-12) Haplotype Frequencies in Cases versus Controls, from the Log-Linear Modeling Analysis
| ConstituentHaplotype | No. of Cases (%) | No. of Controls (%) |
| 3-1 and 14-12 | 32 (11.2) | 5 (2.6) |
| 3-1 and not 14-12 | 29 (10.5) | 10 (5.3) |
| Not 3-1 and 14-12 | 58 (21.0) | 15 (7.9) |
| Not 3-1 and not 14-12 | 157 (56.9) | 160 (84.2) |
For the expected cell frequency, m is the grand mean, dis represents the explanatory variable describing the disease, and h31, h1412, and h1145 represent the three susceptibility haplotypes. The factors separated by “•” represent the interaction terms. Since the haplotypes were treated as factors, it is prudent to fix their margins (Plackett 1981). This is achieved by fitting the h1412*h1145*h31 term in the final model. This term represents the one-way, two-way, and three-way interactions of the haplotypes, independent of disease, and thus absorbs the background LD present in the population. The final model gave a residual deviance of 4.9 with 5 df, which resulted in an excellent fit to the observed haplotype counts (table 3). Dropping either of the two significant haplotype disease interactions resulted in highly significant changes of deviance, ΔD, which is asymptotically distributed χ21. For h1412, ΔD= 24.1 (P=2×10-6) and for h31, ΔD = 9.1 (P=.001). Plotting the residuals using a Q-Q plot indicated that the main features of the data had been adequately captured by the fitted model (data not shown).
Several conclusions can be drawn from the fitted model. First, in the absence of disease status, examination of the one-, two-, and three-way interactions revealed significant interactions between only immediately adjacent haplotypes. The two terms describing the interaction between h31 and h1412 and the three-way interaction with the disease phenotype are not significant (data not shown) and are only included in the model in order to fix the margins. This does not mean that LD does not spread over all three haplotypes; rather, the neighboring haplotypes explain most of the LD in the log-linear model. Log-linear modeling performed with only the two flanking haplotypes (h1412 and h31) showed a significant LD interaction (data not shown). Second, the 11-4-5 haplotype (h1145) is not associated with disease. This suggests that the TNF locus and 14 other expressed class III genes within this haplotype (MHC Consortium 1999) are not associated with MS per se but instead are associated by virtue of their tight LD with the class II–associated susceptibility haplotype, h1412 (DRB1*1501-DQB1*0602). Third, 3-1 (h31) and 14-12 (h1412) show no higher-order interactions with MS, implying that the haplotypes act additively as independent susceptibility factors and therefore do not interact multiplicatively. It is possible that there are higher-order interactions between h31 and h1412; however, because these are not detectable with this data set, these effects would be weak.
Two-Point LD Analysis
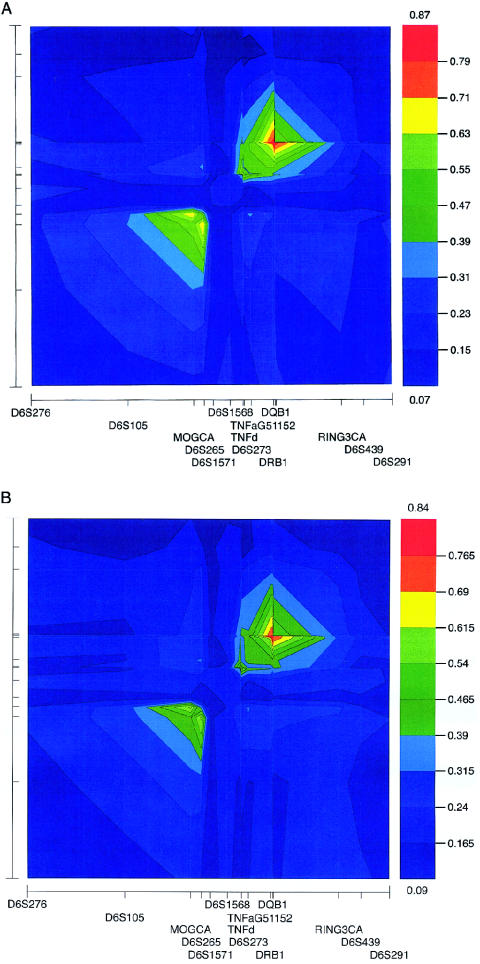
The haplotype data were subjected to two-point LD analysis producing a global overview of LD, using the D′ measure implemented in GOLD (fig. 3) (Abecasis and Cookson 2000). GOLD uses the multiallelic D′ measure, as defined by Hedrick (1987), which is a weighted average of the more familiar biallelic D′ measure. The blocklike structure of LD within HLA is profound for both cases and controls, with two discrete regions displaying elevated levels of suppressed recombination. The LD plots for case (fig. 3A) and control (fig. 3B) data are very similar, with the D′ global maximum only slightly higher in