Abstract
Sarcopenia corresponds to the degenerative loss of skeletal muscle mass, quality, and strength associated with ageing and leads to a progressive impairment of mobility and quality of life. However, the cellular and molecular mechanisms involved in this process are not completely understood. A hallmark of cellular and tissular ageing is the accumulation of oxidatively modified (carbonylated) proteins, leading to a decreased quality of the cellular proteome that could directly impact on normal cellular functions. Although increased oxidative stress has been reported during skeletal muscle ageing, the oxidized protein targets, also referred as to the ‘oxi-proteome’ or ‘carbonylome’, have not been characterized yet. To better understand the mechanisms by which these damaged proteins build up and potentially affect muscle function, proteins targeted by these modifications have been identified in human rectus abdominis muscle obtained from young and old healthy donors using a bi-dimensional gel electrophoresis-based proteomic approach coupled with immunodetection of carbonylated proteins. Among evidenced protein spots, 17 were found as increased carbonylated in biopsies from old donors comparing to young counterparts. These proteins are involved in key cellular functions such as cellular morphology and transport, muscle contraction and energy metabolism. Importantly, impairment of these pathways has been described in skeletal muscle during ageing. Functional decline of these proteins due to irreversible oxidation may therefore impact directly on the above-mentioned pathways, hence contributing to the generation of the sarcopenic phenotype.
Keywords: Ageing, Human rectus abdominis skeletal muscle, Oxidative stress, Protein carbonylation, Proteomics
Abbreviations: 1D, one-dimensional; 2D, bi-dimensional; ATP, adenosine triphosphate; CK, creatine kinase; DNP, 2–4-dinitrophenylhydrazone; DNPH, 2,4-dinitrophenylhydrazine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPD1, Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic; HSP70, Heat shock 70 kDa protein; IEF, Isoelectric focusing; MM-CK, muscle-type creatine kinase; MS, mass spectrometry; MyBPC, myosin-binding protein C; PCr–CK, phosphocreatine–creatine kinase; RA, rectus abdominis; RMI, relative modification index; ROS, reactive oxygen species; ZASP, LIM domain-binding protein 3
Graphical abstract
Highlights
-
•
Oxidative proteome modifications have been analyzed in aged skeletal muscle.
-
•
Proteins targeted by oxidation have been identified in human rectus abdominis muscle.
-
•
Such proteins are involved in muscle contraction, energy metabolism or transduction.
-
•
Functional alterations of these proteins may contribute to the sarcopenic phenotype.
Introduction
Skeletal muscles require dynamic changes in energy supply and oxygen flux for contraction, making it prone to reactive oxygen species (ROS)-mediated damage as a result of an increase in electron flux and leakage from the mitochondrial respiratory chain. Although physiological concentrations of ROS can function as signaling molecules that regulate proliferation, growth, differentiation, and apoptosis [1], when ROS levels overcome the capacity of cellular antioxidant systems, they become toxic, introducing oxidative modifications on cellular macromolecules such as nucleic acids, lipids and, in particular, proteins, inflicting alterations to normal cellular functions. In skeletal muscle, oxidative stress state has negative consequences on action-potential conduction, excitation–contraction coupling, satellite cell differentiation, muscle contraction and mitochondrial respiration [2].
Skeletal muscle ageing is associated with the gradual degenerative loss of skeletal muscle mass, quality, and strength, a condition known as sarcopenia. Oxidative stress contribute at least in part to muscle atrophy [3–6], and previous studies have addressed the extent of protein oxidative damage in the development of sarcopenia in mammalian models [7–10]. Although it is believed that oxidative stress contributes to skeletal muscle dysfunction through macromolecular damage, the molecular mechanisms remain elusive. Protein post-translational modifications induced by ROS are important features of oxidative stress [11]. Among them, protein carbonyl content is by far the most commonly used marker of protein oxidation [12,13]. In fact, protein carbonylation occurs when proteins directly react with ROS, leading to the formation carbonyl groups (aldehydes and ketones) for instance on such amino acids side chains as arginine, lysine, threonine and proline. Introduction of carbonyl groups on proteins can also occur through the reaction of aldehydic products of lipid peroxidation and of dicarbonyl compounds upon glycation and glycoxidation. Since it often leads to a loss of protein function and an increased thermosensitivity or hydrophobicity of the targeted protein [14,15], protein carbonylation has been considered as an indicator of protein damage.
In human skeletal muscle, preliminary studies on vastus lateralis and external intercostal muscles have shown increased accumulation of protein carbonyls during ageing [16–19]. However, in most cases, the protein targets of these oxidative damages and their functional consequences have not been identified. Indeed, this is an essential step to get a complete view of protein oxidative modifications and to understand the mechanisms by which these oxidized proteins potentially contribute to muscle weakness and dysfunction during ageing.
Therefore, proteomic studies, including the analysis of protein abundance as well as protein carbonylation are expected to provide valuable information to unravel the key molecular pathways implicated. In fact, proteomics and in particular bi-dimensional (2D) gels represent appropriate tools for the detection and identification of specific carbonylated proteins in a complex mixture [2,13,20]. The identification of such oxidatively modified proteins (i.e. the oxi-proteome components), can give some insights into the mechanisms by which these damaged proteins accumulate and potentially affect cellular and/or tissular function during ageing or in disease conditions [21]. In this paper, the occurrence and characterization of carbonylated proteins was studied in human rectus abdominis muscle obtained from young and old healthy donors. Although no significant differences in global protein carbonylation was observed at the proteome level, we have used 2D gel electrophoresis based proteomic approaches to improve the resolution of individual proteins for the quantitative analysis of their carbonylation status and further identification of these major skeletal muscle proteins that are targeted by oxidative damage during human rectus abdominis skeletal muscle ageing.
Material and methods
Human biopsies
Human rectus abdominis muscle biopsies were obtained during surgery. Each biopsy used has the written consent of the volunteer donor. A total of 22 human muscle biopsies were used: 11 from healthy men individuals between 0 to 12 years old (named young samples) and 11 from healthy men individuals between 52 and 76 years old (named old samples) (Table 1). All muscle biopsies had an initial wet weight between 15 and 24 mg (Table 1). The study was approved by the Ethical Committee at the Uppsala University Hospital.
Table 1.
