Abstract
Obesity is an increasingly serious health problem in the world. Body mass index (BMI), percentage fat mass, and body fat mass are important indices of obesity. For a sample of pedigrees that contains >10,000 relative pairs (including 1,249 sib pairs) that are useful for linkage analyses, we performed a whole-genome linkage scan, using 380 microsatellite markers to identify genomic regions that may contain quantitative-trait loci (QTLs) for obesity. Each pedigree was ascertained through a proband who has extremely low bone mass, which translates into a low BMI. A major QTL for BMI was identified on 2q14 near the marker D2S347 with a LOD score of 4.04 in two-point analysis and a maximum LOD score (MLS) of 4.44 in multipoint analysis. The genomic region near 2q14 also achieved an MLS >2.0 for percentage of fat mass and body fat mass. For the putative QTL on 2q14, as much as 28.2% of BMI variation (after adjustment for age and sex) may be attributable to this locus. In addition, several other genomic regions that may contain obesity-related QTLs are suggested. For example, 1p36 near the marker D1S468 may contain a QTL for BMI variation, with a LOD score of 2.75 in two-point analysis and an MLS of 2.09 in multipoint analysis. The genomic regions identified in this and earlier reports are compared for further exploration in extension studies that use larger samples and/or denser markers for confirmation and fine-mapping studies, to eventually identify major functional genes involved in obesity.
Introduction
Obesity is a disease condition with excess body fat that adversely affects health (Kopelman 2000). Obesity is associated with many diseases, such as type 2 diabetes mellitus, hypertension, coronary heart disease, and some cancers (Must et al. 1999; Kopelman 2000). A higher BMI (an important measure of obesity that expresses the ratio of weight to the square of height in kilograms per square meter) is associated with higher mortality (Calle et al. 1999). About 250 million adults (7% of the world adult population) are considered obese (defined as having a BMI >30 kg/m2), and two or three times as many may be considered overweight (defined as having a BMI of 25–30 kg/m2) (Kuczmarski et al. 1994; Seidell 1999). In the United States, 59.4% of men and 50.7% of women are overweight or obese (Flegal et al. 1998). The direct costs associated with obesity are estimated to be ∼$100 billion per year, which represents 5.7% of the national health expenditure in the United States in 1995 (Wolf and Colditz 1998). A more recent analysis shows that the health care cost of obesity in the United States is most likely between 0.89% and 4.32% (Allison et al. 1999b).
Obesity is a complex trait that is determined by multiple genetic and environmental factors (including physiological, behavioral, and sociocultural factors) (Comuzzie and Allison 1998; Echwald 1999; Pérusse and Bouchard 1999). Numerous studies have been conducted to characterize the heritability of obesity-related phenotypes. Most studies are based on BMI, but a few are based on fat mass and percentage fat mass (PFM) and a few on lean mass. The estimates of heritability (h2) of BMI ranged from 0.05 to 0.90 (Stunkard et al. 1986, 1990; Bouchard et al. 1988; Selby et al. 1991; Harris et al. 1995; Vogler et al. 1995; Allison et al. 1996; Borecki et al. 1998; Rice et al. 1999). For fat mass, h2 estimates range from 20% to 65% (Bouchard et al. 1988; Comuzzie et al. 1995, 1997; Rice et al. 1997a; Samaras et al. 1997; Nguyen et al. 1998). For PFM, h2 estimates are 62%–80% (Rice et al. 1997b; Sakurai et al. 1997; Faith et al. 1999). In h2 studies of fat mass and PFM, few have employed measurement by dual energy x-ray absorptiometry (DXA) (but see Samaras et al. 1997 and Nguyen et al. 1998), largely because of the relatively recent availability of DXA and its associated relatively large expenses for the measurement. For lean mass, h2 estimates are 0.52%–0.80% (Forbes et al. 1995; Arden and Spector 1997; Nguyen et al. 1998). In our study sample, after adjustment for age, sex, and life style factors, h2, estimates of BMI, fat mass, PFM, and lean mass (the latter three phenotypes measured by DXA) are 0.52%–0.57% (Deng et al. 2001a).
The number and the identity of the genes underlying the susceptibility to obesity have received extensive study (Comuzzie and Allison 1998; Echwald 1999; Pérusse and Bouchard 1999). For the common variation of obesity phenotypes, segregation analyses in several populations suggest that major genes with recessive effects may account for 35%–45% of the variation in obesity-related phenotypes (e.g., BMI, fat mass, and PFM), after adjustment for sex and age (Moll et al. 1991; Rice et al. 1993; Comuzzie et al. 1995). Other segregation analyses did not reveal any major gene effect but inferred the existence of multiple genes, each with minor effects (Lecomte et al. 1997; Borecki et al. 1998). A number of candidate genes have been tested by association and/or linkage for obesity phenotypes (Deng et al. 2000b; Pérusse et al 2001). Genomewide linkage studies have been conducted, and several genomic regions have been reported that may harbor obesity-susceptibility genes (Comuzzie et al. 1997; Hager et al. 1998; Hanson et al. 1998; Norman et al. 1998; Lee et al. 1999; Mitchell et al. 1999; Walder et al. 2000; Watanabe et al. 2000; Hsueh et al. 2001; Feitosa et al. 2002). The genomic regions identified so far seem to be largely unconfirmed (Pérusse et al. 2001), except in a few cases (e.g., a QTL for serum leptin concentration on 2p21 [Comuzzie et al. 1997; Hager et al. 1998] and a locus for obesity on 10p [Hager et al. 1998; Hinney et al. 2000]). Hence, further gene-mapping studies in various samples and populations are warranted. Only by comparing different studies of various population samples and by disentangling biological and statistical causes that underlie the consistent results—as well as the inconsistent ones—may we eventually identify major QTLs for obesity.
In the present study, we genotyped 380 highly heterozygous markers across the whole genome for each of the 630 subjects, who are members of 53 white pedigrees that contain >10,000 relative pairs (including 1,249 sib pairs) that are useful for linkage analyses. We performed a whole-genome linkage scan to identify genomic regions that are potentially important for obesity phenotypes. The study identified a major QTL for BMI and several other genomic regions of potential importance for BMI, fat mass, PFM, and lean mass, and it provides a basis for further extension and confirmation studies for these putative genomic regions and for close examination of the candidate genes identified inside these genomic regions in the present study population.
Subjects and Methods
Subjects
The study was approved by the Creighton University institutional review board. All the study subjects signed informed-consent documents before entering the project. The study subjects came from an expanding database that is being created for studies to search for genes underlying the risk of osteoporosis and obesity that is underway in the Osteoporosis Research Center of Creighton University. Only healthy people (defined by the exclusion criteria that are detailed below) were included in the analysis. All the study subjects were white individuals of European origin, and 53 pedigrees with 630 subjects (248 males and 382 females) from 2–4 generations were analyzed. The pedigrees range from 3 to 99 individuals, with a mean (SE) of 11.7 (2.4). Each pedigree was identified through a proband having bone mineral density (BMD) Z scores less than −1.28 at the hip or spine. BMD values expressed as Z scores adjust for age, sex, and ethnic difference in healthy referent populations. Hence, to identify QTLs for BMD (Deng et al. 2001b), we selected the probands from the lowest 10% of the population BMD variation for the purpose of achieving higher statistical power than would be achieved through random sampling (Risch and Zhang 1995; Deng and Li, in press). Because BMD and BMI are significantly correlated (Deng et al. 2000a), the sampling scheme of our study pedigrees for enhancing the power of linkage analysis to detect genomic regions for BMD variation may also have a similar effect (if any) on BMI and related obesity phenotypes. In fact, the probands tend to have BMI that are lower than the average in their pedigrees (see “Results” section).
