Abstract
The poly(A)-binding protein nuclear 1 is encoded by the PABPN1 gene, whose mutations result in oculopharyngeal muscular dystrophy, a late-onset disorder for which the molecular basis remains unknown. Despite recent studies investigating the functional roles of PABPN1, little is known about its regulation. Here, we show that PABPN1 negatively controls its own expression to maintain homeostatic levels in human cells. Transcription from the PABPN1 gene results in the accumulation of two major isoforms: an unspliced nuclear transcript that retains the 3′-terminal intron and a fully spliced cytoplasmic mRNA. Increased dosage of PABPN1 protein causes a significant decrease in the spliced/unspliced ratio, reducing the levels of endogenous PABPN1 protein. We also show that PABPN1 autoregulation requires inefficient splicing of its 3′-terminal intron. Our data suggest that autoregulation occurs via the binding of PABPN1 to an adenosine (A)-rich region in its 3′ untranslated region, which promotes retention of the 3′-terminal intron and clearance of intron-retained pre-mRNAs by the nuclear exosome. Our findings unveil a mechanism of regulated intron retention coupled to nuclear pre-mRNA decay that functions in the homeostatic control of PABPN1 expression.
INTRODUCTION
A fundamental step in the expression of most protein-coding genes is the addition of a polyadenosine [poly(A)] tail at the 3′ end of mRNAs, which normally promotes gene expression by stimulating nuclear export and translation. A key protein involved in the metabolism of polyadenylated RNAs is the poly(A)-binding protein nuclear 1 (PABPN1). PABPN1 is an evolutionarily conserved protein that is best characterized for its role in mRNA polyadenylation, where it can stimulate poly(A) synthesis in vitro by increasing the processivity of the poly(A) polymerase (1–3). The requirement for PABPN1 during mRNA polyadenylation does not appear to be universal, however. Accordingly, whereas depletion of PABPN1 in primary mouse myoblasts causes poly(A) tail shortening (4), depletion of PABPN1 in human cell lines does not inhibit global gene expression and does not lead to a noticeable impairment in mRNA polyadenylation (5). PABPN1 was also shown to function in gene regulation by influencing the length of 3′ untranslated regions (UTR) of specific genes via the control of polyadenylation signal recognition (6, 7). Furthermore, PABPN1 was found to function in a pathway of RNA decay that targets long noncoding RNAs (lncRNAs) for rapid nuclear turnover and that involves the exosome complex of 3′-5′ exonucleases (5, 8). Therefore, PABPN1 is involved in several important layers of posttranscriptional gene regulation.
Although PABPN1 expression is ubiquitous, its protein levels vary greatly among cell types and tissues (9), suggesting that tight control of PABPN1 expression is important for normal cell and organismal physiology. Notably, mutations in the PABPN1 gene result in oculopharyngeal muscular dystrophy (OPMD), a late-onset disorder that is associated primarily with eyelids drooping, swallowing difficulties, and proximal limb weakness (10). OPMD mutations have been reported in more than 35 countries (11) and result in a PABPN1 protein with a slightly extended polyalanine tract, a consequence of short trinucleotide expansions in the PABPN1 coding sequence (12). Although the mechanism by which short GCG insertions in the PABPN1 gene cause OPMD remains unknown, altered PABPN1 mRNA levels were found in muscle biopsy specimens of OPMD patients relative to those of age-matched controls (13, 14). In addition, reduced expression of PABPN1 mRNA correlates with a poor prognosis in non-small-cell lung cancer (15). As yet, however, little is known about mechanisms that control PABPN1 expression or why it is misregulated in OPMD and cancer.
Besides transcription, cells use multiple layers of posttranscriptional regulation to control gene expression. Alternative splicing is one important mechanism of posttranscriptional gene regulation in which a primary transcript is differentially processed to produce distinct mRNA and protein isoforms. Alternative splicing is controlled by cis-acting elements on pre-mRNAs that are targeted by sequence-specific RNA-binding proteins, which promote or repress productive splicing by the spliceosome (16). Of the different types of alternative splicing in vertebrates, intron retention (IR) is thought to be a rare event; thus, it has remained poorly understood (17). Intron retention is a form of alternative splicing in which one or more introns remain unspliced in a polyadenylated transcript (18). Retained introns often contain premature stop codons, which leads to their cytoplasmic degradation by the nonsense-mediated RNA decay machinery (19–21). Alternatively, translation of an intron-retained transcript can produce a truncated protein, for example, Rem1, which is important for meiotic differentiation in fission yeast (22). Intron retention also can function in gene regulation via nuclear decay of unspliced pre-mRNAs in budding and fission yeasts (23–25). Recently, intron retention was found to control the expression of functionally related genes that are important during the differentiation program of granulocytes (26) and neuronal cells (27), suggesting that intron retention is an underappreciated mechanism of posttranscriptional gene regulation in mammals. Accordingly, recent transcriptome-wide analyses of intron retention across diverse human and mouse cell types indicate that intron retention is far more prevalent than previously appreciated (28, 29). However, compared to the extensive set of mechanisms known to control alternative exon splicing (16, 30), few mechanisms of regulated intron retention have been described.
In this study, we report a mechanism of regulated intron retention coupled to nuclear pre-mRNA decay that functions in the control of PABPN1 homeostasis in human cells. We found that excess levels of PABPN1 protein caused a significant decrease in the spliced/unspliced PABPN1 RNA ratio, thereby reducing endogenous levels of PABPN1 protein. This control required inefficient splicing of the PABPN1 3′-terminal intron via a weak 5′ splice site as well as competitive binding to pre-mRNA between PABPN1 and SRSF10, acting as a splicing activator for PABPN1. Our data suggest that PABPN1 represses splicing of its 3′-terminal intron by binding to an adenosine-rich region in the 3′ UTR of the PABPN1 transcript, thereby promoting pre-mRNA decay by the nuclear exosome. Thus, our findings have identified a mode of posttranscriptional gene regulation in which the control of RNA splicing influences the balance between processing and decay of pre-mRNAs in the nucleus.
MATERIALS AND METHODS
Cell culture.
HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% of tetracycline-free fetal bovine serum (FBS). Inducible expression of green fluorescent protein (GFP)-PABPN1 and GFP-PRMT3 was achieved by flippase-mediated recombination in HEK293FT cells (31). Briefly, PABPN1 and PRMT3 cDNAs were PCR amplified using forward and reverse primers that included attB sequences. PCR products were introduced in pDONR221 using BP clonase II-mediated recombination (Life Technologies). cDNAs then were transferred into pgLAP1 or pgLAP2 plasmids (31) using LR Clonase II (Life Technologies). pgLAP1- and pgLAP2-derived constructs were cotransfected in HEK293FT cells with pOG44, which expresses the Flp recombinase. Positively integrated cells were selected with 150 μg/ml of hygromycin and 15 μg/ml of blasticidin. Positive clones were maintained in DMEM supplemented with 10% tetracycline-free FBS, 75 μg/ml of hygromycin, and 15 μg/ml of blasticidin. Induction of GFP-PABPN1 and GFP-PRMT3 expression was achieved with 500 ng/ml of doxycycline for 48 to 72 h. Short interfering RNAs (siRNAs) were transfected with Lipofectamine 2000 (Life Technologies) at a final concentration of 25 nM for 72 h. Sequences of siRNAs can be found in the supplemental material.
Expression constructs.
Owing to the large number of constructs, the relevant information is provided in the supplemental material.
Protein analyses and antibodies.
Cells were washed two times with phosphate-buffered saline (PBS) and then collected in 1.5-ml tubes. Ninety percent of the cells was kept for RNA extraction, and 10% was used for protein extraction. The cell pellet was resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 2 mM MgCl2, 1 mM dithiothreitol [DTT], and 1× complete protease inhibitor cocktail [Roche]) and incubated at 4°C for 15 min. The mixture was centrifuged for 15 min at 13,000 rpm. The supernatant was transferred into a new tube, loading dye was added to a final 1× concentration (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS [vol/vol], 0.1 M DTT, and bromophenol blue), and the mix was heated for 5 min at 95°C. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting using the following primary antibodies: anti-PABPN1 (Epitomics), antitubulin and antiactin (T5168 and A5441, respectively; Sigma-Aldrich), anti-PRMT3 (A302-526A; Bethyl), anti-GFP (11814460001; Roche), anti-hnRNP A1/A2 and anti-hnRNP H antibodies (32), anti-hnRNP C (PIMA124631; Fisher Scientific), anti-UPF1 (A301-902A; Bethyl), anti-XRN2 (A301-102A; Bethyl), and anti-hRRP40 (A303-909A; Bethyl). Membranes then were probed with a donkey anti-rabbit antibody conjugated to IRDye 800CW (926-32213; LI-COR) and a goat anti-mouse antibody conjugated to Alexa Fluor 680 (A-21057; Life Technologies). Detection of the proteins was performed using an Odyssey infrared imaging system (LI-COR). All of the quantitative protein analyses presented in this study were generated from at least three independent transfection/induction experiments, in which the data and error bars represent averages and standard deviations, respectively.
RNA preparation and analyses.
Total RNA was prepared using TRIzol (Life Technologies) and analyzed by Northern blotting, as previously described (5). To analyze nuclear and cytosolic RNAs, cells were fractionated according to reference 33. For quantitative RT-PCR analyses, cDNA was synthesized with random primers (Omniscript; Qiagen) and analyzed by real-time PCR using position-specific primers as previously described (5). All of the quantitative RT-PCR results presented in this study were generated from at least three independent transfection/induction experiments, in which the data and error bars represent averages and standard deviations, respectively.
RIP assays.