Characteristics of the samples biopsies.
| Sample no | Donor age (years) | Wet weight (mg) | Protein recovery ratio (w/w in %)a |
|---|---|---|---|
| 1 | 0 | 24.00 | 7.0 |
| 2 | 1 | 20.80 | 4.3 |
| 3 | 5 | 20.00 | 7.6 |
| 4 | 9 | 17.00 | 9.3 |
| 5 | 6.5 | 15.00 | 7.1 |
| 6 | 10.75 | 21.90 | 7.6 |
| 7 | 12 | 22.00 | 9.1 |
| 8 | 7.1 | 22.00 | 9.7 |
| 9 | 76 | 20.30 | 9.5 |
| 10 | 70 | 18.00 | 8.3 |
| 11 | 74 | 19.10 | 7.8 |
| 12 | 66 | 19.60 | 8.0 |
| 13 | 74 | 19.20 | 8.4 |
| 14 | 65 | 18.66 | 5.2 |
| 15 | 65 | 18.78 | 6.6 |
| 16 | 71 | 24.00 | 10.8 |
| 17 | 12 | 20.00 | 15.6 |
| 18 | 3 | 20.50 | 3.6 |
| 19 | 9 | 20.38 | 11.0 |
| 20 | 56 | 20.47 | 7.4 |
| 21 | 61 | 20.53 | 17.1 |
| 22 | 52 | 20.13 | 27.2 |
Protein recovery ratio corresponds to the protein amount in mass / biopsy mass.
Protein extraction for proteomics analyses
Proteins extracts from skeletal muscle biopsies were obtained by physical disruption of the sample biopsies using a ULTRA-TURRAX® T25 (IKA®) at 4 °C in a lysis buffer containing 10 mM Tris–HCl (pH 7.4), 8 M urea, 2 M thiourea, 4% CHAPS and 20 mM DTT. After incubation on ice for 20 min, soluble proteins were recovered after clarification by centrifugation for 40 min at 21,000 g. Proteins were further precipitated using the 2D clean-up kit (GE Healthcare) and the resulting pellet was re-suspended into the same lysis buffer. Protein concentrations were determined by the Bradford Method [22] using the Bio-Rad Protein Kit Assay (Bio-Rad).
Protein carbonyl immunodetection after derivatization with DNPH
Carbonylated proteins were derivatized with 2,4-dinitrophenylhydrazine (DNPH) to form 2–4-dinitrophenylhydrazone (DNP) proteins adducts [23]. For total carbonyl quantification, equal quantities of proteins were loaded and separated by SDS-PAGE 12% (v/v). Chemicals for SDS-PAGE were purchased from Bio-Rad. All other chemicals were of analytical grade and obtained from Sigma-Aldrich. For the detection of carbonylated proteins, gels were electrotransferred onto Hybond-C nitrocellulose membranes (GE Healthcare) and incubated with anti-DNP antibodies (1:5000, Sigma-Aldrich). Carbonylated proteins were revealed by a fluorescent anti-rabbit IgG 800CW (1:15,000) polyclonal antibody (LI-COR). Densitometry analyses were performed using NIH ImageJ software and the data are expressed as % volume in pixels.
For 2D gel electrophoresis, derivatization of proteins carbonyls was achieved on IPG strips after isoelectric focusing (IEF), with a 10 mM DNPH, 2 M HCl solution at room temperature (RT) as described previously [24]. 500 µg of protein were diluted on a rehydratation buffer (7 M urea, 2 M thiourea, 1% Amberlite, 4% CHAPS, 1.2% Destreak (GE Healthcare), 0.5% Pharmalyte pH 3–10 (GE Healthcare)) and loaded into 13 cm IPG strips pH 3–10 NL (GE Healthcare). Gel rehydration of the IPG strips was done overnight at RT in an Immobiline DryStrip rewelling tray. IEF was performed using an Ettan™ IPGphor™ 3 Isoelectric Focusing System (GE Healthcare) at 20 °C using the following electrical profile: step, 150 V for 11 h; grad, 1000 V for 3 h; grad, 8000 V for 2 h; step, 8000 V for 1 h. Neutralization step after derivatization with DNPH was performed with 2 M Trizma-Base containing 30% of glycerol (GE Healthcare). Before SDS-PAGE, all IPG strips were equilibrated with a 6 M urea, 10% SDS and 30% Glycerol (GE Healthcare), 0.5 M Tris–HCl (pH 8.8) solution for 2 steps of 15 min: the first one with equilibration solution containing 1% DTT and the second with equilibration solution containing 3% iodoacetamide. SDS-PAGE was carried out using the Protean II system (Bio-Rad). For each sample, two 12% (v/v) polyacrylamide gels were performed in parallel: one for total protein stains with Coomassie Brilliant Blue G-250 (Bio-Rad) for further mass spectrometry analysis; and the other with derivatized protein residues for electrotransfer onto nitrocellulose membranes, where total amount of proteins were stained by a Fast Green solution (Sigma-Aldrich) prior to antibodies incubation for loading control. Membranes were then blocked with Odyssey blocking buffer (LI-COR) overnight at 4 °C. Primary anti-DNP antibodies (1:5000, Sigma-Aldrich) were incubated for 1 h at RT. Revelation was done by a fluorescent anti-rabbit IgG 800CW (1:15,000) polyclonal antibody (LI-COR). Washing steps were done with a PBS/0.1% Tween solution. Carbonylated proteins were revealed by the Odyssey Infrared Imaging System (LI-COR).
Proteomics data acquisition and analysis
Spot detection and quantification were carried out using the image Master 2D Platinum 7 software (GE Healthcare). For comparison, we used the data expressed as spot % volume (%vol) in pixels, which corresponds to a normalized value of the spot volume by considering the total volume of all the spots present in the membrane [25]. To take into account the total amount of loaded protein in each IPG strip, we defined a %vol per carbonylated spot (N%vol) by normalizing the %vol of each anti-DNP incubated membrane with its corresponding %vol on the Fast Green-stationed membrane. The relative modification index ratio (RMI ratio) was obtained by dividing the N%vol of the old group of samples by the N%vol of the young one.
In-gel digestion and mass spectrometry
Spots of interest were manually excised from Coomassie Blue stained 12% (v/v) polyacrylamide gels and were automatically in-gel digested by trypsin [26] using a robot Freedom EVO 100 digester/spotter robot (Tecan). Resulted peptides were then desalted for mass spectrometry (MS/MS) analysis. Results obtained were subjected to a search on the SwissProt database using the MASCOT software (Matrix Science Ltd., London, UK). The software compares the MASCOT peptide sequences derived from spectra with those in libraries for protein identification. Search parameters were as follows: database, SwissProt; taxonomy, all entries or mammalian; enzyme, trypsin; allow up to one missed cleavage; fixed modifications, none; variable modifications, methionine oxidation; peptide mass tolerance, 70 ppm; and fragment mass tolerance, 500 ppm.