The exclusion criteria for the study subjects were a history of (1) serious residual effects of cerebral vascular disease; (2) diabetes mellitus, except for easily controlled, non–insulin-dependent diabetes mellitus (defined as adult asymptomatic hyperglycemia controlled by diet or oral agents); (3) chronic renal disease manifested by serum creatinine >1.9 mg/dl; (4) chronic liver disease or alcoholism; (5) significant chronic lung disease; (6) >6 mo of corticosteroid therapy at pharmacologic levels; (7) >6 mo of treatment with anticonvulsant therapy; (8) evidence of other metabolic or inherited bone disease, such as hyper- or hypoparathyroidism, Paget disease, osteomalacia, or osteogenesis imperfecta; (9) rheumatoid arthritis or collagen disease; (10) recent (within the past year) major gastrointestinal disease, such as peptic ulcer, malabsorption, chronic ulcerative colitis, regional enteritis, or any significant chronic diarrhea state; (11) significant disease of any endocrine organ that would affect bone mass; (12) hyperthyroidism; (13) any neurologic or musculoskeletal condition that would be a nongenetic cause of low bone mass; and (14) any disease, treatment, or condition that would be a nongenetic cause of low bone mass. The exclusion criteria were assessed by nurse-administered questionnaires and/or examination of medical records and were applied most rigorously to potential subjects contacted between the ages of 25 and 50 years. About 5.1% of the total people screened were excluded from our study sample because they met at least one of the exclusion criteria. The exclusion criteria applied might render the allele frequencies and thus the genetic effects of potential obesity QTLs different from those in our general population. This may be the case when some of the above disease states are genetically correlated with obesity, for which there is generally no evidence at present.
Genotyping
For each subject, blood (20 cc) was drawn into EDTA-containing tubes by certified phlebotomists and was stored chilled (∼4°C) until DNA extraction, which was usually completed within the next five calendar days. DNA was extracted by employing a kit (Gentra Systems), according to the procedures detailed in the manufacturer's instructions. DNA was genotyped using fluorescently labeled markers, as we did before (Deng et al. 2000c). The 400 dinucleotide markers with which we started our genotyping are commercially available through Perkin-Elmer Applied Biosystems (ABI Prism Linkage Mapping Sets, version 2). The PCR was performed on PE 9700 thermocyclers (Applied Biosystems). PCR cycling conditions followed those suggested in the ABI Prism Linkage Mapping Sets, version 2. Genotyping was performed using Applied Biosystems automated DNA sequencing systems (models 377 and 310) running the Genescan and Genotyper softwares for allele identification and sizing. A genetic database management system (GenoDB) developed by us (Li et al. 2001) was employed to manage the phenotype and genotype data for linkage analyses. GenoDB is also employed for allele binning (including setting up allele-binning criteria and converting allele sizes to distinct allele numbers), quality control, and formatting of data for PedCheck (O'Connell and Weeks 1998) and for linkage analyses by SOLAR. PedCheck was employed for checking the conformation to Mendelian inheritance patterns at all the marker loci and for checking the relationships of family members within pedigrees. The genotyping error rate, determined by at least three rounds of sample replication in experiments and data analyses by PedCheck, was ∼0.3%. Of the markers, 380 (including 362 on autosomes) were successfully genotyped. These markers have an average population heterozygosity of ∼0.79 and are spaced an average of ∼8.6 cM from adjacent markers throughout the human genome.
Measurement
Fat mass and lean mass were measured by DXA with a Hologic 2000+ or 4500 scanner. Measurements of BMD on the two types of machines agreed within 1%, but the body mass measurements differed in an unsystematic way. For 24 people who underwent scans on the same day on the two types of machines, the difference in whole body mass is ∼3%. All machines were calibrated daily. The body-composition bar was used on every whole-body scan on the Hologic 2000+. On the Hologic 4500, the bar was not needed for the body scans; instead, it was scanned every week. In the early stage of the study, the subjects were recruited for studying osteoporosis only. Thus, we did not install the hardware and software to perform the body composition assessment until later in the recruitment for the project. Therefore, we have data on fat mass, PFM, and lean mass for only 289 subjects from 38 pedigrees. Software and hardware have been kept up to date throughout the implementation of the project. For the Hologic 2000+ scanner that was in service during 1994–1999, the software was updated from version 5.67a to version 5.71a. For the Hologic 4500 scanner that has been in service since 1997, the software has been updated from version 8.19a to version 8.26a. The PFM is the ratio of fat mass divided by body weight (i.e., the sum of fat mass plus lean mass plus bone mineral content). The measurement precision of BMI as reflected by coefficient of variation was 0.2%. The coefficients of variation for fat mass, PFM, and lean mass were 2.2%, 2.2%, and 1.0%, respectively, for measurements obtained on the Hologic 2000+, and were 1.2%, 1.1%, and 0.7%, respectively, for measurements obtained on the Hologic 4500. Members of the same pedigree were usually measured on the same type of machine.
Statistical Analyses
A variance-component linkage analysis (Amos 1994; Amos et al. 1996; Almasy and Blangero 1998) for quantitative traits was performed. The analysis is based on specifying the expected genetic covariances between relatives as a function of the identity by descent at a given marker locus. The analysis considers the phenotypic and genetic information from all pedigree members simultaneously. The analysis assumed joint multivariate normality of phenotypic values, additive genetic effects, and no interaction between genes and the residual. The program employed was SOLAR (Sequential Oligogenic Linkage Analysis Routines) (Almasy and Blangero 1998), which is available on the Internet. The pedigree-ascertainment scheme based on the low BMD values of probands leads to lower-than-average BMI values (see “Results”). The ascertainment scheme was accounted for in the analyses by identifying to the program the probands and their phenotypic values for each pedigree. The built-in modules of the SOLAR program will then be able to account for the ascertainment scheme by using conditional likelihoods in LOD score computation. The statistical properties of the variance-component analyses after the ascertainment correction have been preliminarily investigated (de Andrade et al. 1997; de Andrade and Amos 2000; Sham 2000).
In linkage analysis, age and sex were adjusted as covariates for the obesity phenotypic values, because these generally affect the variation of obesity phenotypes significantly in our study population (Deng et al. 2000b). Analyses were also performed without adjusting for one or both of these covariates. Adjustment for significant covariates in genetic analyses can generally increase the genetic signal-to-noise ratio in linkage detection by decreasing the proportion of the residual phenotypic variation attributable to random environmental factors (Deng et al. 1999). This may thus improve statistical power in our linkage analyses. Comparison of the analyses with and without adjustment for a significant covariate may shed light on the genomic regions identified regarding their direct importance for the trait per se or indirect importance via the influence on the covariate only. The variance-component analyses implemented in SOLAR are generally robust to violations of normality of the data (Williams et al. 1999). However, some types of nonnormality of the data may inflate the type I error rate in excess of specified nominal levels (Allison et al. 1999a). The phenotype data were tested by graphical methods (Sokal and Rohlf 1995) and found not to deviate from normal distributions. Marker-allele frequencies were obtained by maximum likelihood estimation in SOLAR. Hypothesis testing for linkage was conducted by the maximum-likelihood method by investigating the relationship of genetic covariances and the identity-by-descent between relatives. The method compares the maximum likelihoods obtained in the full model (which shows linkage so the locus is a QTL and accounts for some additive genetic variance) with the nested null model (which does not show linkage so the locus is not a QTL). The difference between the two log10 likelihoods yields a LOD score. Twice the difference of the loge of the likelihoods of these two models is asymptotically distributed as a 1/2: 1/2 mixture of a χ2 variable and a point mass at 0 (Almasy and Blangero 1998) with 1 df. Using SOLAR, we performed two-point and multipoint linkage analyses, respectively, for the obesity phenotypes. LOD scores may be converted to approximate P values (prior to accounting for multiple testing), which is commonly employed in statistical testing through a χ2 distribution (Lander and Kruglyak 1995). When a putative QTL is suggested, the proportion of phenotypic variation attributable to this QTL can be estimated by SOLAR. The estimate is usually an upper bound of the genetic effect due to the locus (Göring et al 2001).