A 10-cm dish of HEK293T cells was transfected using Lipofectamine for 48 h with DNA constructs that expressed GFP-SRSF10 and GFP. Cells were washed three times with PBS and incubated 20 min in RNA coimmunoprecipitation (RIP) lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 40 U/ml RNase OUT [Life Technologies], and complete protease inhibitor cocktail) with rocking at 4°C. Lysate was centrifuged at 13,000 rpm for 20 min at 4°C. Ten percent of the supernatant was kept for RNA extraction (input fraction). The rest of the lysate then was incubated with 25 μl of GFP-TRAP beads (ChromoTek) for 4 h at 4°C. The beads subsequently were washed 4 times with RIP lysis buffer. RNA was extracted using TRIzol reagent (Life Technologies) and subsequently analyzed by quantitative reverse transcription-PCR (RT-qPCR).
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed as described previously (7), with a few modifications. Briefly, 67-nucleotide (nt)-long RNA oligonucleotides (sequences are in the supplemental material) corresponding to the A-rich region of the PABPN1 3′ UTR were generated by in vitro transcription using the mirVana miRNA probe construction kit (AM1550; Ambion). Recombinant glutathione S-transferase (GST) or GST-PABPN1 (1.5 to 45 pmol) was added to binding reaction mixtures, including RNA probes (15,000 cpm) in the binding buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.2 mg/ml bovine serum albumin [BSA], 0.01% Nonidet P-40, 0.5 mM DTT, 100 mM KCl, 2 mM EDTA, 2 U/μl RNase OUT) for 15 min at room temperature in a final 15-μl volume. For competition assays, labeled RNA probes were incubated in the presence of 5- and 10-fold molar excesses of cold RNA oligonucleotides. The binding reaction mixtures were analyzed on native PAGE-glycerol gels, dried, and visualized using a Typhoon Trio instrument (GE Healthcare).
RESULTS
Excess PABPN1 protein results in reduced PABPN1 expression via a mechanism that involves intron retention.
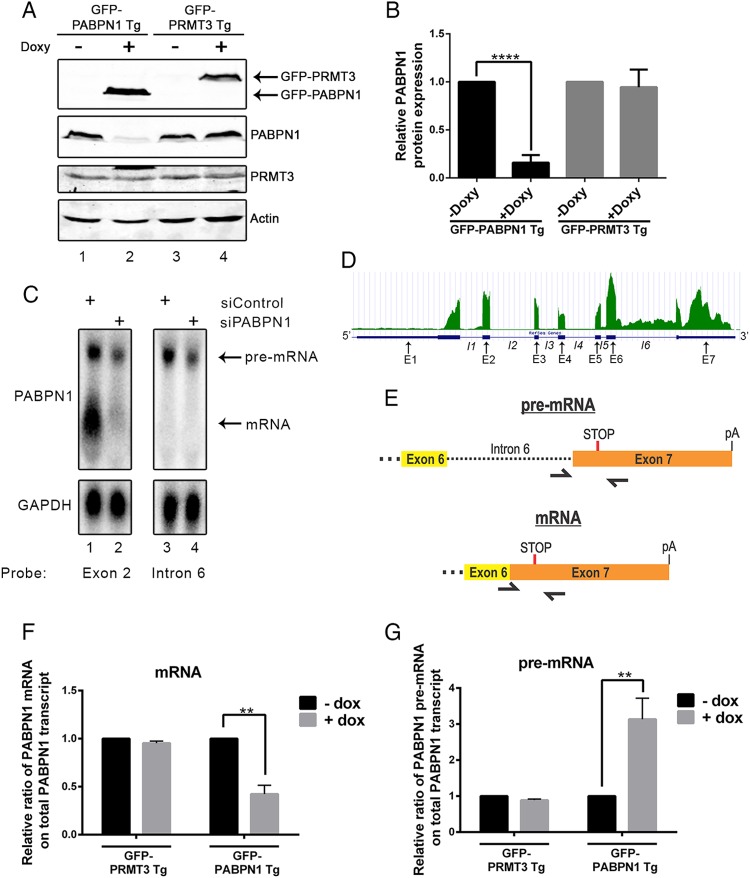
We used a previously described recombination system (31) to generate a human kidney cell line (HEK293) that expresses a single copy of the PABPN1 cDNA under the control of a tetracycline-inducible promoter. The product of the PABPN1 transgene was expressed as a C-terminal fusion to the green fluorescent protein (GFP-PABPN1 Tg) or a FLAG tag (FLAG-PABPN1 Tg). Unexpectedly, the addition of doxycycline to induce GFP-PABPN1 expression resulted in a marked decrease in the levels of endogenous PABPN1 protein (Fig. 1A, compare lanes 1 and 2; quantification is shown in panel B). As a control, the induction of a similarly prepared GFP-PRMT3 transgene did not affect PABPN1 expression (Fig. 1A, compare lanes 3 and 4; quantification is in panel B) and did not result in reduced levels of endogenous PRMT3 (Fig. 1A, lanes 3 and 4). The use of an independent cell line that expresses a FLAG-PABPN1 transgene also showed a significant reduction of endogenous PABPN1 after induction of FLAG-PABPN1 expression (see Fig. S1A and B in the supplemental material). Interestingly, the use of stable cells that conditionally induce normal (10 alanines) and OPMD-like, alanine-expanded (17 alanines) versions of PABPN1 demonstrated comparable negative regulation of endogenous PABPN1 protein (see Fig. S1C and D in the supplemental material). Together, these results support the existence of an autoregulatory feedback loop in which excess PABPN1 protein negatively controls expression from the PABPN1 gene.
FIG 1.
Increased gene dosage of PABPN1 results in reduced levels of endogenous PABPN1 protein and mRNA. (A) Western blot analysis using extracts from untreated (−) or doxycycline-treated (+) transgenic (Tg) cell lines that express GFP-PABPN1 (lanes 1 and 2) and GFP-PRMT3 (lanes 3 and 4). (B) Quantification of endogenous PABPN1 protein in the presence and absence of doxycycline in the indicated cell lines. ****, P < 0.0001 by Student's t test. (C) Northern blot analysis using RNA probes complementary to PABPN1 exon 2 (lanes 1 and 2) and intron 6 (lanes 3 and 4) sequences, using total RNA from HEK293T cells treated with PABPN1-specific (lanes 2 and 4) and control (lanes 1 and 3) siRNAs. (D) RNA-seq read distribution along the PABPN1 gene from human HeLa cells. The bottom row shows the RefSeq gene annotation for the corresponding exons (E1 to E7) and introns (I1 to I6). (E) Schematic of primers used to measure the levels of spliced and unspliced PABPN1 transcripts by RT-qPCR. (F and G) RT-qPCR analysis measuring the relative fraction of total PABPN1 transcript that is spliced mRNA (F) and unspliced pre-mRNA (G) using RNA from the indicated induced (+Doxy) and noninduced (−Doxy) transgenic cell lines. Data are expressed relative to those for the uninduced control (−Doxy). **, P < 0.01 by Student's t test.
To begin to address how increased dosage of PABPN1 protein altered PABPN1 expression, we first examined the levels of endogenous PABPN1 transcripts by Northern blotting. Northern analysis of PABPN1 revealed the presence of two major transcript variants (Fig. 1C, lane 1). Interestingly, siRNA-mediated silencing of PABPN1 expression showed a substantially greater reduction of the short isoform relative to the level of the long isoform (Fig. 1C, compare lanes 1 and 2), suggesting that the long PABPN1 RNA isoform was more resistant to siRNA-mediated silencing. PABPN1 transcript isoforms previously have been reported in mouse cells and attributed to the differential use of polyadenylation signals downstream of the stop codon (9). Therefore, we assessed whether the short and long PABPN1 isoforms detected by northern analysis of total human RNA could be the result of alternative polyadenylation. However, 3′ rapid amplification of cDNA ends (RACE) analyses coupled to DNA sequencing identified a single region of a poly(A) site selection located 886 to 891 nucleotides downstream of the stop codon. Moreover, deep sequencing analysis of poly(A) site mapping by 3′ region extraction and deep sequencing (READS) (34) indicated the use of one main region of polyadenylation site selection ∼900 nt downstream of the stop codon in PABPN1 transcripts (see Fig. S2 in the supplemental material). Thus, we concluded that the two major isoforms of PABPN1 expressed in HEK293T cells are not transcripts with different 3′ UTRs.
To explore the nature of the transcript isoforms produced from the PABPN1 gene, we analyzed data from an RNA-sequencing (RNA-seq) experiment that we previously conducted using HeLa cells (5). The analysis of read coverage along the PABPN1 gene revealed considerably greater signal from the last intron (intron 6) than from the other PABPN1 introns (Fig. 1D), suggesting that a fraction of PABPN1 transcripts retains intron 6. Importantly, the use of an RNA probe specific to intron 6 confirmed that the long PABPN1 isoform corresponds to an unspliced pre-mRNA that failed to splice intron 6 (Fig. 1C, lanes 3 and 4). Quantification of northern data indicated that unspliced PABPN1 transcripts represent almost 20% of the total PABPN1 RNA in HEK293T cells. Using biochemical fractionation to separate nuclear and cytoplasmic RNA populations, we found that the unspliced PABPN1 transcript was detected primarily in the nuclear fraction, whereas the spliced mRNA was mainly cytosolic (see Fig. S3 in the supplemental material).
