Abstract
Lymphocyte subpopulation levels are used for prognosis and monitoring of a variety of human diseases, especially those with an infectious etiology. As a primary step to defining the major gene variation underlying these phenotypes, we conducted the first whole-genome screen for quantitative variation in lymphocyte count, CD4 T cell, CD8 T cell, B cell, and natural killer cell numbers, as well as CD4:CD8 ratio. The screen was performed in 15 of the CEPH families that form the main human genome genetic project mapping resource. Quantitative-trait loci (QTLs) that account for significant proportions of the phenotypic variance of lymphocyte subpopulations were detected on chromosomes 1, 2, 3, 4, 8, 9, 11, 12, and 18. The most significant QTL found was for CD4 levels on chromosome 8 (empirical P=.00005). Two regions of chromosome 4 showed significant linkage to CD4:CD8 ratio (empirical P=.00007 and P=.003). A QTL for the highly correlated measures of CD4 and CD19 levels colocalized at 18q21 (both P=.003). Similarly, a shared region of chromosome 1 was linked to CD8 and CD19 levels (P=.0001 and P=.002, respectively). Several of the identified chromosome regions are likely to harbor polymorphic candidate genes responsible for these important human phenotypes. Their discovery has important implications for understanding the generation of the immune repertoire and understanding immune-system homeostasis. More generally, these data show the power of an integrated human gene–mapping approach for heritable molecular phenotypes, using large pedigrees that have been extensively genotyped.
Introduction
A properly functioning immune system is a necessary component of human survival. Cell populations that are part of the immune system—and are responsible for functional responses—carry surface structures that can be recognized by monoclonal antibodies (Mabs). The major lymphocyte populations identified in this way are CD4 (CD4 antigen [MIM 186940]) and CD8 (CD8 antigen [MIM 186910]) T cells, B cells (CD19 antigen [MIM 107265]), and natural killer (NK) cells. These subsets of lymphocytes play a vital role in defense against tumors, bacteria, viruses, and other parasites, and in the pathology of autoimmune diseases (Panayi et al. 1992; Janeway and Travers 1996; Pardoll 2001). Levels of these cells and derived measures such as CD4:CD8 ratio (CD4:CD8 T cell ratio [MIM 601083]) vary among humans, and a significant proportion of this variation appears to result from genetic differences (Amadori et al. 1995; Evans et al. 1999; Hall et al. 2000). The best example of the clinical relevance of lymphocyte subpopulation variation is in the prognosis and monitoring of the acquired immunodeficiency syndrome (AIDS). Patients with initially high levels of CD4 T cells and high CD4:CD8 T cell ratios show slower progression to AIDS than do patients with lower values (Hersh et al. 1984; Taylor et al. 1989; Fahey et al. 1990; Coates et al. 1992; Saah et al. 1992; Selwyn et al. 1992; Amadori et al. 1996).
As a first step in the identification of candidate genes that specify variation in lymphocyte levels, we determined the degree of genetic control over quantitative differences in six peripheral-blood lymphocyte subpopulations by measuring the heritability of these phenotypes. To identify quantitative-trait loci (QTLs) for the six phenotypes, we performed linkage analysis with genome-scan data at ∼5-cM marker density in a cohort of 15 healthy three-generation families that comprises a maximum of 209 white individuals from Utah. This is the first time that such a comprehensive set of immune-system variables has been analyzed in this way, and our findings should eventually lead to an improved understanding of the genetic factors influencing the development of the immune repertoire and of immune-system homeostasis.
Subjects and Methods
Subjects
Subjects were recruited through the Utah Genetics Reference Project (UGRP) based in Salt Lake City. Institutional review board approval was obtained, and each subject gave informed consent. These subjects are a subset of the CEPH family collection, which is part of the set of families from Utah that were originally collected in the early 1980s. A brief description of these families is given by White and colleagues (1985). The aim was to obtain a set of samples representing large sibships with grandparents. Complete three-generation families were ascertained anecdotally. Originally, White and colleagues sampled 18 families, each with four living grandparents and seven or more children >5 years old, as well as 10 families with similar characteristics. These and other CEPH families have been used to generate linkage maps as part of the CEPH Human Genome Mapping Project (Dib et al. 1996).