Results
The basic characteristics of the study subjects, stratified by age and sex, are summarized in Appendix A (table 3). Some information about the family structure of the study pedigrees is summarized in Appendix B (table 4). Appendix B shows that, for the analyses for BMI, there are >10,000 informative relative pairs (including 1,249 sibling pairs, 1,098 grandparent-grandchild pairs, 1,993 avuncular pairs, and 2,589 first-cousin pairs). For the analyses for fat mass, lean mass and PFM, there are nearly 6,000 informative relative pairs (including 744 sibling pairs, 710 grandparent-grandchild pairs, 1,148 avuncular pairs, and 1,500 first-cousin pairs). These large numbers of informative relative pairs reflect the richness of the genetic information for linkage analyses in our sample. The correlations between BMI and spine and hip BMD are, respectively, 0.27 and 0.44, both significant at P<.01. In our sample, the mean (SE) of BMI, adjusted for age and sex, is 26.45 (0.21) for all the subjects and is 23.97 (0.63) for probands. The BMI value is significantly smaller (P<.01) in the probands, reflecting our sampling scheme through low BMD values.
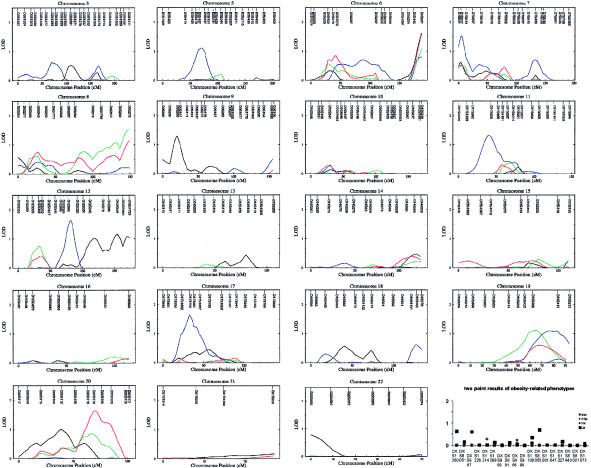
The results for the chromosomes that had a maximum LOD score >2.0 in multipoint linkage analyses for any one of the four obesity phenotypes are summarized in figure 1, the results for the other chromosomes are summarized in figure 2. Because the current version of SOLAR does not handle multipoint linkage analyses for the X chromosome, to convey the linkage signal pattern on the X chromosome, we plotted two-point LOD score results for the X chromosome (fig. 2). For the analysis results presented, obesity phenotypes were adjusted, by use of multiple regression, for significant covariates of age and sex. The results (not shown) of the linkage analyses for obesity phenotypes that were not adjusted for covariates showed a similar pattern in LOD scores and qualitatively the same results, suggesting that any impact of the genomic regions identified here produces a direct effect on obesity phenotypes rather than influencing phenotypes through a covariate.
Figure 1.
Multipoint linkage analysis results for the chromosomes that had an MLS >2.0. Results are shown for BMI (black), PFM (green), fat mass (red), and lean mass (blue).
Figure 2.
Multipoint linkage analysis results for the autosomes that had a maximum LOD score <2.0 and the two-point linkage analysis results for the X chromosome. Results are shown for BMI (black), PFM (green), fat mass (red), and lean mass (blue). Note that the scale of the Y-axis is different from that in figure 1.
Notably (fig. 1 and tables 1 and 2), for BMI, a major QTL on 2q14 is identified with a maximum LOD score (MLS) of 4.44 (P=.000003) at 128 cM from pter on chromosome 2 in multipoint linkage analyses. In two-point linkage analyses, a LOD score of 4.04 (P=.000008) is achieved at the marker D2S347 on 2q14. For this putative major QTL for BMI, as much as 28.2% of BMI variation (after adjustment for age and sex) may be attributable to this locus. In addition, the importance of this genomic region is manifested for PFM and body fat mass. For PFM, 2q14 may contain a QTL with an MLS of 2.10 (P=.00093) at 128 cM from pter on chromosome 2 in multipoint analyses, and a LOD score of 1.91 (P=.0015) is achieved at the marker D2S347 on 2q14 in two-point analyses. For body fat mass, 2q14 achieved an MLS of 2.21 (P=.0007) at 128 cM from pter on chromosome 2 in multipoint analyses, and in two-point analyses, a LOD score of 2.03 (P=.0011) is achieved at D2S347 on 2q14.
Table 1.
Markers and Genomic Regions with LOD Scores >1.5 in Two-Point Linkage Analyses for Obesity Phenotype Variation and the Identified Candidate Genes inside the Genomic Regions
| Phenotypesand Marker | ChromosomeLocation | LODScore | QTLs in Animal Models |
| BMI:a | |||
| D1S468 | 1p36 (4.2 cM from pter) | 2.75 | 10%–13% Abdominal fat (Brockmann et al. 1998) and 16% BMI (Kovacs et al. 1997) |
| D2S347 | 2q14 (131.5 cM from pter) | 4.04 | Adiposity (Pomp 1997) |
| D2S160 | 2q12 (123.0 cM from pter) | 2.56 | 1.9% Late weight gain (Cheverud et al. 1996; Vaughn et al. 1999) |
| D4S1592 | 4q12 (69.5 cM from pter) | 2.29 | 4% 6-wk Weight gain (Keightley et al. 1996) |
| D6S281 | 6q27 (190.1 cM from pter) | 1.77 | 6.1% Adiposity (Taylor et al. 1997) |
| PFM: | |||
| D2S206 | 2q36 (240.8 cM from pter) | 1.95 | Subcutaneous fat (Mehrabian et al. 1998); 10.9% body weight (Hirayama et al. 1999) |
| D2S347 | 2q14 (131.5 cM from pter) | 1.91 | Adiposity (Pomp 1997) |
| Fat Mass: | |||
| D2S347 | 2q14 (131.5 cM from pter) | 2.03 | Adiposity (Pomp 1997) |
| D6S281 | 6q27 (190.1 cM from pter) | 2.02 | 6.1% Adiposity (Taylor et al. 1997) |
| D8S549 | 8p22 (31.7 cM from pter) | 1.95 | 24% Body weight (Gauguier et al. 1996) |
| Lean Mass: | |||
| D12S83 | 12q14 (75.2 cM from pter) | 1.79 | |
| D5S407 | 5q11 (64.7 cM from pter) | 1.59 | Adiposity (Pomp 1997) |
Candidate genes for BMI are as follows: PGD is involved in carbohydrate metabolism. Enzyme catalyzes a step in the pentose pathway and oxidizes glucose-6-phosphate into 6-phosphoglucono-lactone (Wilson et al. 1991). TNFR2 is a polymorphism of the TNFR2 gene, which is associated with obesity, leptin levels, and insulin resistance. (Fernandez-Real et al. 2000). NR0B2 mutations are related to early-onset mild obesity (Nishigori et al. 2001). The candidate genes and their cellular functions (if indicated) in the genomic regions are identified via the human obesity gene map (Pérusse et al. 2001) or Online Mendelian Inheritance in Man. The genetic distances are as indicated by the Marshfield linkage map.