Given that a fraction of PABPN1 transcripts retains the 3′-terminal intron, we next examined whether the induction of GFP-PABPN1 altered the levels of spliced and unspliced PABPN1 isoforms. To detect endogenous PABPN1 transcripts by RT-qPCR and not the mRNA expressed from the GFP-PABPN1 cDNA, we used a reverse primer complementary to a region located in the 3′ UTR of PABPN1 (Fig. 1E). This reverse primer was combined with a forward primer located in intron 6 to detect the unspliced pre-mRNA or with a forward primer across the exon 6-exon 7 junction to detect the spliced PABPN1 mRNA (Fig. 1E). The induction of GFP-PABPN1 by doxycycline caused a significant decrease in the levels of the spliced PABPN1 mRNA (Fig. 1F), which resulted in a 3-fold increase in the percentage of unspliced PABPN1 pre-mRNA relative to that of total PABPN1 RNA (Fig. 1G). As a control, the induction of GFP-PRMT3 did not affect the PABPN1 spliced/unspliced ratio (Fig. 1F to G). These results demonstrate that excess PABPN1 reduces the levels of spliced PABPN1 mRNA and endogenous PABPN1 protein.
Intron 6 of the PABPN1 gene is required for autoregulation.
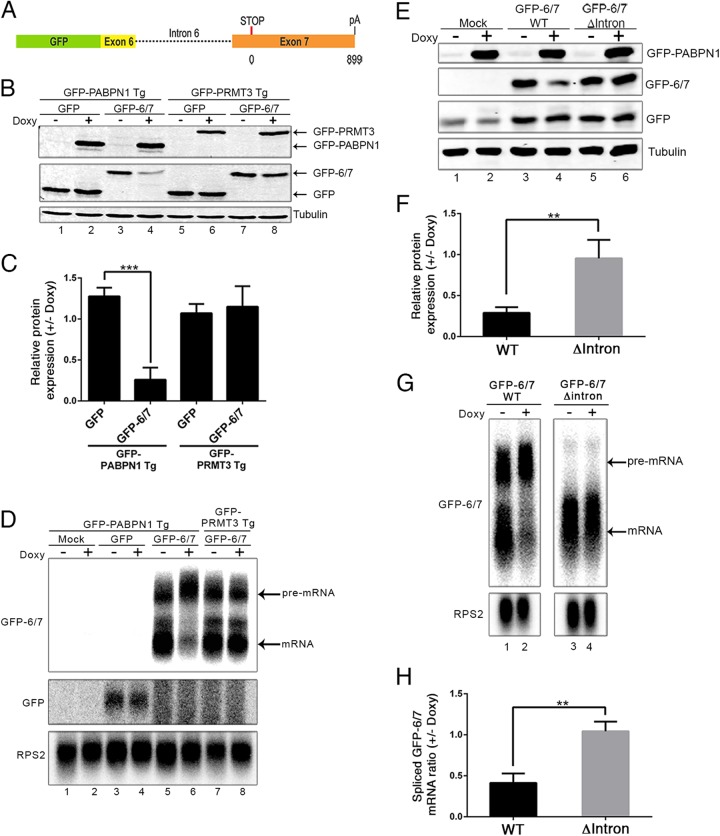
The aforementioned results suggested that PABPN1 controls its own expression by interfering with the splicing of its 3′-terminal intron, resulting in the reduced production of spliced mRNA and, hence, PABPN1 protein. To test this model and get insights into the mechanism of PABPN1 autoregulation, we generated a PABPN1 minigene construct that contained a section of exon 6, intron 6, and exon 7 fused to the 3′ end of the GFP coding sequence (Fig. 2A). Transfection of this minigene construct into the inducible GFP-PABPN1 and GFP-PRMT3 transgenic cell lines resulted in the expression of a protein product in which the last 60 amino acids of PABPN1 were fused to the C terminus of the GFP reporter (GFP-6/7) (Fig. 2B, lanes 3 and 7). The induction of GFP-PABPN1 caused a noticeable decrease in the levels of the GFP-6/7 fusion (Fig. 2B, compare lanes 3 and 4; quantification is shown in panel C), yet no change in GFP-6/7 level was noted after induction of GFP-PRMT3 (Fig. 2B, lanes 7 and 8; quantification in panel C). As an additional control, expression of the wild-type GFP protein was not altered after GFP-PABPN1 induction (Fig. 2B, lanes 1 and 2). We next analyzed the transcript expressed from the minigene construct. To specifically detect the mRNA expressed from the GFP reporter constructs, we designed an antisense RNA probe that is complementary to a section containing plasmid and PABPN1 sequences. Accordingly, this RNA probe did not detect endogenous PABPN1 mRNA and the mRNA expressed from the integrated GFP-PABPN1 transgene (Fig. 2D, lanes 1 and 2). As seen for endogenous PABPN1, northern analysis detected two major transcript isoforms that were expressed from the GFP-6/7 minigene (Fig. 2D, lanes 5 and 7): a short isoform in which intron 6 was spliced out and a long transcript isoform that retained intron 6 (see Fig. S4 in the supplemental material). Importantly, and consistent with our results with endogenous PABPN1, induction of GFP-PABPN1 by doxycycline resulted in reduced levels of spliced mRNA expressed from the GFP-6/7 minigene (Fig. 2D, compare lanes 5 and 6). In contrast, the induction of GFP-PRMT3 did not affect the ratio between spliced and unspliced transcripts expressed from the GFP-6/7 minigene (Fig. 2D, lanes 7 and 8). These results indicate that the GFP-6/7 minigene responds to PABPN1-dependent regulation in a manner similar to that of endogenous PABPN1, indicating that the cis-acting control elements required for the autoregulatory mechanism are present in the minigene.
FIG 2.
3′-Terminal intron of PABPN1 is required for autoregulation. (A) Schematic of the GFP-6/7 minigene construct in which 144 bp of exon 6, intron 6, and exon 7 was fused to the GFP coding sequence. Exon 7 consists of 37 bp of coding sequences, a TAA stop codon, and ∼900 bp of 3′ untranslated region. (B) Western blot analysis using extracts from untreated (−) and doxycycline-treated (+) GFP-PABPN1 (lanes 1 to 4) and GFP-PRMT3 (lanes 5 to 8) transgenic (Tg) cell lines that previously were transfected with the GFP-6/7 (lanes 3 and 4 as well as 7 and 8) or GFP (lanes 1 and 2 as well as 5 and 6) constructs. (C) GFP and GFP-6/7 protein levels were normalized to tubulin and expressed relative to those for the uninduced control (−Doxy). ***, P < 0.001 by Student's t test. (D) Northern blot analysis of the indicated transcripts using total RNA prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 (lanes 1 to 6) and GFP-PRMT3 (lanes 7 and 8) transgenic cell lines that previously were transfected with the GFP-6/7 (lanes 5 to 8) or GFP (lanes 3 and 4) constructs, including a mock transfection control (lanes 1 and 2). (E) Western blot of the indicated proteins using extracts from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic cells that previously were transfected with wild-type (lanes 3 and 4) and intronless (lanes 5 and 6) GFP-6/7 constructs. Cells also were cotransfected with a vector expressing GFP to control for transfection efficiency. (F) GFP-6/7 protein levels were normalized to those for tubulin and GFP (to control for transfection efficiency) and expressed relative to those for the uninduced control (−Doxy). **, P < 0.01 by Student's t test. (G) Northern blot analysis of the indicated transcripts using total RNA prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 Tg cells that previously were transfected with wild-type (lanes 1 and 2) and intronless (lanes 3 and 4) GFP-6/7 constructs. (H) Quantification of spliced GFP-6/7 transcript normalized to RPS2 mRNA and expressed relative to levels for the uninduced control (−Doxy). **, P < 0.01 by Student's t test.
We next addressed whether synthesis of a pre-mRNA containing intron 6 was required for PABPN1-dependent control of GFP-6/7 expression by generating an intronless version of the minigene. Transfection of the intronless and intron-containing constructs into the GFP-PABPN1 transgenic cell line resulted in similar levels of GFP-6/7 protein (Fig. 2E, compare lanes 3 and 5). However, whereas the induction of GFP-PABPN1 resulted in decreased levels of GFP-6/7 protein expressed from the intron-containing minigene (Fig. 2E, lanes 3 and 4), no change in GFP-6/7 levels was observed after GFP-PABPN1 induction in cells expressing the intronless version of the minigene (Fig. 2E, lanes 5 and 6; quantifications are in panel F). In agreement with this protein analysis, northern data indicated that GFP-PABPN1 induction did not affect the levels of GFP-6/7 mRNA expressed from the intronless construct (Fig. 2G, lanes 3 and 4; quantifications in panel H). These results indicate that PABPN1 autoregulation occurs at the level of unspliced pre-mRNAs that retain intron 6.
Poor splicing efficiency is required for PABPN1-dependent regulation.
