Abstract
TANK-binding kinase 1 (TBK1)-mediated induction of type I interferon (IFN) plays a critical role in host antiviral responses and immune homeostasis. The negative regulation of TBK1 activity is largely unknown. We report that suppressor of cytokine signaling 3 (SOCS3) inhibits the IFN-β signaling pathway by promoting proteasomal degradation of TBK1. Overexpression and knockdown experiments indicated that SOCS3 is a negative regulator of IFN regulatory factor 3 (IRF3) phosphorylation and IFN-β transcription. Moreover, SOCS3 directly associates with TBK1, and they colocalize in the cytoplasm. SOCS3 catalyzes K48-linked polyubiquitination of TBK1 at Lys341 and Lys344 and promotes subsequent TBK1 degradation. On the contrary, SOCS3 knockdown markedly increases the abundance of TBK1. Interestingly, both the BOX domain of SOCS3 and Ser172 phosphorylation of TBK1 are indispensable for the processes of ubiquitination and degradation. Ectopic expression of SOCS3 significantly inhibits vesicular stomatitis virus (VSV) and influenza A virus strain A/WSN/33 (WSN)-induced IRF3 phosphorylation and facilitates the replication of WSN virus by detecting the transcription of its viral RNA (vRNA). Knockdown of SOCS3 represses WSN replication. Collectively, these results demonstrate that SOCS3 acts as a negative regulator of IFN-β signal by ubiquitinating and degrading TBK1, shed light on the understanding of antiviral innate immunity, and provide a potential target for developing antiviral agents.
INTRODUCTION
Innate immunity is the first barrier to protect host cells from microbial infection, which is a great threat to health and even lives of humans. RIG-I-like receptors (RLRs), such as RIG-I and MDA5, can bind to viral RNA through its C-terminal regulatory domain (RD) and interact with the caspase activation and recruitment domain (CARD) of the mitochondrial adaptor protein MAVS by its N-terminal CARDs (1–4). Subsequently, MAVS activates downstream kinases TANK-binding kinase 1 (TBK1)/IKKε and IKKα/β, leading to the activation of transcription factors interferon regulatory factors 3 and 7 (IRF3/7) and NF-κB, respectively. As a consequence, these factors are translocated into the nucleus and initiate transcription of type I interferon (IFN) and other proinflammatory cytokines (5). IFN-α/β bind to the cell surface receptors and activate the Janus tyrosine kinase (JAK) family, which in turn provokes JAK and signal transducer and activator of transcription (JAK/STAT) signal transduction cascades. The ultimate products, IFN-stimulated genes (ISGs), antagonize the replication of various pathogens (6).
Suppressor of cytokine signaling (SOCS) is another important protein family stimulated by JAK/STAT. The SOCS proteins comprise eight members, including cytokine-inducible Src homology 2 (SH2) domain-containing protein (CIS) and SOCS1 to -7 (7). They share several domains: a central SH2 domain that participates in associations with substrates through recognition of the phosphorylated tyrosine residues; a C-terminal SOCS BOX domain that is involved in the formation of the E3 ligase complex with elongins B and C, cullin5, and Rbx-1 (8, 9); and an N-terminal extended SH2 subdomain (N-ESS) that contributes to substrate association (10). In addition, the unique kinase inhibitory regions (KIRs) of SOCS1 and SOCS3 inhibit JAKs by acting as their pseudosubstrate (11, 12). SOCS3 binds to and inhibits JAKs through its SH2 domain and N-terminal KIR (11).
In addition to the feedback inhibition of JAK/STAT signaling, it has been reported that SOCS3 can be induced via infection by several viruses (13–16) and represses innate immune responses, including NF-κB (17) and the interferon signal pathway (13, 14). For example, SOCS3 is upregulated by influenza virus (13, 14), herpes simplex virus 1 (15), and HIV (16) and blocks the expression of type I interferon, resulting in the augmentation of virus replication; SOCS3 interacts with TRAF6 and attenuates its ubiquitination, thus hindering the formation of the TRAF6/TAK1 complex and reducing the kinase activity of TAK1 (17). However, the molecular mechanism of SOCS3-mediated inhibition of IFN-β expression remains largely unknown.
TBK1 plays critical roles in RIG-I signaling. Activated TBK1 phosphorylates transcription factor IRF3 to transcribe type I interferon following viral stimulation. The activity of TBK1 is regulated in multiple ways, including phosphorylation, ubiquitination, kinase activity modulation, and complex disruption (18). It has been reported that many proteins promote or inhibit Lys63 (K63)-linked polyubiquitination of TBK1, such as Nrdp1 (19), MIB1/2 (20), CYLD (21), and A20-TAX1BP1-ABIN1 (22). On the other hand, E3 ubiquitin ligases DTX4 and TRAF-interacting protein (TRIP) also target TBK1 for K48-linked ubiquitination and degradation and attenuate IFN-β production (23, 24). However, the negative regulation of TBK1 activity is not well understood.
In this study, we elucidated the mechanism of SOCS3 in negative regulation of the IFN-β signal pathway by targeting TBK1 for proteasome-dependent degradation. SOCS3 forms a complex with TBK1 and intensifies K48-linked polyubiquitination of TBK1 at Lys341 and Lys344. Knockdown of SOCS3 significantly enhances activation of the IFN-β signal and abrogates degradation of TBK1. Moreover, the replication of influenza A virus (IAV) can be effectively augmented by SOCS3. Collectively, SOCS3 drives proteasomal degradation of TBK1 and negatively regulates antiviral innate immune responses.
MATERIALS AND METHODS
Plasmids and viruses.
Flag-TBK1 S172A was kindly provided by Hongbin Shu (Wuhan University, Wuhan, China). RIG-I, MAVS (also termed VISA), TBK1, TRAF3, IRF3, and IFN-stimulated response element (ISRE) reporter plasmids were gifts from Xin Ye at the Institute of Microbiology, Chinese Academy of Sciences (Beijing, China). Plasmids harboring genes for IKKε and TANK were kindly provided by Dongyan Jin (The University of Hong Kong, Hong Kong, China). Expression vectors for hemagglutinin (HA)-tagged wild-type Ub, K48-Ub, and K63-Ub were from Feng Shao (NIBS, China). His-tagged wild-type (WT) Ub was obtained from Eli Song at the Institute of Biophysics, Academy of Sciences in Beijing, China. TBK1 mutants (KD, KU, ΔULD, CC, K341R, K344R, K323R, K372R, K341/344R, and K38A) were generated by cloning corresponding cDNAs into a pcDNA3.0-Flag vector (Invitrogen). Similarly, SOCS3 truncations [Δ(N+KIR), ΔBOX, ΔSH2, Δ(ESS+SH2), and Δ(PEST+BOX)] were constructed by cloning the respective cDNA sequences into pcDNA4-Myc/His or pGEX4T-1 expression vectors (Invitrogen), and the Myc-SOCS3 rescue plasmid was constructed according to the sequence of SOCS3-specific small interfering RNA 1 (siRNA 1). The reverse genetic system of IAV A/WSN/33 (WSN) virus was provided by George F. Gao at Institute of Microbiology, Chinese Academy of Sciences. Vesicular stomatitis virus (VSV) was purchased from the National Institutes for Food and Drug Control.
Reagents.
The rabbit antibodies (Abs) against phospho-IRF3 (Ser396; 4947), IRF3 (4302), p-TBK1 (5483), TBK1 (3013), and SOCS3 (2923) were purchased from Cell Signaling Technology (Boston, MA). The mouse antibodies specific for Myc (sc-40), ubiquitin (sc-8017), and β-actin (47778) were from Santa Cruz Biotechnology (Santa Cruz, CA). The mouse anti-Flag (F3165) antibody was obtained from Sigma (USA). Rabbit Myc (A00172) and goat HA (A00168) antibodies were bought from Genscript (Nanjing, China). The mouse anti-His Ab was purchased from ZSGB-BIO (Beijing, China). Poly(I·C) was obtained from Sigma and used at a final concentration of 1 μg/ml for 293T cells and mouse embryonic fibroblasts (MEF). MG132 (from Santa Cruz Biotechnology) and protein G beads and glutathione S-transferase (GST) beads (GE Healthcare, USA) were obtained. A 4′,6-diamidino-2-phenylindole (DAPI) staining kit (KGA511) was bought from KeyGENBioTECH (Nanjing, China). A TNT T7-coupled reticulocyte lysate system and dual-luciferase reporter assay system were obtained from Promega (Madison, WI). An enzyme-linked immunosorbent assay (ELISA) kit was purchased from USCN Life Science (SEA222Mu).
Cell culture and transfection.
293T, HeLa, RAW 264.7, and A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, USA) at 37°C in 5% CO2. MEF were prepared from embryos at day 15 and cultured in DMEM supplemented with 10% FBS. Indicated plasmids or siRNAs were transfected into cells with Entranster-H (Engreen, Beijing, China) or Lipofectamine 2000 (Invitrogen).
Dual-luciferase reporter assays.