Table 2.
Markers and Genomic Regions with LOD Scores >1.5 in Multipoint Linkage Analyses for Obesity Phenotype Variation and the Identified Candidate Genes inside the Genomic Regions
| Phenotype and Chromosome Location | LODScore | QTLs in Animal Models |
| BMI:a | ||
| 1p36 (0 cM from pter) | 2.09 | See table 1 |
| 2q14 (128 cM from pter) | 4.44 | See table 1 |
| 4q12 (68 cM from pter) | 2.09 | See table 1 |
| 6q27 (188 cM from pter) | 1.61 | See table 1 |
| PFM: | ||
| 2q14 (128 cM from pter) | 2.10 | 1.9% Late weight gain (Cheverud et al. 1996; Vaughn et al. 1999) |
| 8q24 (149 cM from pter) | 1.54 | 14.4% 14-wk % Fat (Horvat et al. 2000); 24% body weight (Gauguier et al. 1996) |
| Fat mass:b | ||
| 2q14 (128 cM from pter) | 2.21 | 1.9% Late weight gain (Cheverud et al. 1996); Vaughn et al. 1999) |
| 6q27 (188 cM from pter) | 1.59 | See table 1 |
| 20q13 (69 cM from pter) | 1.64 | |
| Lean mass: | ||
| 12q14 (82 cM from pter) | 1.64 | |
| 17p12 (33 cM from pter) | 1.64 | |
| 7p22 (5 cM from pter) | 1.52 | 4.9% 14-wk % Fat (Horvat et al. 2000); 5.4% weight (Chung et al. 1997) |
PGD, TNFR2, and NR0B2 are candidate genes for BMI. See table 1, footnote a.
See “Discussion” section.
In addition to the genomic region 2q14, a few other genomic regions were identified that had LOD scores >1.5 and that may harbor QTLs for the obesity phenotypes under study (fig. 1 and table 1). For BMI, four additional genomic regions were suggested. Region 1p36 may contain a QTL with an MLS of 2.09 (P=.00095) at the terminal end of short arm on chromosome 1 in multipoint analyses. In two-point analyses, a LOD score of 2.75 (P=.00019) is achieved at D1S468 on 1p36. Region 4q12 may contain a QTL with an MLS of 2.09 (P=.00096) at 68 cM from pter on chromosome 4 in multipoint analyses. In two-point analyses, a LOD score of 2.29 (P=.00058) is achieved at D4S1592 on 4q12. Two genomic regions with LOD scores >1.50 in either multi- or two-point linkage analyses (fig. 2; table 1) are also noticeable. The genetic signals manifested in these genomic regions may deserve further close examination in extended samples and/or denser molecular markers. These noteworthy genomic regions include 6q27 with an MLS of 1.61 (P=.0032) in multipoint analyses at 188 cM from pter on chromosome 6 in multipoint analyses, and those at the marker D6S281 on 6q27 with a LOD score of 1.77 (P=.0021), the marker D2S160 on 2q12 with a LOD score of 2.56 (P=.00030), in two-point analyses. For PFM, in two-point analyses, a LOD score of 1.95 (P=.0014) is achieved at D2S206 on 2q36; 8q24 achieved an MLS of 1.54 (P=.0039) in multipoint analyses.
For fat mass, 6q27 achieved an MLS of 1.59 (P=.0034) at 188 cM from pter on chromosome 6 in multipoint analyses. In two-point analyses, a LOD score of 2.02 (P=.0011) is achieved at D6S281 on 6q27. The other noteworthy genomic regions are 20q13, which achieved an MLS of 1.64 (P=.0030) at 69 cM from the pter in multipoint analyses, and the marker D8S549 on 8p22, which achieved a LOD score of 1.95 (P=.0014) in two-point analyses.
For lean mass, we did not detect any genomic region showing a LOD score >2.0 in multipoint or two-point linkage analyses. Some genomic regions with LOD scores >1.50 are noteworthy. In multipoint analyses, these genomic regions include 12q14 with an MLS of 1.64 (P=.0030) at 82 cM from pter on chromosome 12, 17p12 with an MLS of 1.63 (P=.0031) at 33 cM from the pter on chromosome 17, and 7p22 with an MLS of 1.52 (P=.0041) at 5 cM from pter on chromosome 7. In two-point analyses, a LOD score of 1.79 (P=.0020) is achieved at D12S83 on 12q14, and a LOD score of 1.59 is achieved at D5S407 on 5q11.
Because the pedigree size varies dramatically (from 3 to 99), we performed additional linkage analyses for the largest pedigree and for the rest of the pedigrees. We found that the evidence from the two independent subsamples generally provide support for each other for the major significant results presented for the combined sample, although the significance is weaker in the subsamples.
Discussion
Various phenotypes have been employed for obesity research. The World Health Organization proposed BMI as a simple measure of obesity. BMI has been used widely, because it can be measured conveniently at low cost for large samples (Comuzzie and Allison 1998). However, BMI cannot distinguish fat mass from lean mass and thus may not always be appropriate (Allison and Saunders 2000). For example, athletes often have a BMI >25 kg/m2, along with only 10%–15% body fat (Allison and Saunders 2000). Body fat mass and PFM are considered phenotypically more homogeneous than BMI and are more appropriate as phenotypes for obesity research (Lecomte et al. 1997). However, because fat mass and PFM cannot be measured easily and inexpensively, they have not been used as widely as BMI. It may also be important to study body lean mass, because excess body weight consists of not only fat mass but also lean mass (Chagnon et al. 2000).
Our purpose is to identify, through a whole-genome scan and by comparison with earlier studies, genomic regions that may contain QTLs underlying the variation of obesity phenotypes. For a sample of pedigrees that contains >10,000 useful relative pairs, we performed a whole-genome linkage scan, using 380 microsatellite markers (with an average heterozygosity of ∼0.79 and spaced ∼8.6 cM apart throughout the whole human genome). For whole-genome linkage studies, phenotypic characteristics and measurement quality of phenotypes—as reflected by coefficient of variation—are often summarized. However, the quality of genotyping data, an important aspect of whole-genome linkage studies and genetic studies involving genotyping in general (Gordon et al. 2001), has seldom been reported. The genotyping quality should also be reported, because it affects the assessment of the quality of data and thus the robustness of results in linkage studies (Terwilliger et al. 1990). The rate of absent or erroneous genotyping data is estimated to be quite low (0.3%) in our study, a fact that is expected to contribute to the strength of our linkage results.