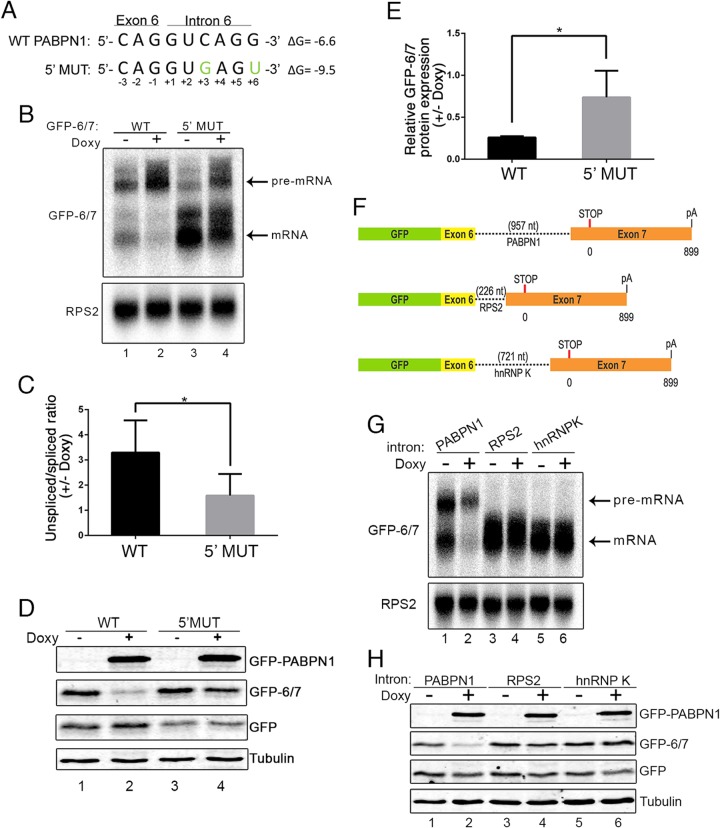
The observation that roughly a fifth of PABPN1 cellular transcripts represent unspliced pre-mRNAs suggested that the PABPN1 3′-terminal intron is inefficiently spliced. 5′ Splice site (5′ ss) selection by the U1 snRNP is a critical step that determines the efficiency of intron splicing. Consensus 5′ ss nucleotides that favor base pairing with the U1 snRNA tend to show higher splicing efficiency (35). Analysis of sequences at the exon-intron junction of intron 6 in the PABPN1 transcript revealed a relatively weak 5′ ss due to the presence of a cytidine (C) at position +3 (Fig. 3A), which is either an adenine (A) or a guanine (G) in >90% of human 5′ ss (35). Moreover, a G is at position +6 of the intron (Fig. 3A), whereas a uracil (U) is present in ∼50% of the human 5′ ss (35). To test whether splicing efficiency influences PABPN1 autoregulation, we created a 5′ ss mutant (5′ MUT) that introduced a C-to-G mutation at position +3 and a G-to-U mutation at position +6, which are expected to favor base pairing with the U1 snRNA and, as a result, increase splicing efficiency. Consistent with this prediction, expression from the minigene construct with the 5′ ss mutations robustly increased splicing, as demonstrated by the increased level of spliced mRNA and decreased level of unspliced pre-mRNA relative to the level for the wild-type intron (Fig. 3B, compare lane 3 to lane 1). Importantly, the 5′ MUT showed reduced sensitivity to PABPN1-dependent regulation compared to that of the minigene with the wild-type intron (Fig. 3B, compare lanes 1 and 2 to lanes 3 and 4, and C). Western blot analysis also revealed reduced PABPN1-dependent regulation for the 5′ ss mutations, as seen by the reduced downregulation of GFP-6/7 protein in the 5′ MUT compared to that of the wild-type construct (Fig. 3D and E). These results show that increasing the efficiency of intron 6 splicing reduces the capacity of PABPN1 to negatively control GFP-6/7 expression.
FIG 3.
Inefficient splicing of the PAPBN1 terminal intron is necessary for PABPN1 autoregulation. (A) The 5′ ss sequences of wild-type (WT) and mutated (5′ MUT) PABPN1 intron 6 are shown. Nucleotide changes introduced in intron 6 sequence to enhance splicing efficiency are shown in green. Nucleotide positions are numbered and indicated under each nucleotide. ΔG, predicted free energy of 5′ ss. (B) Northern blot analysis using total RNA prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic (Tg) cells that previously were transfected with wild-type (lanes 1 and 2) and 5′ ss mutant (lanes 3 and 4) GFP-6/7 constructs. (C) Quantification of GFP-6/7 unspliced/spliced ratios from Northern blot data and expressed relative to those for the uninduced control (−Doxy). *, P < 0.05 by Student's t test. (D) Western blot analysis of the indicated proteins using extracts from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic cells that previously were transfected with wild-type (lanes 1 and 2) and 5′ ss mutant (lanes 3 and 4) GFP-6/7 constructs. (E) GFP-6/7 protein levels were normalized to those for tubulin and GFP (to control for transfection efficiency) and expressed relative to those for the uninduced control (−Doxy). *, P < 0.05 by Student's t tests. (F) Schematic of GFP-6/7 constructs with PABPN1, RPS2, and HNRNPK 3′-terminal introns used to study PABPN1-dependent regulation. (G and H) Northern blot (G) and Western blot (H) analyses of extracts prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic cells that previously were transfected with GFP-6/7 constructs containing the PABPN1 (lanes 1 and 2), RPS2 (lanes 3 and 4), and HNRNPK (lanes 5 and 6) introns. Cells also were cotransfected with a vector expressing GFP to control for transfection efficiency.
To further support the idea that PABPN1 autoregulation depends on intron 6 and splicing efficiency, we replaced the inefficiently spliced 3′-terminal intron of PABPN1 with the efficiently spliced 3′-terminal introns of the RPS2 and HNRNPK genes (Fig. 3F). As expected for efficiently spliced introns, unspliced GFP-6/7 pre-mRNA was not detected when the minigene construct contained the RPS2 and HNRNPK introns (Fig. 3G, lanes 3 and 5). Importantly, minigenes with RPS2 and HNRNPK introns were resistant to PABPN1-dependent regulation, as analyzed at the RNA (Fig. 3G, lanes 3 to 6) and protein (Fig. 3H, lanes 3 to 6) levels. Moreover, we could engineer the PABPN1-insensitive RPS2 intron to become sensitive to GFP-PABPN1 induction by introducing mutations in the 5′ ss of the RPS2 intron that disfavor base pairing to the U1 snRNA (see Fig. S5 in the supplemental material). From these data, we conclude that the poor splicing efficiency of the PABPN1 3′-terminal intron is important for autoregulation.
3′-end polyadenylation is not required for PABPN1 autoregulation.
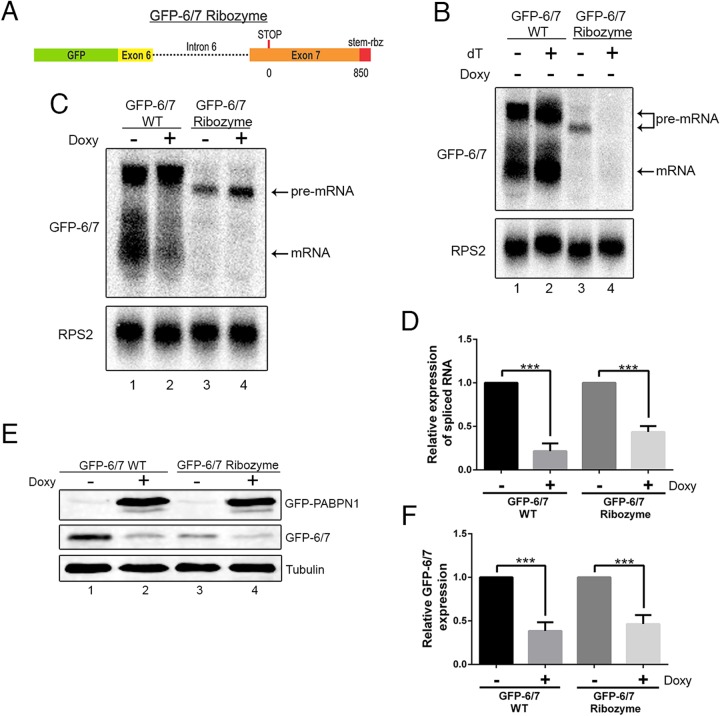
Autoregulation by PABPN1 at the level of the unspliced pre-mRNA predicts a mechanism that involves binding to the 3′-end poly(A) tail. To test whether polyadenylation is required for PABPN1 self-regulation, we generated a minigene construct in which a variant of the hepatitis Delta ribozyme was inserted instead of the PABPN1 polyadenylation signal (Fig. 4A). We chose the hepatitis Delta ribozyme because it was shown previously to cleave efficiently in human cells (36). A hairpin loop derived from the 3′ end of human histone mRNA also was inserted 5′ of the ribozyme cleavage site to stabilize the upstream transcript (36). Wild-type and ribozyme constructs were transfected into HEK293T cells, and expression of the GFP-6/7-derived transcripts was analyzed by Northern blotting. As can be seen in Fig. 4B, the majority of the transcripts produced from the ribozyme construct were unspliced pre-mRNAs that retained intron 6 (lane 3). This observation is consistent with results showing that poly(A) site recognition by the 3′-end processing machinery facilitates the splicing of 3′-terminal introns (37, 38). Although not readily detectable by northern analysis, RT-PCR using a primer that spans the exon-exon junction indicated that spliced transcripts were produced from the ribozyme construct (see Fig. S6 in the supplemental material). We next verified that the GFP-6/7 ribozyme construct expressed nonpolyadenylated transcripts by selection of poly(A)+ RNAs using oligo(dT)-based purification. Northern analysis of oligo(dT)-selected RNA expressed from the wild-type GFP-6/7 construct detected both spliced and unspliced transcripts (Fig. 4B, lane 2). In contrast, RNAs produced from the ribozyme construct were not detected in the oligo(dT)-bound fraction (Fig. 4B, lane 4). As a control, equal levels of polyadenylated RPS2 mRNA were recovered by oligo(dT) purification using total RNA prepared from cells transfected with wild-type and ribozyme constructs (Fig. 4B, compare lanes 2 and 4), confirming that poly(A) RNA selection efficiency was similar for both samples. From these results, we conclude that the GFP-6/7 transcripts expressed from the ribozyme construct are not polyadenylated.
FIG 4.