cases (0.87) than controls (0.84). Given the differences in sample size, small differences of this nature would be expected. The global and intrablock patterns of LD are virtually identical in cases and controls, which is a predictable result, given that we did not examine LD surrounding particular haplotypes but rather overall LD in the two samples. The distal block of LD in case haplotypes extends ∼3 Mb centromeric of D6S105 (extended class I), to D6S1571 (classical class I) (fig. 1). The local D′ maximum (0.63–0.71) in the region is located between MOGCA and D6S265. Centromeric of D6S1571, a region of reduced LD extends to the TNF locus. The level of LD then gradually increases from TNFa to the DRB1-DQB1-G51152 haplotype, which defines the location of the global and local D′ maximum of 0.87 in case and control haplotypes. Centromeric of this haplotype towards RING3CA, the level of LD decreases; however, even at this marker, D′ is 0.31–0.39.
Figure 3.
Two-point LD analysis, using GOLD, for case (A) and control (B) haplotypes. The relative location of markers used to construct the haplotypes is represented on the X-axis but is more clearly depicted in figure 1. The X-axis has been rotated left through 90° for the Y-axis, with the ticks identifying the relative marker location for both axes. The LD measure, D′, is graphically represented adjacent to each GOLD plot. For cases, D′ values are 0.07–0.87, and, for controls, they are 0.09–0.84. Between the top and bottom D′ value, the range has been divided into 10 intervals, each represented by a colored box. The D′ value delineating each interval is indicated.
Discussion
In the absence of genotypes from isolated chromosomes, one relies on algorithms to infer haplotypes. These can either derive probability distributions for potential haplotypes, such as the expectation-maximization algorithm (Slatkin and Excoffier 1996), or treat the haplotypes as variables to be estimated using the genotypic data from family members. One example of the latter method is an application of the Hidden Markov Model (HMM) that is known as the Lander-Green algorithm, in the GENEHUNTER program (Kruglyak et al. 1996). We have used this method for inferring haplotypes; however, a caveat when using haplotypes inferred by GENEHUNTER is that the program returns only the haplotype that maximizes the likelihood of the observed data, given a specific marker map. It may be the case that there are several possible haplotypes of equal or very close probability. Other algorithms—for example, a Monte Carlo Markov chain approach, as implemented in SIMWALK—may yield different haplotypes. To date, we have used only GENEHUNTER, which uses the Viterbi algorithm from the HMM methodology (Rabiner 1989) to reconstruct haplotypes; however, we anticipate that by genotyping a constellation of close relatives, we can obtain phase information that would be sufficient to permit alternate algorithms to infer identical haplotypes in most families. In the future we plan to investigate haplotype reconstruction qualitatively for these different algorithms on genomewide data from this sample.
We have found susceptibility and protective haplotypes in the class I region independent of the class II susceptibility haplotype, DRB1*1501-DQB1*0602, which replicated the findings of a recent study in different populations of northern European ancestry (Fogdell-Hahn et al. 2000). In addition, we have shown that the TNF locus and 14 other genes delineated by a 150-kb haplotype (TNFa-D6S273) in the class III region are not involved in susceptibility to MS in this sample.