Our sample was recruited for the purpose of efficiently locating genes that underlie BMD (Deng et al. 2001b), and BMD and body weight are known to be correlated (Deng et al. 2000a). Because bone mass is a part of body mass, BMD and BMI may also share some genes underlying their variation, although the significance of the shared genetic determination is unknown and is not revealed by comparison of our results in the present study with those from another of our other studies (H.W.D., G.H.X., Q.Y.H., H.S., H.Y.D., Y.J.L., T.C., J.I.L., H.T.Z., K.M.D., and R.R.R., unpublished data). The ascertainment of our multigeneration pedigrees through probands with extremely low BMD values translates into lower-than-average BMI values of the probands, a practice that may yield improved power if any effect is present (Risch and Zhang 1995; Deng and Li, in press) in our sample. The strategy for identifying genes for obesity is, essentially, to search for genes underlying the variation of obesity phenotypes. The recruitment of pedigrees via probands with low BMI and the use of genetic studies of thinness may yield important and complementary findings to genetic studies of obesity conducted by ascertainment through obese subjects (Bulik and Allison 2001). The loci identified via the ascertainment of low or high extreme phenotype distribution should all be those important for the variation of obesity phenotypes and thus are important for obesity research. The efficacy of gene identification via ascertainment through probands of extremely low or extremely high phenotypic values has been demonstrated (e.g., Risch and Zhang 1995; Deng and Li, in press). In our study, the larger sample size for the analyses of BMI yields higher statistical power than that for fat mass, lean mass and PFM. The total sample in our study is in the intermediate range compared with earlier genomewide scan studies for obesity (Comuzzie et al. 1997; Hager et al. 1998; Hanson et al. 1998; Norman et al. 1998; Lee et al. 1999; Mitchell et al. 1999; Walder et al. 2000; Watanabe et al. 2000; Hsueh et al. 2001; Feitosa et al. 2002). The exact power of each study may depend not only on the total sample size but also on a number of other factors, such as pedigree structure, study design and analyses, genotyping quality and molecular markers employed, specific phenotypes under study, potential population-specific genetic effects of QTLs, phenotype distribution within pedigrees or relative pairs, and ascertainment scheme. It would be interesting, in the future, to compare the linkage results obtained in reference to the relative power under various conditions for various studies involving linkage scans.
Several genomic regions that may contain QTLs for the variation of the obesity phenotypes studied were identified. Notably, the genomic region near the marker D2S347 on chromosome 2 may contain a QTL for BMI variation. We estimate that this QTL may explain as much as 28.2% of BMI variation (after adjustment for age and sex). In addition, there are several genomic regions identified for the obesity phenotypes studied with LOD scores >1.5 in two- and/or multipoint linkage analyses. Several of the genomic regions identified/suggested coincide with earlier QTL-mapping studies from animal models and harbor several candidate genes (table 1). The importance of chromosome 2 has been revealed for serum leptin concentration and/or fat mass (Comuzzie et al. 1997; Hager et al. 1998). The exact positions of the peak LOD scores (and thus the QTLs involved) reported by Comuzzie et al. (1997), Hager et al. (1998), and ourselves are different on chromosome 2 (∼80 cM apart on the Marshfield linkage map). However, the importance of chromosome 2 in obesity etiology seems to be corroborated by a putative QTL region we identified on this chromosome. Several mouse studies have suggested that genes influencing body weight and fatness may be located on chromosome 2, in regions homologous to human chromosome 2q (Pomp 1997), which further strengthens the importance of chromosome 2 for obesity. The peak on chromosome 1 (1p36) coincided with the region containing the gene TNFR2 (tumor necrosis factor-α receptor 2), whose polymorphism is associated with obesity, leptin levels, and insulin resistance (Fernandez-Real et al. 2000). In addition, mutations of NR0B2 (nuclear receptor subfamily 0, group B, member 2) were reported to be associated with early-onset mild obesity (Nishigori et al. 2001). Several of the other genomic regions identified/suggested, such as 4q12 and 6q27, coincide with earlier QTL-mapping studies from animal models and several candidate genes are suggested (Keightley et al. 1996; Taylor et al. 1997).
On chromosome 20q13, Lee et al. (1999) demonstrated a QTL for BMI and PFM; the LOD scores of BMI, PFM, and fat mass in our pedigrees are, respectively, 0.52 (P=.061), 0.85 (P=.024) and 1.64 (P=.003) on 20q13. Considering that the strength of evidence required for replication of a previously found linkage is different from that in a whole-genome linkage analysis (Lander and Kruglyak 1995), our results lend support to the finding by Lee et al. (1999) that chromosome 20 may harbor a QTL affecting obesity phenotypes. The chromosome region 20q11-13 contains several putative candidate genes for obesity, as outlined below. Agouti-signaling protein (ASIP), whose gene is located on 20q11-13, is a potent inhibitor of α-melanocyte-stimulating hormone receptors 3 and 4 (MC3R and MC4R) (Fong et al. 1997), and the lack of functional MC4R leads to obesity in mice (Huszar et al. 1997). Mutations of agouti gene on 20q11-13 lead to obesity in mice (Miller et al. 1993). CEBPB (CAAT/enhancer-binding protein β) is related to adipocyte differentiation and is considered one of the candidate genes (Yeh et al. 1995). Mutations of GNAS1 (guanine nucleotide-binding protein, alpha stimulating activity polypeptide 1) in 20q11-13 is associated with Albright hereditary oteodystrophy, which is partly characterized by obesity (Gunay-Aygen et al. 1997).
Our data show that, except for the body lean mass and fat mass, the traits analyzed are generally correlated significantly. The phenotypic correlations are, respectively, 0.83 (between BMI and fat mass), 0.33 (between BMI and lean mass), 0.51 (between BMI and PFM), 0.03 (between fat mass and lean mass), 0.84 (between body fat mass and PFM), and −0.48 (between body lean mass and PFM). However, a significant and high phenotypic correlation does not necessarily imply a significant and high genetic correlation (Deng et al. 2002), which indexes the degree of shared genetic determination of two phenotypes. Hence, it is not surprising that, although some loci identified in this study show linkage signal to multiple phenotypes (e.g., 2q14 for BMI, PFM, and body fat mass), other loci identified are somewhat more phenotype specific. The polygenic nature of inheritance (Deng et al. 2001b) and the different sample sizes for the analyses of BMI and other phenotypes may contribute to some of the different findings for various phenotypes. When multiple correlated phenotypes are employed in analyses, LOD scores need to be judged in consideration of multiple testing. We performed our analyses for individual phenotypes in this study; alternatively, multivariate analyses may be performed (Allison et al. 1998). In another report (Deng et al., in press), we used factor scores of the principal component of various bone mass phenotypes in complex segregation analyses. However, when the same strategy of multivariate analyses is employed in our linkage analyses for bone sizes at different skeletal sites, multivariate analysis is less sensitive individual univariate analyses in detecting linkage signals (Q.Y.H., F.H.X., H.S., H.Y.D., T.C., Y.J.L., Y.Z.L., J.L.L., K.M.D., R.R.R., and H.W.D., unpublished data). This may be due to heterogeneity in genetic determination of individual loci to different phenotypes involved, a topic that we wish to pursue elsewhere.
The genomic regions identified in this and earlier studies need to be subject to extension studies with more samples and denser markers for confirmation. Those genomic regions and chromosomes that are cross-supported by several types of study (e.g., linkage analyses in humans, QTL mapping in animal models, and candidate genes identified from molecular and cellular studies) may deserve particular attention in our future endeavors. Once the genomic regions are confirmed, subsequent fine-mapping studies need to be pursued to pinpoint the QTLs to genomic regions <1 cM, for eventual identification of the genes and functional mutations involved in the variation of the obesity phenotypes. The candidate genes identified in genomic regions linked to obesity may be subject to association studies and/or transmission-disequilibrium studies (Spielman et al. 1993; Allison 1997) for confirmation of linkage and association. Efforts from multiple approaches and from various groups for the study of different populations should eventually unravel the QTLs that are important for obesity.