For dual-luciferase reporter assays, cells were transfected with plasmids carrying an IFN-β or ISRE luciferase reporter gene together with pRL-TK and other plasmids. Twenty-four hours after transfection, cells were collected and lysed. Subsequently, luciferase activity was measured with the dual-luciferase reporter assay system (Promega) according to the manufacturer's protocols. Data were normalized by the ratio of firefly luciferase activity to Renilla luciferase activity.
RNA interference (RNAi) and quantitative reverse transcription-PCR (qRT-PCR).
siRNA oligonucleotides were transfected into 293T or HeLa cells by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The sequences for SOCS3 siRNAs were si-1, 5′-GACCCAGUCUGGGACCAAGdTdT-3′; si-2, 5′-GAAGAGCCUAUUACAUCUAdTdT-3′; and si-3, 5′-CACCUGGACUCCUAUGAGA dTdT-3′. These siRNAs were all synthesized in RiboBio (Guangzhou, China).
Total RNA was extracted with TRNzol (Tiangen, Beijing, China) according to the manufacturer's protocols. cDNA was synthesized using a reverse transcription kit (Tiangen, Beijing, China). A SYBR RT-PCR kit (TaKaRa, Dalian, China) was used for quantitative real-time PCR assays. The primers used for RT-PCR were as follows: h-SOCS3 forward, 5′-AGCAGATGGAGGGTTCTGCTTTGT-3′, and reverse, 5′-ATTGGCTGTGTTTGGCTCCTTGTG-3′; h-IFN-β forward, 5′-GTCAGAGTGGAAATCCTAAG-3′, and reverse, 5′-ACAGCATCTGCTGGTTGAAG-3′; h-GAPDH forward, 5′-GGAGAAACCTGCCAAGTATG-3′, and reverse, 5′-TTACTCCTTGGAGGCCATGTAG-3′; NA forward, 5′-ATTCAAGGGGGACCTTTAAGGACAG-3′, and reverse, 5′-CTGACCAAGCAACCGATTCAA ACCT-3′; NP forward, 5′-GATCTGGCACTCCAATTTGAATGAT-3′, and reverse, 5′-CTAGGGAGGGTTGAA CCCTGCATCA-3′; PA forward, 5′-AGAGGACCTGAAAATCGAAACAAAC-3′, and reverse, 5′-TATTGACTCGCCTTGCTCATCGATG-3′; M1 forward, 5′-ACAGAGACTTGAAGATGTCTTTGCA-3′, and reverse, 5′-CTAAAATCCCCTTAGTCAGAGGTGA-3′; 18SrRNA forward, 5′-GTAACCCGTTGAACCCCATT-3′, and reverse, 5′-CCATCCAATCGGTAGTAGCG-3′.
Immunoprecipitation and immunoblotting.
Cells were collected and lysed with 1× lysis buffer (Cell Signaling Technology), and then lysates were incubated with appropriate antibodies and protein G beads (Santa Cruz Biotechnology) at 4°C overnight. The beads were washed three times with immunoprecipitation (IP) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 1% Nonidet P-40), followed by immunoblotting (IB) analysis. For the immunoblotting, cells were harvested and lysed with 1× RIPA buffer (Cell Signaling Technology), and then samples were boiled for 5 min together with 2× loading buffer for SDS-PAGE. Proteins were then transferred onto nitrocellulose membranes (Bio-Rad). The membrane was sealed with 5% fat-free milk in Tris-buffered saline with Tween (TBS-T) for 2 h at room temperature and incubated with the appropriate primary antibody at 4°C overnight. Then, the membrane was washed three times with TBS-T and incubated with a horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. After washing three times with TBS-T, the membrane was flushed with enhanced chemiluminescence (ECL) reagent (Applygen, Beijing, China). The bands were detected via X-ray film (Fuji, Japan) exposed to developer and fixing solutions.
Ubiquitination assays.
For ubiquitination assays, cells were transfected with the indicated plasmids, followed by collecting and lysing cells using 1× cell lysis buffer (Cell Signaling Technology). Then, cell supernatant was incubated with protein G beads and appropriate antibodies at 4°C overnight. Beads were washed three times with IP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 1% Nonidet P-40). The samples were boiled for 5 min together with 2× loading buffer and then SDS-PAGE was performed, followed by transfer onto nitrocellulose membranes, which were sealed with 5% milk and incubated with primary and secondary antibodies. The final results were obtained via ECL (Applygen, Beijing, China), and film was exposed to developer and fixing solution.
GST pulldown assay.
GST and GST-fused proteins were expressed and purified from Escherichia coli strain BL21 Star. The indicated plasmids were transfected into 293T cells for 24 h and then collected, and cells were lysed by using lysis buffer. The lysates were incubated with prepared GST or GST-fused proteins, including GST-fused SOCS3 protein and its mutant proteins, at 4°C overnight. The beads were washed three times with phosphate-buffered saline (PBS) and further boiled for 5 min with loading buffer. Prepared samples were analyzed by IB.
In vitro translation assays.
Flag-tagged TBK1 truncations (KU and CC) were translated in vitro with a TNT T7-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. The in vitro-translated products were incubated with GST or GST-fused SOCS3 protein at 4°C overnight. Then, beads were washed three times and boiled to be used for IB.
Immunofluorescence microscopy.
For immunofluorescence microscopy, cells were transfected with Flag-TBK1 and Myc-SOCS3 plasmids for 24 h. Following elution, fixing, and blocking with 5% bovine serum albumin (BSA), cells were incubated with primary antibodies (mouse anti-Flag and rabbit anti-Myc Abs) and secondary antibodies (fluorescein isothiocyanate-conjugated anti-mouse IgG Ab and tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG Ab). The nuclei were stained with DAPI (KeyGENBioTECH). The final result was observed by using laser confocal fluorescence microscopy (Leica TCS SP2, Germany).
Generation and infection of viruses.
Generation of IAV was performed as previously described (25). A series of vRNA expression plasmids were first transfected into 293T cells for 3 days, and then the supernatants were harvested and used to propagate IAV in MDCK cells.
For the antivirus assays, two kinds of viruses, including IAV and VSV, were used to infect 293T cells or A549 cells. First of all, the cells were washed three times with PBS, followed by infection with the virus (diluted in DMEM containing 2.5 μg/ml trypsin) for 1 h. Then, the cells were washed three times with PBS and cultured in DMEM containing 2.5 μg/ml trypsin for the indicated times. Finally, the cells were collected and used to perform IB analysis or qRT-PCR.
ELISA.
RAW 264.7 or primary MEF cells were stimulated by poly(I·C) at a final concentration of 10 μg/ml or 1 μg/ml, respectively. Then, the culture supernatants were harvested and used to measure the concentrations of IFN-β according to the manufacturer's instructions.
Statistical analysis.
All experiments were performed for more than three replicates, and a two-tailed Student t test with a P value of <0.05 was used to identify the significance of the data.
RESULTS
SOCS3 negatively regulates the IFN-β signaling pathway.
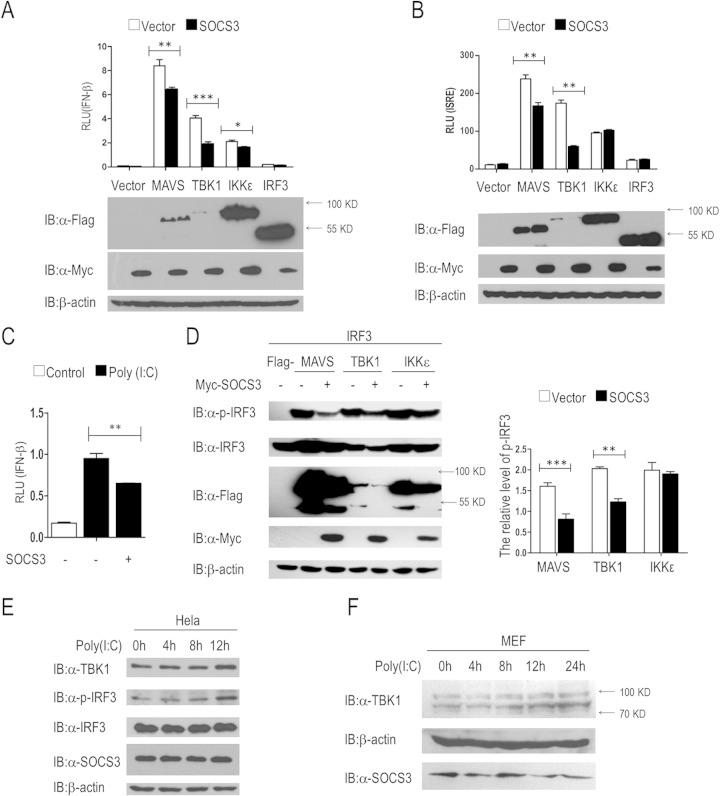
We performed dual-luciferase reporter screening to identify candidate regulators of the RIG-I signal pathway (26), and the results revealed that SOCS3 is an inhibitor of this signal axis (data not shown). In order to further verify the regulation, a dual-luciferase reporter assay was performed by transfecting HEK293T human embryonic kidney cells (293T cells) with plasmids carrying the IFN-β promoter luciferase reporter, an internal control Renilla luciferase, and a RIG-I signaling adaptor (MAVS, TBK1, IKKε, or IRF3) in the absence or presence of the SOCS3 expression vector. The data showed that SOCS3 overexpression significantly attenuated the activation of the IFN-β promoter induced by MAVS, TBK1, and IKKε but not that of IRF3 (Fig. 1A). These results suggested that SOCS3 acts upstream of IRF3, and TBK1 is a candidate target. To evaluate whether SOCS3 directly represses the IFN-β promoter, we detected the effects of this protein on TBK1-induced activation of an ISRE luciferase reporter. The results suggested that MAVS or TBK1, but not IKKε or IRF3, mediated ISRE activation that was markedly reduced by SOCS3 overexpression (Fig. 1B). In addition, we found that SOCS3 potently lowered poly(I·C)-stimulated IFN-β expression in 293T cells (Fig. 1C).
