Abstract
The nonhomologous end-joining (NHEJ) pathway is essential for the preservation of genome integrity, as it efficiently repairs DNA double-strand breaks (DSBs). Previous biochemical and genetic investigations have indicated that, despite the importance of this pathway, the entire complement of genes regulating NHEJ remains unknown. To address this, we employed a plasmid-based NHEJ DNA repair screen in budding yeast (Saccharomyces cerevisiae) using 369 putative nonessential DNA repair-related components as queries. Among the newly identified genes associated with NHEJ deficiency upon disruption are two spindle assembly checkpoint kinases, Bub1 and Bub2. Both observation of resulting phenotypes and chromatin immunoprecipitation demonstrated that Bub1 and -2, either alone or in combination with cell cycle regulators, are recruited near the DSB, where phosphorylated Rad53 or H2A accumulates. Large-scale proteomic analysis of Bub kinases phosphorylated in response to DNA damage identified previously unknown kinase substrates on Tel1 S/T-Q sites. Moreover, Bub1 NHEJ function appears to be conserved in mammalian cells. 53BP1, which influences DSB repair by NHEJ, colocalizes with human BUB1 and is recruited to the break sites. Thus, while Bub is not a core component of NHEJ machinery, our data support its dual role in mitotic exit and promotion of NHEJ repair in yeast and mammals.
INTRODUCTION
The repair of DNA double-strand breaks (DSBs) is an essential process required for the preservation of genome integrity and the normal functioning of the cell (1). These cytotoxic lesions are repaired by major DSB repair pathways, including the homologous recombination (HR) (2) and nonhomologous end-joining (NHEJ) systems (1). While the former is the prevalent pathway in the unicellular budding yeast Saccharomyces cerevisiae (3), the latter is more prevalent in mammalian cells, especially those that are quiescent (4), and can repair DNA lesions even if there is no homologous strand (5). Notably, the impairment of NHEJ in mammalian cells is frequently linked to genomic instability, cancer, and lymphoid V(D)J (i.e., variable, diversity, and joining gene segments) recombination defects. Therefore, a detailed molecular understanding of this pathway would provide critical insight into the genetic risk factors related to carcinogenesis or immunological disorders (6).
As in mammalian cells, the core components of the classical NHEJ pathway in S. cerevisiae depends on three major complexes, YKu (Ku), MRX, and DNL4, which are rapidly recruited to DSBs (7). Initially, the yeast Ku heterodimer (Ku70/80) binds to each end of a DSB, serving as an anchor for protein complexes involved in securing and annealing the break, also suppressing the competing HR pathway (8). After this, the DSB processing complex MRX (Mre11-Rad50-Xrs2), which acts at an early stage of both the NHEJ and HR repair pathways (9), spans the lesions so that the DNA ligase complex, DNL4 (i.e., Dnl4-Lif1-Nej1), can rejoin the DSB ends (10).
While the actions of these core protein complexes in yeast have long been elucidated, efficient DNA repair by NHEJ also depends on a wide array of other coordinated cellular processes. For example, the yeast chromatin structure remodeling (11) and histone acetyltransferase (12) complexes have well-described roles in NHEJ. Additionally, phosphorylation of yeast H2A (γ-H2A) histone and the H2AX histone variant (referred to as γ-H2AX) in mammals by ATM (ataxia telangiectasia mutated)/ATR (ATM and Rad3-related)-like kinases has been shown to promote efficient NHEJ (13). Also, both the homologous repair and nonhomologous repair of DSBs depend on the DNA damage response (DDR) pathway, which detects DNA lesions via recognition factors of the MRX complex (14) and regulates the activities of protein kinases, including Rad9, Rad53, and the checkpoint effector protein Chk1 (15). These kinases induce cell cycle arrest and the upregulation of DNA repair systems, leading to profound alterations in chromatin structure, dynamics, and gene expression (15).
Aside from these processes, DNA damage has also been shown to trigger the spindle assembly checkpoint (SAC) pathway in yeast, preventing cell cycle progression prior to cell commitment to anaphase (16). In particular, the SAC kinase protein, Bub1, in conjunction with other kinetochore factors essential for the SAC pathway (e.g., Mad1, Mad2, Mad3, and Bub3), mediates cell cycle arrest in the event of spindle fiber damage (16). Another SAC pathway-associated component, Bub2, localizes to the spindle pole body and forms an association with its GTPase-activating cocomplex member, Bfa1, to trigger a delay in anaphase progression or mitotic exit (16). Both the Bub1 and Bub2 (herein referred as “Bub”) branches of the SAC pathway also limit cell cycle progression by preventing the activation of the anaphase promoting complex (APC), an essential, multisubunit, E3 ubiquitin-protein ligase that depends on the coactivators Cdc20 and Cdh1 to target anaphase-inhibitory substrates, such as B-type cyclins (16). These B-type cyclins act as key regulators of cell cycle proteins (e.g., activation of cyclin-dependent protein kinase, Cdc28 by the mitotic cyclin Clb2) and are tightly regulated by major transcription factors, such as the Swi4-Swi6 cell cycle box binding factor, a component of the SCB-binding factor (SBF) complex that regulates the expression of proteins involved in budding, spindle pole formation, cell wall biogenesis, and DNA synthesis (16). Notably, genome-wide pooled short hairpin RNA (shRNA) screens in different cancer cell lines have identified components of the SAC, APC, and cyclins to be essential for proliferation in certain cancer-specific cells (e.g., breast and pancreatic cancer cells) (17), suggesting involvement of this mitotic checkpoint abnormality in tumor progression beyond DSB repair.
While Bub1 and Bub2 activate the SAC via distinct pathways (18), they both work in concert with the major targeting components of the APC, cyclins, and SBFs in cell cycle control (19). Moreover, the ability of these Bub proteins to interchangeably stimulate the Rad9 and Rad53 DNA damage checkpoint kinases suggests that the Bub pathways provide robust and redundant protection against spindle damage (20) and also implies a likely role for these proteins in response to mitotic DSBs. In fact, recent observations in mammalian cells have indicated that Bub1 functions in the DDR signaling pathway (21), but its role in supporting NHEJ efficiency remains unclear.
In this study, we posit that in addition to the previously known components of the NHEJ pathway discovered from large-scale genetic screens (11, 22, 23), there are yet unidentified proteins conserved in yeast and humans that promote end-joining repair of DNA DSBs. To determine this, we employed a variant of a comprehensive plasmid-based DNA repair screening approach specific to NHEJ in S. cerevisiae. Among the newly identified NHEJ factors, we identified Bub1, Bub2, and several other nonessential components involved in cell cycle regulation. Further investigations into the effects of Bub proteins on NHEJ revealed that they are efficiently recruited to DSBs at later times and affect the fidelity of repair at the cleavage site, as well as the process of DNA end resection after DSB induction. Loss of bub further confirmed that the observed retention of Rad53 and γ-H2A phosphorylation in the vicinity of the DNA break is due to a delay in DNA damage repair. Phosphoproteomic screening of bub mutant strains in response to DNA damage implicated several yeast kinase substrates on Tel1 S/T-Q sites. NHEJ defects observed in BUB1-depleted mammalian cells suggest a conserved role for BUB proteins in yeast and humans. Our findings also reveal that the Bub-mediated branches of the SAC cooperate with cell cycle regulatory proteins (Apc9, Clb2, and Swi4) to alter NHEJ, overlapping with the DDR pathway that uses Rad53 kinase and phosphorylated H2A at the DNA break. Altogether, the findings confirm that these systems are functionally reliant on one another, and perturbations to any of them can impair normal DDR and negatively impact cell survival.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
The yeast strains, plasmids, and primers used in this study are listed in Table S1 in the supplemental material. Mutant strains in JKM139 were generated by lithium acetate transformation with a PCR product containing the NATMX6 (nourseothricin resistance) cassette flanked by regions homologous to the gene of interest. Nonessential haploid MATa kanamycin-marked deletion mutant strains were obtained from the yeast deletion library (24). Wild-type and mutant strains were cultured in YPD (1% yeast extract, 2% Bacto peptone, 2% glucose), except as noted.
Compilation of DNA repair-related target set.
An exhaustive review of literature studies, gene ontology (GO) annotations, and public database surveys (25, 26) was used to manually curate 510 genes that are of relevance to DNA-repair related processes. In certain cases, gene candidates were included based on their epistatic associations with known NHEJ factors (24, 25) or when they displayed hypersensitivity to DNA damage-inducing agents targeting the NHEJ pathway, as assessed in large-scale chemical-genetic screens (27). The target index also includes some key NHEJ factors as positive controls, to assess the reliability of our assay. By using the NHEJ assay (see below), we screened 369 (of the 510) DNA repair-related nonessential genes, while the remainder either failed or were not available in the deletion mutant library.
Plasmid-based NHEJ or HR and chromosomal DSB repair assays in yeast.
The plasmid end-joining or repair assay for NHEJ in yeast was performed essentially as previously described (12). Colony formation for linear/circular transformations for mutant replicates over linear/circular for wild-type replicates handled on the same day were used to estimate NHEJ repair efficiency. Conversely, HR efficiency in yeast was assayed essentially as described previously (28) with the following modifications. About 200 ng of the purified LacZ amplicon (PCR amplified from pGV255-LIVE plasmid using LacZ forward and reverse primers) was cotransformed into the relevant yeast mutant strain together with 10 ng of the BglII-digested linear pGV256-DEAD plasmid by a standard transformation method. The quantitative X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) colony-lift filter assay was then used to monitor the activity of the LacZ reporter gene by transferring the transformants onto a nitrocellulose membrane. The membrane subjected to X-Gal was incubated for roughly 4 h at 30°C, until the blue colonies were detected. HR efficiency was calculated as the number of blue (i.e., LacZ-producing) colonies over a parallel circular plasmid transformation (i.e., with uncut pGV256 plasmid) and the tested mutant strains were normalized to the wild type.