The 15 families we phenotyped and analyzed in the present study were CEPH pedigrees 1350, 1377, 1362, 1418, 1408, 1345, 1340, 1477, 1349, 1421, 1346, 1334, 1424, 1375, and 1358. Each family visited a clinic at the University of Utah Medical Center, where phenotypic data were gathered and a thorough clinical examination was performed. Follow-up phenotyping was not possible in every case, either because of deaths during the intervening 17 years or because original siblings were unavailable. The health status of the subjects was evaluated through a full physical examination and interview, and families were included for study without any selection for segregation of disease.
Flow Cytometry
Fasting venous blood samples (5 ml) were drawn into glass Vacutainer tubes containing sterile preservative-free heparin (Becton Dickinson) and were used within 2 h. Samples were taken between 7:00 and 8:00 am, to minimize any circadian fluctuation in leukocyte population levels.
Fluorescent-conjugated antibody mixtures were added to 100 ml of whole blood, were incubated at 22°C for 15 min, and were then processed using the ImmunoPrep reagent system (Beckman Coulter), which includes a 1% paraformaldehyde fixation step. The cells then remain suitable for flow cytometry for as long as 5 d, when kept at 4°C. All Mabs used were directly conjugated antihuman mouse IgG1 and were IgG1-FITC 679.1Mc7, IgG1-PE 679.1Mc7, IgG1-Cy5 679.1Mc7, CD3-Cy5 UCHT1, and CD45-Cy5 J.33 (all Immunotech/Beckman Coulter), as well as CD4-FITC SK3, CD8-PE SK1, CD16FITC NKP15, CD19PE 4G7, and CD56PE MY31 (Becton Dickinson).
Flow cytometry was performed within 40 h of cell fixation and was performed using a Coulter EPICS-XL I set up for three-color detection and XL system II software (Beckman Coulter). The cytometer was calibrated daily, using ImmunoCheck beads (Beckman Coulter), to maintain a half-peak coefficient of variation of <2 for each channel. Lymphocytes were gated according to their forward- and side-scatter properties, and 5,000 events were recorded. The lymphocyte purity of this region was calculated using a CD45-specific Mab and was >99% in each case. Positive thresholds were defined by quadrant regions, which enclosed the negative control population on each axis. FlowCount (Beckman Coulter) beads were used to estimate the concentration of lymphocytes. Absolute lymphocyte numbers, in millions of cells per milliliter of blood, were calculated using the mean of quadruplicate samples. If one of the four samples could not be analyzed for technical reasons, the mean was calculated from the remaining three samples. When two of the samples could not be analyzed, the data point was excluded from the study. Absolute numbers of lymphocyte subsets were determined as a percentage of the total number of lymphocytes.
STR Marker Genotyping
Genotype data for the 15 extended pedigrees used in the present study were available from the CEPH database. Genetic markers were largely autosomal STRs, and positions were assigned using the Marshfield map (Broman et al. 1998). The average intermarker distance was ∼5 cM. A detailed list of the 655 genetic markers used is available from the authors.
Heritability Analysis
Table 1 summarizes the number of spouse-spouse, parent-offspring, and sibling-sibling pairs available for analysis of immune phenotypes. Estimates of familial correlations and their standard deviations were obtained after traits were adjusted, according to the covariates age and sex, using REGC. Heritability was calculated from the familial correlations, using ASSOC2 within SAGE (Statistical Analysis for Genetic Epidemiology), under the assumption that parent-offspring and sibling-sibling correlations are the same (SAGE 2001a).
Table 1.