Results and discussion
Initial screening looking at changes at the global proteome level of carbonylated proteins was performed after derivatization of protein carbonyls with DNPH followed by immunodetection of DNP protein adducts after SDS-PAGE (Fig. 1A). Densitometry analysis was performed and normalized by total protein content for each sample (Fig. 1B and C). No significant differences at the global level in protein carbonyl content was detected between groups, in agreement with what was previously reported by Marzani et al. who did not find a statistical difference on protein carbonyl content during ageing in both rectus abdominis and vastus lateralis human muscles [18]. More recently, Fanò et al., using skeletal muscle biopsy samples obtained from vastus lateralis, found that protein carbonyls, showed a significant increase during ageing. However, this difference was not significant after splitting by gender [27]. Importantly, immunodetection of carbonylated proteins after one-dimensional (1D) electrophoresis separation has serious limitations for the resolution of single protein bands and can provide only restricted information.
Fig. 1.
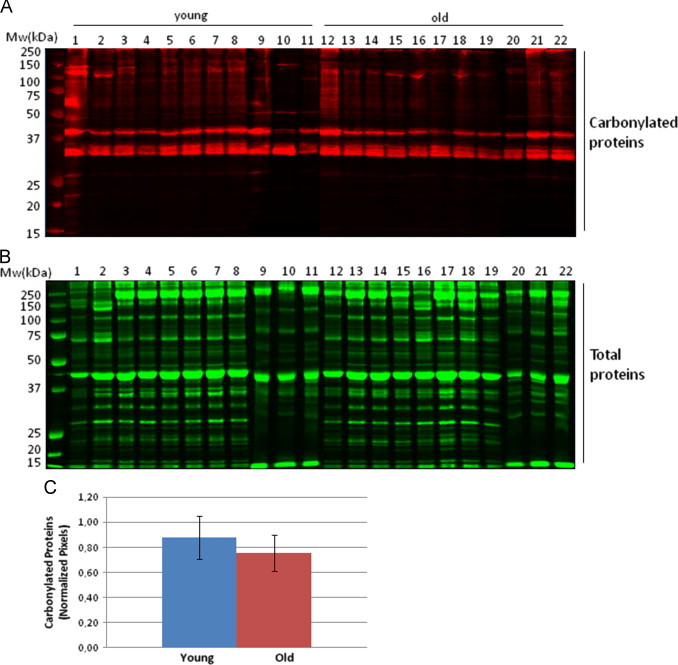
Analysis of total carbonylated proteins of human rectus abdominis muscle biopsies. (A) Carbonylated protein profiles from young and old human biopsies after 1D gel electrophoresis. Protein carbonyl were detected by Western-blotting against protein-DNP derivatives and monitored by fluorescent secondary antibody hybridation. (B) Total protein profiles stained with colloidal Coomassie brilliant blue G after 1D gel electrophoresis. (C) Densitometric analysis of carbonylated protein western-blots. Semi-quantitative assessment of modified proteins was done using total protein staining for normalization. Relative values are expressed as mean±S.D. (n=11) and no significant difference was found between the young and old groups.
To further analyze the occurrence of protein carbonylation at the single protein level, 2D gel electrophoresis separation of protein extracts was performed prior to immunodetection of carbonylated proteins. 2D gels are very appropriate to investigate protein isoforms since many post-translational modifications (such as carbonylation of arginine or lysine residues) often leads to changes in the isoelectric point of proteins, and thus shift the position of the protein in 2D gels. After electrotransfer onto nitrocellulose membranes total protein profiles were obtained by fast-green staining (Fig. 2). All the analyzed samples displayed a similar protein migration pattern, suggesting no drastic shifts in the protein profiles at the expression level (Fig. 2, right panels) between the two experimental groups. Immunodetection of carbonylated proteins was performed after their derivatization by DNPH as described in Material and methods. Interestingly, the pattern of the modified proteins was not superimposable with the pattern obtained for the total protein staining, indicating that certain proteins represent preferential targets for these deleterious oxidative modifications. Several proteins of high molecular weight were found preferentially carbonylated (Fig. 2, left panel) while certain proteins with an isoelectric point higher than 5, also appeared to be preferentially carbonylated.
Fig. 2.
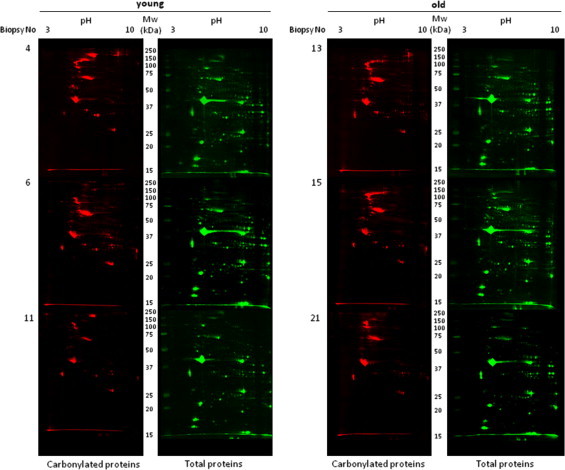
Oxi-proteome analysis of young and old human skeletal muscle samples. Protein extracts from young (n=3) and old (n=3) human skeletal muscle biopsies were separated by 2D gel electrophoresis. After the second dimension, gels were electrotransferred onto nitrocellulose membranes for subsequent immune detection of DPNH-derivatized carbonylated proteins (left panels). Densitometry analysis was done by image Master 2D software (GE Healthcare) using total protein staining (right panels) as loading control.
A relative modification index (RMI) per spot was calculated in order to evidence differentially carbonylated proteins between the two experimental groups taking into account their protein expression levels. Seventeen protein spots exhibited an RMI ratio consistently higher than 1.3 (increasingly carbonylated) in all biopsies from the aged group as compared with the young one. On the other hand, fourteen protein spots showed decreased carbonylation in the old group (RMI<0.7). Protein spots evidenced as increasingly carbonylated were excised from Coomassie Blue stained 2D gels (Fig. 3) and analyzed by MS/MS for protein identification. Protein spots that were identified in the aged group are listed in Table 2. Among the identified proteins, 8 are muscle specific, 4 are ubiquitous in different tissues and organ systems, and 2 proteins belong from plasma (Table 2).