Acknowledgments
Investigators of this work were partially supported by grants from Health Future Foundation, National Institutes of Health grants K01 AR02170-01, R01 AR45349-01, R01 GM60402-01A1, P01 DC01813-07), State of Nebraska Cancer and Smoking Related Disease Research Program grant LB 595, State of Nebraska Tobacco Settlement Fund grant LB692, US Department of Energy grant DE-FG03-00ER63000/A00, Creighton University, grants from National Science Foundation of China, and grants from Hunan Normal University and the Ministry of Education of China. We thank Hai-Tao Zhang, Jian Li, Xuan Zhang, Dong-Bon Lai, Xu-Tao Deng, and Yan Zhou for their extensive genotyping effort for this project. Assistance in DNA extraction by the technicians of the Osteoporosis Research Center of Creighton University is appreciated. We thank two anonymous reviewers for their constructive comments that helped to improve the manuscript.
Appendix A
Table A1.
Basic Characteristics of the Study Subjects for BMI, Fat Mass, Lean Mass, and PFM Stratified by Sex and Age
|
Mean (SD) [Sample Size] for |
||||
| Age Range (years)and Sex | BMIa(kg/m2) | Fat Mass(kg) | Lean Mass(kg) | PFM |
| 20–29: | ||||
| M | 24.93 (3.07) [23] | 16.26 (9.16) [14] | 61.21 (7.14) [14] | .19 (.09) [14] |
| F | 24.63 (5.32) [40] | 23.65 (11.69) [33] | 41.51 (4.82) [33] | .34 (.08) [33] |
| 30–39: | ||||
| M | 26.94 (4.00) [69] | 19.72 (9.67) [35] | 60.55 (7.16) [35] | .23 (.08) [35] |
| F | 25.00 (4.86) [101] | 24.7 (8.10) [61] | 41.31 (5.21) [35] | .35 (.07) [35] |
| 40–49: | ||||
| M | 27.12 (3.47) [65] | 23.15 (7.89) [29] | 59.88 (6.76) [29] | .26 (.05) [29] |
| F | 26.75 (6.31) [110] | 27.73 (11.79) [60] | 40.89 (6.08) [60] | .38 (.08) [60] |
| 50–59: | ||||
| M | 28.02 (3.36) [32] | 24.07 (7.57) [13] | 59.11 (4.7) [13] | .28 (.05) [13] |
| F | 26.15 (5.41) [55] | 28.39 (11.33) [16] | 39.02 (5.69) [16] | .40 (.07) [16] |
| 60–69: | ||||
| M | 28.43 (4.93) [33] | 20.56 (4.62) [5] | 60.78 (9.73) [5] | .24 (.02) [5] |
| F | 27.2 (5.93) [58] | 29.95 (9.21) [13] | 36.68 (4.53) [13] | .43 (.06) [13] |
| ⩾70: | ||||
| M | 27.74 (3.72) [30] | 21.84 (6.90) [6] | 55.26 (3.05) [6] | .27 (.05) [6] |
| F | 25.87 (3.36) [42] | 23.57 (7.22) [4] | 35.15 (2.59) [4] | .39 (.07) [4] |
For BMI, the total sample size shown is 658. The number is greater than the 630 subjects genotyped, because markers of 28 subjects could not be amplified (usually as a result of poor quality or degradation of the DNA) or because our archive contained only phenotype data. However, because the genotypes of these 28 subjects can often be unambiguously inferred from their relatives, their phenotype data are useful in linkage analyses. Therefore, their phenotype data are also included in the summary for the above table.
Appendix B
Table B1.
Relationships Used in Analyses by SOLAR
|
No. of Pairs Measured for |
||
| Relative Pair | BMI | FM and LMa |
| Parent-offspring | 1,380 | 892 |
| Sibling | 1,249 | 744 |
| Grandparent-grandchild | 1,098 | 710 |
| Avuncular | 1,993 | 1,148 |
| Half sibling | 23 | 22 |
| Great grandparent–grandchild | 356 | 222 |
| Grand avuncular | 445 | 147 |
| Half avuncular | 23 | 23 |
| Great grand avuncular | 3 | 3 |
| Great great grandparent-grandchild | 6 | 6 |
| First cousin | 2,589 | 1,500 |
| First cousin, once removed | 1,607 | 528 |
| First cousin, twice removed | 3 | 3 |
| Half first cousin | 13 | 13 |
| Second cousin | 1,971 | 755 |
| Second cousin, once removed | 12 |
12 |
| Total | 12,771 | 6,728 |
FM = fat mass; LM = lean mass.
Electronic-Database Information
URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Division of Statistical Genetics, http://watson.hgen.pitt.edu/register/soft_doc.html (for PedCheck) [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- SOLAR, http://www.sfbr.org/sfbr/public/software/solar/index.html [Google Scholar]
References
- Allison DB (1997) Transmission-disequilibrium tests for quantitative traits. Am J Hum Genet 60:676–690 [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Kaprio J, Korkelia M, Koskenvuo M, Neale MC, Hayakawa K (1996) The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord 20:501–506 [PubMed] [Google Scholar]
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999a) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Saunders ES (2000) Obesity in North America. Med Clin North Am 84:305–332 [DOI] [PubMed] [Google Scholar]
- Allison DB, Thiel B, Jean PS, Elston RC, Infante MC, Schork NJ (1998) Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet 63:1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Zannolli R, Narayan KMV (1999b) The direct health care costs of obesity in the United States. Am J Public Health 89:1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Zhu DK, Boerwinkle E (1996) Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet 60:143–160 [DOI] [PubMed] [Google Scholar]
- Arden NK, Spector TD (1997) Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res 12:2076–2081 [DOI] [PubMed] [Google Scholar]
- Borecki IB, Higgins M, Schreiner PJ, Arnett DK, Mayer Davis E, Hunt SC, Province MA (1998) Evidence for multiple determinants of the body mass index: the National Heart, Lung, and Blood Institute Family Heart Study. Obes Res 6:107–114 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Pérusse L, Leblanc C, Tremblay A, Thériault G (1988) Inheritance of the amount and distribution of human body fat. Int J Obes 12:205–215 [PubMed] [Google Scholar]
- Brockmann GA, Haley CS, Renne U, Knott SA, Schwerin M (1998) Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics 150:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik CM, Allison DB (2001) The genetic epidemiology of thinness. Obes Rev 2:107–116 [DOI] [PubMed] [Google Scholar]
- Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW Jr (1999) Body-mass index and mortality in a prospective cohort of US adults. N Engl J Med 341:1097–1105 [DOI] [PubMed] [Google Scholar]
- Chagnon YC, Borecki IB, Pérusse L, Roy S, Lacaille M, Chagnon M, Ho-Kim MA, Rice T, Province MA, Rao DC, Bouchard C (2000) Genome-wide search for genes related to the fat-free body mass in the Quebec Family Study. Metabolism 49:203–207 [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ, Duarte FA, van Swinderen B, Cothrqan K, Perel C (1996) Quantitative trait loci for murine growth. Genetics 142:1305–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WK, Zheng M, Chua M, Kershaw E, Power-Kehoe L, Tsuji M, Wu-Peng XS, Williams J, Chua SC Jr, Leibel RL (1997) Genetic modifiers of leprfa associated with variability in insulin production and susceptibility to NIDDM. Genomics 41:332–344 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Allison DB (1998) The search for human obesity genes. Science 280:1374–1377 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Blangero J, Mahaney MC, Mitchell BD, Hixson JE, Samollow PB, Stern MP, MacCluer JW (1995) Major gene with sex-specific effects influences fat mass in Mexican Americans. Genet Epidemiol 12:475–488 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitchell BD, Mahancy MC, Dyer TD, Stern MP, MacCluer, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass in located on human chromosome 2. Nat Genet 15:273–276 [DOI] [PubMed] [Google Scholar]