The efficiencies of chromosomal DSBs in yeast for the selected set of strains derived from JKM139 were assayed essentially as described elsewhere (12). Briefly, overnight cultures of wild-type or mutant strains were grown in fresh YPD or YP-raffinose (1% yeast extract, 2% Bacto peptone, 2% raffinose) at 30°C to an optical density at 600 nm (OD600) of ∼0.5. Harvested cells were washed extensively in sterile water to remove residual glucose, which may otherwise interfere with the expression of the galactose promoter. To induce HO (homothallic switching endonuclease), serial dilutions of the mid-log phase of the wild-type or mutant cells were plated on YP medium supplemented with 2% galactose. For comparison, cultures from the same set of strains were plated on YP medium with 2% glucose. Colony formation was measured after 2 to 3 days of incubation at 30°C. Chromosomal DSB efficiency was computed as the number of wild-type or mutant colonies that survived in galactose medium over the number that survived in glucose medium. Each measurement was repeated independently a minimum of three times.
DNA end resection and sequencing.
The wild-type (JKM139) and derivative mutant strains were grown in YP medium containing raffinose (20 g/liter) as a carbon source to the mid-exponential phase. To induce DNA breaks, galactose (20 g/liter) was added to the culture. Samples taken at the indicated time points were briefly centrifuged (3,000 × g) for 3 min, and the cell pellets were resuspended in an equal volume of phosphate-buffered saline (PBS) and frozen in liquid nitrogen until the pellets from all time points were collected. Thawed frozen cells gently disrupted by vortexing with glass beads for 5 min were used to isolate the chromosomal DNA through a standard phenol-chloroform extraction method. The DNA was then digested with restriction enzymes (BssSαI and PvuII) and separated on neutral agarose gels. Dot blotting was carried out as described previously (29), except that biotinylated DNA probes were used for the labeling reaction. The spot intensity of each of the tested strains at various time points from the blots was quantified by image analysis in Adobe Photoshop and corrected to their corresponding spots from uninduced cells.
For sequencing analysis, individual wild-type and mutant colonies from the plasmid repair assay (which received cleaved p416 plasmid) grown in 5 ml minimal medium lacking uracil were used to reisolate plasmids, which were then purified and subjected to standard Sanger sequencing (TCAG; Hospital for Sick Children). The resulting sequences were quantified to assess the frequency and extent of mutation events measured at the cleavage site by characterizing them into three groups. These include sequences with (i) no mutation at the break site, (ii) small sequence loss of overhangs (<4 bp), and (iii) large sequence loss (>4 bp).
Mammalian cell line, RNAi/CRISPR constructs, and transfection.
The mammalian cell line, RNA interference (RNAi), and CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9) reagents used in this study are listed in Table S1 in the supplemental material. Human osteosarcoma U2OS cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. The shRNA construct specific for BUB1 was purchased from the RNAi consortium lentiviral library available from Open Biosystems, whereas the on-target human siRNA specific for TBC1D1 knockdown (SMARTpools) was synthesized by Dharmacon. shRNAs and siRNAs were introduced in U2OS cells using lentivirus-mediated transfection to create stable cells.
A BUB1 chromosomal gene knockout was generated by cloning the chemically synthesized single guide RNA (sgRNA)-encoding oligonucleotides (see Table S1 in the supplemental material) targeting BUB1 (designed using the CRISPR design tool), into the lentiCRISPR v2 (Addgene) vector backbone. Briefly, the sequence-verified plasmid DNA (i.e., lentiCRISPR v2 containing BUB1 sgRNA) was transfected into the cultured U2OS cells in 6-well plates, following the standard transfection procedure with Lipofectamine LTX with Plus reagent.
NHEJ and HR reporter assays in mammalian cells.
The purified product of the reporter construct (pEGFP-Pem1-Ad2; a gift from V. Gorbunova) digested with I-SceI at 37°C was transfected either with empty vector lentiCRISPR v2 or BUB1-ligated lentiCRISPR v2 into the U2OS cells by transfection with Lipofectamine LTX with Plus reagent. After 72 h posttransfection, cells were harvested and fixed for fluorescence-activated cell sorting (FACS) analysis. For HR reporter assays, pDRGFP (Addgene plasmid 26475) and pCBASceI (Addgene plasmid 26477) constructs (gifts from Maria Jasin [30, 31]), expressing I-SceI endonuclease from a mammalian promoter that introduces DSBs at genomic I-SceI sites (30), were transfected with empty vector lentiCRISPR v2 or BUB1-ligated lentiCRISPR v2 into the U2OS cells, followed by FACS analysis at 72 h posttransfection. The percentages of green fluorescent protein (GFP)-positive cells in the control and knockout were counted to measure NHEJ or HR repair efficiency.
Microscopic, cell morphology, and immunofluorescence analyses.
To examine microcolony formation, the mid-log phase of the wild-type JKM139 strain and isogenic mutant cells were washed in sterile water and plated on YP plates containing 2% galactose. The plates were incubated at 30°C, and at least 100 cells with microcolonies (i.e., colonies of 1, 2, 4, or more than 4 cells) were counted for each time point over a period of 24 h under an Olympus stereomicroscope.
Cell morphology inspected in harvested cells from yeast wild-type and mutant cultures were fixed in 70% ethanol and washed several times with phosphate-buffered saline (PBS). Cells stained with DAPI (4′,6-diamidino-2-phenylindole) were visualized for large-budded cells or elongated nuclei using a Zeiss Observer Z1 inverted microscope with Colibri 2 epifluorescence.
γ-H2A phosphorylation on DSB sites was visualized by staining yeast cells as previously described (32), with some modifications. Cells from cultures grown overnight were subcultured in YPD to the mid-log phase (OD600 of ∼0.5), washed in sterile water, and then grown in YP medium supplemented with 2% galactose for 1 h to induce DNA damage. Approximately 1 ml of the induced cells was removed at the indicated time points and fixed with 1% formaldehyde. Cells were washed, treated with zymolyase (10 μg/ml), spotted onto poly-l-lysine-coated slides, and stained with anti-phospho-histone H2A (Ser129) primary antibody (1:1,000; Abcam) for 1 h prior to washing of the cells with PBS and Alexa Fluor 488-conjugated secondary antibody (1:500) for 30 min. To visualize nuclei, cells were subsequently treated with RNase A (0.2 mg/ml) and stained with propidium iodide (2 μg/ml) for 1 h prior to visualization. The γ-H2A foci were visualized using a Zeiss epifluorescence microscope, and at least 50 cells were analyzed per time point. Images were acquired with ZEN software supplied with the Zeiss epifluorescence microscope and processed further with Adobe Photoshop CS5.
To assess 53BP1 (p53-binding protein 1) focus formation, U2OS cells were transfected with the BUB1 shRNAs, along with a nontargeting shRNA control. After 48 h of puromycin selection, cells were seeded onto coverslips and fixed with 4% paraformaldehyde in PBS for 15 min, followed by three washes with PBS. Cells were permeabilized with ∼0.3% Triton X-100 for 10 min and blocked in PBS containing 10% FBS, ∼0.3% Tween 20, and 0.3 M glycine for 30 min. Cells were then immunostained with a rabbit anti-53BP1 antibody (Abcam) at 1:500 for 1 h followed by three washes in PBS and incubation with a goat anti-rabbit IgG Alexa Fluor 488-conjugated secondary antibody at 1:1,000 for 1 h. For proper visualization of 53BP1 foci, nuclei were counterstained with DAPI (1 μg/ml), with images acquired using a Zeiss epifluorescence microscope. A similar procedure was adapted for TBC1D1, except that siRNA targeting TBC1D1, along with a scrambled siRNA control were transfected into U2OS cells seeded onto a sterile coverslip placed into a 6-well dish containing the appropriate culture medium.
Colocalization (53BP1 and BUB1) experiments were performed by growing U2OS cells on fibronectin-coated acid-washed glass coverslips. Briefly, cells were rinsed in PBS and then fixed in 4% acid-free formaldehyde for 10 min at room temperature, followed by permeabilization (0.25% Triton X-100 in PBS) and blocking in 10% FBS in PBS containing 0.1% Tween 20 and 0.3 M glycine. Cells were incubated overnight with rabbit polyclonal anti-53BP1 and mouse monoclonal anti-BUB1 antibodies (Abcam) in PBS solution. After subsequent washing, cells were incubated with goat polyclonal anti-rabbit antibody conjugated with Alexa Fluor 488 (1:300; Abcam) and donkey anti-mouse IgG conjugated with Alexa Fluor 647 (1:300; Abcam) in the same buffer as the primary antibody. Cells were briefly washed in PBS and water, mounted on glass slides in DAPI-containing mounting medium, and visualized.
Cell synchronization and flow cytometry.
Wild-type or mutant cells in the exponential phase were synchronized in YPD plus 20 μg/ml nocodazole for 2 h at 30°C and subsequently treated with 40 μg/ml bleomycin (BLM) for 30 min. Cells were released from nocodazole by two washes with YPD, and samples aliquoted at each indicated time point were fixed in 70% ethanol and analyzed by flow cytometry.
Immunoblotting and immunoprecipitation.
To assess Rad53 or γ-H2A phosphorylation, cells from wild-type and mutant cells were harvested at the indicated time points, and extracts were prepared essentially as described above. Proteins were separated on 10% SDS–polyacrylamide gels and transferred onto nitrocellulose membranes using the manufacturer's instructions. Membranes were probed with anti-Rad53 (1:5,000) or anti-phospho-histone H2A (1:5,000; Ser129) primary antibody, followed by goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (1:5,000). Immunoblots were visualized using chemiluminescence.
BUB1 immunoprecipitation coupled with mass spectrometry (MS) was performed by lysing U2OS cells cross-linked with dithiobis succinimidyl propionate in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1% NP-40, 1 mM EDTA). About 10 mg of protein incubated with protein G-magnetic microbeads and BUB1 antibody at 4°C for ∼4 h was purified using a magnetic column, followed by three washes with RIPA buffer containing detergent and the last two washes with detergent-free buffer. Trypsin-digested purified protein with proteolytic digestion mixture (2 M urea, 50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol, 5 μg/ml immobilized trypsin) was eluted using a buffer containing 2 M urea, 50 mM Tris-HCl (pH 7.5), and 5 mM choloroacetamide. The peptide mixtures were then subjected to shotgun peptide sequencing using gel-free liquid chromatography-tandem mass spectrometry (MS/MS) to identify the interacting proteins, essentially as previously described (33).