FIG 1.
SOCS3 inhibits IFN-β signaling pathway. (A to C) Luciferase assays revealed SOCS3-mediated suppression of RIG-I signaling. 293T cells were transfected with plasmids harboring IFN-β promoter genes or with the ISRE promoter together with the indicated plasmids (A and B), with or without poly(I·C) cotransfection (1 μg/ml) (C). At 24 h after transfection, the cells were lysed and the luciferase activities were monitored. (D) SOCS3 negatively regulates induced IRF3 phosphorylation. 293T cells were transfected with Flag-tagged MAVS, TBK1, or IKKε vectors and IRF3 plasmid (50 ng/ml) with or without a Myc-SOCS3 expression vector. Immunoblotting was used to analyze the levels of IRF3 phosphorylation and the expression of total IRF3. The intensity of p-IRF3 in Western blotting bands was measured with ImageJ software. (E and F) Immunoblot analysis in HeLa cells or primary MEF cells after poly(I·C) treatment at a final concentration of 1 μg/ml. α, antibody. Error bars indicate standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To reveal the mechanism of SOCS3-mediated inhibition of the IFN-β promoter, we assessed the phosphorylation of IRF3. We transfected a SOCS3 expression vector together with MAVS, TBK1, or IKKε plasmid into 293T cells and found that SOCS3 substantially attenuated the IRF3 phosphorylation induced by MAVS and TBK1 but not that by IKKε (Fig. 1D). These data suggested that SOCS3 negatively regulates the type I interferon signal pathway upstream of IRF3.
To evaluate endogenous p-IRF3 and TBK1 under physiologic conditions, poly(I·C) was used to stimulate HeLa cells and primary MEF cells for different times, respectively. Within 12 h, TBK1 and IRF3 phosphorylation expression levels were all upregulated in a time-dependent manner after poly(I·C) stimulation (Fig. 1E). A similar result was obtained in primary MEF cells (Fig. 1F).
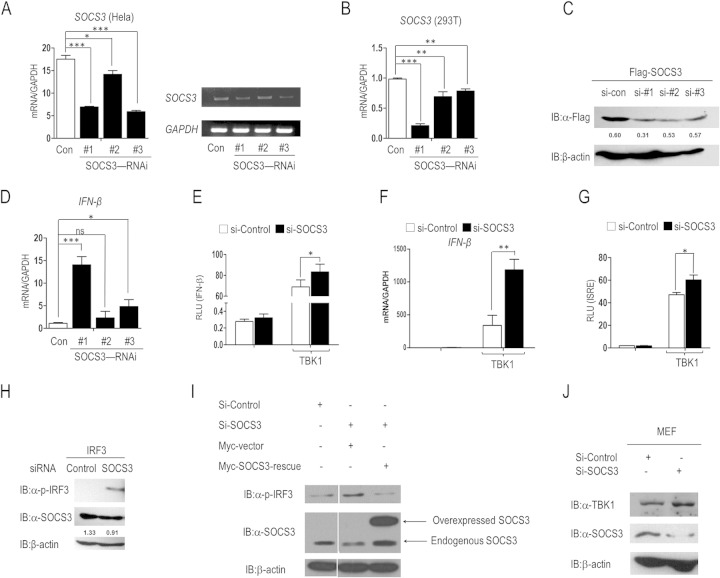
Knockdown of SOCS3 heightens IFN-β transcription.
We next sought to demonstrate whether knockdown of SOCS3 could enhance IFN-β response. Three SOCS3-specific siRNAs (named si-1, si-2, and si-3) were used to verify the hypothesis. We first evaluated the knockdown efficiency of these siRNAs at the mRNA level. As shown in Fig. 2A and B, si-1 and si-3 significantly reduced the abundance of endogenous SOCS3 mRNA in HeLa cells, whereas si-1 markedly reduced SOCS3 expression in 293T cells. In addition, we assessed the effects of these siRNAs by Western blotting. We cotransfected a Flag-SOCS3 plasmid with an siRNA control or SOCS3-specific siRNAs into 293T cells, and the results confirmed the knockdown (Fig. 2C). Furthermore, transfection of SOCS3 targeting siRNA resulted in higher IFN-β production at the mRNA level (Fig. 2D). Collectively, si-1 had the best knockdown efficiency in both HeLa and 293T cells (Fig. 2A, B, and C) and resulted in the highest IFN-β expression (Fig. 2D). This siRNA was used in the following experiments.
FIG 2.
Knockdown of SOCS3 activates the IFN-β signaling pathway. (A) qRT-PCR (left) and RT-PCR (right) analysis of SOCS3 knockdown efficiency at the mRNA level in HeLa cells. (B) qRT-PCR analysis of SOCS3 knockdown efficiency at the mRNA level in 293T cells. (C) Immunoblot analysis of the knockdown of exogenous SOCS3 in 293T cells transfected with Flag-tagged SOCS3 plasmid and siRNA control or SOCS3-specific siRNAs. (D) Real-time PCR analysis of IFN-β mRNA in 293T cells transfected with control siRNA or siRNA against SOCS3. (E) 293T cells were transfected with siRNA control (si-control) or si-1 for 36 h and further transfected with plasmids harboring genes for the IFN-β promoter and TK reporter genes, with or without a TBK1 expression vector. The activity of the IFN-β promoter was analyzed in dual-luciferase assays. (F) HeLa cells were transfected with si-control or si-1 and further transfected with or without TBK1 plasmid. The transcription of IFN-β mRNA was evaluated by real-time PCR. (G) Thirty-six hours after transfection with siRNAs, 293T cells were transfected with the indicated plasmids. Luciferase assays were performed to evaluate the activity of the ISRE reporter. (H) Immunoblot analysis of IRF3 phosphorylation in 293T cells transfected with IRF3 plasmid (50 ng/ml) and si-control or si-1. (I) Immunoblot analysis of IRF3 phosphorylation in 293T cells transfected with the indicated siRNA or SOCS3 rescue plasmid. Overexpressed and endogenous SOCS3 is indicated with an arrow. (J) Primary MEF cells were transfected with si-control or si-1. The expression levels of TBK1 and SOCS3 were evaluated by IB analysis. Error bars indicate standard deviations. *, P < 0.05; ** P < 0.01; ***, P < 0.001; ns, nonsignificant.
Subsequently, we detected the effects of SOCS3 knockdown on RIG-I signaling. As shown in Fig. 2E and F, transfection of SOCS3 siRNA markedly increased TBK1-induced activation of the IFN-β promoter and IFN-β transcription. Consistent with these observations, TBK1-mediated ISRE reporter activation was also upregulated in cells transfected with SOCS3 siRNA (Fig. 2G). Furthermore, knockdown of SOCS3 considerably enhanced the abundance of IRF3 phosphorylation (Fig. 2H). These results were consistent with those for SOCS3 overexpression.
To exclude off-target effects and validate the RNAi data, we included a rescue control. As shown in Fig. 2I, the IRF3 phosphorylation level was significantly upregulated by SOCS3 knockdown and further repressed by cotransfection of the SOCS3 rescue plasmid.
In addition, we checked the effect of SOCS3 knockdown in primary cells. MEF cells were isolated from embryos of BALB/c mice at day 15 and were cultured in DMEM supplemented with 10% FBS. We transfected prepared MEF cells with control siRNA or SOCS3-specific siRNA. The result suggested that SOCS3 knockdown significantly increased the abundance of TBK1 (Fig. 2J). These data further confirmed that SOCS3 is a negative regulator of type I interferon signal transduction.