Fig. 3.
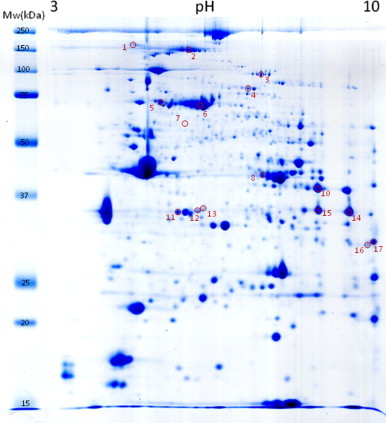
Coomassie blue staining of one representative 2D gel. The numbers and positions of the 17 selected spots identified by MS/MS correspond to those found as consistently increased on the old group (RMI>1.3).
Table 2.
Localization of carbonylated proteins identified from old skeletal muscle biopsies.
| Protein name and localization | Swiss-Prot accession no | Protein spot noa | Mascot scoreb | Sequence coverage (%) | No. of sequenced peptides | Theoretical protein mass (kDa) | Theoretical PI | RMI ratioc |
|---|---|---|---|---|---|---|---|---|
| Ubiquitous proteins: | ||||||||
| Collagen alpha-1(VI) chain | CO6A1_HUMAN | 1 | 205 | 7 | 5 | 108 | 5.3 | 4.34 |
| Heat shock cognate 71 kDa protein | HSP7C_HUMAN | 5 | 256 | 15 | 5 | 71 | 5.4 | 1.63 |
| Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic (GPD1) | GPDA_HUMAN | 13 | 214 | 22 | 4 | 38 | 5.8 | 1.52 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | G3P_HUMAN | 14 | 243 | 12 | 3 | 36 | 8.6 | 1.67 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | G3P_HUMAN | 15 | 424 | 25 | 5 | 36 | 8.6 | 1.68 |
| Voltage-dependent anion-selective channel protein 1 | VDAC1_HUMAN | 16 | 685 | 39 | 8 | 31 | 8.6 | 1.88 |
| Muscle specific proteins: | ||||||||
| Myosin-binding protein C, slow-type (MyBPC) | MYPC1_HUMAN | 2 | 119 | 3 | 3 | 128 | 5.8 | 1.54 |
| Glycogen phosphorylase, muscle form | PYGM_HUMAN | 3 | 882 | 22 | 13 | 97 | 6.6 | 1.67 |
| Myosin-7 | MYH7_HUMAN | 7 | 63 | – | 1 | 223 | 5.6 | 3.12 |
| Creatine kinase M-type | KCRM_HUMAN | 8 | 206 | 9 | 3 | 43 | 6.8 | 1.44 |
| Creatine kinase M-type | KCRM_HUMAN | 9 | 416 | 20 | 6 | 43 | 6.8 | 1.54 |
| Fructose-bisphosphate aldolase A | ALDOA_HUMAN | 10 | 611 | 31 | 7 | 39 | 8.3 | 1.65 |
| Troponin T, slow skeletal muscle | TNNT1_HUMAN | 11 | 512 | 22 | 6 | 33 | 5.9 | 1.87 |
| Troponin T, slow skeletal muscle | TNNT1_HUMAN | 12 | 244 | 22 | 5 | 33 | 5.9 | 1.42 |
| LIM domain-binding protein 3 (ZASP) | LDB3_HUMAN | 17 | 206 | 8 | 4 | 77 | 8.5 | 1.70 |
| Plasma proteins: | ||||||||
| Serotransferrin | TRFE_HUMAN | 4 | 399 | 17 | 10 | 77 | 6.8 | 1.31 |
| Serum albumin | ALBU_HUMAN | 6 | 986 | 29 | 13 | 69 | 5.9 | 1.33 |
Spots of interest were identified by MALDI-TO–FTOF-MS as described under Material and methods. For each spot, different parameters clarifying protein identification by MS are indicated.
Protein spot number refers to the numbered spots in Fig. 3.
Mascot protein scores greater than 56 are significant (P<0.05).
RMI ratio represents the Relative Modification Index ratio.
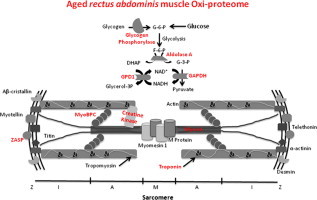
The identified skeletal muscle proteins were then analyzed and grouped by metabolic pathways and cellular functions. Major biological functions include muscle contraction, energy transduction and energy metabolism (Fig. 4). Among proteins involved in muscle contraction, we have found that myosin 7, troponin T, myosin-binding protein C (MyBPC), and LIM domain-binding protein 3 (ZASP) are increasingly carbonylated in aged muscle. Interestingly, the decreased speed of contraction observed in old age at both the muscle and motor protein levels is a hallmark of skeletal muscle ageing [28–31]. Myosin is a highly conserved protein that converts chemical energy into mechanical force and a key protein for muscle contraction. Previous studies have shown increased glycation of myosin in both fiber types of aged rats [32]. In addition, the carbonylated residues have been identified [33]. Interactions of myosin with cytoskeletal proteins such as titin, myomesin/M protein and MyBPC play an important role in thick filaments physiology. MyBPC contributes to the assembly and stabilization of thick filaments and modulates the formation of actomyosin cross-bridges, via direct interactions with both thick myosin and thin actin filaments [34,35]. The importance of MyBPC to muscle contraction is further emphasized by the discovery that mutations in genes encoding MyBPC cause myopathies in both skeletal [36,37] and cardiac muscles [38–40]. Increased oxidation of MyBPC, together with the previously reported age-dependent decrease of myosin isoforms and regulatory proteins like myosin binding proteins C and H [41–44], may contribute to the destabilization of muscle fibers. In addition, perturbations in the thin sarcomere fillaments of muscle fibers have also been associated with differently expression of actin and its regulatory proteins such as troponin and tropomyosin [42,43,45,46]. Here we found increased carbonylation levels of troponin T in the elderly. Troponin inhibits the actomyosin Mg2+-ATPase and the Ca2+ release from the sarcoplasmic reticulum inducing a change in the troponin–tropomyosin conformation that exposes myosin binding sites on the actin filament and activates the myosin ATPase, thereby allowing muscle contraction [47,48]. Since muscle contraction depends on myofibrillar thin filament interactions [49], the oxidative damage to these proteins may well compromise this interaction and hence muscle contraction.