- de Andrade M, Amos CI (2000) Ascertainment issues in variance components models. Genet Epidemiol 19:333–344 [DOI] [PubMed] [Google Scholar]
- de Andrade M, Thiel TJ, Yu L, Amos CI (1997) Assessing linkage on chromosome 5 using components of variance approach: univariate versus multivariate. Genet Epidemiol 14:773–778 [DOI] [PubMed] [Google Scholar]
- Deng HW, Chen WM, Conway T, Zhou Y, Davies KM, Stegman MR, Deng HY, Recker RR (2000a) Determination of bone mineral density of the hip and spine in human pedigrees by genetic and life-style factors. Genet Epidemiol 19:160–177 [DOI] [PubMed] [Google Scholar]
- Deng HW, Lai DB, Li J, Conway T, Jing Li, Xu FH, Davies KM, Recker RR (2001a) Characterization of genetic and life-style factors for determining variation of BMI, fat mass, percentage fat mass, and lean mass. J Clin Densitom 4:353–362 [DOI] [PubMed] [Google Scholar]
- Deng HW, Li J. The effect of selective sampling on transmission disequilibrium test of a QTL. Genet Res (in press) [DOI] [PubMed] [Google Scholar]
- Deng HW, Li J, Li JL, Johnson M, Dowd R, Gong G, Deng H, Recker RR (2000b) Association of estrogen receptor-α (ER) genotypes with body mass index in normal healthy postmenopausal Caucasian women. J Clin Endocrinol Metab 85:2748–2751 [DOI] [PubMed] [Google Scholar]
- Deng HW, Li J, Li JL, Johnson M, Recker RR (1999) Association of VDR and ER genotypes with bone mass in postmenopausal women: different conclusions with different analyses. Osteoporos Int 9:499–507 [DOI] [PubMed] [Google Scholar]
- Deng HW, Livhshits G, Xu FH, Conway T, Davies KM, Deng HY, Recker RR. Evidence of a major gene for bone mineral density/content in human pedigrees identified via probands with extreme bone mineral density. Ann Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Deng HW, Mahaney MC, Williams J, Li J, Conway T, Davies KM, Li JL, Deng HY, Recker RR (2002) Relevance of the genes for bone mass variation to susceptibility to osteoporotic fractures and its implications to gene search for complex human diseases. Genet Epidemiol 22:12–25 [DOI] [PubMed] [Google Scholar]
- Deng HW, Xu FH, Conway T, Deng XT, Davies KM, Li JL, Deng HY, Recker RR (2001b) Is population BMD variation linked to the marker d11s987 on chromosome 11q12–13. J Clin Endocrinol Metab 86:3735–3741 [DOI] [PubMed] [Google Scholar]
- Deng HW, Zhou Y, Johnson M, Li J, Recker RR (2000c) Product difference of multiplex and singleplex PCR and its practical implications for human genetic studies. Biotechniques 29:298–308 [DOI] [PubMed] [Google Scholar]
- Echwald SM (1999) Genetics of human obesity: lessons from mouse models and candidate genes. J Intern Med 245:653–666 [DOI] [PubMed] [Google Scholar]
- Faith MS, Pietrobelli A, Nuñez C, Heo M, Heymsfield SB, Allison DB (1999) Evidence for independent genetic influence on fat mass and body mass in a pediatric twin sample. Pediatrics 104:61–67 [DOI] [PubMed] [Google Scholar]
- Feitosa MF, Borecki IB, Rich SS, Arnett DK, Sholinsky P, Myers RH, Leppert M, Province MA (2002) Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet 70:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Real JM, Vendrell J, Ricart W, Broch M, Gutierrez C, Casamitjana R, Oriola J, Richart C (2000) Polymorphism of the tumor necrosis factor-α receptor 2 gene is associated with obesity, leptin levels, and insulin resistance in young subjects and diet-treated type 2 diabetic patients. Diabetes Care 23:831–837 [DOI] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kuczmarski RJ, Johnson CI (1998) Overweight and obesity in the United States: prevalence and trends, 1960–1994. Int J Obes Relat Metab Disord 22:39–47 [DOI] [PubMed] [Google Scholar]
- Fong TM, Mao C, MacNeil T, Kalyani R, Smith T, Weinberg D, Tota MR, Van der Ploeg LH (1997) ART (protein product of agouti-related transcript) as an antagonist of MC-3 and MC-4 receptors. Biochem Biophys Res Commun 237:629–631 [DOI] [PubMed] [Google Scholar]
- Forbes GB, Sauer EP, Weitkamp LR (1995) Lean body mass in twins. Metabolism 44:1442–1446 [DOI] [PubMed] [Google Scholar]
- Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, Penicaud L, Lathrop M, Ktorza A (1996) Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet 12:38–43 [DOI] [PubMed] [Google Scholar]
- Gordon D, Simon C, Heath C, Liu X, Ott J (2001) A transmission disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am J Hum Genet 69:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunay-Aygun M, Cassidy S, Nicholls R (1997) Prader-Willi and other syndromes associated with obesity and mental retardation. Behav Genet 27:307–324 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR, Tambs K, Magnus P (1995) Sex-specific effects for body mass index in the new Norwegian twin panel. Genet Epidemiol 12:251–265 [DOI] [PubMed] [Google Scholar]
- Hinney A, Ziegler A, Oeffner F, Wedewardt C, Vogel M, Wulftange H, Geller F, Stubing K, Siegfried W, Goldschmidt HP, Remschmidt H, Hebebrand J (2000) Independent confirmation of a major locus for obesity on chromosome 10. J Clin Endocrinol Metab 85:2962–2965 [DOI] [PubMed] [Google Scholar]
- Hirayama I, Yi Z, Izumi S, Arai I, Suzuki W, Nagamachi Y, Kuwano H, Takeuchi T, Izumi T (1999) Genetic analysis of obese diabetes in the TSOD mouse. Diabetes 48:1183–1191 [DOI] [PubMed] [Google Scholar]
- Horvat S, Bunger L, Falconer VM, Mackay P, Law A, Bulfield G, Keightley PD (2000) Mapping of obesity QTLs in a cross between mouse lines divergently selected on fat content. Mamm Genome 11:2–7 [DOI] [PubMed] [Google Scholar]
- Hsueh WC, Mitchell BD, Schneider JL, St Jean PL, Pollin TI, Ehm MG, Wagner MJ, Burns DK, Sakul H, Bell CJ, Shuldiner AR (2001) Genome-wide scan of obesity in the old order Amish. J Clin Endocrinol Metab 86:1199–1205 [DOI] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F (1997) Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88:131–141 [DOI] [PubMed] [Google Scholar]
- Keightley PD, Hardge T, May L, Bulfield G (1996) A genetic map of quantitative trait loci for body weight in the mouse. Genetics 142:227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman PG (2000) Obesity as a medical problem. Nature 404:635–643 [DOI] [PubMed] [Google Scholar]
- Kovacs P, Voigt B, Berg S, Vogt L, Kloting I (1997) WOK. 1W rats: a potential animal model of the insulin resistance syndrome. Ann NY Acad Sci 827:94–99 [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Flegal KM, Campbell SM, Johnson CL (1994) Increasing prevalence of overweight among US adults: the National Health and Nutrition Examination Surveys—1960 to 1991. JAMA 272:205–211 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting the reporting results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lecomte E, Herbeth B, Nicaud V, Rakotovao R, Artur Y, Tiret L (1997) Segregation analysis of fat mass and fat-free mass with age- and sex-dependent effects: the Stanislas Family Study. Genet Epidemiol 14:51–62 [DOI] [PubMed] [Google Scholar]