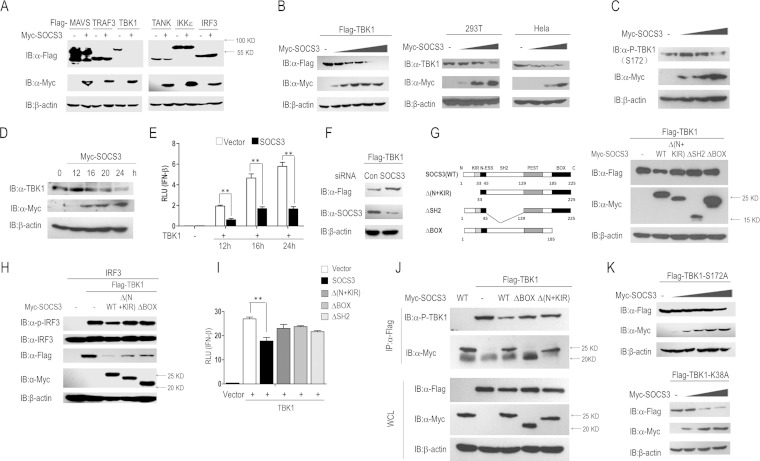
SOCS3 targets TBK1 for degradation.
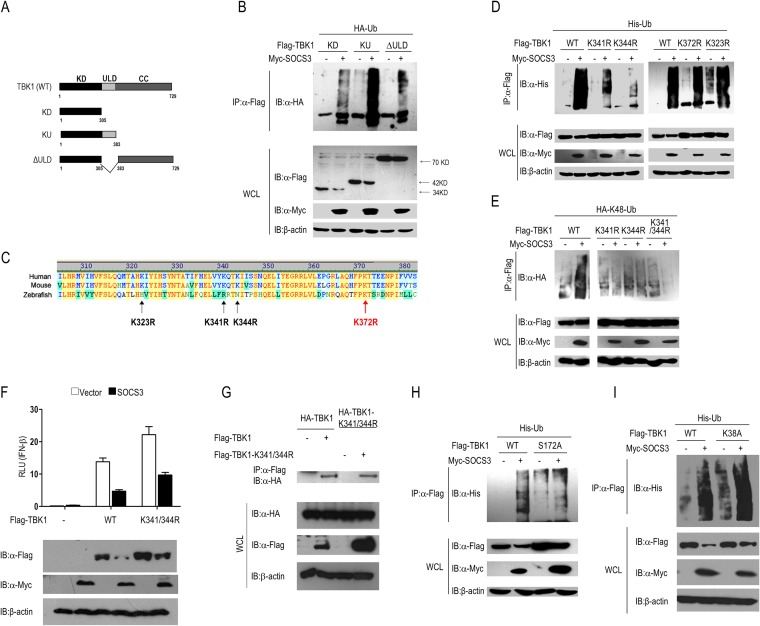
We observed that SOCS3 overexpression reduces the abundance of TBK1 at the protein level (Fig. 1A, B, and D); the data from the dual-luciferase reporter assay (Fig. 1A and B) and IRF3 phosphorylation assay (Fig. 1D) suggested that SOCS3 acts upstream of IRF3 and downstream of TBK1, and so we doubt whether TBK1 is a target of SOCS3. We transfected a SOCS3 expression vector together with a plasmid carrying the gene for a RIG-I signal adaptor (MAVS, TRAF3, TBK1, TANK, IKKε, or IRF3) into 293T cells. The results indicated that TBK1, but not other adaptors, was considerably decreased by SOCS3 ectopic expression (Fig. 3A). Subsequently, we found that SOCS3 overexpression induced degradation of ectopic and endogenous TBK1 in a dose-dependent manner (Fig. 3B). Besides, the phosphorylation level of endogenous TBK1 was significantly inhibited by SOCS3 overexpression (Fig. 3C). In addition, we performed the time course experiments of SOCS3-mediated degradation of TBK1 and the inhibition of IFN-β activity. As shown in Fig. 3D and E, endogenous TBK1 degradation induced by SOCS3 overexpression started at 16 h after transfection, whereas SOCS3-mediated inhibition of TBK1-stimulated IFN-β transcription started at 12 h. Furthermore, knockdown of SOCS3 increased the level of ectopic expressed TBK1 protein (Fig. 3F). All these data demonstrated that the stability of TBK1 is regulated by SOCS3.
FIG 3.
SOCS3 targets TBK1 for degradation. (A) Plasmids carrying genes for adaptors of the RIG-I signal were separately transfected into 293T cells with or without Myc-SOCS3 cotransfection. IB was performed to identify the abundance of these adaptors (Flag-tagged MAVS, TRAF3, TBK1, TANK, IKKε, and IRF3). (B and C) 293T or HeLa cells were transfected with the indicated plasmids. The expression levels of TBK1 and p-TBK1 (S172) were evaluated by Western blotting. (D and E) The time courses of SOCS3-mediated degradation of TBK1 and inhibition of IFN-β activity were evaluated by Western blotting and a dual-luciferase assay in 293T cells, respectively. (F) 293T cells were transfected with control siRNA (si-control) or si-1 (SOCS3-specific siRNA) by using Lipofectamine 2000 for 36 h, followed by the transfection with Flag-TBK1. IB analysis showed the expression of TBK1 and SOCS3 proteins. (G) Constructs of Myc-tagged wild-type SOCS3 and its truncations (left). Flag-tagged TBK1 and wild-type or mutated Myc-SOCS3 were transfected into 293T cells for 24 h, and then IB for TBK1 expression levels (right). (H) 293T cells were transfected with IRF3 (50 ng/ml), Flag-TBK1, and Myc-SOCS3 wild type or truncations for 24 h, and IRF3 phosphorylation and total IRF3 were analyzed by IB. (I) A dual-luciferase assay was performed to identify the activation of the IFN-β reporter in 293T cells transfected with the indicated plasmids. (J) 293T cells were transfected with the indicated plasmids for 24 h. Cells lysates were incubated with protein G beads plus anti-Flag Ab at 4°C overnight, followed by immunoblot analysis with anti-p-TBK1 Ab. (K) 293T cells were transfected with increasing amounts of SOCS3 plasmid and Flag-TBK1-S172A mutant or Flag-TBK1-K38A mutant for 24 h. The expression of the two mutants was evaluated by Western blotting. Error bars indicate standard deviations. **, P < 0.01.
To further evaluate which domain of SOCS3 is responsible for TBK1 degradation, we generated a series of SOCS3 mutants, including Δ(N+KIR), ΔSH2, and ΔBOX (Fig. 3G). Overexpression of the wild-type SOCS3 but not the Δ(N+KIR), ΔBOX, or ΔSH2 mutated forms of SOCS3 reduced the level of ectopically expressed Flag-TBK1 (Fig. 3G). In another assay, compared with wild-type SOCS3, truncations such as Δ(N+KIR) or ΔBOX restored the expression of TBK1 as well as TBK1-induced phosphorylation of IRF3 in 293T cells (Fig. 3H). Interestingly, the dual-luciferase assay showed that these SOCS3 mutants failed to repress TBK1-induced activation of the IFN-β promoter (Fig. 3I). Meanwhile, we also checked the effect of wild-type SOCS3 or its mutants on TBK1 phosphorylation by IP and subsequent Western blotting assays. 293T cells were transfected with the indicated plasmids (Fig. 3J), and TBK1 protein was purified by IP with anti-Flag antibody and evaluated in an IB assay with anti-p-TBK1. The results suggested that TBK1 phosphorylation was significantly reduced by wild-type SOCS3 but not its mutants [ΔBOX and Δ(N+KIR)] (Fig. 3J). This result was consistent with the result that wild-type SOCS3 but not its mutants induced the degradation of TBK1 (Fig. 3G).
Interestingly, SOCS3 does not have obvious effects on the stability of TBK1-S172A, a mutated form of TBK1 when the autophosphorylation site serine 172, which is critical for virus-stimulated IFN-β induction and IRF3 activation (28), is mutated to alanine (Fig. 3K). For comparison, SOCS3 promoted the degradation of the TBK1 K38A mutant, a kinase-inactive mutant in which the ATP-binding residue Lys38 was mutated to alanine (Fig. 3K). It is reasonable to conclude that the process of SOCS3 reducing the abundance of TBK1 is dependent on TBK1 serine 172 but not lysine 38. It has been reported that some kinases, such as glycogen synthase kinase 3β (GSK3β), promote TBK1 self-association and autophosphorylation at Ser172 (28), which may participate in the process of SOCS3-mediated TBK1 degradation (29).
Taken together, SOCS3 negatively regulates IFN-β signal transduction cascades by targeting TBK1 for degradation, and all fragments detected (N+KIR, SH2, and BOX) are indispensable for this process.