HLA-A typing of a number of affected individuals provided strong evidence that the significantly associated class I susceptibility haplotype (3-1) was defined by the presence of the A*0301 allele. However, we found that the class I susceptibility haplotype had no higher order interactions with the class II region but yielded an additive effect on risk, in contrast to recent findings (Fogdell-Hahn et al. 2000). Nevertheless, these results do not provide sufficient resolution to identify the HLA-A*0301 allele as the causative susceptibility determinant in this region of the HLA complex. Indeed, speculating on the involvement of the HLA-A*0301 gene product on the basis of these results and allelic typing of the HLA-A gene would be premature.
Lie and colleagues (1999) recently reported the mapping of an unidentified gene in the extended class I region that modulates susceptibility to type 1 diabetes and celiac disease. We also identified a protective influence of the class I region, defined by a MOGCA-D6S265 haplotype (1-6). It is unclear at present whether this haplotype corresponds to the HLA-A*0201 allele described recently in Swedish and Norwegian patients with MS (Fogdell-Hahn et al. 2000). We did not analyze this haplotype for interactions with the class II region, because it was only weakly associated after correction for multiple testing. Nevertheless, it remains to be recognized that while genetic variants conferring protection and susceptibility can be detected by association with allelic variants of the HLA-A gene and the encompassing 400-kb haplotype, it is impossible to say at this stage if allelic heterogeneity at the same locus is responsible. Moreover, it is not even clear if these variants reside within the MOGCA-D6S265 haplotype. In addition, there were other MOGCA-D6S265 haplotypes showing marginal evidence for association prior to correction for multiple testing. It is possible that these marginally associated haplotypes would, with a larger data set, reach significance. Hence, it is intriguing to speculate that this region of the HLA complex contains multiple genetic determinants that play a role in MS.
By comparing two-point LD analyses of case and controls haplotypes across the HLA region, a consistent block-like pattern of LD was observed. These findings correspond well with previous studies of recombination rates in the HLA region (Martin et al. 1995; Cullen et al. 1997; Bugawan et al. 2000). The centromeric block of LD spanning class III and class II has long provided a source of confusion for single-marker association studies that use polymorphic markers in close proximity to the TNF locus. In the present study we have been able to dissect away a region of class III, including the TNF locus, from involvement in MS. Recent studies of recombination hot spots (Jeffreys et al. 2000, 2001) in the class II region are supported by the erosion of LD centromeric of the DRB1 gene. Efforts aimed at cloning the gene responsible for hemochromatosis (Feder et al. 1996) in 83% of patients were hindered by the strong LD extending almost 3.8 Mb telomeric of the HLA-A gene.
The HLA region contains genes that contribute to susceptibility and provide protection from MS. In order to conduct a genetic dissection of the HLA region that will help to identify the causative variants, it will be necessary to employ approaches that utilize haplotype information. Testing haplotypes rather than single loci provides the advantage of being able to test genomic segments encoding multiple genes, which represents a way to help distinguish true association from the confounding effects of strong LD. Interaction between HLA genes in trans is documented for type 1 diabetes and is therefore possible in MS. Hence, although the present article addresses haplotype interactions in cis, analyses of interactions in trans are warranted, but we recognize that a larger data set may be required to afford sufficient power for such a study. By analyzing haplotypes through the use of the approaches outlined here, we should gain a better understanding of the way in which the HLA region modulates disease.
Acknowledgments
First, our thanks go to the people with MS and their families, who have shown tremendous enthusiasm towards the Tasmanian MS Program. Thanks also to the control persons and their relatives. We are indebted to Michele Brown for DNA extraction. Thanks to the research assistants Jane Pittaway, Lyn Hall, Kristen Hazelwood, Rhonda McCoy, Deirbhile O’Byrne, and John Cary for their rigorous pursuit of biological samples. Our gratitude to Maree Ring and Annette Banks for genealogical research, to Sue Sawbridge and Tim Albion for database management, to Joan Clough for data entry, and to Natasha Newton and Emma Stubbs for administrative assistance. Thanks to Dr. Stan Sjeica, Dr. Andrew Hughes, and Bozidar Drulovic for clinical diagnosis, and Peter Duigan for HLA-A typing. J.P.R. and M.B. are postdoctoral fellows supported by The Cooperative Research Centre for Discovery of Genes for Common Human Diseases (Gene-CRC). T.J.K. is a Viertel Fellow. T.P.S. and S.J.F. are fellows of the National Health and Medical Research Council of Australia. All genotyping was conducted at the Australian Genome Research Facility. The work contained in the present article was funded by The Gene-CRC, and the National MS Society of the United States of America. The Gene-CRC is established and supported by the Australian Government's Co-operative Research Centre's Scheme.