Fig. 4.
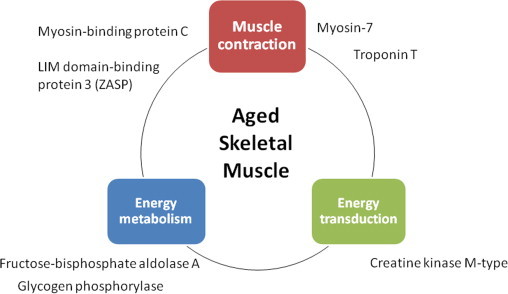
Functional grouping of muscle proteins increasingly oxidized with age. Increasingly oxidized muscle-specific proteins identified in aged rectus abdominis biopsies were grouped in three functional categories: muscle contraction, energy metabolism and energy transduction.
The motor functions of striated muscle crucially depend on the highly ordered arrays of thick myosin and thin actin filaments in sarcomeres. Although its detailed role has not been elucidated yet, the LIM domain-binding protein 3 (ZASP), found as increasingly carbonylated in aged rectus abdominis muscle, functions as an adapter to couple protein kinase C-mediated signaling to the cytoskeleton and to maintain Z-disc stability as well as cytoskeletal ultrastructure during contraction. Accumulation of this protein in its oxidized form may lead to protein aggregates formation and myofibril disintegration that may result in muscle weakness.
Muscle contraction depends also in high-energy fluxes where creatine kinase plays a central role. During muscle contraction, myosin hydrolyzes ATP into ADP upon filament sliding. The renewal of ATP is achieved mainly by the phosphocreatine–creatine kinase (PCr–CK) system. Among the creatine kinase (CK) isoenzymes, the muscle-type CK (MM-CK) specifically binds to the myofibril M-line and is associated with the action-activated myosin ATPase as an intramyofibrillar ATP regenerator [50–52]. Since this MM-CK was found irreversibly oxidized in old rectus abdominis muscle, this could alter muscle metabolism and compromise muscle contraction in the elderly. Besides the PCr–CK system, the glycolytic network and its closer interaction between mitochondria and organelles also provide energy within muscle cells [53]. In this study, 3 glycolytic enzymes appear as highly carbonylated in old slow oxidative skeletal rectus abdominis muscle: the fructose-bisphosphate aldolase A, the glycerol-3-phosphate dehydrogenase (GPD1) and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Table 2). Interestingly, several of these enzymes involved in anaerobic metabolism have been found decreased with age in murine and human skeletal muscle [54,55].
Although slow skeletal muscles uses mainly oxidative mitochondrial processes to generate the levels of ATP needed to maintain contractile activity for long time without showing fatigue [53], they also rely in glycolysis for their energy production [56]. For this reason, muscle metabolism and contraction in aged skeletal muscle would be affected by irreversible carbonylation on glycolytic proteins, as previously reported in certain diseases when these glycolytic enzymes are not functional [57–62]. Defects in the muscle form of glycogen phosphorylase, another protein found increased carbonylated in old rectus abdominis muscle, are involved in type 5 glycogen storage disease, also known as McArdle disease, a myopathy also characterized by exercise intolerance, such as easy fatigability, muscle cramps and contractures as well as muscle weakness [63,64]. Intramuscular glycogen acts as a readily available source of glucose-6-phosphate for glycolysis within skeletal muscle and glycogen phosphorylase catalyses the rate-limiting step in glycogenolysis [65]. Therefore, a deficiency in glycogen phosphorylase may result in the inability to mobilize muscle glycogen during anaerobic metabolism [66].
Although it is well recognized that ageing causes changes in the proteome, the nature and targets of these changes, their consequences on skeletal muscle function and how they may contribute to sarcopenia have not yet been completely elucidated. Our results suggest that oxidative stress during skeletal muscle ageing targets the contractile machinery, but also structural and regulatory proteins. In addition, the main mechanism of energy production, the phosphocreatine kinase system was affected by carbonylation in the elderly while other energetic metabolic pathways such as glycolysis appear to be also affected. Finally, the heat shock 70 kDa protein (HSP70), a key player in protein quality control with different identified roles in skeletal muscle [67], such as protection against oxidative stress, was also found highly carbonylated in old skeletal muscle. Up-regulation of heat shock proteins is a well know feature of muscle ageing [46,68,69]. Further studies should address the functional status of the identified carbonylated proteins and related cellular pathways associated with muscle dysfunction in order to reveal their role in the development of the ageing phenotype.
Acknowledgments
The authors wish to acknowledge the FP7 EU-funded MyoAge Project (No. 223576), and COST Action CM1001, coordinated by Dr. Gillian Butler-Browne and Dr. Tilman Grune, respectively, as well as the AFM-Téléthon for their support. In addition, they are very grateful to M. Cédric Broussard at the Plate-forme Protéomique Université Paris-Descartes 3P5 for performing the mass spectrometry analyses.