The interaction between 53BP1 and BUB1 was verified by coimmunoprecipitation using protein A/G-magnetic microbeads and the rabbit polyclonal anti-BUB1 antibody (Abcam). γ-H2AX phosphorylation levels in control- and BUB1-depleted U2OS cells were assessed by treating cells with and without the DNA-damaging agents for 48 h. Protein extracts from samples were subjected to 10% SDS-PAGE, and the blots were probed with anti-phospho-histone H2AX (Ser139) antibody (Abcam) to assess γ-H2AX phosphorylation levels in depleted cells. Immunoblots were visualized using chemiluminescence.
Phenotypic and clonogenic cell survival assays.
Exponentially growing cultures of the single and double deletion yeast strains were serially diluted and spotted onto YPD plates in the presence or absence of hydroxyurea (50 mM). The sensitivity of the wild-type or mutant strains to these agents was examined after 3 days of incubation at 30°C.
U2OS cells transduced with BUB1 and 53BP1 shRNAs and scrambled control shRNA, via lentivirus, were plated in triplicate on 24-well plates (i.e., with 5 × 103 cells/well, 25 ml virus/well, and 8 mg/ml Polybrene). After 24 h of infection, cells were seeded into new 24-well plates for expansion into fresh medium containing puromycin (2 μg/ml) and incubated for an additional 48 h. Fresh cells were then treated with various concentrations of cisplatin. After 48 h of incubation, cells were washed with warm PBS, and the number of viable cells harvested by trypsinization at 37°C was quantified using a cell counter.
Analysis of cleavage site and ChIP by qRT-PCR.
HO cleavage and chromatin immunoprecipitation (ChIP) analysis were performed essentially as previously described (13), with the following modifications. To measure the amount of intact HO cleavage site DNA, ∼5 ml of cells from overnight YPD cultures was grown in fresh YPD medium or washed and transferred to YP medium containing 2% galactose to induce HO cleavage for 1 h and then washed and resuspended in YPD and sampled by recovering the cells at the indicated time points. Chromosomal DNA was extracted from cells via a phenol-chloroform procedure and was used to measure HO cleavage levels by comparing quantitative real-time PCR (qRT-PCR) amplicons of the induced samples against the corresponding uninduced (glucose-treated) control cells. The analysis was performed using SYBR green to measure the number of amplicons on an iCycler real-time PCR system (Bio-Rad). The primers (see Table S1 in the supplemental material) used for the HO cleavage site at the MATa locus were constructed essentially as described previously (13).
In γ-H2A ChIP experiments, cell samples prepared as described above were cross-linked to DNA using formaldehyde and resuspended in ChIP buffer. The cells used for Bub1 and Bub2 ChIP analysis were washed and resuspended in YP medium containing 2% galactose for 1 h and then used directly without a further wash or recovery period in YPD. The cell pellets were disrupted using glass beads, and the extracts were subsequently sonicated using a Branson Sonifier. Immunoprecipitation was performed using protein G beads and an anti-H2A phosphor-S129 antibody (Abcam) or anti-Bub1/Bub2 or the control anti-Ku70 antibodies (Santa Cruz). The qRT-PCR primers (see Table S1 in the supplemental material) used to measure the DNA amplicons adjacent to the HO endonuclease break site were constructed essentially as previously described (13).
Phosphoproteomic analysis by IMAC and MS.
Nuclei from wild-type (JKM139) and mutant yeast cells grown in the absence (YPD with 2% glucose) and presence of HO endonuclease-induced DSB (YP medium with 2% galactose) were isolated using a glass bead cell disruption-based method. Yeast nuclei were resuspended in ice-cold extraction buffer (20 mM HEPES [pH 8.0], 8 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate, 1 mM sodium fluoride, 1% Triton X-100, 1:200 [vol/vol] dilution of protease inhibitor cocktail), sonicated, and centrifuged for 10 min at 15,000 × g to separate soluble proteins from the debris.
Next, samples were diluted four times with 20 mM HEPES (pH 8.0) and reduced by the addition of 5 mM Tris (2-carboxyethyl) phosphine (TCEP) for 1 h at room temperature, followed by alkylation with 15 mM iodoacetamide for 60 min in the dark. Proteolytic digestion was performed overnight by the addition of TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone)-treated trypsin (Worthington) in a 1:100 enzyme/protein ratio. Approximately 150 μg of the digested mixture (reaction stopped by adding 1% formic acid) was desalted on disposable TopTip C18 columns (Glygen). After lyophilization, the sample was subjected to phosphopeptide enrichment using immobilized metal affinity chromatography (IMAC) as per the manufacturer's instructions (Pierce Fe-nitrilotriacetic acid [NTA] phosphopeptide enrichment kit; Thermo Scientific). The samples were then acidified by adding 1% trifluoroacetic acid, followed by desalting (C18 columns) and drying in vacuum evaporator.
Chromatographic separation of the phosphopeptide mixture was performed on a Proxeon EASY-nanoLC 1000 liquid chromatograph (Thermo Scientific) equipped with a Thermo Scientific Acclaim PepMap C18 nano column (15 cm by 50 μm inside diameter [i.d.], 2 μm, 100 Å) and water-acetonitrile-0.1% formic acid gradient. Samples were loaded onto the column for 100 min at a flow rate of 0.30 μl/min. Peptides were separated with a gradient of 2 to 6% acetonitrile for 1 min, followed by a linear gradient increase from 6 to 24% for 75 min, 24 to 100% for 14 min, and then a 100% acetonitrile wash for 10 min. Eluted peptides were then sprayed into an Orbitrap Elite mass spectrometer (Thermo Scientific) using positive electrospray ionization at an ion source temperature of 250°C and an ion spray voltage of 2.1 kV. Full-scan MS spectra (m/z 350 to 2,000) were acquired in the Orbitrap at a resolution of 60,000 (m/z 400).
Raw MS/MS files were converted to mzXML format and submitted for database searching using SEQUEST-PVM v.27 (rev. 9) under standard workflow conditions and with a nonredundant yeast protein sequence FASTA file from the SGD database (January 2012). Search parameters were set to allow for dynamic modification of methionine oxidation and phosphorylation of STY and one fixed modification of cysteine carbamidomethylation using precursor ion tolerances of 20 ppm. A stringent false-discovery rate of 1% (P ≤ 0.01) was used to filter candidate peptide, protein, and phospho site identifications. To determine the phosphorylation sites that were dependent on Bub, we removed those phospho sites present only under glucose conditions from the corresponding samples subjected to DNA damage. Additionally, we retained substrates that were identified by mass spectrometry at a confidence of 90% or more, calculated essentially as previously described (33), in either the wild type or Bub mutants.
Bioinformatic analyses.
Orthological relationships of annotated and putative DNA repair-related genes were evaluated using a set of 120 eukaryotic species via PhyloPro. The domain homology search for yeast Bub2 in humans was performed by querying the full-length yeast Bub2 protein coding target sequence against the nonredundant human reference protein sequence database. Iterative searches were specifically carried out using a domain-enhanced, lookup-time-accelerated BLAST (DELTA-BLAST) search engine from NCBI. Enrichment analysis of gene ontology (GO) annotation terms was performed using BiNGO (ver. 3.02), a Cytoscape (ver. 3.1.1) plug-in (34), where proteins significantly enriched for a specific GO biological process were evaluated using a hypergeometric test corrected for multiple hypothesis testing (P ≤ 0.05) using a Benjamini-Hochberg false discovery rate correction. Significantly enriched processes were then visualized using the Enrichment Map (ver. 2.0) (35) Cytoscape plug-in.
RESULTS
Large-scale plasmid-based DSB screening reveals new putative NHEJ repair genes.
To systematically screen genes for involvement in NHEJ (Fig. 1A), we generated a candidate gene list through exhaustive surveys of the literature and public databases (25). This resulted in 510 known and putative DNA repair-related genes (see Table S2, sheet 1, in the supplemental material), from which we successfully screened the majority (n = 369) of the nonessential genes as queries in a plasmid NHEJ assay (see Materials and Methods) (Fig. 1A; see Table S2, sheet 2). These 369 genes included those known to participate in NHEJ (n = 10) and those involved in DNA-related repair (n = 57), response to stress or DNA damage (n = 20), DNA replication (n = 18), cell cycle or division (n = 64), histone or chromatin modification (n = 90), and cell maintenance and structure (n = 24), as well as genes with unclear functions (n = 36) (Fig. 1B). Many of these genes are evolutionarily conserved, consistent with their fundamental roles in DNA repair (36) (see Fig. S1A and B and Table S3, sheet 1, in the supplemental material).
FIG 1.
NHEJ targets and overview of the plasmid-based DSB repair screening strategy. (A) Schematic summary of the plasmid-based repair screening methodology (12) and characterization of Bub factors in NHEJ by independent assays for chromosomal DSB repair, γ-H2A focus formation, drug sensitivity, phospho-proteomics, and several others. (B) Functional distribution of known and putative DNA repair-related target genes grouped into 11 broadly representative bioprocesses. (C) Distribution of NHEJ efficiencies for previously known DNA repair-related mutants compared to the wild type, with the selected threshold indicated. (D) Venn diagram showing the overlap of candidate genes (total numbers of genes from the indicated sources are in parentheses) identified from this study versus those from previously published small- and large-scale (11, 22, 23) genetic screens.
To identify the genes related to the NHEJ pathway, we used an established in vitro plasmid-based DSB repair assay (12) in which the linearized p416 plasmid containing the URA3 selection marker was introduced into a yeast array of target single-gene deletion mutants. The plasmid cleavage site (i.e., an induced DSB) was contained in a region with no homologous sequence available in the genome to repair (Fig. 1A). Mutant strains that display less colony growth from the linearized plasmid relative to those from a circularized control plasmid were taken to be potentially involved in NHEJ (see Materials and Methods).
As an independent assessment of the accuracy of our screening approach, we compared two biological replicates of each mutant screen and found that in the majority (>90%) of the screens, there is a consistent effect on NHEJ efficiency (i.e., either with reduction or no reduction) between replicates (see Fig. S1C in the supplemental material). (Inconsistent mutant screens demarcated as outliers are shown in Table S2, sheet 2, in the supplemental material.) We next applied a statistical framework to define an appropriate threshold to derive biologically meaningful information about genes involved in NHEJ repair. Specifically, we tested 29 genes reported previously to have an effect on NHEJ and found that 28 of them reproducibly displayed ≤70% NHEJ efficiency, reflecting the overall sensitivity of our method. In contrast, only 4 of the 19 independent replicate screens of the same wild-type strain resulted in ≤70% NHEJ efficiency (Fig. 1C), suggesting a false-positive error rate of ∼21%. The candidate gene set with NHEJ impairment was categorized based on efficiency values into very severe (<5% NHEJ efficiency), severe (5 to 20%), moderate (21 to 50%), mild (51 to 70%), and having no effect (>70%) (see Fig. S2 and Table S2, sheet 2, in the supplemental material).