Appendix
Table A1.
Allele-Coding Information for Seven Markers in the HLA Region[Note]
| Allele | MOGCA | D6S265 | TNFa | TNFd | D6S273 | DRB1* | DQB1* |
| 1 | 121 | 174 | 104 | 129 | 146 | 01/0103 | 0301 |
| 2 | 127 | 176 | 106 | 131 | 149 | 02 | 0302 |
| 3 | 129 | 178 | 108 | 133 | 151 | 03 | 0303 |
| 4 | 131 | 180 | 110 | 135 | 152 | 04 | 0304 |
| 5 | 133 | 182 | 112 | 137 | 154 | 06 | 0305 |
| 6 | 135 | 184 | 114 | 139 | 156 | 07 | 0402 |
| 7 | 137 | 186 | 116 | 141 | 158 | 08 | 0501 |
| 8 | 139 | 190 | 118 | 143 | 160 | 09 | 0502 |
| 9 | 141 | 192 | 120 | … | … | 10 | 0503 |
| 10 | 143 | 194 | 122 | … | … | 11 | 0504 |
| 11 | 146 | 196 | 124 | … | … | 12 | 0601 |
| 12 | 148 | … | 126 | … | … | 13 | 0602 |
| 13 | 150 | … | 128 | … | … | 14 | 0603 |
| 14 | 152 | … | 130 | … | … | 1501 | 0604 |
| 15 | … | … | … | … | … | 1502 | 0201/0202 |
| 16 | … | … | … | … | … | 1601 | 0605/0609 |
Note.— Except for allelic designation for the DRB1 and DQB1 genes, all “raw” allele data for SSLP markers (MOGCA, D6S265, TNFa, TNFd, and D6S273) are in base pairs. The degree to which allelic typing of the DRB1 and DQB1 genes was resolved was not uniform over all alleles, because prior knowledge existed for association. Hence, we pursued the resolution of DRB1*15, DRB1*16 and DQB1*06 genotypes. For instance, the DRB1*01 (allele 1) typing corresponds to multiple alleles, including DRB1*0103, which have not been resolved. When the population frequency of a particular allele was not split and was not strongly biased (>90%) in favor of one allele, both alleles have been recorded as possible (e.g, 0201/0202 and 0605/0609).
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Australian Genome Research Facility, http://www.agrf.org.au (for genotyping)
- Genome Database, http://www.gdb.org (for primer sequences)
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/genome/guide/human/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MS [MIM 126200])
- R Project for Statistical Computing, http://www.R-project.org/
- University of California, Santa Cruz, Human Genome Project Working Draft, http://genome.ucsc.edu/ (for genetic and physical map information)
- Australian Bureau of Statistics, http://www.abs.gov.au (for 1996 census data)
References
- Abecasis GR, Cookson WO (2000) GOLD: graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Allcock RJ, de la Concha EG, Fernandez-Arquero M, Vigil P, Conejero L, Arroyo R, Price P (1999) Susceptibility to multiple sclerosis mediated by HLA-DRB1 is influenced by a second gene telomeric of the TNF cluster. Hum Immunol 60:1266–1273 [DOI] [PubMed] [Google Scholar]
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148:249–258 [PubMed] [Google Scholar]
- Bugawan TL, Klitz W, Blair A, Erlich HA (2000) High-resolution HLA class I typing in the CEPH families: analysis of linkage disequilibrium among HLA loci. Tissue Antigens 56:392–404 [DOI] [PubMed] [Google Scholar]
- Celius EG, Harbo HF, Egeland T, Vartdal F, Vandvik B, Spurkiand A (2000) Sex and age at diagnosis are correlated with the HLA-DR2, DQ6 haplotype in multiple sclerosis. J Neurol Sci 178:132–135 [DOI] [PubMed] [Google Scholar]
- Chataway J, Feakes R, Coraddu F, Gray J, Deans J, Fraser M, Robertson N, Broadley, S Jones H, Clayton D, Goodfellow PN, Sawcer S, Compston A (1998) The genetics of multiple sclerosis: principles, background and updated results of the United Kingdom systematic genome screen. Brain 121:1869–1887 [DOI] [PubMed] [Google Scholar]
- Coraddu F, Sawcer S, Feakes R, Chataway J, Broadley S, Jones HB, Clayton D, Gray J, Smith S, Taylor C, Goodfellow PN, Compston, A. (1998) HLA typing in the United Kingdom multiple sclerosis genome screen. Neurogenetics 2:24–33 [DOI] [PubMed] [Google Scholar]