PABPN1 autoregulation does not require canonical 3′-end processing and polyadenylation. (A) Schematic diagram of the GFP-6/7 ribozyme construct. (B) Northern blot analysis of total (lanes 1 and 3) and oligo(dT)-selected (lanes 2 and 4) RNA prepared from cells transfected with DNA constructs that express wild-type (lanes 1 and 2) and ribozyme-processed (lanes 3 and 4) GFP-6/7 transcript. (C) Northern blot analysis of total RNA prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic (Tg) cells that previously were transfected with wild-type (lanes 1 and 2) and ribozyme-processed (lanes 3 and 4) GFP-6/7 constructs. (D) Quantitative RT-PCR analysis of spliced GFP-6/7 mRNA produced from wild-type and ribozyme constructs. GFP-6/7 mRNA levels were normalized to RPS2 mRNA and expressed relative to those for the uninduced control (−Doxy). ***, P < 0.001 by Student's t test. (E) Western blot analysis using extracts from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic cells that previously were transfected with wild-type (lanes 1 and 2) and ribozyme (lanes 3 and 4) GFP-6/7 constructs. (F) GFP-6/7 protein levels were normalized to those for tubulin and expressed relative to those for the uninduced control (−Doxy). ***, P < 0.001 by Student's t test.
We next transfected the GFP-PABPN1-inducible cell line with wild-type and ribozyme constructs to address the importance of 3′-end polyadenylation in the mechanism of PABPN1 autoregulation. Consistent with our previous results using the wild-type construct, reduced levels of spliced GFP-6/7 mRNA were observed after GFP-PABPN1 induction (Fig. 4C, lanes 1 and 2). Interestingly, transcripts expressed from the ribozyme constructs still were sensitive to excess PABPN1, as indicated by the accumulation of unspliced pre-mRNA (Fig. 4C, lanes 3 and 4) and reduced levels of spliced mRNA (Fig. 4D) after doxycycline induction. Because spliced transcripts produced from a self-cleaving ribozyme previously were shown to be competent for nuclear export in mammalian cells (36), we also investigated the sensitivity of the ribozyme construct to PABPN1-dependent regulation by measuring GFP-6/7 protein levels. As expected, lower levels of GFP-6/7 proteins were detected in cells transfected with the ribozyme construct than in cells that expressed the wild-type minigene (Fig. 4E, compare lanes 1 and 3). Importantly, GFP-PABPN1 induction caused similar downregulation of GFP-6/7 protein for both wild-type and ribozyme constructs (Fig. 4E and F), consistent with the RNA analyses (Fig. 4C and D). Together, these data argue for a mechanism of PABPN1 autoregulation that is independent of 3′-end polyadenylation.
An adenosine-rich region in the 3′ UTR of the PABPN1 transcript is important for autoregulation.
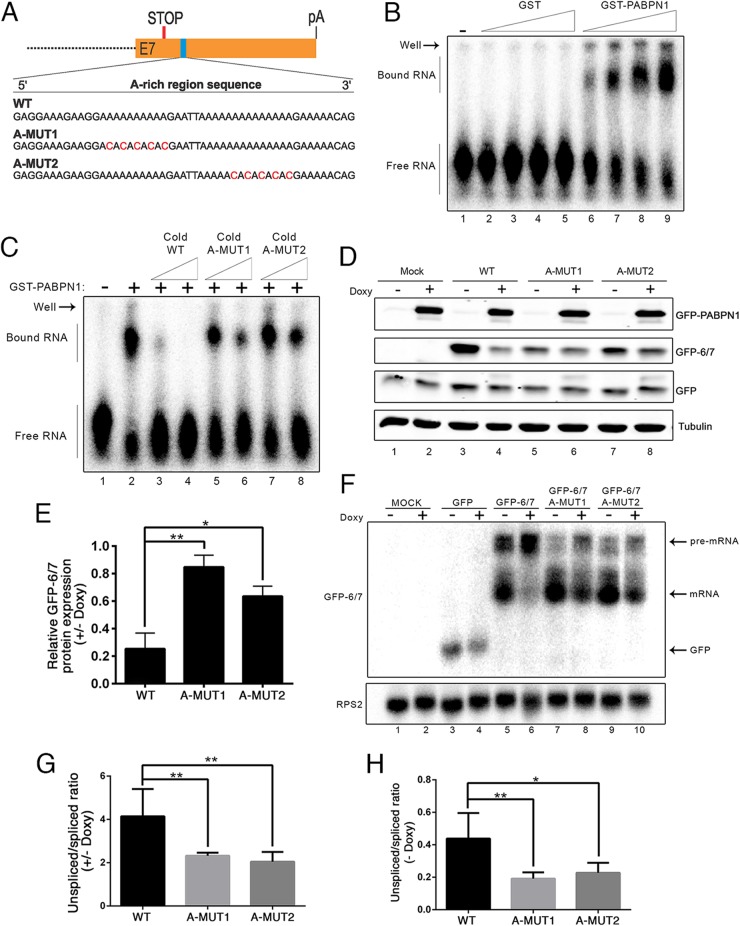
Given results indicating that a 3′ poly(A) tail was not required for PABPN1 self-regulation, we searched for potential PABPN1 binding sites in the PABPN1 transcript. Notably, an A-rich region was apparent in the PABPN1 3′ UTR ∼70-nt downstream from the 3′ ss of the terminal intron (Fig. 5A). Interestingly, this A-rich region is strongly conserved among vertebrate species (see Fig. S7 in the supplemental material). To test whether PABPN1 binds to the A-rich region within its 3′ UTR, we used an RNA gel shift assay to examine binding of recombinant PABPN1 to a 67-nt-long RNA probe that corresponds to PABPN1 3′ UTR sequences containing the A-rich region. As can be seen in Fig. 5B, GST-PABPN1 robustly bound the 3′ UTR RNA probe (lanes 6 to 9), whereas GST alone did not (lanes 2 to 5). Importantly, binding of PABPN1 to this region of its 3′ UTR was largely dependent on the integrity of the A-rich elements, as disruption of the poly(A) stretches with cytidine (A-Mut1 and A-Mut2) (Fig. 5A) clearly reduced PABPN1 binding to this region, as determined by competition assays (Fig. 5C, compare lanes 5 to 8 to lanes 3 and 4). These results suggest direct binding of PABPN1 to the A-rich region of its 3′ UTR.
FIG 5.
Adenosine-rich region in the 3′ UTR of the PABPN1 transcript is important for PABPN1 autoregulation. (A) A 50-nt, adenosine-rich region from the human PABPN1 mRNA is shown. C-to-A changes introduced in adenosine repeats are shown in red and are referred to as A-MUT1 and A-MUT2. (B) Increasing amounts of GST (lanes 2 to 5) and GST-PABPN1 (lanes 6 to 9) were incubated with a 67-nt RNA probe that includes the A repeats of the PABPN1 3′ UTR. The position of free probe and PABPN1-bound complexes is shown on the left. (C) Equal amounts of GST-PABPN1 were incubated with the A-rich 3′ UTR probe in the presence of increasing amounts of cold wild-type (WT, lanes 3 and 4) and C-to-A mutated (A-MUT1, lanes 5 and 6; A-MUT2, lanes 7 and 8) 3′ UTR probes or without any competing nucleic acid (lane 2). (D) Western blot analysis using extracts from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic (Tg) cells that previously were transfected with wild-type (lanes 3 and 4) and C-to-A mutated (lanes 5 to 8) GFP-6/7 constructs. (E) GFP-6/7 protein levels were normalized to those of tubulin and GFP (to control for transfection efficiency) and expressed relative to those for the uninduced control (−Doxy). P < 0.01 (**) and P < 0.05 (*) by Student's t tests. (F) Northern blot analysis using total RNA prepared from untreated (−) and doxycycline-treated (+) GFP-PABPN1 transgenic cells that previously were transfected with wild-type (lanes 5 and 6) and C-to-A mutated (lanes 7 to 10) GFP-6/7 constructs. (G) RT-PCR analysis of unspliced/spliced ratios for wild-type and C-to-A mutant constructs expressed relative to those for the uninduced control (−Doxy). **, P < 0.01 by Student's t test. (H) RT-PCR analysis of unspliced/spliced ratios for wild-type and C-to-A mutant constructs expressed under uninduced conditions (−Doxy). P < 0.01 (**) and P < 0.05 (*) by Student's t tests.
We next examined whether disruption of the adenosine repeats in the PABPN1 3′ UTR affected PABPN1 autoregulation by introducing the A-to-C mutations that decreased PABPN1 binding (Fig. 5A to C) in the GFP-6/7 minigene construct. Analysis of GFP-6/7 protein levels before and after GFP-PABPN1 induction revealed a significant decrease of PABPN1-mediated regulation in cells that expressed constructs containing mutations in the A-rich regions (Fig. 5D, compare lanes 5 to 8 to lanes 3 and 4; quantification is shown in panel E). This observation, obtained by measuring GFP-6/7 protein, was supported by RNA analyses, which indicated that the ability of PABPN1 to decrease spliced mRNA levels was reduced in cells that expressed constructs harboring the A-to-C mutations (Fig. 5F, compare lanes 7 to 10 to lanes 5 and 6). Accordingly, A-to-C mutants significantly reduced the increase in the unspliced/spliced ratio after GFP-PABPN1 induction (Fig. 5G). Northern analysis further revealed that the mutations introduced in the A-rich region stimulated intron 6 splicing, as demonstrated by the significant decrease in the unspliced/spliced ratio relative to that of the wild-type construct under normal uninduced conditions (Fig. 5F, compare lanes 7 and 9 to lane 5; quantification in panel H). Collectively, the data presented in Fig. 5 suggest that PABPN1 autoregulation occurs via binding to the A-rich region of its 3′ UTR, which interferes with the splicing of intron 6, thereby decreasing the levels of spliced mRNA.
Splicing of the PABPN1 3′-terminal intron is controlled by SRSF10, hnRNP H, and hnRNP A2/B1.