As with the previously reported genetic studies (11, 22, 23), our assay correctly captured (at ≤70% NHEJ efficiency) virtually all of the well-studied genes associated with NHEJ (Fig. 1D; see Table S2, sheet 2, in the supplemental material). These include the components of the yeast Ku complex and strains depleted for subunits of the Dnl4 complex (11, 22, 23). Strains lacking the NHEJ factor mre11, defective for DSB repair (11), and with loss of chromatin assembly and silencing factor genes sir2, sir3, and sir4 also showed reduced NHEJ efficiency, emphasizing the direct or indirect functional roles of these components in NHEJ (23). Conversely, the mutant strains corresponding to several new candidates (see Fig. S2 and Table S2, sheet 2, in the supplemental material), including those with the SAC-encoding genes (bub1 and bub2) that displayed moderate reductions in NHEJ efficiency may have been missed in previous screens (11, 22, 23) due to differences in strain backgrounds and experimental methodology or reduced severity of NHEJ impairment compared to that of canonical NHEJ mutants.
Both Bub1 and Bub2 function as initial sensors of DNA damage during mitosis (20), and previous biochemical and genetic investigations have indicated a strong functional coupling between the SAC and the DDR, with DNA lesions being capable of causing cell cycle arrest through a SAC-dependent mechanism (20). In accordance with this notion, but in contrast to other known SAC factors (bfa1, bub3, mad1, and mad2), the reduced colony formation (<50%) of yeast cells due to the loss of bub1 and bub2 was similar to the reductions reported previously for chromatin modification or DNA damage recognition proteins (11, 12, 23) that were suggested to have a likely role in the NHEJ pathway of DSB repair. Consistent with this, the NHEJ defects of bub1 and bub2 (see Fig. S3B in the supplemental material) appeared to be ∼1.2- to 1.5-fold lower than those in the corresponding mutant strains deleted for HR activity (see Fig. S3A), which is comparable to the fold estimates for NHEJ-defective mutants described hitherto (37, 38). While this modest difference probably reflects the relatively minor role of Bub in NHEJ, we chose to characterize the effects of two branches of the SAC checkpoint on NHEJ and its recruitment to DSBs in more detail.
Prior to in-depth investigation of Bub's cellular role in DSB repair by NHEJ, we conducted a focused plasmid end-joining assay of Bub with more replicates and compared its repair efficiency to that of the canonical NHEJ core factor Ku70 to ascertain if the bub NHEJ mutants from the primary DSB repair screens are genuine and to what degree these mutant defects are relative to well-established NHEJ factors (see Fig. S3B in the supplemental material). Consistent with the expectation from the original screen (see Fig. S2 in the supplemental material), the bub mutants showed moderate (or partial) defects in NHEJ, which were much less severe than that observed for the ku70-deficient mutant. However, the strong NHEJ repair defects in ku70 mutants were not further impaired by the deletion of bub (see Fig. S3B in the supplemental material). This result implies that while Bub cannot repair the NHEJ defect at a higher frequency like the core NHEJ factors, it may have an indirect role in the end-joining repair of DNA DSBs by functioning cooperatively with Ku70.
Bub1 and Bub2 promote chromosomal DSB repair by NHEJ.
Since plasmid-based DSBs, as employed in this study for assessing NHEJ repair, may be functionally distinct from chromosomal DSB's, we reexamined the bub deletion mutant strains for chromosomal DSB repair by NHEJ. To do so, we constructed the bub deletion mutants in a yeast JKM139 strain expressing an inducible site-specific HO endonuclease from a galactose promoter. This results in cleavage of a restriction site at the MAT locus and produces an in vivo chromosomal DSB (13). Notably, the parental JKM139 strain that we used lacks the two silent copies of the MAT locus (i.e., the HML and HMR loci located at the left and right ends of chromosome III, respectively) required for the repair of HO-induced DSBs by HR (Fig. 2A). Efficient repair of this break will depend solely on the NHEJ process to ensure the cell's survival (13).
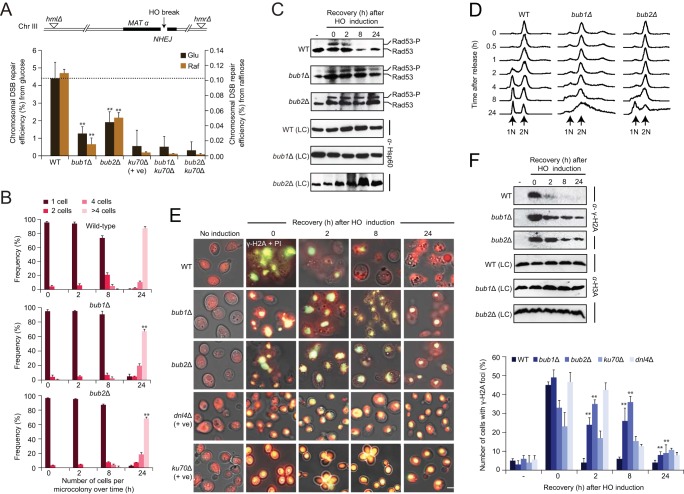
FIG 2.
Bub1 and Bub2 contribute to NHEJ, Rad53 checkpoint activation, mitotic delay, and γ-H2A phosphorylation and focus formation. (A) Schematic illustration (top) of the haploid yeast JKM139 strain on chromosome III expressing galactose-inducible HO endonuclease at the MAT locus, bearing the deletions of HML and HMR donor loci. Shown are the repair efficiencies of HO-induced chromosomal DSBs (bottom) of the indicated mutant and wild-type (WT) strains grown in glucose (Glu) and raffinose (Raf) carbon sources. Efficiency was determined by counting the surviving colonies on galactose versus glucose medium. Note that in the NHEJ assay (using the JKM139 strain background) because there is an extended HO signal, it is likely that there is an artificial inflation of the effects of nonmutagenic NHEJ due to repeated cleavage at an intact breakage site. (B) “Frequency” refers to the proportion of cells (i.e., 1, 2, 4, or >4 cells) per 100 microcolonies in the indicated strains under HO-induced DSBs. (C) Immunoblots show Rad53 activity of WT and mutant cells recovered at the indicated time points after HO induction. Rad53 activation was analyzed by probing with anti-Rad53 antibody. The housekeeping heat shock protein (Hsp60) probed with anti-Hsp60 antibody was used as a loading control (LC). (D) Cell cycle profiles of WT and mutant strains treated with BLM. Cells from the indicated strains were arrested with nocodazole in G2/M (t = 0) and released synchronously into YPD medium containing BLM (40 μg/ml) for 30 min prior to washing of the cells with fresh BLM-free YPD medium. Samples were harvested at the indicated times, and the DNA contents were analyzed by flow cytometry. (E) Accumulation (left) and quantification (right) of γ-H2A focus formation in WT and mutant cells after recovery from HO induction at the indicated time points were analyzed by immunofluorescence with anti-γ-H2A antibody to visualize focus formation (green) and with propidium iodide (PI) to stain nuclei (red). The scale bar equals 5 μm. (F) H2A phosphorylation for WT and mutant strains at the indicated time points was analyzed by Western blotting with anti-γ-H2A antibody, and anti-H3A antibody (probed against histone H3 protein) was used as the loading control (LC). Values indicate means ± standard errors of the means (SEM) from three independent experiments; P ≤ 0.05 (indicated with asterisk), calculated using Student's t test. −, assessment of Rad53 activity (C) and γ-H2A (F) phosphorylation levels prior to HO induction.
In comparison to the wild-type JKM139 cells, both the isogenic bub1 and bub2 mutant strains displayed significantly reduced (P ≤ 0.05) colony formation 48 to 72 h after cleavage was induced by treatment with galactose (Fig. 2A). However, we noticed that the reported survival frequency for the wild-type cells was higher (∼4%) than that in previously published (∼0.1%) studies of JKM139 (39). This discrepancy is due to the growth of the wild-type or mutant strains tested under glucose conditions (prior to transfer to galactose medium) that repressed the galactose-inducible promoter (40, 41), resulting in lower HO cleavage efficiency and increased colony survival. Therefore, we performed the chromosomal DSB assays for wild-type and mutant (bub and ku70) strains in nonrepressing raffinose as a carbon source. In addition to the colony survival rate for wild-type cells in raffinose that was in agreement with the previously reported literature (39), the overall pattern of reduced survival frequency for bub mutants and further reductions for ku70, as expected, remain consistent under both glucose and raffinose conditions (Fig. 2A), although the NHEJ efficiency in the raffinose-cultured cells showed a more robust HO cleavage effect.
To test if the observed reduction in cell survival of the bub mutants was due to defects in DNA break repair, we sequenced the joints from 40 or more repaired plasmids from independent clones. We found that compared to wild-type cells, the deletion of bub1 or bub2 exhibited an increase (from 33% in the wild type to 47% and 57%, respectively) of defective repairs involving small (<4-bp) deletions. In contrast, while bub1 or bub2 mutant cells showed little increase (18% and 7%, respectively) in the repair products with large (>4-bp) deletions, ku70 mutant cells favored repairs with significantly large deletions (66%) at the break site (see Fig. S3C in the supplemental material). This indicates that the bub mutants have a loss of fidelity in NHEJ, but not to the same degree as in strains lacking ku70, suggesting less efficient NHEJ or increased end processing in DSB repair.
Furthermore, after a release from G1 arrest, induction of HO endonuclease in the bub mutant cells generated fewer microcolonies (i.e., those with four or more cells) at 24 h compared to the wild-type strain (Fig. 2B). This delayed recovery of bub mutants is consistent with altered DNA damage checkpoint activation, perhaps via single-stranded DNA (ssDNA) generated by resection (42).
Bub1 and Bub2 contribute to Rad53 activation and mitotic delay.