- Cullen M, Noble J, Erlich H, Thorpe K, Beck S, Klitz W, Trowsdale J, Carrington M (1997) Characterization of recombination in the HLA class II region. Am J Hum Genet 60:397–407 [PMC free article] [PubMed] [Google Scholar]
- Ebers GC, Bulman DE, Sadovnick AD, Paty DW, Warren S, Hader W, Murray TJ, Seland TP, Duquette P, Grey T (1986) A population-based study of multiple sclerosis in twins. N Engl J Med 315:1638–1642 [DOI] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Risch NJ, The Canadian Collaborative Study Group (1995) A genetic basis for familial aggregation in multiple sclerosis. Nature 377:150–151 [DOI] [PubMed] [Google Scholar]
- Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, Foote SJ (2000) Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet 67:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Wolff RK et al. (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13:399–408 [DOI] [PubMed] [Google Scholar]
- Fernandez-Arquero M, Arroyo R, Rubio A, Martin C, Vigil P, Conejero L, Figueredo MA, de al Concha EG (1999) Primary association of a TNF gene polymorphism with susceptibility to multiple sclerosis. Neurology 53:1361–1363 [DOI] [PubMed] [Google Scholar]
- Fogdell-Hahn A, Ligers A, Gronning M, Hillert J, Olerup O (2000) Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens 55:140–148 [DOI] [PubMed] [Google Scholar]
- Foissac A, Salhi M, Cambon-Thomsen A (2000) Microsatellites in the HLA region: 1999 update. Tissue Antigens 55:477–509 [DOI] [PubMed] [Google Scholar]
- Francis DA, Thompson AJ, Brookes P, Davey N, Lechler RI, McDonald WI, Batchelor JR (1991) Multiple sclerosis and HLA: is the susceptibility gene really HLA-DR or -DQ? Hum Immunol 32:119–124 [DOI] [PubMed] [Google Scholar]
- Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117:331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillert J, Olerup O (1993) Multiple sclerosis is associated with genes within or close to the HLA-DR-DQ subregion on a normal DR15,DQ6,Dw2 haplotype. Neurology 43:163–168 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Ritchie A, Neumann R (2000) High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet 9:725–733 [DOI] [PubMed] [Google Scholar]
- Jenisch S, Henseler T, Nair RP, Guo SW, Westphal E, Stuart P, Kronke M, Voorhees JJ, Christophers E, Elder JT (1998) Linkage analysis of human leukocyte antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/-C region. Am J Hum Genet 63:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jersild C, Svejgaard A, Fog T (1972) HL-A antigens and multiple sclerosis. Lancet 1:1240–1124 [DOI] [PubMed] [Google Scholar]
- Jongeneel CV, Briant L, Udalova IA, Sevin A, Nedospasov SA, Cambon-Thomsen A (1991) Extensive genetic polymorphism in the human tumor necrosis factor region and relation to extended HLA haplotypes. Proc Natl Acad Sci USA 88:9717–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Sasasuki T (1991) In HLA 1991, Proceedings of the 11th International Histocompatibility Workshop and Conference. In: Tsuji K, Aizawa M, Sasazuki T (eds) Eleventh International Histocompatibility Workshop reference protocol for the HLA DNA-typing technique. Vol 1. Oxford University Press, Tokyo, pp 397–419 [Google Scholar]
- Kruglyak L, Daly MJ, Reeve Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kwok WW, Kovats S, Thurtle P, Nepom GT (1993) HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J Immunol 150:2263–2272 [PubMed] [Google Scholar]