- Lee JH, Reed DR, Li WD, Xu W, Joo EJ, Kilker RL, Nanthakumar E, North M, Sakul H, Bell C, Price RA (1999) Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet 64:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Deng HY, Lai DB, Recker RR, Deng HW (2001) Towards high-throughput genotyping: a dynamic and automatic software for manipulating large-scale genotype data using fluorescently labeled dinucleotide markers. Genome Res 11:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M, Wen P, Fisler J, Davis R, Lusis A (1998) Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J Clin Invest 101:2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, Duhl DM, Vrieling H, Cordes SP, Ollmann MM, Winkes BM, Barsh GS (1993) Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev 7:454–467 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Comuzzie AG, Almasy L, Blangero J, MacCluer JW, Hixson JE (1999) A quantitative trait locus influencing BMI maps to the region of the β-3 adrenergic receptor. Diabetes 48:1863–1867 [DOI] [PubMed] [Google Scholar]
- Moll PP, Burns TL, Lauer RM (1991) The genetic and environmental sources of body mass index variability: the Muscatine Ponderosity Family Study. Am J Hum Genet 49:1243–1255 [PMC free article] [PubMed] [Google Scholar]
- Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH (1999) The disease burden associated with overweight and obesity. JAMA 282:1523–1529 [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Howard GM, Keely PJ, Eisman JA (1998) Bone mass, lean mass, and fat mass: same genes or same environments? Am J Epidemiol 147:3–16 [DOI] [PubMed] [Google Scholar]
- Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, Kohama T, Yamagata K, Yang Q, Matsuzawa Y, Miki T, Seino S, Kim MY, Choi HS, Lee YK, Moore DD, Takeda J (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci USA 98:575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérusse L, Bouchard C (1999) Genotype-environment interaction in human obesity. Nutr Rev 57:S31–S37 [DOI] [PubMed] [Google Scholar]
- Pérusse L, Chagnon YC, Weisnagel J, Rankinen T, Snyder E, Sands E, Bouchard C (2001) The human obesity gene map: the 2000 update. Obes Res 9:135–169 [DOI] [PubMed] [Google Scholar]
- Pomp D (1997) Genetic dissection of obesity in polygenic animal models. Behav Genet 27:285–306 [DOI] [PubMed] [Google Scholar]
- Rice T, Borecki IB, Bouchard C, Rao DC (1993) Segregation analysis of fat mass and other body composition measures derived from underwater weighing. Am J Hum Genet 52:967–973 [PMC free article] [PubMed] [Google Scholar]
- Rice T, Daw EW, Gagnon J, Bouchard C, Leon AS, Skinner JS, Wilmore JH, Rao DC (1997) Familial resemblance for body composition measures: the HERITAGE family study. Obes Res 5:557–562 [PubMed] [Google Scholar]
- Rice T, Després JP, Daw EW, Gagnon J, Borecki IB, Pérusse L, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C (1997) Familial resemblance for abdominal visceral fat: the HERITAGE family study. Int J Obes Relat Metab Disord 21:1024–1031 [DOI] [PubMed] [Google Scholar]
- Rice T, Pérusse L, Bouchard C, Rao DC (1999) Familial aggregation of body mass index and subcutaneous fat measures in the longitudinal Quebec family study. Genet Epidemiol 16:316–334 [DOI] [PubMed] [Google Scholar]
- Risch N, Zhang H (1995) Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science 268:1584–1589 [DOI] [PubMed] [Google Scholar]
- Sakurai Y, Umeda T, Shinichi K, Honjo S, Wakabayashi K, Todoroki I, Nishikawa H, Ogawa S, Katsurada M (1997) Relation of total and beverage-specific alcohol intake to body mass index and waist-to-hip ratio: a study of self-defense officials in Japan. Eur J Epidemiol 13:893–898 [DOI] [PubMed] [Google Scholar]
- Samaras K, Spector TD, Nguyen TV, Baan K, Campbell LV, Kelly PJ (1997) Independent genetic factors determine the amount and distribution of fat in women after the menopause. J Clin Endocrinol Metab 82:781–785 [DOI] [PubMed] [Google Scholar]
- Seidell JC (1999) Obesity: a growing problem. Acta Paediatr Suppl 88:46–50 [DOI] [PubMed] [Google Scholar]
- Selby JV, Reed T, Newman B, Fabsitz RR, Carmelli D (1991) Effects of selective on estimates of heritability for body mass index in the national heart, lung, and blood institute twin study. Genet Epidemiol 8:371–380 [DOI] [PubMed] [Google Scholar]
- Sham PK, Zhao JH, Cherny SS, Hewitt JK (2000) Variance components QTL linkage analysis of selected and non-normal samples: conditioning on trait values. Genet Epidemiol 19:S22-S28 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry, 3d ed. W H Freeman, New York [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Foch TT, Hrubec Z (1986) A twin study of human obesity. JAMA 256:51–54 [PubMed] [Google Scholar]
- Stunkard AJ, Harris JR, Pedersen NL, McClearn GE (1990) The body-mass index of twins who have been reared apart. N Engl J Med 322:1483–1487 [DOI] [PubMed] [Google Scholar]
- Taylor BA, Phillips SJ (1997) Obesity QTLs on mouse chromosome 2 and 17. Genomics 43:249–257 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Weeks DE, Ott J (1990) Laboratory errors in the reading of marker alleles cause massive reduction in LOD score and lead to gross overestimates of recombination fraction. Am J Hum Genet Suppl 47:A201 [Google Scholar]
- Vaughn TT, Pletscher LS, Peripato A, King-Ellison K, Adams E, Erikson C, Cheverud JM (1999) Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet Res 74:313–322 [DOI] [PubMed] [Google Scholar]
- Vogler GP, Sorensen TI, Stunkard AJ, Srinivasan MR, Rao DC (1995) Influences of genes and shared family environment on adult body mass index assessed in an adoption study by a comprehensive path model. Int J Obes Relat Metab Disord 19:40–45 [PubMed] [Google Scholar]
- Walder K, Hanson RL, Kobes S, Knowler WC, Ravussin E (2000) An autosomal genomic scan for loci linked to plasma leptin concentration in Pima Indians. Int J Obes Relat Metab Disord 24:559–565 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL et al. (2000) The Finland–United States Investigation of Non–Insulin-Dependent Diabetes Mellitus Genetics study. II. An autosomal genome scan for diabetes-related quantitative–trait loci. Am J Hum Genet 67:1186–1200 [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J (1999) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet 65:1134–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AF, Elston RC, Tran LD, Siervogel RM (1991) Use of the robust sib-pair method to screen for single-locus, multiple-locus, and pleiotropic effects: application to traits related to hypertension. Am J Hum Genet 48:862–872 [PMC free article] [PubMed] [Google Scholar]
- Wolf AM, Colditz GA (1998) Current estimates of the economic cost of obesity in the United States. Obes Res 6:97–106 [DOI] [PubMed] [Google Scholar]
- Yeh W, Cao Z, Classon M, McKnight S (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9:168–181 [DOI] [PubMed] [Google Scholar]