Activation of the Rad53 checkpoint kinase in response to DNA damage can be assessed by a phosphorylation-dependent electrophoretic shift in Rad53 mobility (43). This has been widely used to assay for DDR-related checkpoint protein kinases, which are activated by Rad53 subsequent to DNA damage (43). We therefore monitored the activation of Rad53 in wild-type and bub1 and bub2 mutant strains during their recovery from HO-induced damage. We found that Rad53 in wild-type cells was completely dephosphorylated 8 h post-HO-induced DNA damage, whereas in bub mutants, Rad53 remained hyperphosphorylated beyond 8 h (Fig. 2C). This suggests a requirement of Bub1 and Bub2 for Rad53 dephosphorylation during DNA damage recovery.
Accumulation of large-budded cells with elongated single nuclei spanning the bud neck during recovery from DNA damage is indicative of cells being present in and progressing through the G2/M phases of the cell cycle (44). Consistent with this, examination on the nuclear morphology of wild-type or bub1 and bub2 mutant cells stained with DAPI showed that both wild-type and mutant strains accumulated large-budded cells and elongated nuclei within 30 min of removal of BLM (a radiomimetic agent that produces DSBs) (see Fig. S3D in the supplemental material). However, this effect was not sustained for long in the wild-type or NHEJ dnl4 mutant strains as the large-budded cells and elongated nuclei decreased 1 h after BLM removal, possibly due to efficient HR-mediated repair in dnl4 mutants, whereas the bub mutants continued to delay progression through the G2/M phase, exhibiting elongated nuclei for up to 8 h after removal of the drug. Overall, this delayed progression through the G2/M phase indicates that the tendency of these checkpoint mutants to accumulate in anaphase during recovery is similar to a phenotype previously noticed upon deletion of the DNA-repair scaffolding gene rtt107 (44), consistent with a role for Bub in DNA break repair.
Flow cytometry analysis confirmed that subsequent to BLM-induced DNA damage, the bub1 and bub2 mutant cells arrested in the G2/M (2 N DNA content) phase of the cell cycle for up to 24 h, at which point cell cycle progression began to resume. In contrast, wild-type cells completed cell division and reentered G1 at 2 h (Fig. 2D). Moreover, the cells lacking bub1 and bub2 progress more rapidly from G1 to S phase than wild-type cells, resulting in the reduction of G1 accumulation and an increase in the number of cells with intermediate amounts of DNA between 1 and 2 N. These effects were specific, as the prolonged G2/M delay was not observed without BLM treatment (data not shown), indicating that the delay in the reentry into the cell cycle in the bub mutants is due to BLM-induced damage at the DSB sites.
Recruitment of histone H2A to the sites of Bub1 and Bub2 at DNA breaks.
One of the early responses to DNA damage in mammals is phosphorylation of the H2AX histone variant at serine-139 to form γ-H2AX foci (13), whereas in yeast, the phosphorylation of histone H2A (γ-H2A) occurs in a DNA damage-dependent manner at serine-129 (13). These phosphorylated forms of γ-H2A and γ-H2AX have been used as molecular markers for monitoring DSBs (13), and we therefore sought to compare the localization of γ-H2A in yeast bub mutant and wild-type strains subsequent to HO induction (Fig. 2E). While the wild-type cells showed a sharp increase in the formation of γ-H2A foci, they returned to a normal state 2 h after HO induction and were rarely visible at 24 h, indicating rapid repair of the lesions (Fig. 2E). Conversely, the γ-H2A foci remained visible even after 24 h of DNA damage induction in both the bub1 and bub2 mutants, similar to that of the ku70 and dnl4 mutant strains. Notably, these observations are in agreement with the phosphorylation levels of γ-H2A detected by immunoblotting in mutant cells (Fig. 2F), suggesting a delayed response of these strains in repairing DSB sites.
Bub1 and Bub2 bind near sites of DNA damage and facilitate ssDNA formation by resection.
To ascertain if Bub1 or Bub2 was directly bound to DSBs, we performed ChIP with anti-Bub1 and -Bub2 antibodies in wild-type strains followed by qRT-PCR at the HO-induced break site. Both Bub1 and 2 were found to be significantly enriched at the cleavage site compared to a LEU2 control locus after break induction, with Bub1 maximally enriched (∼63-fold) at 30 min and Bub2 maximally enriched at 1 h (∼55-fold enrichment) and with both disassociating gradually for at least 2 h after damage induction (Fig. 3A). Alternately, Ku70 rapidly accumulated (∼60-fold) at the breakage site immediately following the DNA damage and decreased much faster than bub mutants (Fig. 3A), signifying its involvement in the early recognition and recruitment to the sites of DNA damage for efficient repairing (11).
FIG 3.
Bub1 and Bub2 are recruited to the site of DNA damage. (A) A schematic diagram of the HO restriction endonuclease recognition site on chromosome III of the haploid yeast JKM139 strain is shown with the location of primer pair sets that were used to amplify regions, either at or near the HO-induced DSB (I) or at a control locus (II), by qRT-PCR and the resulting relative fold enrichment of Bub1, Bub2, and Ku70 (control) ChIPs near the DSB sites using the ChIP 1 primer pair at the specified time points after HO induction (III). (B) Relative fold enrichment of Bub1 and Bub2 ChIP at the HO cleavage site using the ChIP 1 primer pair at the indicated hours after HO induction in ku70 mutant strain background. (C) Relative fold enrichment of Bub and Ku70 ChIP near the HO cleavage site (distances of 1.4 and 5 kb) using the ChIP 2 and ChIP 3 primer pairs at the indicated time points after HO induction. (D) Resection-mediated ssDNA formation in the indicated strains at various times points after HO induction at the MAT locus, 5 kb downstream of the MAT or LEU2 control locus (for neutral blots, see Fig. S3E in the supplemental material). Results are shown as means ± standard deviations (SD) (n = 3). (E) Serial cell dilution assay showing the growth inhibition of the indicated strains exposed to hydroxyurea (HU; 50 mM).
To test whether recruitment of Bub to the DSB depends on Ku70, we performed additional ChIP assays with anti-Bub1 and -Bub2 antibodies in ku70 mutant cells at various time points after HO induction. Notably, while Bub recruitment to the break site was observed at later times (Fig. 3A), there is a relative reduction of Bub binding to a DSB in ku70 mutants at 1 h post-DSB induction (Fig. 3B), indicating that Ku70 can be functioning only indirectly in the recruitment of Bub to the sites near the break as any genuine Ku-dependent NHEJ factors do not accumulate at DSB in the absence of Ku (45). Recruitment of both Bub1 and Bub2 was also detected at 0.3 kb and 1.4 kb from the cleavage sites as early as 30 min to 1 h after HO induction. However, by 2 h after cleavage, the recruitment of both Bub1 and Bub2 decreased significantly. Bub recruitment was significantly less prevalent at all time points in regions up to 5 kb distant from the DSB (Fig. 3A and C), suggesting that the Bub is highly localized near the break site.
Since end processing factors have been implicated in 5′-to-3′ HO-induced DSB resection (46), we monitored alterations in DNA-end resection profiles at the HO-induced DSBs in bub1 and bub2 mutant cells. This was done using a neutral dot blot approach (29) with probes designed adjacent to the HO cleavage site at the MAT locus, 5 kb distal from the DSB site, and at a control LEU2 locus (∼100 kb distal from the HO site) (see Fig. S3E in the supplemental material). Our quantitative results (Fig. 3D) showed that in contrast to ku70, cells bearing the bub1 mutation exhibited a steady increase in 3′-end ssDNA resection 2 to 8 h after HO induction, at sites close to the DSB, whereas the resection in cells with the bub2 mutation appeared as early as 30 min after HO induction. However, when assayed at 5 kb away from the break site, the resection was still noticeable in bub mutants 4 h after HO induction, at which time point the ku70 mutant showed resection almost as efficiently as the wild-type strain, suggesting that the Bub and Ku70 process resection, albeit to various degrees.
Bub1 and Bub2 are required for increased cellular resistance to DNA damage.
Since genotoxic agents induce single-and double-strand breaks (11), we compared the sensitivity of the yeast bub1 and bub2 mutant strains to the exogenous genotoxic agent hydroxyurea (HU), which depletes 2′-deoxynucleoside 5′-triphosphate pools to initiate chromosomal breaks (13). Like the ku70 strain that has aberrant NHEJ (13), bub mutants showed more resistance to the cytotoxic effect of HU than wild-type cells. Furthermore, deletion of dnl4 or ku70 in bub mutant strains caused similar levels of resistance to HU (Fig. 3E), suggesting that the observed increase in resistance of bub mutants to DSBs in the absence of ku70 or dnl4 is due to enhanced HR.
A phosphoproteomic screen identifies in vivo substrates of Bub1 and Bub2.
While ATM/ATR substrates for several SAC proteins, including Bub1, were identified previously by large-scale proteomics (47), it is still unclear what in vivo substrates are targeted by the Bub1 and Bub2 checkpoint kinases in response to HO-induced DNA damage. To address this, we used IMAC coupled with MS to screen for Bub1/Bub2-dependent or -independent phosphorylation in HO-induced DSBs. The numbers of peptides phosphorylated in bub mutants, wild-type cells, or both following HO-induced DSBs were quantified by measuring the ion abundance in the mass spectra (see Materials and Methods) (Fig. 4A).
FIG 4.
Phosphoproteomic analysis of bub mutants after HO-induced DSB at the MAT locus. (A) Strategy to identify phosphoprotein substrates that are dependent (case 1) or independent (cases 2 and 3) of Bub1/Bub2 in response to DSB induced by HO endonuclease. (B) The distribution of protein copy number per cell gathered from Ghaemmaghami et al. (60) is shown for phosphoproteins identified in this study versus the total yeast cellular proteome. (C) The number and percentage of Bub-dependent or -independent kinase and nonkinase phosphorylation sites identified by MS at high confidence (left), and the relative ion abundances (right) are shown for two representative Bub-dependent kinase-phosphorylated substrates. (D) Comparative analysis on the frequency of amino acids at the +1 position of phosphorylated serine or threonine for Bub kinase-dependent phosphorylation (denoted as a hydrophobic or hydrophilic residue) against all identified phosphopeptides. (E) Motif analysis showing the distribution of amino acid residues surrounding the phospho-SQ sites on the Bub-dependent substrates after HO induction. (F) Network showing the connectivity between Bub and phosphorylated kinase substrates from various processes in response to DSB induction. (G) Immunoblot analyses of HO-induced DSB with cells expressing a hemagglutinin (HA)-tagged Akl1 and Ste20 Bub-dependent kinase substrates in the anti-HA immunoprecipitates using anti-phospho-S/T-Q antibody. Anti-HA antibody was used as the loading control (LC).