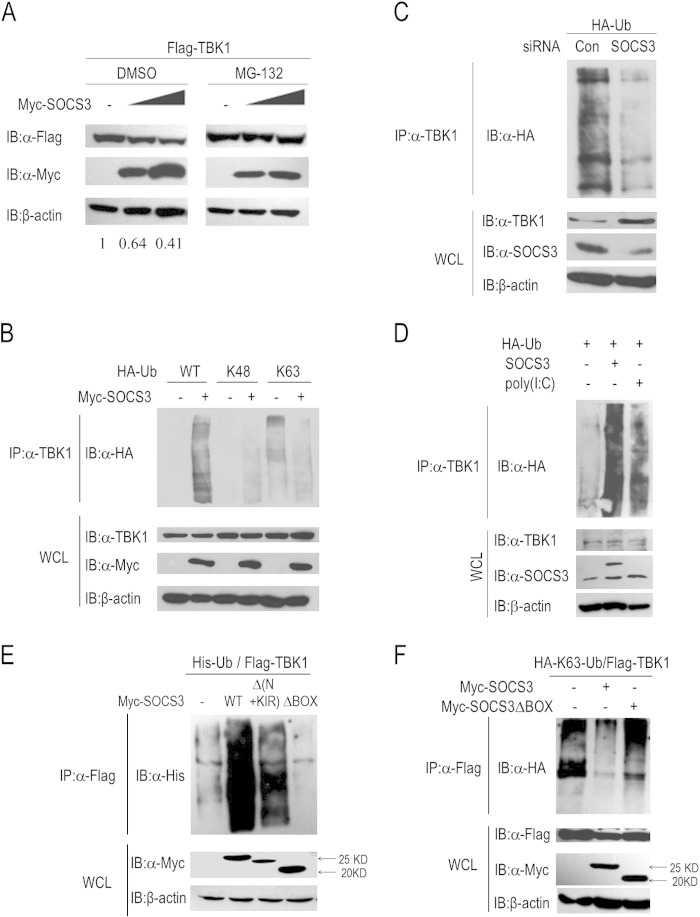
SOCS3 enhances K48-linked polyubiquitination of TBK1.
Several studies have suggested that SOCS3 functions as an E3 ligase by forming complexes with elongins B and C, cullin5, and Rbx-1 in many physiological processes (8, 9). Intriguingly, the accumulation and function of TBK1 is also regulated by ubiquitination, including K48-linked and K63-linked types. In view of the facts, we determined whether SOCS3 lowered the abundance of TBK1 protein by the ubiquitin-proteasome pathway. Initially, we evaluated whether SOCS3-induced TBK1 degradation occurs via the proteasomal pathway. We transfected 293T cells with the indicated plasmids and treated them with dimethyl sulfoxide (DMSO) or MG132 for 6 h. Immunoblot analysis suggested that loss of TBK1 protein induced by SOCS3 was blocked by the proteasome inhibitor MG132 (Fig. 4A). Furthermore, ubiquitination of endogenous TBK1 was enhanced in 293T cells with coexpression of SOCS3 (Fig. 4B, left two panels). To determine the specific ubiquitination type of TBK1, we further included two ubiquitin mutants, K48 and K63, in the experiments, in which all lysine residues were replaced by arginine residues except for at site 48 or 63, respectively. The data suggested that SOCS3 promotes more K48-linked ubiquitination of TBK1 whereas it attenuates its K63-linked ubiquitination (Fig. 4B). As K48-linked polyubiquitination is accompanied by proteasomal degradation of target proteins, this phenomenon is consistent with the result of SOCS3-mediated TBK1 degradation. Besides, we included TBK1 ubiquitination assays after knockdown of SOCS3 to validate the result shown in Fig. 4B. As shown in Fig. 4C, knockdown of SOCS3 significantly attenuated TBK1 ubiquitination. In addition, the endogenous TBK1 ubiquitination level was tested with different treatments. We found that SOCS3 overexpression or poly(I·C) stimulation dramatically promoted the endogenous ubiquitination of TBK1 (Fig. 4D).
FIG 4.
SOCS3 increases K48-linked ubiquitination of TBK1. (A) Immunoblot analysis of TBK1 expression in 293T cells with DMSO or MG132 treatment for 6 h after transfection for 24 h. (B) 293T cells were transfected with plasmids harboring genes for wild-type or mutated HA-Ub with or without Myc-SOCS3 cotransfection for 24 h. Cell lysates were incubated with protein G beads plus anti-TBK1 Ab at 4°C overnight, followed by immunoblot analysis with anti-HA Ab. (C) 293T cells were transfected with control siRNA (si-control) or si-1 for 24 h, followed by further transfection with HA-Ub plasmids for 24 h. Cell lysates were used to perform ubiquitination assays. (D) Ubiquitination assays of endogenous TBK1. 293T cells were transfected with HA-Ub and SOCS3 plasmid or poly(I·C), and then the cell lysates were incubated with protein G beads plus anti-TBK1 Ab at 4°C overnight, followed by immunoblot analysis with anti-HA Ab. (E) 293T cells were transfected with plasmids harboring genes for Flag-TBK1, His-Ub, and Myc-tagged wild-type or truncated SOCS3 for 24 h, followed by immunoprecipitation with protein G beads plus anti-Flag Ab and immunoblot analysis with anti-His Ab. (F) 293T cells were transfected with Flag-TBK1, HA-Ub(K63), or Myc-SOCS3 wild type or ΔBOX for 24 h, and then lysates were analyzed in ubiquitination assays.
In addition, we evaluated domains of SOCS3 that play crucial roles in the process of ubiquitination and degradation of TBK1. Compared to wild-type SOCS3, the ubiquitination level of TBK1 was considerably diminished by the ΔBOX truncation but not the mutant with the N-terminal residues and the KIR domain deleted (Fig. 4E). These results indicated that the BOX domain of SOCS3 participates in the process of TBK1 ubiquitination and degradation. Finally, we found that the BOX domain of SOCS3 is indispensable for SOCS3 to attenuate K63-linked ubiquitination of TBK1, which is important for its activity, and the mechanism remains to be revealed (Fig. 4F). Therefore, SOCS3 reduces the abundance of TBK1 by enhancing its K48-linked ubiquitination.
K341 and K344 of TBK1 are pivotal sites for ubiquitination.
To determine the sites responsible for SOCS3-induced polyubiquitination, we constructed three truncations of TBK1 (Fig. 5A), including KD (kinase domain), KU (lacking the coiled-coil domain), and ΔULD (lacking the ubiquitin-like domain). Subsequently, these three mutants were separately transfected into 293T cells together with an HA-tagged ubiquitin vector in the absence or presence of SOCS3 plasmid. Compared to KU and ΔULD, the KU truncation led to the largest amount of polyubiquitin upon SOCS3 overexpression (Fig. 5B), which indicated that the ubiquitin-like domain (ULD) contains potential ubiquitination sites. In this region, we identified four lysine residues (K323, K341, K344, and K372) which are conserved in human and mouse but not in zebrafish (Fig. 5C). We mutated these sites (K323R, K341R, K344R, and K372R) separately and transfected each of them or the wild-type TBK1 into 293T cells with His-Ub, accompanied or not by Myc-SOCS3 cotransfection. Compared to wild-type TBK1, mutation of K341 or K344 but not K323 or K372 significantly decreased SOCS3-induced TBK1 ubiquitination (Fig. 5D). Consistent with this observation, there was no obvious SOCS3-induced K48-linked polyubiquitination of mutants with K341R, K344R, or K341/344R (lysine residues of both sites replaced by arginine) (Fig. 5E). These results demonstrated that Lys341 and Lys344 in TBK1 are two major sites responsible for SOCS3-stimulated ubiquitination. However, the double mutant of TBK1 failed to rescue IFN-β activity inhibited by SOCS3, which indicated that another mechanism may be also involved in SOCS3-mediated TBK1 degradation (Fig. 5F). Meanwhile, co-IP experiments with Flag-tagged and HA-tagged wild-type or the double mutant TBK1 showed that the K341/344R mutation does not interfere in the process of TBK1 self-association (Fig. 5G).
FIG 5.
Identification of ubiquitination sites on TBK1. (A) Constructs of wild-type TBK1 and its truncations. (B) 293T cells were transfected with plasmids carrying genes for HA-Ub and TBK1 truncations with or without Myc-SOCS3 cotransfection, and then cell extracts were used to perform ubiquitination assays. (C) Identification of four lysines in the ULD domain of TBK1. (D) 293T cells were transfected with plasmids harboring genes for His-Ub and TBK1 mutations (K341R, K344R, K372R, or K323R) with or without Myc-SOCS3 cotransfection, and then cell extracts were used to perform ubiquitination assays. (E) 293T cells were transfected with plasmids carrying genes for HA-Ub (K48) and TBK1 mutations (K341R, K344R, or K341/344R) with or without Myc-SOCS3 cotransfection, and then extracts were used to perform ubiquitination assays. (F) Dual-luciferase assay results for SOCS3-mediated suppression of IFN-β activation induced by wild-type or the K341/344R mutant of TBK1 and Western blot assay results for the expression of these proteins. (G) Coimmunoprecipitation results, to check TBK1 dimerization in 293T cells transfected with different tagged TBK1 wild-type or K341/344R mutant plasmids. (H and I) Lysates of 293T cells transfected with His-Ub and Flag-TBK1, Flag-TBK1-S172A, or Flag-TBK1-K38A, with or without Myc-SOCS3 cotransfection were analyzed in ubiquitination assays.