References
- 1.Barbieri E., Sestili P. Reactive oxygen species in skeletal muscle signaling. J. Signal Trans. 2012;2012:982794. doi: 10.1155/2012/982794. 22175016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baraibar M.A., Gueugneau M., Duguez S., Butler-Browne G., Bechet D., Friguet B. Expression and modification proteomics during skeletal muscle ageing. Biogerontology. 2013;14(3):339–352. doi: 10.1007/s10522-013-9426-7. 23624703 [DOI] [PubMed] [Google Scholar]
- 3.Powers S.K., Kavazis A.N., McClung J.M. Oxidative stress and disuse muscle atrophy. J. Appl. Physiol. 2007;102(6):2389–2397. doi: 10.1152/japplphysiol.01202.2006. 17289908 [DOI] [PubMed] [Google Scholar]
- 4.Pellegrino M.A., Desaphy J.-F., Brocca L., Pierno S., Camerino D.C., Bottinelli R. Redox homeostasis, oxidative stress and disuse muscle atrophy. J. Physiol. 2011;589(9):2147–2160. doi: 10.1113/jphysiol.2010.203232. 21320887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers S.K., Smuder A.J., Criswell D.S. Mechanistic links between oxidative stress and disuse muscle atrophy. Antioxid. Redox Signal. 2011;15(9):2519–2528. doi: 10.1089/ars.2011.3973. 21457104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers S.K., Smuder A.J., Judge A.R. Oxidative stress and disuse muscle atrophy: cause or consequence? Curr. Opin. Clinical Nutr. Metab. Care. 2012;15(3):240–245. doi: 10.1097/MCO.0b013e328352b4c2. 22466926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson D.C., Betzenhauser M.J., Reiken S., Meli A.C., Umanskaya A., Xie W., Shiomi T., Zalk R., Lacampagne A., Marks A.R. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 2011;14(2):196–207. doi: 10.1016/j.cmet.2011.05.014. 21803290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choksi K.B., Nuss J.E., Deford J.H., Papaconstantinou J. Age-related alterations in oxidatively damaged proteins of mouse skeletal muscle mitochondrial electron transport chain complexes. Free Radic. Biol. Med. 2008;45(6):826–838. doi: 10.1016/j.freeradbiomed.2008.06.006. 18598756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snow L.M., Fugere N.A., Thompson L.V. Advanced glycation end-product accumulation and associated protein modification in Type II skeletal muscle with aging. J. Gerontol. A: Biol. Sci. Med. Sci. 2007;62(11):1204–1210. doi: 10.1093/gerona/62.11.1204. 18000139 [DOI] [PubMed] [Google Scholar]
- 10.Thompson L.V., Durand D., Fugere N.A., Ferrington D.A. Myosin and actin expression and oxidation in aging muscle. J. Appl. Physiol. 2006;101(6):1581–1587. doi: 10.1152/japplphysiol.00426.2006. 16840579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breusing N., Grune T. Biomarkers of protein oxidation from a chemical, biological and medical point of view. Exp. Gerontol. 2010;45(10):733–737. doi: 10.1016/j.exger.2010.04.004. 20403419 [DOI] [PubMed] [Google Scholar]
- 12.Baraibar M.A., Friguet B. Oxidative proteome modifications target specific cellular pathways during oxidative stress, cellular senescence and aging. Exp. Gerontol. 2013;48(7):620–625. doi: 10.1016/j.exger.2012.10.007. 23127722 [DOI] [PubMed] [Google Scholar]
- 13.Baraibar M.A., Ladouce R., Friguet B. Proteomic quantification and identification of carbonylated proteins upon oxidative stress and during cellular aging. J. Proteomics. 2013;92:63–70. doi: 10.1016/j.jprot.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Grune T., Shringarpure R., Sitte N., Davies K. Age-related changes in protein oxidation and proteolysis in mammalian cells. J. Gerontol. A: Biol. Sci. Med. Sci. 2001;56(11):B459–B467. doi: 10.1093/gerona/56.11.b459. 11682566 [DOI] [PubMed] [Google Scholar]
- 15.Baraibar M.A., Barbeito A.G., Muhoberac B.B., Vidal R. A mutant light-chain ferritin that causes neurodegeneration has enhanced propensity toward oxidative damage. Free Radic. Biol. Med. 2012;52(9):1692–1697. doi: 10.1016/j.freeradbiomed.2012.02.015. 22348978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianni P., Jan K.J., Douglas M.J., Stuart P.M., Tarnopolsky M.A. Oxidative stress and the mitochondrial theory of aging in human skeletal muscle. Exp. Gerontol. 2004;39(9):1391–1400. doi: 10.1016/j.exger.2004.06.002. 15489062 [DOI] [PubMed] [Google Scholar]
- 17.Pansarasa O., Castagna L., Colombi B., Vecchiet J., Felzani G., Marzatico F. Age and sex differences in human skeletal muscle: role of reactive oxygen species. Free Radic. Res. 2000;33(3):287–293. doi: 10.1080/10715760000301451. 10993482 [DOI] [PubMed] [Google Scholar]
- 18.Marzani B., Felzani G., Bellomo R.G., Vecchiet J., Marzatico F. Human muscle aging: ROS-mediated alterations in rectus abdominis and vastus lateralis muscles. Exp. Gerontol. 2005;40(12):959–965. doi: 10.1016/j.exger.2005.08.010. 16213688 [DOI] [PubMed] [Google Scholar]
- 19.Barreiro E., Coronell C., Laviña B., Ramírez-Sarmiento A., Orozco-Levi M., Gea J., PENAM Project Aging, sex differences, and oxidative stress in human respiratory and limb muscles. Free Radic. Biol. Med. 2006;41(5):797–809. doi: 10.1016/j.freeradbiomed.2006.05.027. 16895800 [DOI] [PubMed] [Google Scholar]
- 20.Rogowska-Wrzesinska A., Le Bihan M.-C., Thaysen-Andersen M., Roepstorff P. 2D gels still have a niche in proteomics. J. Proteomics. 2013;88:4–13. doi: 10.1016/j.jprot.2013.01.010. 23353020 [DOI] [PubMed] [Google Scholar]
- 21.Baraibar M.A., Liu L., Ahmed E.K., Friguet B. Protein oxidative damage at the crossroads of cellular senescence, aging, and age-related diseases. Oxid. Med. Cell. Longev. 2012;2012:1–8. doi: 10.1155/2012/919832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. 942051 [DOI] [PubMed] [Google Scholar]
- 23.Levine R.L., Garland D., Oliver C.N., Amici A., Climent I., Lenz A.G., Ahn B.W., Shaltiel S., Stadtman E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. 1978225 [DOI] [PubMed] [Google Scholar]