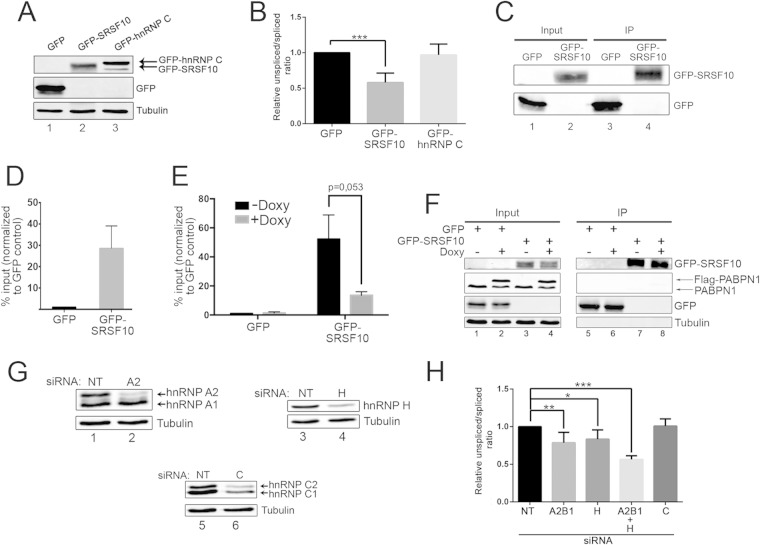
The data described above suggest that PABPN1 self-regulation occurs at the level of the unspliced pre-mRNA. Because introns with weak splice sites frequently are substrates for regulation by trans-acting factors, we searched for proteins contributing to the control of PABPN1 intron 6 splicing. Examination of the nucleotide sequence of the two exons flanking PABPN1 intron 6 revealed a conserved GA-rich region located at the beginning of the downstream exon and adjacent to the A-rich region bound by PABPN1 (see Fig. S7 in the supplemental material). GA-rich sequences are a hallmark of serine-arginine-rich splicing factor 10 (SRSF10) binding, as determined by SELEX (39) and RNAcompete (40), and have been shown to be overrepresented in the vicinity of SRSF10-activated exons (41). To test whether SRSF10 regulates PABPN1 splicing, we overexpressed a GFP-tagged version of SRSF10 in HEK293T cells (Fig. 6A) and analyzed the levels of spliced and unspliced PABPN1 transcripts. Overexpression of GFP-SRSF10 significantly promoted PABPN1 splicing, as demonstrated by the reduced unspliced/spliced ratio (Fig. 6B). In contrast, overexpression of GFP-hnRNP C (Fig. 6A) did not affect the ratio between spliced and unspliced transcript (Fig. 6B).
FIG 6.
SRSF10, hnRNP H, and hnRNP A2/B1 modulate PABPN1 intron 6 splicing. (A) Western blot analysis using total extracts prepared from HEK293T cells that were transfected with constructs expressing GFP (lane 1), GFP-SRSF10 (lane 2), and GFP-hnRNP C (lane 3). (B) RT-qPCR analysis of the endogenous PABPN1 unspliced/spliced ratio using total RNA prepared from HEK293T cells that previously were transfected with constructs expressing GFP, GFP-SRSF10, and GFP-hnRNP C. ***, P < 0.001 by Student's t test. (C and D) RIP assays were performed using GFP and GFP-SRSF10. Pre-mRNA association (IP/input ratio) for GFP and GFP-SRSF10 was analyzed by RT-qPCR, and the values then were set to 1.0 for the control GFP purification. (E) RT-qPCR analysis of RIP assays using GFP and GFP-SRSF10 that were expressed in HEK293T cells that conditionally express Flag-PABPN1 using a doxycycline (Doxy)-sensitive promoter. PABPN1 pre-mRNA association was measured as described for panel D, and the values were sent to 1.0 for the GFP control that was purified from uninduced cells (−Doxy). The P value was calculated using Student's t test. (F) Western blot validation of a RIP assay, as described for panel E, for GFP immunoprecipitations (IP; lanes 5 to 8) and whole-cell extracts (input; lanes 1 to 4). (G) Western blot validation of RNAi-mediated depletion of hnRNP A2/B1 (lane 2), hnRNP H (lane 4), and hnRNP C (lane 6). (H) RT-qPCR analysis of PABPN1 unspliced/spliced ratios using total RNA prepared from HEK293T cells treated with siRNAs specific to hnRNP H, hnRNP A2/B1, and hnRNP C, as well as a nontarget control siRNA. P < 0.001 (***), P < 0.01 (**), and P < 0.05 (*) by Student's t test.
We next addressed whether SRSF10 directly regulates the splicing of PABPN1. RNA coimmunoprecipitation (RIP) experiments were performed to examine whether unspliced PABPN1 transcripts copurify with SRSF10. We purified GFP-SRSF10 (Fig. 6C) and measured the association of unspliced PABPN1 transcripts by RT-PCR relative to a control GFP purification. A clear enrichment of unspliced PABPN1 transcript was found in the GFP-SRSF10-bound fraction relative to the control GFP purification (Fig. 6D), a result consistent with the idea that stimulation of PABPN1 splicing by SRSF10 (Fig. 6B) is a consequence of binding to the unspliced PABPN1 pre-mRNA. Given the close proximity between the GA-rich and A-rich regions at the beginning of exon 7 of the PABPN1 transcript (see Fig. S7 in the supplemental material), we next tested whether binding of PABPN1 to the A-rich region affected the association between SRSF10 and the PABPN1 pre-mRNA. For this, we used RIP assays to examine the impact of Flag-PABPN1 overexpression on GFP-SRSF10 binding to endogenous PABPN1 pre-mRNAs. As can be seen in Fig. 6E, the level of PABPN1 pre-mRNA associated with SRSF10 was substantially reduced in cells in which Flag-PABPN1 was induced (+Doxy) compared to levels in noninduced cells (−Doxy), despite the purification of similar levels of GFP-SRSF10 from both induced and noninduced cells (Fig. 6F, lanes 7 and 8). These data suggest that PABPN1 binding to the A-rich region of its exon 7 negatively affects the association between the splicing factor, SRSF10, and the PABPN1 pre-mRNA.
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are another important class of RNA-binding proteins that function in the regulation of alternative splicing, frequently in the control of the alternative splicing of cassette exons (42). Recently, transcriptome-wide binding of several hnRNPs was determined by cross-linking immunoprecipitation followed by sequencing (CLIP-seq) analysis (43). hnRNP H and hnRNP A2/B1 were found to bind intron 6 of PABPN1. To examine the functional significance of this binding to the splicing of the PABPN1 3′-terminal intron, we used siRNAs to efficiently deplete hnRNP H and hnRNP A2/B1 from HEK293T cells (Fig. 6G) and measured the levels of spliced and unspliced PABPN1 transcripts by quantitative RT-PCR. As can be seen in Fig. 6H, the individual depletion of hnRNP H and hnRNP A2/B1 resulted in a slight decrease in the levels of unspliced pre-mRNA relative to those of spliced mRNA and compared to cells treated with control siRNAs. In contrast, the single depletion of hnRNP C did not perturb the unspliced/spliced PABPN1 RNA ratio. Interestingly, codepletion of hnRNP H and hnRNP A2/B1 showed an additive effect in the ratio of unspliced/spliced transcripts relative to those for either single depletion (Fig. 6H), consistent with the observation that hnRNP H and hnRNP A2/B1 bind different regions of PABPN1 intron 6 (43). However, the induction of GFP-PABPN1 in cells previously depleted of both hnRNP H and hnRNP A2/B1 did not significantly alter the autoregulation capacity of PABPN1 (data not shown). Together, these results suggest that hnRNP H and hnRNP A2/B1 function as splicing repressors for PABPN1 intron 6 but are not directly involved in the mechanism of PABPN1 autoregulation.
PABPN1 autoregulation requires the nuclear RNA exosome but not nonsense-mediated RNA decay.
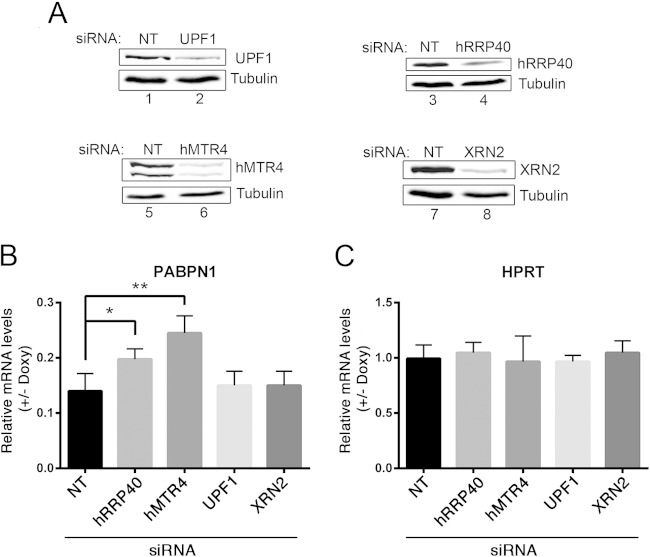
The data presented above are consistent with a model in which PABPN1-dependent intron retention promotes the rapid degradation of unspliced pre-mRNAs in the nucleus, thereby reducing the cytoplasmic levels of spliced mRNA. To examine whether nuclear RNA decay by the exosome was involved in PABPN1 autoregulation, we used siRNAs to knock down hRRP40 and hMTR4 (Fig. 7A, lanes 4 and 6), two central components of human exosome ribonucleolytic activity (44). The GFP-PABPN1 inducible cell line first was transfected with gene-specific siRNAs and cultured in the absence or presence of doxycycline to induce GFP-PABPN1. Total RNA was subsequently prepared from induced and noninduced cells, and quantitative RT-PCR was used to examine the impact of exosome deficiency on PABPN1 autoregulation by measuring the levels of PABPN1 mRNA. Consistent with our previous results, induction of GFP-PABPN1 resulted in a robust ∼85% decrease in PABPN1 mRNA in cells treated with control siRNAs (Fig. 7B). Strikingly, the most prominent defect in PABPN1 autoregulation was observed in hMTR4-depleted cells, in which PABPN1 mRNA levels were significantly increased following GFP-PABPN1 induction relative to those in cells treated with control siRNAs (Fig. 7B). PABPN1 autoregulation also was reduced in hRRP40-depleted cells (Fig. 7B). No significant changes in PABPN1 mRNA levels were detected in cells deficient for a key component of nonsense-mediated RNA decay, UPF1, and for the nuclear 5′-3′ exonuclease, XRN2 (Fig. 7A and B). Importantly, the doxycycline-dependent changes observed in hMTR4- and hRRP40-depleted cells were specific to the PABPN1 mRNA, as levels of the control HPRT mRNA were not altered by exosome deficiency after GFP-PABPN1 induction (Fig. 7C). These results indicate that the nuclear exosome contributes to PABPN1 autoregulation.