After elimination of those phosphopeptides present under non-DNA-damaging conditions (see Materials and Methods), a total of 476 high-confidence (≥90%) nonredundant phosphorylation sites on 164 proteins were identified after induction of DSBs with HO (see Table S3, sheet 2, in the supplemental material). Nearly 60% (285 of 476) of the Bub1/Bub2-dependent or -independent phosphorylation occurred primarily on serine residues (see Fig. S4A in the supplemental material). This is consistent with earlier reports of serine phosphorylation in response to DNA damage (48). Of these, nearly 46% (75 of 164) of the identified substrates have human orthologues (see Table S3, sheet 2), and more than half (85 of 164) are involved in transport, metabolism, transcription, regulation, DDR, signaling, and DNA replication, repair, or recombination (see Fig. S4B). GO enrichment analysis of the phosphoproteins further indicated a significant enrichment (P ≤ 1.82 × 10−7) for proteins functioning in regulation, transport, and signaling (see Fig. S4C and Table S3, sheet 3) that have roles in DDR-related pathways (47).
Analysis of phosphoprotein abundance showed an obvious bias toward highly abundant proteins (>105 copies per cell), with phosphorylation detected even among proteins of low abundance (<103 copies per cell) (Fig. 4B). The majority (150 of 164) of the HO-induced bub1 and bub2 kinase substrates were distinct from each other (see Fig. S4D in the supplemental material), consistent with the independent functions of these two proteins (18). In addition, about 69% (328 of 476) of the nonkinase (e.g., Cdc48 and Kog1) and ∼5% (22 of 476) of the kinase (e.g., Pkc1 and Pfk2) phosphorylated sites containing substrates were dependent on bub1 or bub2 (Fig. 4C; see Table S3, sheet 2, in the supplemental material).
Because DNA damage checkpoint kinases such as Mec1 (the yeast homolog of human ATR) and Tel1 (the yeast homolog of the human ATM) phosphorylate DDR proteins at S/T-Q consensus sites (47), we examined the frequency of amino acid residues immediately adjacent to the +1 position of the phosphorylated serine (S) or threonine (T) followed by a glutamine (Q) for all identified phosphorylation sites. Phosphorylation of the S/T-Q motif accounted for nearly 5% (22 of 476) of all phosphorylation sites identified, with ∼77% (17 of 22) of the phosphorylation being Bub1/Bub2 dependent (Fig. 4D). By comparison, the hydrophobic amino acid leucine, which is found around SQ or TQ sequences (Fig. 4E) and is phosphorylated by ATM (49), appears to be the most commonly phosphorylated residue in Bub1/Bub2-dependent phosphorylation, accounting for ∼22% (4 of 18) of these phosphopeptides.
While our approach detected many Bub-dependent and -independent substrates, we focused specifically on 5 high-confidence kinase substrates that are directly dependent on Bub1/Bub2 after DSB induction (Fig. 4F; see Table S3, sheet 2, in the supplemental material). These include kinases involved in metabolism (Pfk2), cell cycle, cytoskeleton, and signal transduction (Pkc1, Akl1, and Ste20) and one of unknown function (Ypk3). Some of these kinase substrates were further confirmed by Western blotting using an antibody that specifically recognizes the phosphorylated serine or threonine motifs (Fig. 4G). Taken together, these results suggest that Bub1 and Bub2 phosphorylate many proteins from diverse pathways in response to DNA damage and are important regulators of the DDR.
Human BUB1 binds near the sites of DNA damage and interacts with factors regulating NHEJ.
The conserved domains of Bub1 and Bub2 (Fig. 5A) encouraged us to examine if RNAi-mediated knockdown of these genes in human U2OS osteosarcoma cells (an in vitro model cell line for DDR studies) can result in the formation of nuclear foci at the DSB sites, similar to formation of γ-H2A foci in bub1 and bub2 yeast mutants. After induction of DSBs, the mediator proteins involved in end-joining repair (50), such as BRCA1, the MRE11-NBS1 (XRS2)-RAD50 nuclease complexes, and 53BP1, accumulate in nuclear foci, where the retention of these proteins at DNA breaks requires phosphorylated H2AX (50). Consistent with γ-H2A localization in yeast bub mutants, shRNA knockdowns of human BUB1 (ortholog of yeast Bub1) in U2OS cells caused an accumulation of spontaneous 53BP1 foci (Fig. 5B). To rule out the possibility that this effect was due solely to a G2/M-phase checkpoint defect, BUB1 knockdowns were arrested in G2/M phase by nocodazole treatment (data not shown) and were found to display a significant (P ≤ 0.05) 11-fold reduction in focus formation, indicating that the 53BP1 focus formation was not due only to cell cycle checkpoint arrest.
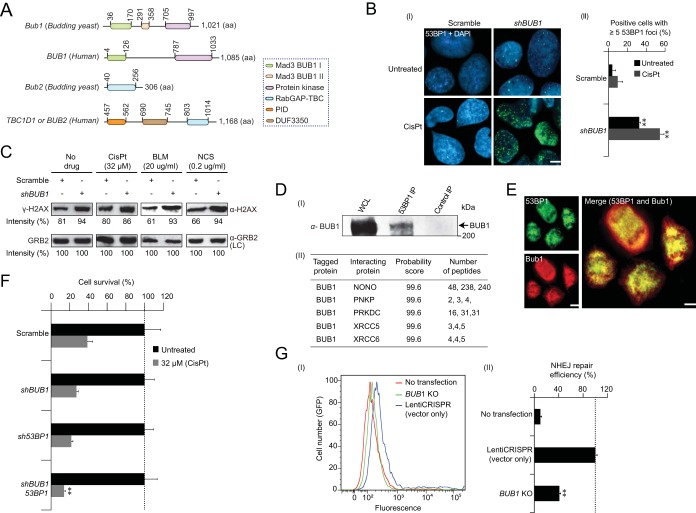
FIG 5.
Bub1 NHEJ is conserved in mammalian cells. (A) Schematic showing the conserved domain organization and length (amino acids) of Bub1 and Bub2 in budding yeast and humans. Colored boxes indicate the homology of the domain regions. (B) Accumulation (I) and quantitative (II) analysis of 53BP1 foci (green) formation in U2OS cells transfected with BUB1 shRNA in the presence and absence of cisplatin (CisPt). DAPI (blue) was used for nuclear staining and visualization of 53BP1 foci. The scale bar equals 5 μm. (C) Immunoblot analysis of H2AX phosphorylation levels in shBUB1 and control-depleted cells in the presence and absence of the indicated concentration of cisplatin (CisPt), bleomycin (BLM), or neocarzostatin (NCS) using an anti-γ-H2AX antibody. Epidermal growth factor receptor-binding protein (GRB2) probed with an anti-GRB2 antibody was used as the loading control (LC). (D) Immunoblot analyses (I) of 53BP1 in the input whole-cell lysate (WCL) and anti-53BP1 immunoprecipitates (IP) using anti-BUB1 antibody in human U2OS cells. The molecular mass of the marker protein by SDS-PAGE is indicated. NHEJ proteins copurified from the endogenous BUB1 (II) using anti-BUB1 antibody (peptides from three independent replicates shown) in human U2OS cells were identified at high confidence (99%) by MS/MS. (E) Immunofluorescent images showing the 53BP1 (green) colocalization with BUB1 (red) in the human U2OS cells. Cells were immunostained with anti-53BP1 and anti-BUB1 antibodies. The scale bar equals 8 μm. (F) Clonogenic survival of control shRNA and BUB1- and 53BP1-depleted U2OS cells treated with cisplatin (CisPt; 32 μM). Asterisks indicate significantly reduced (P ≤ 0.05, Student's t test) cell survival of BUB1 53BP1 double knockdowns compared to their corresponding single knockdowns. (G) FACS-sorted positive GFP cells (I), indicating NHEJ repair efficiency (II) in U2OS cells transfected with the empty LentiCRISPR vector (blue) or the BUB1 CRISPR knockout (KO [green]). U2OS cells not transfected with the indicated knockout or vector (red) served as a control. Fluorescence values are in arbitrary units. Data (B and G) are represented as the mean ± SEM (n ≥ 3); asterisks indicate significant (P ≤ 0.05, Student's t test) difference between the indicated knockdowns or knockouts versus scrambled shRNA or LentiCRISPR control.
Conversely, compared to the untreated BUB1 RNAi cells, the number of nuclear foci increased nearly 2-fold following treatment with the DNA cross-linking agent cisplatin, which impairs the cellular NHEJ pathway (51). This elevated 53BP1 focus formation correlated with levels of γ-H2AX phosphorylation in BUB1-depleted cells (Fig. 5C), demonstrating that BUB1 defects can result in increased DNA damage or defects in DNA repair and suggesting a conserved role for this SAC gene in NHEJ between yeast and humans.
These observations and the known requirement of mammalian ATM kinase in 53BP1 formation (52) prompted us to examine if BUB1 function is dependent on a physical interaction with 53BP1. While affinity purification coupled with mass spectrometry (AP-MS) of endogenous BUB1 in U2OS cells failed to identify 53BP1, BUB1 efficiently coprecipitated with native 53BP1 in solubilized U2OS cell lysates (Fig. 5D). BUB1 also colocalized with 53BP1 in the dividing U2OS cells (Fig. 5E), consistent with the known localization of 53BP1 in the kinetochore of mitotic cells (53). Other cellular proteins that were consistently captured using AP-MS include NHEJ proteins required for DSB and V(D)J recombination (e.g., PRKDC, NONO, PNKP, and XRCC6 [yeast Ku70]-XRCC5 [yeast Ku80] dimer) (Fig. 5D) and proteins involved in DNA repair-related functions, regulation of DDR, and epigenetic control (e.g., histone or chromatin assembly) (see Fig. S5A and Table S3, sheet 4, in the supplemental material). Thus, there is a stable physical association of human BUB1 with proteins implicated in NHEJ and DNA repair-related processes.