It has been suggested that SOCS3 fails to degrade TBK1-S172A but not TBK1-K38A (Fig. 3J), so we further investigated whether SOCS3 is responsible for their ubiquitination. SOCS3 significantly increased polyubiquitination of the wild type or K38A mutant but not the S172A mutant of TBK1 in 293T cells (Fig. 5H and I), indicating that SOCS3-mediated TBK1 ubiquitination and degradation require serine 172 but not lysine 38 of TBK1.
Therefore, K341 and K344 of TBK1 are pivotal sites for ubiquitination induced by SOCS3, and the process is dependent on TBK1 serine 172 but not lysine 38.
SOCS3 forms a complex with TBK1.
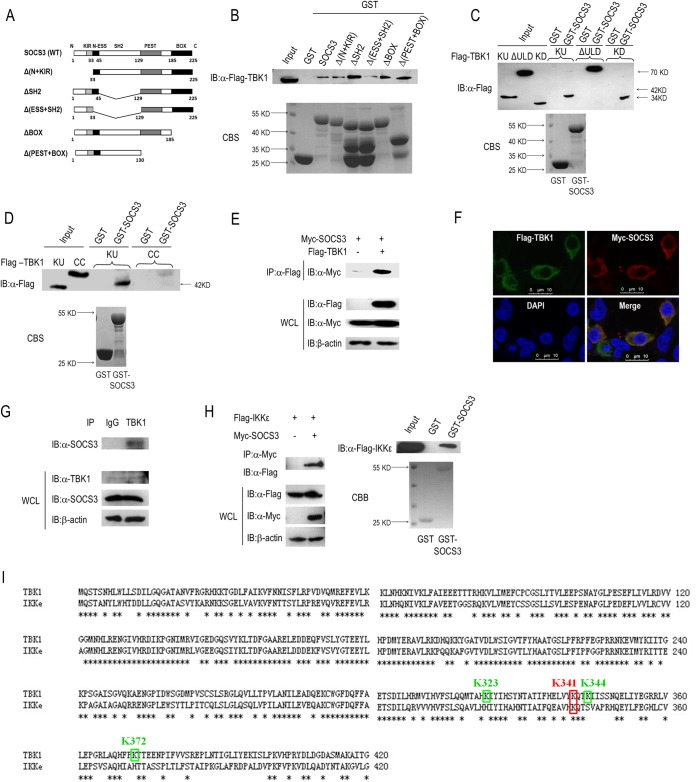
Our studies suggested that SOCS3 negatively regulates IFN-β activation by enhancing K48-linked ubiquitination and degradation of TBK1. Then, we asked whether SOCS3 could directly interact with TBK1. GST-fused SOCS3 protein was expressed and purified from E. coli and incubated with lysates from 293T cells transfected with Flag-tagged TBK1 expression vector. An IB assay suggested that SOCS3 directly associates with TBK1 (Fig. 6B). To determine regions of SOCS3 that participate in the interaction with TBK1, we expressed and purified GST-fused SOCS3 truncations (termed Δ(N+KIR), ΔSH2, Δ(ESS+SH2), ΔBOX, and Δ(PEST+BOX)) from E. coli (Fig. 6A and B). Then, these GST-fused proteins were incubated with lysates from Flag-TBK1-overexpressing 293T cells. The results of the GST pulldown assay showed that there was less association between TBK1 and the Δ(ESS+SH2) mutant of SOCS3 among all the interactions (Fig. 6B), indicating that the N-terminal ESS domain is responsible for SOCS3 forming a protein complex with TBK1.
FIG 6.
SOCS3 and TBK1 form complexes in cells. (A) Constructs of GST-tagged wild-type SOCS3 and its truncations. (B) GST or GST-fused proteins were incubated with lysates of 293T cells transfected with Flag-tagged TBK1 at 4°C overnight. Immunoblotting was used to analyze the association between TBK1 and wild-type or mutated SOCS3. (C) GST or GST-fused SOCS3 protein was incubated with lysates of 293T cells transfected with Flag-tagged TBK1 truncations (KU, ΔULD, and KD) at 4°C overnight. Immunoblotting was used to analyze the associations between SOCS3 and TBK1 mutants. (D) GST or GST-fused SOCS3 protein was incubated with in vitro-translated KU or CC protein produced via a reticulocyte lysate system, and the interactions between SOCS3 and TBK1 mutants were identified via IB. (E) Coimmunoprecipitation was used to evaluate the association between SOCS3 and TBK1 in 293T cells transfected with Myc-SOCS3 and Flag-3.0 or Flag-TBK1. (F) Immunofluorescence microscopy detected the colocalization of Flag-TBK1 and Myc-SOCS3 in 293T cells. (G) Results of co-IP assays of endogenous interactions between SOCS3 and TBK1 with rabbit IgG or TBK1 antibody, followed by IB analysis with anti-SOCS3 Ab. (H) Results of co-IP and GST pulldown analyses of the association between SOCS3 and IKKε. (I) Identification of ubiquitinated lysine residues between TBK1 and IKKε. Bars, 10 μm. CBS, Coomassie blue stain.
Reciprocally, we performed a GST pulldown assay to determine the regions of TBK1 required for its interaction with SOCS3. We incubated GST-fused SOCS3 protein with lysates from 293T cells transfected with Flag-tagged TBK1 truncations (KD, KU, and ΔULD). All these TBK1 mutants harbor the kinase domain, and Western blotting revealed that all of them interact with SOCS3 (Fig. 6C). These results suggested the kinase domain of TBK1 is sufficient for the interaction. In addition, we incubated GST-fused SOCS3 protein with products from in vitro-translated KU or CC proteins by using a reticulocyte lysate system (Fig. 6D). The results showed that SOCS3 interacts with KU but not CC (Fig. 6D), suggesting that the CC domain does not participate in the formation of the SOCS3-TBK1 complex. Theoretically, the TBK1-KU protein (containing 383 amino acids) is larger than the TBK1-CC protein (containing 346 amino acids). In fact, we detected that the TBK1-CC protein has a higher molecular weight than the TBK1-KU protein (Fig. 6D), which is consistent with the report of Cui et al. (23). We also performed co-IP in 293T cells transfected with Myc-SOCS3 and Flag-3.0 or Flag-TBK1, which further indicated that SOCS3 directly associates with TBK1 (Fig. 6E). To check the endogenous association between TBK1 and SOCS3, we also performed co-IP in 293T cells with IgG or TBK1 antibody. IB analysis suggested that SOCS3 forms complex with TBK1 under physiological conditions (Fig. 6G). Finally, we performed an immunofluorescence microscope assay and confirmed the colocalization of SOCS3 and TBK1 in the cytoplasm (Fig. 6F). Taken together, SOCS3 directly binds to TBK1 to promote ubiquitination and degradation of this protein, and the N-terminal ESS domain of SOCS3 and the kinase domain of TBK1 are indispensable for their physical interaction.
Though IKKε-induced IFN-β activity, ISRE activity, and IRF3 phosphorylation were less obviously inhibited by ectopic expression of SOCS3 than that induced by TBK1 (Fig. 1A, B and D), we also checked whether SOCS3 interacts with IKKε, which is structurally similar to TBK1 (another IKK-related kinase). Co-IP and GST pulldown assays showed that SOCS3 associated with IKKε (Fig. 6H). In addition, we determined whether the ubiquitinated lysine residues were conserved between TBK1 and IKKε. As shown in Fig. 6I, the lysine residue 341 of TBK1 is not only involved in the process of SOCS3-mediated TBK1 polyubiquitination (Fig. 5D and E) but also conserved between the two kinases.
SOCS3 facilitates viral replication.