FIG 7.
PABPN1 autoregulation requires the nuclear RNA exosome. (A) Western blot validations of RNAi-mediated depletion of UPF1 (lane 2), hRRP40 (lane 4), hMTR4 (lane 6), and XRN2 (lane 8). (B and C) Quantitative RT-PCR analysis of spliced PABPN1 mRNA (B) and HPRT mRNA (C) levels using total RNA prepared from HEK293T cells treated with siRNAs specific to hRRP40, hMTR4, UPF1, and XRN2, as well as a nontarget (NT) control siRNA. To specifically assay for PABPN1-dependent effects, data were normalized to those for RPS2 mRNA and expressed relative to those for the uninduced conditions (−Doxy). P < 0.01 (**) and P < 0.05 (*) by Student's t tests.
DISCUSSION
Autoregulation is a mechanism for the homeostatic control of gene expression that becomes particularly important in cases in which a deficiency or a surplus of protein affects cell functions. PABPN1 is an evolutionarily conserved RNA-binding protein with multiple roles in posttranscriptional gene regulation. Accordingly, studies have demonstrated that a deficiency in PABPN1 alters the expression of specific genes (5, 7, 23) and causes proliferation defects (4). Conversely, studies also have shown that overexpression of Pab2, the yeast homolog of PABPN1, results in growth inhibition in Schizosaccharomyces pombe (45), while the muscle-specific overexpression of wild-type mammalian PABPN1 in Drosophila results in muscle defects (46). In oculopharyngeal muscular dystrophy (OPMD), mutations in the PABPN1 coding sequence affect mRNA levels (13, 14). Thus, the autoregulatory mechanism disclosed in this study supports the idea that PABPN1 expression needs to be strictly controlled for normal cell physiology. Importantly, we provide mechanistic insights into how this autoregulatory pathway involves PABPN1 binding to an adenosine-rich region in its 3′ UTR, which promotes the degradation of intron-retained PABPN1 pre-mRNA in the nucleus.
Gene regulation by nuclear pre-mRNA decay.
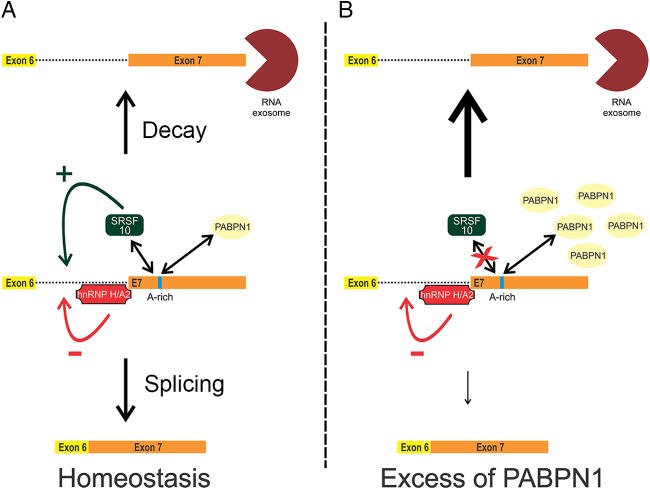
In this study, we report on the ability of PABPN1 to control its steady-state protein level via a negative feedback loop that results in reduced mRNA levels. Using a minigene construct that recapitulated PABPN1-dependent regulation, we showed that autoregulation occurs at the level of unspliced pre-mRNA (Fig. 2) and requires inefficient splicing of the 3′-terminal intron (Fig. 3). In addition, we provide evidence that PABPN1 autoregulation depends on the activity of the nuclear RNA exosome, as a deficiency in exosome function reduced the ability of excess PABPN1 to negatively control its endogenous mRNA level (Fig. 7). Collectively, our results are consistent with a model of PABPN1 homeostatic expression in which pre-mRNA decay by the nuclear exosome competes with splicing of the PABPN1 3′-terminal intron by the spliceosome (Fig. 8A). The equilibrium between decay and splicing of unspliced PABPN1 pre-mRNAs is influenced by PABPN1 protein levels (Fig. 8A and B). Accordingly, our in vitro and in vivo data suggest that PABPN1 directly interacts with a highly conserved adenosine-rich region in its 3′ UTR, and by this means it negatively affects the splicing of the 3′-terminal intron. Therefore, at high concentrations, PABPN1 can more effectively interfere with splicing of its terminal intron and influence the equilibrium toward pre-mRNA decay by the exosome, providing a mechanism for self-regulation (Fig. 8B). Although most of the results presented in the current study support an autoregulatory mechanism that acts primarily at the RNA level, we do not exclude that posttranslational mechanisms, such as the formation of insoluble nuclear PABPN1 aggregates (47), also contribute to control PABPN1 protein levels.
FIG 8.
Model for PABPN1 autoregulation. (A) The homeostatic control of PABPN1 expression involves competition between pre-mRNA decay by the exosome and splicing of the 3′-terminal intron by the spliceosome. Splicing regulators and PABPN1 influence the equilibrium between pre-mRNA splicing and decay. (B) When the nuclear concentration of PABPN1 is high, binding of PABPN1 to an adenosine (A)-rich region in its 3′ UTR interferes with splicing of the 3′-terminal intron, which promotes pre-mRNA decay by the exosome.
Despite evidence that supports the contribution of exosome-dependent nuclear RNA decay in the mechanism of PABPN1 autoregulation (Fig. 7), the fact that intron-retained PABPN1 pre-mRNAs are readily detected at steady state (Fig. 1) suggests that a pool of polyadenylated unspliced PABPN1 transcript is relatively stable in the nucleus. This observation is consistent with recent findings from Boutz et al., which show that thousands of intron-retained transcripts stably accumulate in the nucleus and are not subject to unproductive splicing by nonsense-mediated decay (28). Given that excess levels of PABPN1 caused a strong (80 to 90%) reduction in PABPN1 mRNA levels yet only marginal (5 to 10%) accumulation of intron-retained pre-mRNA, we propose that PABPN1 autoregulation mainly targets cotranscriptional splicing of intron 6, rather than the pool of intron-retained PABPN1 transcripts that accumulate in the nucleus. PABPN1 autoregulation at the level of cotranscriptional splicing would also explain why a deficiency in nuclear exosome did not affect the level of intron-retained pre-mRNA after GFP-PABPN1 induction (data not shown) but increased PABPN1 mRNA levels (Fig. 7): reduced competition from nuclear pre-mRNA decay increases the opportunity for productive cotranscriptional splicing of the 3′ terminal intron, resulting in the observed increase of PABPN1 mRNA in exosome-deficient cells.
PABPN1 did not affect the expression of intronless and intron-swap constructs (Fig. 2 and 3), despite preserving the adenosine-rich tract in their 3′ UTR. These data argue against the idea that the mechanism of PABPN1 autoregulation proceeds mainly by inhibiting the export of bound transcripts. Rather, our results are more consistent with a role of PABPN1 in the splicing modulation of its 3′-terminal intron. The exact mechanism underlying PABPN1-mediated splicing regulation still needs investigation, but it may include an influence on SRSF10 recruitment (Fig. 8). Accordingly, the results from our RIP assays (Fig. 6) suggest that binding of SRSF10 and PABPN1 to the flanking GA-rich and A-rich regions of the PABPN1 3′ UTR, respectively, are mutually exclusive. Indeed, the preferred RNA motif recognized by PABPN1 in vitro is A-G/A-A-A-G/A-A, whereas it is A-G-A-G-A-G/A-G/A for SRSF10 (40). Such sequence similarity could involve competitive binding to the PABPN1 pre-mRNA, whereby excess PABPN1 would interfere with SRSF10 recruitment, preventing the activator role of SRSF10 in the splicing of the PABPN1 3′-terminal intron (Fig. 8B). A model in which PABPN1 competes with RNA processing was proposed as the mechanism whereby PABPN1 controls alternative polyadenylation (APA) of specific genes, most likely by competing with the 3′-end processing machinery (6, 7). The nature of the gene specificity for PABPN1-regulated APA remains unclear, however. The results presented in this study raise the possibility that PABPN1 as well as other poly(A)-binding proteins, in addition to their general roles in RNA metabolism, also function in gene-specific regulation through binding to internal adenosine-rich tracts. In that respect, the major human PABP in the cytoplasm, PABPC1, binds to an adenosine-rich region in the 5′ UTR of its own mRNA, thereby negatively repressing translation (48), while the major nuclear PABP in S. cerevisiae, Nab2, binds to an internal poly(A) stretch in its mRNA to repress 3′-end processing (49). Because genome-encoded internal poly(A) tracts are relatively common in human genes, with the majority of ≥12-adenosine stretches located in introns and 3′ UTRs (50), it is tempting to speculate that these template-encoded poly(A) tracts function in gene-specific regulation via PABPs.