As 53BP1 physically binds and colocalizes with BUB1 (Fig. 5D and E), we examined the effects of BUB1 and 53BP1 knockdowns on the survival of U2OS cells in response to cisplatin-induced DNA damage. While knockdowns of both 53BP1 and BUB1 did not impair the survival of U2OS cells in the absence of DNA damage, their combined knockdown effect showed significantly (P ≤ 0.05) reduced survival of cisplatin-treated U2OS cells relative to the corresponding single knockdowns (Fig. 5F), suggesting participation of BUB1 and 53BP1 in DDR.
We next investigated if, like yeast Bub1 (Fig. 2A), the mammalian BUB1 orthologue has similar NHEJ activity in vitro in the U2OS cells. To test this, we employed an established GFP reporter system (54), where the GFP coding sequence is intruded upon by inserting an adenoviral exon flanked by recognition sequences for I-SceI endonuclease. Upon I-SceI digestion, DSBs are generated, and only successful repair of DSBs by NHEJ will reconstitute the functional GFP. The resulting GFP-expressing positive cells were quantified by FACS (see Fig. S5B in the supplemental material). Notably, compared with cells infected with empty vector, the efficiency of NHEJ was significantly (P ≤ 7 × 10−9) reduced in BUB1 knockout cells (Fig. 5G) and was 2-fold lower than the corresponding knockout in U2OS cells with HR activity deleted (see Fig. S5B and C in the supplemental material), suggesting a conserved functional role of BUB1 in repairing DSBs, potentially through stimulation of the robust NHEJ pathway.
Conversely, although yeast bub2 shares a conserved domain (the Tre1/Bub2/Cdc16 [TBC] Rab-binding domain; E value, ≤2e−54) with human TBC1D1 (Fig. 5A; see Fig. S6 in the supplemental material), siRNA knockdown of TBC1D1 cells did not cause 53BP1 focus formation (data not shown), suggesting Bub2 function is not conserved in TBC1D1. Indeed, iterative similarity searches by BLASTP with the full-length sequence of yeast Bub2 failed to identify TBC1D1 as a potential orthologue.
Bub1 and Bub2 cooperate with the Apc9, Clb2, and Swi4 factors in yeast to mediate NHEJ repair.
Cell cycle arrest by the SAC hinders the activation of the APC, which regulates cyclin-dependent kinases, such as Cdc28 and the SBF transcription factors (16). This system integration is evident from the analysis of previously published interaction data (see Table S3, sheet 5, in the supplemental material), which showed extensive physical and functional dependencies between these processes (see Fig. S7A in the supplemental material). While mutations of some APCs (e.g., apc9 and cdc26), cyclins (e.g., clb2 and clb5), and SBF components (e.g., swi4 and swi6) that are not essential for viability exhibited only moderate or mild reduction in NHEJ efficiency (see Fig. S2 in the supplemental material), we chose apc9, clb2, and swi4 for further investigation, as loss of function of these genes in our DSB repair screens showed a 40 to 50% reduction in NHEJ (see Table S2, sheet 2, in the supplemental material).
Using a recent multicondition differential epistatic map (55), we found that a subset of APC mutants had genetic interaction profiles that were highly correlated with bub1 and bub2 mutant strains under three DSB-inducing conditions (Fig. 6A; see Table S3, sheet 6, in the supplemental material). Indeed, in both plasmid-borne (Fig. 6B) and HO-induced chromosomal (Fig. 6C) DSB assays, we found that like the bub ku70 double mutant strains, the deletion of bub with any of the apc9, clb2, or swi4 genes showed no further decrease in DSB repair efficiency compared to that in constitutive single-gene mutants. This suggests a cooperative role for Bub with Apc9, Clb2, and Swi4 in NHEJ repair. However, the efficiency of DSB repair was considerably (P ≤ 0.05) reduced in bub ku70 mutants compared to the loss of function of both bub and one of the apc9, clb2, or swi4 alleles. The delayed recruitment of Bub checkpoint proteins to the DNA break site in the absence of ku70 (Fig. 3B) is consistent with the participation of Bub proteins with Ku70 in NHEJ of DSBs.
FIG 6.
Bub1 and Bub2 functions jointly with APC, cyclins, and SBF to promote NHEJ. (A) Heat map showing clusters of similar correlated GI profiles (blue, negative correlation; yellow, positive correlation) between the bub and APC double mutants (except Swi4 and Clb2 as they were not tested) in response to genotoxic agents: methyl methanesulfonate (MMS), 0.01%; camptothecin (CPT), 5 μg/ml; Zeocin (Zeo), 75 μM. Pearson's correlation coefficient (PCC) was computed for each BUB-APC gene pair across other genes tested using the epistatic interaction data derived from Guénolé et al. (55). (B and C) NHEJ (B) and chromosomal DSB (C) repair efficiency of the indicated bub double mutants and their corresponding single-mutant strains grown in the presence of glucose (Glu) and raffinose (Raf) carbon sources. (D) Schematic diagram of the HO restriction endonuclease recognition site on the chromosome III of the haploid yeast JKM139 strain is shown with the location of primer pair sets that were used to amplify regions on the HO-induced DSB, or at a control locus, by qRT-PCR for measurement of cleavage efficiency. (E) Amounts of intact cut site PCR signal (quantified using qRT-PCR) upon HO induction in the indicated single (left) and double mutants (right) are shown over time. The HO1 and HO2 primers were used to assess the HO-induced DSB site. Intact cut site PCR signal was computed as the ratio of HO signals in induced cells relative to signal in the control locus. (F) Tentative model describing the role of Bub1 and Bub2 on NHEJ. See main text for details. (B, C, and E) Data are represented as means ± SEM (n = 3). Asterisks indicate significant (P ≤ 0.05 [Student's t test]) differences between single mutants versus the wild type (WT).
Bub1 and Bub2 act with Apc9, Clb2, and Swi4 to mediate NHEJ at the site of DSB repair.
Functional cooperation of Bub with Apc9, Clb2, and Swi4 (Fig. 6B) prompted us to assess if Bub1 and Bub2 act together with any of these factors directly at the site of HO-induced DSBs in vivo. We therefore probed their effect on DSBs by recovering chromosomal DNA from the single and double deletion strains after HO induction, and the efficiency of HO cleavage repair was measured (see Materials and Methods) (Fig. 6D). qRT-PCR analysis across the HO recognition site showed that the initial 80% cleavage (20% PCR signal) at the HO site was mostly repaired within 30 min (Fig. 6E). In contrast, bub1 and bub2 mutants, as well as the apc9, clb2, and swi4 mutants, either alone or in combination, showed reduced repair (with ∼40 to 80% PCR signals) even 2 h post-HO induction, suggesting a delay in processing DSB repair.
We next sought to determine if, like bub1 and bub2 (Fig. 2F), the apc9, clb2, and swi4 components regulated the recruitment of H2A near the DSB induced at the MAT locus, either alone or in combination with the bub mutants. To verify this, we performed additional ChIP followed by qRT-PCR in these mutant strains with an anti-phospho-histone-specific H2A antibody. Consistent with our other observations, H2A is phosphorylated to its maximum of ∼12-fold enrichment in wild-type cells as early as 30 min post-HO induction (see Fig. S7B in the supplemental material) and decreases thereafter to ∼2-fold enrichment at the later time point (i.e., after 1 h). In contrast, the levels of H2A phosphorylation in both bub1 and bub2 as well as the apc9, clb2, and swi4 single and double mutants displayed only ∼2- to 5-fold enrichment, even 2 h after HO induction. These observations correlate strongly with the formation of γ-H2A foci in apc9, clb2, and swi4 mutants, either alone or in combination with bub1 and bub2 after HO induction (see Fig. S7C in the supplemental material [representative bub1 or bub2 apc9 double mutant shown]). Taken together, the recruitment of Bub proteins and Apc9, Clb2, and Swi4 components to sites near a DSB in vivo underscores an intriguing functional link of these processes in the mechanism of NHEJ, either directly or indirectly, to sustain genome integrity.
DISCUSSION
Understanding how cells respond to and repair DNA damage through DSB recognition and repair pathways is of fundamental importance, as incorrect repair of the DSBs may lead to genomic instability, cancers, and cell death (6). In this study, using a comprehensive plasmid-based DSB repair screening approach, we identify loss-of-function mutants exhibiting moderate to mild NHEJ defects in S. cerevisiae. We characterize in further detail two SAC kinases, Bub1 and Bub2, in the NHEJ process of DSB repair.
Consistent with the critical roles of the SAC and DDR pathway proteins in checkpoint activation (21), our data indicate that phosphorylation of both Rad53 and γ-H2A is continuously altered in bub mutants in response to HO-induced DNA damage, whereas these DSB signatures are rapidly diminished in wild-type cells. This suggests DSB repair and resumption of the cell cycle are much quicker in wild-type cells than in the bub mutants, which display these signatures for much longer periods. This is consistent with Bub1 and Bub2 actively functioning in efficient DSB repair. Likewise, increases in ssDNA generation in bub mutants in response to DSBs indicate that NHEJ failure due to bub dysfunction can lead to increased end processing. ChIP experiments further confirmed that both Bub1 and Bub2 are recruited efficiently at or near the sites of DNA damage, albeit to a lesser extent in the absence of Ku70. Given the observed physical and functional connectivity of Bub with Ku70 (mammalian XRCC6), it is possible that Ku assists in recruiting Bub to the break site to encourage DDR.
Because ATM/ATR substrates involved in DNA damage signaling stimulate kinases, including Rad53 (47), we conducted an unbiased phosphoproteomics screen to identify potential targets of Bub proteins in HO-induced DSBs. The identified Bub phosphorylation sites on Tel1 S/T-Q sites were involved in DDR, signaling, transport, replication, recombination, and repair, consistent with the finding that DNA damage-activated kinases phosphorylate a diverse network of nuclear proteins and pathways (47). Nevertheless, many new Bub substrates identified in this study that were phosphorylated in response to DNA damage warrant further functional characterization to understand how kinase/nonkinase Bub protein substrates coordinate DDR and phosphorylation-mediated signaling pathways.