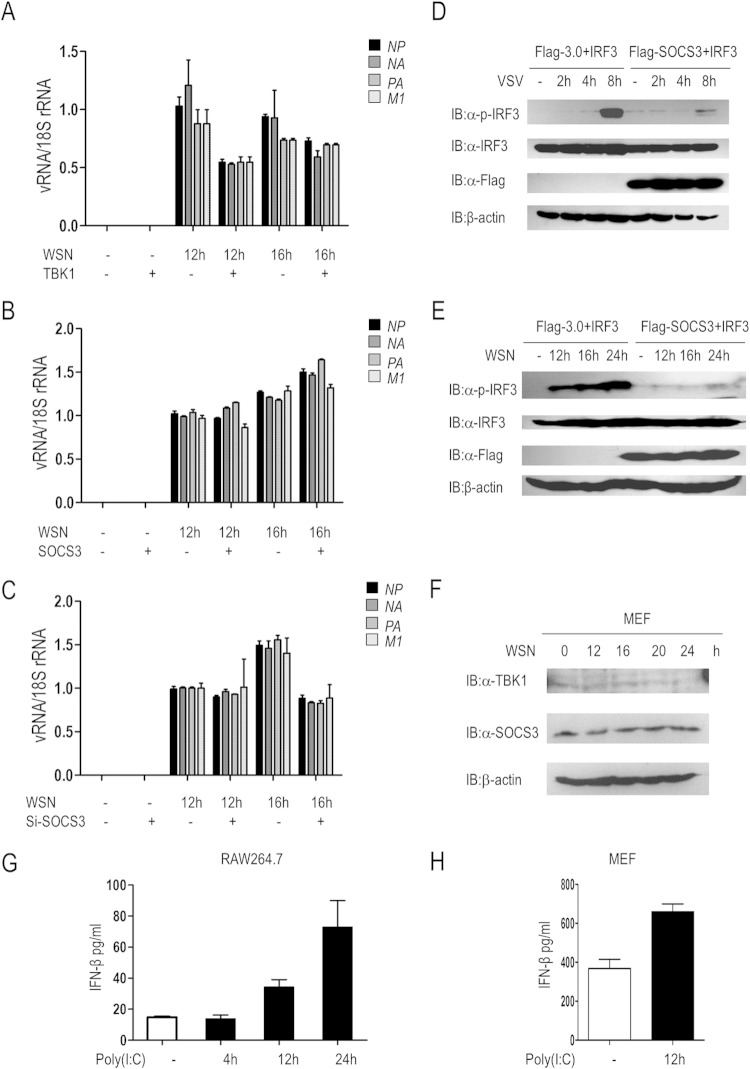
Type I interferon is an important barrier to defend against viral infection, and so we further investigated the effects of SOCS3 on viral replication. Cells were transfected with TBK1 or SOCS3 for 24 h, followed by WSN stimulation for 0 h, 12 h, or 16 h. Subsequently, qRT-PCR was performed to detect the abundance of vRNAs (NP, NA, PA, and M1) in cells. The results showed that the transcription of these vRNAs was significantly inhibited by TBK1 but increased by SOCS3 (Fig. 7A and B). Conversely, knockdown of SOCS3 obviously reduced their abundance (Fig. 7C).
FIG 7.
SOCS3 facilitates viral replication. (A) 293T cells were transfected with or without TBK1 for 24 h, followed by WSN infection for 0 h, 12 h, or 16 h. The transcription of vRNAs (NP, NA, PA, and M1) was evaluated by qRT-PCR. (B) 293T cells were transfected with or without SOCS3 for 24 h, followed by WSN infection for 0 h, 12 h, or 16 h. The transcription of vRNAs (NP, NA, PA, and M1) was evaluated by qRT-PCR. (C) Control siRNA or SOCS3-specific siRNA (si-1) was transfected into A549 cells for 48 h, followed by WSN infection for 0 h, 12 h, or 16 h. qRT-PCR was performed to detect the expression of vRNAs (NP, NA, PA, and M1). (D) IRF3 phosphorylation was evaluated in 293T cells transfected with IRF3 (50 ng/ml) and Flag-3.0 or Flag-SOCS3 for 24 h followed by VSV infection for the indicated times. (E) 293T cells were transfected with the indicated plasmids for 24 h, and then cells were infected with WSN for an additional 0 h, 12 h, 16 h, or 24 h. IB was used for analysis of the phosphorylation of IRF3 and total IRF3. (F) Immunoblot analysis results showing the expression of TBK1 and SOCS3 in primary MEF cells with WSN virus infection. (G and H) ELISA results for IFN-β production at the protein level in RAW 264.7 cells or primary MEF cells with 10 μg/ml or 1 μg/ml poly(I·C) stimulation, respectively. Error bars indicate standard deviations.
In addition to the transcription of WSN vRNAs, we also investigated the potential role of SOCS3 on RNA virus-stimulated downstream signaling cascades. We evaluated IRF3 phosphorylation in 293T cells transfected with Flag-tagged 3.0 or SOCS3 and subsequently infected the cells with VSV for 0 h, 2 h, 4 h, or 8 h. As shown in Fig. 7D, phosphorylation of IRF3 was markedly increased in Flag-3.0-transfected cells upon VSV infection, which was significantly reduced by SOCS3 overexpression. Consistent with this result, ectopic expression of SOCS3 significantly inhibited IRF3 phosphorylation induced by WSN infection for 12 h, 16 h, or 24 h (Fig. 7E). Besides, we performed the WSN infection experiment in mouse primary MEF cells. As shown in Fig. 7F, WSN virus activated the highest TBK1 protein level at 16 h after infection, and then the protein abundance was downregulated upon upregulation of SOCS3 protein expression. Furthermore, we performed ELISAs to check poly(I·C)-induced IFN-β production in RAW 264.7 cells and primary MEF cells. In addition, the data indicated that poly(I·C) stimulation resulted in more IFN-β production at a protein level (Fig. 7G and H). Collectively, SOCS3 blocks IRF3 signaling and facilitates the replication of virus.
DISCUSSION
Innate immunity is a forceful weapon to defend against viruses. The RIG-I signaling pathway is triggered through recognition of RNA viruses, followed by signal cascades that drive expression of type I interferons and other proinflammatory cytokines (5). Subsequently, a JAK/STAT signal is further provoked by IFN-α/β to stimulate the transcription of antiviral genes as well as SOCS3, which plays an important role in the regulation of NF-κB and JAK/STAT signal pathways (11, 17). Though it has been reported that SOCS3 lowers the expression of IFN-β (13), the molecular mechanism remains largely unknown.
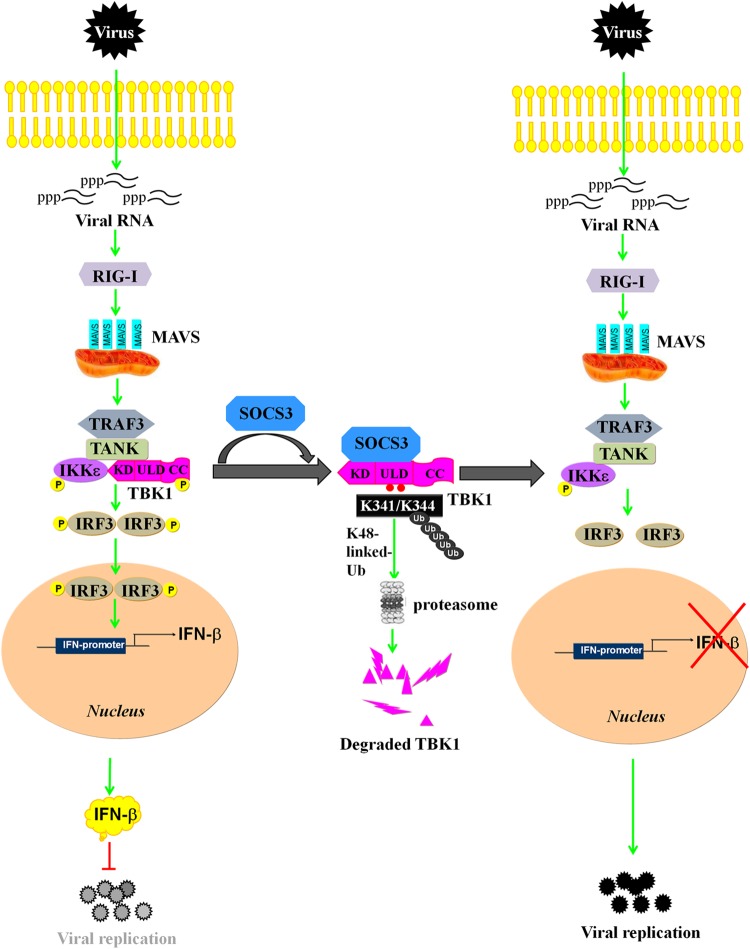
In this study, we found that SOCS3 negatively regulates the IFN-β signaling pathway through promoting K48-linked polyubiquitination of TBK1 at Lys341 and Lys344 sites and, subsequently, its degradation (Fig. 8). We further identified that ectopic expression of SOCS3 inhibits IFN-β activation induced by multiple adaptors except for IRF3, whereas knockdown of SOCS3 enhances IFN-β signaling. It has been reported that the SOCS3 BOX domain is involved in the formation of the E3 ligase complex by interaction with elongins B and C, cullin5, and Rbx-1 (8, 9). As a member of the complex, SOCS3 participates in the process of degrading some proteins, such as focal adhesion kinase (FAK) (27) and CD33-related Siglec 7 (30). Although the (N+KIR) and BOX domains of SOCS3 are indispensable for the degradation of TBK1, they may be involved in different mechanisms. Here, we found that the BOX domain is responsible for ubiquitination of TBK1, which results in its subsequent degradation (Fig. 4E). Traditionally, the SOCS3 SH2 domain mediates the association with substrate by recognition of phosphorylated tyrosine residues (9), whereas the N-ESS domain facilitates substrate association (10). In this study, we found that the N-ESS domain is required in the process of SOCS3-TBK1 complex formation (Fig. 6B). Our study revealed TBK1 acts as a target protein of SOCS3, and this new findings broadens our understanding of the function of SOCS3 as an E3 ligase via complex formation with other proteins.
FIG 8.