- 24.Baraibar M.A., Hyzewicz J., Rogowska-Wrzesinska A., Ladouce R., Roepstorff P., Mouly V., Friguet B. Oxidative stress-induced proteome alterations target different cellular pathways in human myoblasts. Free Radic. Biol. Med. 2011;51(8):1522–1532. doi: 10.1016/j.freeradbiomed.2011.06.032. 21810466 [DOI] [PubMed] [Google Scholar]
- 25.Ahmed E.K., Rogowska-Wrzesinska A., Roepstorff P., Bulteau A.-L., Friguet B. Protein modification and replicative senescence of WI-38 human embryonic fibroblasts. Aging Cell. 2010;9(2):252–272. doi: 10.1111/j.1474-9726.2010.00555.x. 20102351 [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. 17406544 [DOI] [PubMed] [Google Scholar]
- 27.Fanò G., Mecocci P., Vecchiet J., Belia S., Fulle S., Polidori M.C., Felzani G., Senin U., Vecchiet L., Beal M.F. Age and sex influence on oxidative damage and functional status in human skeletal muscle. J. Muscle Res. Cell Mot. 2001;22(4):345–351. doi: 10.1023/a:1013122805060. 11808774 [DOI] [PubMed] [Google Scholar]
- 28.Höök P., Li X., Sleep J., Hughes S., Larsson L. In vitro motility speed of slow myosin extracted from single soleus fibres from young and old rats. J. Physiol. 1999;520(2):463–471. doi: 10.1111/j.1469-7793.1999.00463.x. 10523415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Larsson L. Maximum shortening velocity and myosin isoforms in single muscle fibers from young and old rats. Am. J. Physiol.—Cell Physiol. 1996;270:C352–C360. doi: 10.1152/ajpcell.1996.270.1.C352. [DOI] [PubMed] [Google Scholar]
- 30.Larsson L., Li X., Frontera W.R. Effects of aging on shortening velocity and myosin isoform composition in single human skeletal muscle cells. Am. J. Physiol. 1997;272(2 1):C638–C649. doi: 10.1152/ajpcell.1997.272.2.C638. 9124308 [DOI] [PubMed] [Google Scholar]
- 31.Höök P., Sriramoju V., Larsson L. Effects of aging on actin sliding speed on myosin from single skeletal muscle cells of mice, rats, and humans. Am. J. Physiol.—Cell Physiol. 2001;280(4):C782–C788. doi: 10.1152/ajpcell.2001.280.4.C782. 11245594 [DOI] [PubMed] [Google Scholar]
- 32.Ramamurthy B., Larsson L. Detection of an aging-related increase in advanced glycation end products in fast- and slow-twitch skeletal muscles in the rat. Biogerontology. 2013;14(3):293–301. doi: 10.1007/s10522-013-9430-y. 23681254 [DOI] [PubMed] [Google Scholar]
- 33.Li M., Ogilvie H., Ochala J., Artemenko K., Iwamoto H., Yagi N., Bergquist J., Larsson L. Aberrant post-translational modifications compromise human myosin motor function in old age. Aging Cell. 2015;14(2):228–235. doi: 10.1111/acel.12307. 25645586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackermann M.A., Kontrogianni-Konstantopoulos A. Myosin binding protein-C: a regulator of actomyosin interaction in striated muscle. J. Biomed. Biotechnol. 2011;2011:636403. doi: 10.1155/2011/636403. 22028592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Dijk S.J., Bezold K.L., Harris S.P. Earning stripes: myosin binding protein-C interactions with actin. Pflugers Arch. 2014;466(3):445–450. doi: 10.1007/s00424-013-1432-8. 24442149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markus B., Narkis G., Landau D., Birk R.Z., Cohen I., Birk O.S. Autosomal recessive lethal congenital contractural syndrome type 4 (LCCS4) caused by a mutation in MYBPC1. Hum. Mutat. 2012;33(10):1435–1438. doi: 10.1002/humu.22122. 22610851 [DOI] [PubMed] [Google Scholar]
- 37.Ackermann M.A., Patel P.D., Valenti J., Takagi Y., Homsher E., Sellers J.R., Kontrogianni-Konstantopoulos A. Loss of actomyosin regulation in distal arthrogryposis myopathy due to mutant myosin binding protein-C slow. FASEB J. 2013;27(8):3217–3228. doi: 10.1096/fj.13-228882. 23657818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlossarek S., Mearini G., Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J. Mol. Cell. Cardiol. 2011;50(4):613–620. doi: 10.1016/j.yjmcc.2011.01.014. 21291890 [DOI] [PubMed] [Google Scholar]
- 39.Bonne G., Carrier L., Richard P., Hainque B., Schwartz K. Familial hypertrophic cardiomyopathy: from mutations to functional defects. Circ. Res. 1998;83(6):580–593. doi: 10.1161/01.res.83.6.580. 9742053 [DOI] [PubMed] [Google Scholar]
- 40.Harris S.P., Lyons R.G., Bezold K.L. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 2011;108(6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. 21415409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capitanio D., Vasso M., Fania C., Moriggi M., Viganò A., Procacci P., Magnaghi V., Gelfi C. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics. 2009;9(7):2004–2020. doi: 10.1002/pmic.200701162. 19333999 [DOI] [PubMed] [Google Scholar]
- 42.Gannon J., Doran P., Kirwan A., Ohlendieck K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 2009;88(11):685–700. doi: 10.1016/j.ejcb.2009.06.004. 19616867 [DOI] [PubMed] [Google Scholar]
- 43.Gelfi C., Vigano A., Ripamonti M., Pontoglio A., Begum S., Pellegrino M.A., Grassi B., Bottinelli R., Wait R., Cerretelli P. The human muscle proteome in aging. J. Proteome Res. 2006;5(6):1344–1353. doi: 10.1021/pr050414x. 16739986 [DOI] [PubMed] [Google Scholar]
- 44.Thompson L.V., Durand D., Fugere N.A., Ferrington D.A. Myosin and actin expression and oxidation in aging muscle. J. Appl. Physiol. 2006;101(6):1581–1587. doi: 10.1152/japplphysiol.00426.2006. 16840579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doran P., O’Connell K., Gannon J., Kavanagh M., Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8(2):364–377. doi: 10.1002/pmic.200700475. 18050275 [DOI] [PubMed] [Google Scholar]
- 46.Staunton L., Zweyer M., Swandulla D., Ohlendieck K. Mass spectrometry-based proteomic analysis of middle-aged vs. aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int. J. Mol. Med. 2012;30(4):723–733. doi: 10.3892/ijmm.2012.1056. 22797148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei B., Jin J.-P. Troponin T isoforms and posttranscriptional modifications: evolution, regulation and function. Arch. Biochem. Biophys. 2011;505(2):144–154. doi: 10.1016/j.abb.2010.10.013. 20965144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes A.V., Potter J.D., Szczesna-cordary D. The role of troponins in muscle contraction. I. UBMB Life. 2002;54(6):323–333. doi: 10.1080/15216540216037. 12665242 [DOI] [PubMed] [Google Scholar]