- Lie BA, Sollid LM, Ascher H, Ek J, Akselsen HE, Ronningen KS, Thorsby E, Undlien DE (1999) A gene telomeric of the HLA class I region is involved in predisposition to both type 1 diabetes and coeliac disease. Tissue Antigens 54:162–168 [DOI] [PubMed] [Google Scholar]
- Martin M, Mann D, Carrington M (1995) Recombination rates across the HLA complex: use of microsatellites as a rapid screen for recombinant chromosomes. Hum Mol Genet 4:423–428 [DOI] [PubMed] [Google Scholar]
- Martin MP, Harding A, Chadwick R, Kronick M, Cullen M, Lin L, Mignot E, Carrington M (1998) Characterization of 12 microsatellite loci of the human MHC in a panel of reference cell lines. Immunogenetics 47:503 [DOI] [PubMed] [Google Scholar]
- Masterman T, Ligers A, Olsson T, Andersson M, Olerup O, Hillert J (2000) HLA-DR15 is associated with lower age at onset in multiple sclerosis. Ann Neurol 48:211–219 [PubMed] [Google Scholar]
- McDonnell GV, Mawhinney H, Graham CA, Hawkins SA, Middleton D (1999) A study of the HLA-DR region in clinical subgroups of multiple sclerosis and its influence on prognosis. J Neurol Sci 165:77–83 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium, The (1999) Complete sequence and gene map of the human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Mumford CJ, Wood NW, Kellar-Wood H, Thorpe JW, Miller DH, Compston DA (1994) The British Isles survey of multiple sclerosis in twins. Neurology 44:11–15 [DOI] [PubMed] [Google Scholar]
- Nair RP, Stuart P, Henseler T, Jenisch S, Chia NV, Westphal E, Schork NJ, Kim J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2000) Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet 66:1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olerup O, Hillert J (1991) HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue Antigens 38:1–15 [DOI] [PubMed] [Google Scholar]
- Ota M, Mizuki N, Katsuyama Y, Tamiya G, Shiina T, Oka A, Ando H, Kimura M, Goto K, Ohno S, Inoko H (1999) The critical region for Behçet disease in the human major histocompatibility complex is reduced to a 46-kb segment centromeric of HLA-B, by association analysis using refined microsatellite mapping. Am J Hum Genet 64:1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paty DW, Oger JJ, Kastrukoff LF, Hashimoto SA, Hooge JP, Eisen AA, Eisen KA (1988) MRI in the diagnosis of MS: a prospective study with comparison of clinical evaluation, evoked potentials, oligoclonal banding, and CT. Neurology 38:180–185 [DOI] [PubMed] [Google Scholar]
- Plackett RL (1981) The analysis of categorical data, 2d ed. Charles Griffin, London [Google Scholar]
- Poser CM, Paty DW, Scheinberg L, McDonald WI, Davis FA, Ebers GC, Johnson KP, Sibley WA, Silberberg DH, Tourtellotte WW (1983) New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol 13:227–231 [DOI] [PubMed] [Google Scholar]
- Pridmore SA (1990) The large Huntington's disease family of Tasmania. Med J Aust 153:593–595 [DOI] [PubMed] [Google Scholar]
- Rabiner LR (1989) A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE 77:257–286 [Google Scholar]
- Roth MP, Nogueira L, Coppin H, Clanet M, Clayton J, Cambon-Thomsen A (1994) Tumor necrosis factor polymorphism in multiple sclerosis: no additional association independent of HLA. J Neuroimmunol 51:93–99 [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59:97–105 [DOI] [PubMed] [Google Scholar]
- Slatkin M, Excoffier L (1996) Testing for linkage disequilibrium in genotypic data using the expectation-maximization algorithm. Heredity 76:377–383 [DOI] [PubMed] [Google Scholar]
- Udalova IA, Nedospasov SA, Webb GC, Chaplin DD, Turetskaya RL (1993) Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics 16:180–186 [DOI] [PubMed] [Google Scholar]