Next, as in yeast, we demonstrate in mammalian cells that BUB1 has a conserved role in NHEJ. BUB1 participates with proteins acting in DNA repair-related pathways, including principal NHEJ factors, and DSBs in BUB1-depleted cells induce γ-H2AX phosphorylation and 53BP1 focus formation. The latter finding and the presence of Bub-phosphorylated substrates on Tel1 S/T-Q sites reinforce the finding that ATM-mediated BUB1-dependent phosphorylation is likely to activate the DDR (21) and accumulate γ-H2AX and 53BP1 focus formation in a BUB1 binding- and ATM-dependent manner (56).
Previous observations have indicated that the SAC pathway operates through the regulation of the APC (Fig. 6F), which in association with Cdc20 and under the tight regulation of SBF transcription factors, ubiquitinates and targets mitotic cyclins for degradation (16). Our findings indeed indicate that the bub mutants also harboring a deletion of apc9, clb2, or swi4 have no additional loss of NHEJ efficiency compared to respective single mutants. Similarly, the mutants with apc9, clb2, and swi4 mutations, alone or in combination with bub, showed similar dynamics in the formation of γ-H2A foci as the bub single mutants. These results are consistent with a broad cellular role for Bub in DDR that is highly connected with other pathways through DSB repair.
While the results presented in this study primarily emphasize the digenic combination of bub with apc9, clb2, and swi4 in NHEJ repair, additional relationships may have been overlooked, as we did not test the role of Bub proteins with all of the components of the APC and all cyclins. Additionally, there were nonessential components of the APC and cyclins that were tested and failed to show a reduction in NHEJ efficiency, suggesting that only certain components of the APC and cyclins tend to function specifically in NHEJ by cooperating with Bub, while others may have either a role in an unrelated process or even an opposing (i.e., antagonistic) function with Bub. This is indeed supported by the findings from a recent bub1 and bub2 genetic screen under DNA-damaging conditions (55), which showed positively and negatively correlated genetic interaction profiles with components of the APC.
Altogether, our results support a model in which Bub proteins act as a part of the DNA repair machinery, not necessarily as a core component of the NHEJ process, because some of our findings undoubtedly showed that Bub is distinct from NHEJ factors (Fig. 6F). For example, BLM results (see Fig. S3D in the supplemental material) showed a DSB response not shared by NHEJ mutants. Likewise, the Bub DSB ChIP profile (Fig. 3) is similar to those of histone methylation (H3K9Me2) and other DDR proteins at later stages of DSB repair (57, 58) than NHEJ, as evident by contrasting it to the Ku NHEJ protein, where Bub shows a much longer persistence at breaks. As well, the reported delay in division for bub mutants that is consistent with an alteration of checkpoint activation perhaps occurs via ssDNA generated by resection, as opposed to functioning directly in the NHEJ pathway, because both bub and ku70 mutants resect the ssDNA at various degrees. Nonetheless, based on the other characterizations of Bub's broader cellular role in DSB repair, it is likely that Bub is involved in promoting NHEJ activity in response to DNA damage, which is consistent with the synergistic interactions previously observed between DSB repair and SAC genes (59), as well as with the known role of Bub1 in DDR signaling (21).
Prolonged recruitment of γ-H2A and Mec1/Tel1-dependent Rad9 phosphorylation of Rad53 in bub mutant cells further signify the involvement of Bub in DDR as a consequence of DNA damage. Finally, like phosphorylated γ-H2A in yeast, the chromatin-associated factor 53BP1 has been shown to promote DSB repair via NHEJ in mammalian cells (50), which is in general agreement with our observation that 53BP1 colocalizes with human BUB1 in dividing cells and is recruited at the DSB sites. As phosphorylated γ-H2AX and 53BP1 accumulate in human BUB1-depleted cells, this indicates that, like Bub1 in yeast, human BUB1 functions in NHEJ and in protection from spontaneous genome instability (Fig. 6F).
In addition to the role played by Bub proteins in activating the mitotic checkpoint (16), they appear to function in concert with a suite of APC proteins, cyclins, and SBF factors to promote the repair of DNA breaks in NHEJ. While further mechanistic work is required to understand how these various components contribute to canonical NHEJ (Fig. 6F), the results presented here elucidate a fundamental role for Bub1 and Bub2 in the cellular response to DSBs mediated by the NHEJ repair pathway. In this work, we have generated a valuable resource by identifying a set of subtle novel candidates governing the NHEJ repair pathway. Conservation of these genes in humans and their relationship to cancerogenesis (17) further supports a broad applicability of our findings to inform future studies aimed at characterizing the mechanistic function for these variants in human tumors and in the development of target- and mechanism-based therapeutics.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Vera Gorbunova (University of Rochester) for providing the NHEJ reporter system and Tina Singh from Grant Brown's laboratory (University of Toronto) for performing flow cytometry analysis. We thank the members of M. Babu's laboratory for technical assistance and Gabe Musso (Harvard Medical School, MA) and the anonymous reviewers of the manuscript for constructive suggestions, helpful discussions, and comments.
This research was supported by grants from the Natural Sciences and Engineering Research Council to A.G. and the Canadian Institutes of Health Research to M.B. (MOP-125952). M.B. holds a CIHR New Investigator award. M.J. and J.V. are recipients of a Saskatchewan Health Research Foundation Postdoctoral Fellowship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00007-15.
REFERENCES
- 1.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Heyer WD. 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudas A, Chovanec M. 2004. DNA double-strand break repair by homologous recombination. Mutat Res 566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Yano K, Morotomi-Yano K, Adachi N, Akiyama H. 2009. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res 50:97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
- 5.Lisby M, Rothstein R. 2009. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gent DC, Hoeijmakers JH, Kanaar R. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 7.Daley JM, Palmbos PL, Wu D, Wilson TE. 2005. Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. 2006. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8:1105–1115. doi: 10.1016/S1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 10.Palmbos PL, Wu D, Daley JM, Wilson TE. 2008. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics 180:1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol 25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jessulat M, Alamgir M, Salsali H, Greenblatt J, Xu J, Golshani A. 2008. Interacting proteins Rtt109 and Vps75 affect the efficiency of non-homologous end-joining in Saccharomyces cerevisiae. Arch Biochem Biophys 469:157–164. doi: 10.1016/j.abb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 13.van Attikum H, Fritsch O, Hohn B, Gasser SM. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Paull TT. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 15.Stracker TH, Usui T, Petrini JH. 2009. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom J, Cross FR. 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol 8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 17.Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JL, van Dyk D, Fedyshyn B, Luhova M, Brito GC, Vizeacoumar FJ, Vizeacoumar FS, Datti A, Kasimer D, Buzina A, Mero P, Misquitta C, Normand J, Haider M, Ketela T, Wrana JL, Rottapel R, Neel BG, Moffat J. 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraschini R, Formenti E, Lucchini G, Piatti S. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol 145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zich J, Hardwick KG. 2010. Getting down to the phosphorylated ‘nuts and bolts’ of spindle checkpoint signalling. Trends Biochem Sci 35:18–27. doi: 10.1016/j.tibs.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Kim EM, Burke DJ. 2008. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet 4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Wang H, Xu Y, Brinkman KL, Ishiyama H, Wong ST, Xu B. 2012. The kinetochore protein Bub1 participates in the DNA damage response. DNA Repair (Amst) 11:185–191. doi: 10.1016/j.dnarep.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson TE. 2002. A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics 162:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooi SL, Shoemaker DD, Boeke JD. 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 24.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. 2010. The genetic landscape of a cell. Science 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O'Donnell L, Reguly T, Breitkreutz A, Sellam A, Chen D, Chang C, Rust J, Livstone M, Oughtred R, Dolinski K, Tyers M. 2013. The BioGRID interaction database: 2013 update. Nucleic Acids Res 41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erdemir T, Bilican B, Cagatay T, Goding CR, Yavuzer U. 2002. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol Microbiol 46:947–957. doi: 10.1046/j.1365-2958.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- 29.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson C, Moynahan ME, Jasin M. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev 12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. 2013. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res 41:5784–5798. doi: 10.1093/nar/gkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babu M, Krogan NJ, Awrey DE, Emili A, Greenblatt JF. 2009. Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry. Methods Mol Biol 548:187–207. doi: 10.1007/978-1-59745-540-4_11. [DOI] [PubMed] [Google Scholar]
- 34.Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 35.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosti E, Katakowski JA, Schaetzlein S, Kim HS, Ryan CJ, Shales M, Roguev A, Krogan NJ, Palliser D, Keogh MC, Edelmann W. 2014. Evolutionarily conserved genetic interactions with budding and fission yeast MutS identify orthologous relationships in mismatch repair-deficient cancer cells. Genome Med 6:68. doi: 10.1186/s13073-014-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clikeman JA, Khalsa GJ, Barton SL, Nickoloff JA. 2001. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics 157:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Frank-Vaillant M, Marcand S. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev 15:3005–3012. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston M, Flick JS, Pexton T. 1994. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol 14:3834–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flick JS, Johnston M. 1990. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol 10:4757–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison JC, Haber JE. 2006. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 43.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW. 2006. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell 17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo K, Vega-Palas MA, Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 48.van Attikum H, Gasser SM. 2005. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol 6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 49.Kim ST, Lim DS, Canman CE, Kastan MB. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 50.Hartlerode AJ, Scully R. 2009. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sears CR, Turchi JJ. 2012. Complex cisplatin-double strand break (DSB) lesions directly impair cellular non-homologous end-joining (NHEJ) independent of downstream damage response (DDR) pathways. J Biol Chem 287:24263–24272. doi: 10.1074/jbc.M112.344911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rappold I, Iwabuchi K, Date T, Chen J. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol 153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jullien D, Vagnarelli P, Earnshaw WC, Adachi Y. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J Cell Sci 115:71–79. [DOI] [PubMed] [Google Scholar]
- 54.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. 2004. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A 101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guenole A, Srivas R, Vreeken K, Wang ZZ, Wang S, Krogan NJ, Ideker T, van Attikum H. 2013. Dissection of DNA damage responses using multiconditional genetic interaction maps. Mol Cell 49:346–358. doi: 10.1016/j.molcel.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hein J, Boichuk S, Wu J, Cheng Y, Freire R, Jat PS, Roberts TM, Gjoerup OV. 2009. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J Virol 83:117–127. doi: 10.1128/JVI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. 2011. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A 108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. 2010. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol 20:328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.