- 49.Scott W., Stevens J., Binder-Macleod S.A. Human skeletal muscle fiber type classifications. Phys. Ther. 2001;81:1810–1816. 13510739 [PubMed] [Google Scholar]
- 50.Wallimann T., Schlösser T., Eppenberger H.M. Function of M-line-bound creatine kinase as Intramyofibrillar ATP regenerator at the receiving end of the phosphorylcreatine Shuttle in muscle. J. Biol. Chem. 1984;259(8):5238–5246. 6143755 [PubMed] [Google Scholar]
- 51.Hornemann T., Kempa S., Himmel M., Hayess K., Fürst D.O., Wallimann T. Muscle-type creatine kinase interacts with central domains of the M-band proteins myomesin and M-protein. J. Mol. Biol. 2003;332(4):877–887. doi: 10.1016/s0022-2836(03)00921-5. 12972258 [DOI] [PubMed] [Google Scholar]
- 52.Hornemann T., Stolz M., Wallimann T. Isoenzyme-specific interaction of muscle-type creatine kinase with the sarcomeric M-line is mediated by NH(2)-terminal lysine charge-clamps. J. Cell Biol. 2000;149(6):1225–1234. doi: 10.1083/jcb.149.6.1225. 10851020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. 22013216 [DOI] [PubMed] [Google Scholar]
- 54.Piec I., Listrat A., Alliot J., Chambon C., Taylor R.G., Bechet D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005;19(9):1143–1145. doi: 10.1096/fj.04-3084fje. 15831715 [DOI] [PubMed] [Google Scholar]
- 55.Gueugneau M., Coudy-Gandilhon C., Gourbeyre O., Chambon C., Combaret L., Polge C., Taillandier D., Attaix D., Friguet B., Maier A.B., Butler-Browne G., Béchet D. Proteomics of muscle chronological ageing in post-menopausal women. BMC Genomics. 2014;15(1):1165. doi: 10.1186/1471-2164-15-1165. 25532418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halseth A.E., Bracy D.P., Wasserman D.H. Functional limitations to glucose uptake in muscles comprised of different fiber types. Am. J. Physiol.—Endocrinol. Metab. 2001;280(6):E994–E999. doi: 10.1152/ajpendo.2001.280.6.E994. 11350781 [DOI] [PubMed] [Google Scholar]
- 57.Kishi H., Mukai T., Hirono A., Fujii H., Miwa S., Hori K. Human aldolase A deficiency associated with a hemolytic anemia: thermolabile aldolase due to a single base mutation. Proc. Natl. Acad. Sci. USA. 1987;84(23):8623–8627. doi: 10.1073/pnas.84.23.8623. 2825199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kreuder J., Borkhardt A., Repp R., Pekrun A., Göttsche B., Gottschalk U., Reichmann H., Schachenmayr W., Schlegel K., Lampert F. Brief report: inherited metabolic myopathy and hemolysis due to a mutation in aldolase A. N. Engl. J. Med. 1996;334(17):1100–1104. doi: 10.1056/NEJM199604253341705. 8598869 [DOI] [PubMed] [Google Scholar]
- 59.Nakajima H., Amano W., Kubo T., Fukuhara A., Ihara H., Azuma Y.-T., Tajima H., Inui T., Sawa A., Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J. Biol. Chem. 2009;284(49):34331–34341. doi: 10.1074/jbc.M109.027698. 19837666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato T., Morita A., Mori N., Miura S. Glycerol 3-phosphate dehydrogenase 1 deficiency enhances exercise capacity due to increased lipid oxidation during strenuous exercise. Biochem. Biophys. Res. Commun. 2015;457(4):1–6. doi: 10.1016/j.bbrc.2015.01.043. 25603051 [DOI] [PubMed] [Google Scholar]
- 61.Vigelsø A., Dybboe R., Hansen C.N., Dela F., Helge J.W., Guadalupe Grau A. GAPDH and β-actin protein decreases with aging, making stain-Free technology a superior loading control in western blotting of human skeletal muscle. J. Appl. Physiol. 2015;118(3):386–394. doi: 10.1152/japplphysiol.00840.2014. 25429098 [DOI] [PubMed] [Google Scholar]
- 62.MacDonald M.J., Marshall L.K. Mouse lacking NAD+-linked glycerol phosphate dehydrogenase has normal pancreatic beta cell function but abnormal metabolite pattern in skeletal muscle. Arch. Biochem. Biophys. 2000;384(1):143–153. doi: 10.1006/abbi.2000.2107. 11147825 [DOI] [PubMed] [Google Scholar]
- 63.Kitaoka Y., Ogborn D.I., Nilsson M.I., Mocellin N.J., MacNeil L.G., Tarnopolsky M.A. Oxidative stress and Nrf2 signaling in McArdle disease. Mol. Genet. Metab. 2013;110(3):297–302. doi: 10.1016/j.ymgme.2013.06.022. 23906480 [DOI] [PubMed] [Google Scholar]
- 64.Kohn T.A., Noakes T.D., Rae D.E., Rubio J.C., Santalla A., Nogales-Gadea G., Pinós T., Martín M.A., Arenas J., Lucia A. McArdle disease does not affect skeletal muscle fibre type profiles in humans. Biol. Open. 2014;3(12):1224–1227. doi: 10.1242/bio.20149548. 25432515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katz A., Westerblad H. Regulation of glycogen breakdown and its consequences for skeletal muscle function after training. Mamm. Genome. 2014;25(9–10):464–472. doi: 10.1007/s00335-014-9519-x. 24777203 [DOI] [PubMed] [Google Scholar]
- 66.Lucia A., Nogales-Gadea G., Pérez M., Martín M.A., Andreu A.L., Arenas J. McArdle disease: what do neurologists need to know? Nat. Clin. Pract. Neurol. 2008;4(10):568–577. doi: 10.1038/ncpneuro0913. 18833216 [DOI] [PubMed] [Google Scholar]
- 67.Liu Y., Gampert L., Nething K., Steinacker J.M. Response and function of skeletal muscle heat shock protein 70. Front. Biosci. 2006;11:2802–2827. doi: 10.2741/2011. 16720354 [DOI] [PubMed] [Google Scholar]
- 68.Doran P., Gannon J., O’Connell K., Ohlendieck K. Aging skeletal muscle shows a drastic increase in the small heat shock proteins alphaB-crystallin/HspB5 and cvHsp/HspB7. Eur. J. Cell Biol. 2007;86(10):629–640. doi: 10.1016/j.ejcb.2007.07.003. 17761354 [DOI] [PubMed] [Google Scholar]
- 69.Lombardi A., Silvestri E., Cioffi F., Senese R., Lanni A., Goglia F., de Lange P., Moreno M. Defining the transcriptomic and proteomic profiles of rat ageing skeletal muscle by the use of a cDNA array, 2D- and blue native-PAGE approach. J. Proteomics. 2009;72(4):708–721. doi: 10.1016/j.jprot.2009.02.007. 19268720 [DOI] [PubMed] [Google Scholar]