Model of SOCS3-mediated inhibition of the IFN-β signaling cascade via promotion of ubiquitination and degradation of TBK1. Upon RNA virus infection, RIG-I recognizes 5′-triphosphate viral RNA and activates MAVS, which further recruits TBK1 and IKKε to phosphorylate IRF3. IRF3 undergoes phosphorylation and dimerization to stimulate the production of IFN-β, which activates a downstream signal to suppress viral replication. However, SOCS3 inhibits IFN-β activity by targeting TBK1 for K48-linked ubiquitination on the K341 and K344 sites and subsequent degradation, thus facilitating viral replication.
Obviously, SOCS3 is a negative regulator of RIG-I signaling. It functions as a member of the E3 ligase complex both in the NF-κB signaling pathway and the IFN-β signaling pathway. It has been reported that SOCS3 suppresses ubiquitination of TRAF6, thus hindering the formation of the TRAF6/TAK1 complex and TAK1 kinase activity, which results in the inhibition of the NF-κB signal (17). In addition, we found that SOCS3 negatively regulates the IFN-β signal by promoting the ubiquitination and degradation of TBK1 (Fig. 8).
TBK1, a key member of the IFN-β signaling pathway, is activated by the mitochondrial adaptor protein MAVS, which further phosphorylates transcription factor IRF3 to transcribe type I interferon (29). It has been reported that GSK3β phosphorylates TBK1 and promotes its self-association and autophosphorylation at Ser172, which is important for its activity (28). In addition to phosphorylation, TBK1 can be regulated by K63-linked and K48-linked ubiquitination (19–21, 23, 24, 31, 32). Here, we found that SOCS3 promoted ubiquitination and degradation of TBK1 (Fig. 3A and B and 4A and B), which led to inhibition of IFN-β activation (Fig. 1). TBK1 was degraded in a SOCS3 dose-dependent manner (Fig. 3B); however, it can avoid being ubiquitinated and degraded by SOCS3 if the serine at residue 172 of TBK1 is mutated to alanine (Fig. 3K and 5H). In addition, the kinase domain of TBK1 is also responsible for SOCS3-TBK1 complex formation. Consequently, Ser172 of TBK1, as an autophosphorylation site (28), is indispensable for SOCS3-mediated inhibition of IFN-β. The ULD of TBK1 plays important roles in the regulation of IRF3 activation (33). We found that ULD is mainly responsible for SOCS3-induced polyubiquitination of TBK1, and two critical ubiquitination sites (K341 and K344) of TBK1 were identified in the process of SOCS3-mediated regulation (Fig. 5D and E). Similar to SOCS3, NLRP4 and TRIP also negatively regulate the IFN-β signaling cascade by targeting TBK1 for Lys48 (K48)-linked polyubiquitination and degradation. However, different from SOCS3, as a member of the E3 ligase complex, NLRP4 functions by recruiting the E3 ubiquitin ligase DTX4 to TBK1, whereas TRIP is an E3 ligase (23, 24).
The cross talk between the RIG-I and JAK/STAT signaling pathways has been known for years. Viruses or cytoplasmic nucleic acids activate RIG-I signaling and recruit STAT6, an important transcription factor of the JAK/STAT pathway, to the endoplasmic reticulum, where it is phosphorylated by TBK1 on Ser407, independently of JAKs. The phosphorylated STAT6 then undergoes dimerization and translocates from the cytoplasm to the nucleus and initiates the transcription of specific targets responsible for immune cell homing (34). It still remains a mystery how JAK/STAT axis feedback regulates RIG-I signaling. It has been reported that infection by some RNA viruses, such as influenza A virus and HIV, upregulates SOCS3 expression via the JAK/STAT and NF-κB pathways (14, 16). We found that SOCS3 negatively regulated RIG-I signaling by targeting TBK1 for proteasome-mediated degradation. As a consequence, ectopic expression of SOCS3 significantly increased the replication of WSN (Fig. 7B), whereas knockdown of SOCS3 greatly lowered vRNA abundance (Fig. 7C).
Taken together, we have demonstrated a new mechanism of immunologic escape. SOCS3 negatively regulates cellular antiviral responses through promoting K48-linked ubiquitination and proteasomal degradation of TBK1. These findings provide new insights into antiviral innate immunity and identify a potential target for the development of antiviral drugs.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 Program; 2012CB518900, 2011CB504706, 2011CB504805, and 2011CB504304), the National Natural Science Foundation of China (81171572), and the Guangdong Innovative Research Team Program (2009010058).
We are grateful to Lanqing Ma (Institute of Microbiology, Chinese Academy of Sciences) for her technical assistance.
We declare no competing financial interests.
REFERENCES
- 1.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, Endres S, Hartmann G. 2006. 5′-triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 2.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Sousa CRE. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 3.Seth RB, Sun LJ, Ea CK, Chen ZJJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Xu LG, Wang YY, Han KJ, Li LY, Zhai ZH, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 5.McWhirter SM, ten Oever BR, Maniatis T. 2005. Connecting mitochondria and innate immunity. Cell 122:645–647. doi: 10.1016/j.cell.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 7.Krebs DL, Hilton DJ. 2001. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SBH, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A 96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piessevaux J, Lavens D, Peelman F, Tavernier J. 2008. The many faces of the SOCS box. Cytokine Growth Factor Rev 19:371–381. doi: 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Babon JJ, McManus EJ, Yao SG, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS. 2006. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell 22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. 1999. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 12.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. 1999. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J 18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pothlichet J, Chignard M, Si-Tahar M. 2008. Innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNARI-dependent pathway. J Immunol 180:2034–2038. doi: 10.4049/jimmunol.180.4.2034. [DOI] [PubMed] [Google Scholar]
- 14.Pauli E-K, Schmolke M, Wolff T, Viemann D, Roth J, Bode JG, Ludwig S. 2008. Influenza A virus inhibits type I IFN signaling via NF-κB-dependent induction of SOCS-3 expression. PLoS Pathog 4:e1000196. doi: 10.1371/journal.ppat.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Miura S, Jimbow K, Fujii N. 2004. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J Virol 78:6282–6286. doi: 10.1128/JVI.78.12.6282-6286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhtar LN, Qin H, Muldowney MT, Yanagisawa LL, Kutsch O, Clements JE, Benveniste EN. 2010. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J Immunol 185:2393–2404. doi: 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frobose H, Ronn SG, Heding PE, Mendoza H, Cohen P, Mandrup-Poulsen T, Billestrup N. 2006. Suppressor of cytokine signaling-3 inhibits interleukin-1 signaling by targeting the TRAF-6/TAK1 complex. Mol Endocrinol 20:1587–1596. doi: 10.1210/me.2005-0301. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W. 2013. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett 587:542–548. doi: 10.1016/j.febslet.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. 2009. The E3 ubiquitin ligase Nrdp1 ‘preferentially' promotes TLR-mediated production of type I interferon. Nat Immunol 10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Wang L, Berman M, Kong Y-Y, Dorf ME. 2011. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, Xavier R, Ting AT. 2008. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep 9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao L, Coope H, Grant S, Ma A, Ley SC, Harhaj EW. 2011. ABIN1 protein cooperates with TAX1BP1 and A20 proteins to inhibit antiviral signaling. J Biol Chem 286:36592–36602. doi: 10.1074/jbc.M111.283762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui J, Li Y, Zhu L, Liu D, Zhou S, Wang HY, Wang R-F. 2012. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Wang L, Zhao X, Zhao K, Meng H, Zhao W, Gao C. 2012. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med 209:1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, Wang Q, Kang T, Chen S, Huang W. 2012. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol 14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 26.Chen S, Sheng C, Liu D, Yao C, Gao S, Song L, Jiang W, Li J, Huang W. 2013. Enhancer of Zeste homolog 2 is a negative regulator of mitochondria-mediated innate immune responses. J Immunol 191:2614–2623. doi: 10.4049/jimmunol.1203143. [DOI] [PubMed] [Google Scholar]
- 27.Liu E, Cote JF, Vuori K. 2003. Negative regulation of FAK signaling by SOCS proteins. EMBO J 22:5036–5046. doi: 10.1093/emboj/cdg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei C-Q, Zhong B, Zhang Y, Zhang J, Wang S, Shu H-B. 2010. Glycogen synthase kinase 3 beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 33:878–889. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Orr SJ, Morgan NM, Buick RJ, Boyd CR, Elliott J, Burrows JF, Jefferies CA, Crocker PR, Johnston JA. 2007. SOCS3 targets Siglec 7 for proteasomal degradation and blocks Siglec 7-mediated responses. J Biol Chem 282:3418–3422. doi: 10.1074/jbc.C600216200. [DOI] [PubMed] [Google Scholar]
- 31.Parvatiyar K, Barber GN, Harhaj EW. 2010. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J Biol Chem 285:14999–15009. doi: 10.1074/jbc.M110.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charoenthongtrakul S, Gao L, Parvatiyar K, Lee D, Harhaj EW. 2013. RING finger protein 11 targets TBK1/IKKi kinases to inhibit antiviral signaling. PLoS One 8:e53717. doi: 10.1371/journal.pone.0053717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, Hofmann K, Akira S, Doetsch V, Dikic I. 2007. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J 26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]