The PABPN1-dependent mechanism of gene regulation identified in this study is reminiscent of how S. pombe Pab2 controls the expression of specific intron-containing genes via exosome-mediated pre-mRNA decay (23). One of the important similarities between these two nuclear pre-mRNA decay pathways is that they both depend on poor splicing efficiency. Accordingly, the inefficient splicing required by these pre-mRNA decay pathways likely increases the window of opportunity for PABPN1/Pab2 and the exosome to compete with RNA splicing.
Intron retention as a mechanism for PABPN1 gene regulation.
Although RNA splicing generally is thought to occur cotranscriptionally (51), our data suggest that the 3′-terminal intron of PABPN1 frequently is spliced posttranscriptionally. Importantly, our findings revealed that intron retention (IR) is a mechanism allowing PABPN1 to control its own expression. Previous studies have shown that IR is used as a mechanism for the self-control of certain RNA-binding proteins, most frequently splicing factors (19, 52, 53). In these cases, as well as for most IR-dependent mechanisms of gene regulation, NMD appears to be the primary pathway used to achieve negative control of mRNA levels (29, 54). In contrast, our results argue that NMD is not involved in PABPN1 autoregulation, as depletion of UPF1 did not impair the ability of GFP-PABPN1 to repress endogenous PABPN1 mRNA levels (Fig. 7). Rather, our data are consistent with a mechanism of IR-mediated nuclear retention coupled to pre-mRNA decay by the exosome.
In general, IR is considered an uncommon mechanism of alternative splicing. However, recent studies indicate that IR is widespread in mammalian transcriptomes (29) and important for the regulation of functionally related groups of genes (26, 27). Mechanistically, our results indicated that a weak 5′ ss and splicing repressors, hnRNP H and hnRNP A2/B1, contribute to the degree of PABPN1 IR. Future investigations will be required to examine whether RNA polymerase II-pausing (29) and/or chromatin-associated proteins (55) also influence retention of the PABPN1 3′-terminal intron. It also will be interesting to determine the subnuclear localization of intron-retained PABPN1 transcripts, as retention at or near the site of transcription is distinctive for defective RNAs that are directed to exosome-mediated decay (56). Alternatively, unspliced PABPN1 transcripts could accumulate in a specific nuclear environment, functioning as a reservoir for rapid posttranscriptional splicing in response to cellular cues or homeostatic regulation.
Relatively few data are available on the patterns of PABPN1 expression and regulation in humans. In mice, PABPN1 protein levels vary substantially across tissues as a result of the differential expression of two major mRNA isoforms that have been proposed to result from the alternative use of poly(A) sites (9). However, it should be noted that the proximal poly(A) site identified for the mouse PABPN1 mRNA is 66 bp downstream of the stop codon (9), which maps precisely inside the conserved adenosine-rich region in the 3′ UTR, raising the possibility of mispriming by oligo(dT)-based reverse transcription. In fact, we detected intron-retained PABPN1 transcripts in the C2C12 mouse cell line (see Fig. S8 in the supplemental material), suggesting that PABPN1 IR is conserved in mice. Thus, in addition to functioning in autoregulation, the IR-mediated mechanism of gene regulation described in this study could be used to control PABPN1 levels in different cell types and tissues. For instance, by altering the concentration and/or activity of splicing regulators, such as hnRNP H, hnRNP A2/B1, and SRSF10, cells could control PABPN1 mRNA levels, as demonstrated by our data.
In summary, we have uncovered a mechanism of regulated intron retention coupled to pre-mRNA decay that controls PABPN1 expression in human cells. Our data did not reveal any significant differences in autoregulation capacity between normal and alanine-expanded versions of PABPN1 (see Fig. S1 in the supplemental material). However, it remains possible that the negative autoregulatory loop described in this study becomes sufficiently impaired during the 30- to 50-year period needed to develop the first OPMD symptoms to account for the altered levels of PABPN1 mRNA expressed from OPMD alleles (13, 14), potentially resulting in the accumulation of aberrant PABPN1 protein.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bin Tian (Rutgers New Jersey Medical School) for PABPN1 3′ READS data and Roscoe Klinck for critical reading of the manuscript. We thank Anne-Marie Landry-Voyer for generating the cell lines that conditionally express normal and alanine-expanded versions of PABPN1.
This work was supported by the Canadian Institutes of Health Research grant MOP-106595 to F.B. F.B. is a Canada Research Chair in the Quality Control of Gene Expression.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00070-15.
REFERENCES
- 1.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J 22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahle E. 1991. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66:759–768. doi: 10.1016/0092-8674(91)90119-J. [DOI] [PubMed] [Google Scholar]
- 4.Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. 2010. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum Mol Genet 19:1058–1065. doi: 10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet 8:e1003078. doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, den Dunnen JT, van der Maarel SM, Raz V, ‘t Hoen PA. 2012. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res 40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Bresson SM, Conrad NK. 2013. The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet 9:e1003893. doi: 10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apponi LH, Corbett AH, Pavlath GK. 2013. Control of mRNA stability contributes to low levels of nuclear poly(A) binding protein 1 (PABPN1) in skeletal muscle. Skelet Muscle 3:23. doi: 10.1186/2044-5040-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee A, Apponi LH, Pavlath GK, Corbett AH. 2013. PABPN1: molecular function and muscle disease. FEBS J 280:4230–4250. doi: 10.1111/febs.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brais B. 2009. Oculopharyngeal muscular dystrophy: a polyalanine myopathy. Curr Neurol Neurosci Rep 9:76–82. doi: 10.1007/s11910-009-0012-y. [DOI] [PubMed] [Google Scholar]
- 12.Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korczyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA. 1998. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 13.Anvar SY, Raz Y, Verway N, van der Sluijs B, Venema A, Goeman JJ, Vissing J, van der Maarel SM, ‘t Hoen PA, van Engelen BG, Raz V. 2013. A decline in PABPN1 induces progressive muscle weakness in oculopharyngeal muscle dystrophy and in muscle aging. Aging 5:412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroder JM, Klossok T, Weis J. 2011. Oculopharyngeal muscle dystrophy: fine structure and mRNA expression levels of PABPN1. Clin Neuropathol 30:94–103. doi: 10.5414/NPP30094. [DOI] [PubMed] [Google Scholar]
- 15.Ichinose J, Watanabe K, Sano A, Nagase T, Nakajima J, Fukayama M, Yatomi Y, Ohishi N, Takai D. 2014. Alternative polyadenylation is associated with lower expression of PABPN1 and poor prognosis in non-small cell lung cancer. Cancer Sci 105:1135–1141. doi: 10.1111/cas.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 18.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. 2007. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 20.Lejeune F, Maquat LE. 2005. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. 2008. Promoter-driven splicing regulation in fission yeast. Nature 455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- 23.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Elela SA. 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 25.Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. 2012. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol Cell 47:267–280. doi: 10.1016/j.molcel.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, Thoeng A, Khoo TL, Bailey CG, Holst J, Rasko JE. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 27.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boutz PL, Bhutkar A, Sharp PA. 2015. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witten JT, Ule J. 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres JZ, Miller JJ, Jackson PK. 2009. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9:2888–2891. doi: 10.1002/pmic.200800873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, Lucier JF, Gervais-Bird J, Tremblay K, Prinos P, Klinck R, Elela SA, Chabot B. 2008. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol 28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu W, Levy DE. 2006. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods 39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10:133–139. doi: 10.1038/nchembio.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roca X, Krainer AR, Eperon IC. 2013. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev 27:129–144. doi: 10.1101/gad.209759.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. 2005. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol Cell 20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. 2006. An interaction between U2AF 65 and CF I (m) links the splicing and 3′ end processing machineries. EMBO J 25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niwa M, Berget SM. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 39.Shin C, Manley JL. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407–417. doi: 10.1016/S0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 40.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Wu W, Li H, Cheng Y, Wei N, Zong J, Feng X, Xie Z, Chen D, Manley JL, Wang H, Feng Y. 2014. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res 42:4019–4030. doi: 10.1093/nar/gkt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han SP, Tang YH, Smith R. 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 43.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M Jr, Yeo GW. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 45.Perreault A, Lemieux C, Bachand F. 2007. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J Biol Chem 282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 46.Chartier A, Benoit B, Simonelig M. 2006. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J 25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raz V, Routledge S, Venema A, Buijze H, van der Wal E, Anvar S, Straasheijm KR, Klooster R, Antoniou M, van der Maarel SM. 2011. Modeling oculopharyngeal muscular dystrophy in myotube cultures reveals reduced accumulation of soluble mutant PABPN1 protein. Am J Pathol 179:1988–2000. doi: 10.1016/j.ajpath.2011.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Bag J. 1998. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J Biol Chem 273:34535–34542. doi: 10.1074/jbc.273.51.34535. [DOI] [PubMed] [Google Scholar]
- 49.Roth KM, Wolf MK, Rossi M, Butler JS. 2005. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol Cell Biol 25:1577–1585. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH. 2014. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip Rev RNA 5:601–622. doi: 10.1002/wrna.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. 2010. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge Y, Porse BT. 2014. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays 36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- 55.Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, Xu W, Mu S, Wen H, Qiu J, Wang Z, Yang P, Wu F, Hui J, Fu X, Shi X, Shi YG, Xing Y, Lan F, Shi Y. 2014. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 56:298–310. doi: 10.1016/j.molcel.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. 2011. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol 18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.