Summary of Utah Genetic Reference Project (CEPH) Families Used for Linkage Analysis
|
No. of |
|||
| Trait | Spouse Pairs | Sibling Pairs | Parent-Offspring Pairs |
| Lymphocyte count, CD4 T cell level, CD8 T cell level, CD4:CD8 ratio | 18 | 385 | 225 |
| CD19 B cell level | 17 | 374 | 211 |
| NK cell level | 7 | 209 | 119 |
QTL Analysis
QTL analysis was performed using the SIBPAL2 program implemented in the SAGE package (SAGE 2001). The W2 variation of the revised Haseman-Elston regression approach was followed (Haseman and Elston 1972; Shete et al. 1999; Xu et al. 2000; SAGE 2001c). The W2 method uses a weighted combination of squared trait difference and squared mean–corrected trait sum and is similar to the method described by Xu and colleagues (2000) and, with modifications, to the method described by Shete and colleagues (1999). Weights are chosen proportional to the inverse residual variance of the squared differences and sums. The method for combining pairs within multiple sibships is fully described by Xu et al. (2000). In our data set, the W2 product and difference variants of the revised Haseman-Elston method gave comparable results (data not shown) (Haseman and Elston 1972; Elston et al. 2000; Xu et al. 2000). W2 results are reported. Age and sex were included as covariates in the regression models. Neither covariate was significant in most of the analyses.
Simulation of Empirical P Values
The method is described in the SAGE 4.0 manual (SAGE 2001). Nominal P values were calculated assuming the W2 test statistic follows a t distribution that is asymptotically correct. However, because it was unclear whether the sample size was sufficient to ensure asymptopia, we also calculated the P values empirically, using a Monte Carlo permutation method. We permuted the allele sharing within families so that the correlation structure among relatives remained the same. The test statistics were then computed for each multipoint marker position across the chromosome; 10,000 simulations were performed. The true test statistic was then compared to the distribution of the permuted test statistics (at each point) to derive the empirical P value at each point.
Quantitative Transmission/Disequilibrium Test (QTDT) Analysis
The method proposed by George et al. (1999) was used to conduct QTDT analysis. This test detects association in the presence of linkage, does not require independence of observations, and allows for analysis of pedigree data and adjustment for covariates. The maximum-likelihood estimates of the parameters and the standard errors of the estimates are computed by numerical methods. These procedures are implemented in the program ASSOC2 of the SAGE software package (SAGE 2001a).
Results
Distributions and Covariates
Table 2 shows distributions of values including numbers studied, means, standard deviations, and maximum and minimum values for the six lymphocyte-subpopulation parameters studied in the 15 CEPH pedigrees. All values were within the ranges expected on the basis of previously published data. Mean age of subjects was 42.5 years (SD 18.7 years; range 18–90 years). Variation in several of the traits under study is highly correlated, and Pearson correlation coefficients are summarized in table 3. Relationships may be divided into trivial and material. For example, the lymphocyte count variable is largely additively composed of CD4 T cell, CD8 T cell, CD19 B cell, and NK cell numbers. This leads to highly significant correlations (each P=.0001). Other correlations, however, such as CD19 B cells with both CD4 and CD8 T cells (0.57 and 0.44, both P=.0001) and NK cells with CD4 and CD8 T cells (0.26 and 0.57, P=.01 and P=.0001, respectively) are not so easily explained away and may reflect shared environmental and/or genetic factors that impact upon multiple phenotypes. Age was negatively correlated with CD19 B cell level (P=.002) and nonsignificantly related to lymphocyte count but had no significant effect on the other phenotypes in these families.
Table 2.
Summary Statistics on Variables Measured in 15 Utah Genetic Reference Project (CEPH) Families Used for Linkage Analysis[Note]
| Variable | No. | Mean | SD | MinimumValue | MaximumValue |
| Lymphocyte count | 142 | 1.29 | .45 | .18 | 2.63 |
| CD19 level | 138 | .16 | .08 | .02 | .41 |
| NK level | 83 | .06 | .04 | .001 | .21 |
| CD4 level | 142 | .59 | .24 | .08 | 1.4 |
| CD8 level | 142 | .36 | .16 | .05 | .94 |
| CD4:CD8 ratio | 142 | 1.81 | .83 | .19 | 6.25 |
| Age | 100 | 42.54 | 18.76 | 18 | 90 |
Note.— With the exception of CD4:CD8 ratio and age, values are expressed in millions per milliliter.
Table 3.
Variables Measured in 15 Utah Genetic Reference Project (CEPH) families Used for Linkage Analysis
|
Correlation Coefficients (P) of |
||||||
| Variable | CD19 | NK | CD4 | CD8 | CD4:CD8Ratio | Age |
| Lymphocyte count | .69 (.0001) | .45 (.0001) | .87 (.0001) | .80 (.0001) | .02 (.80) | −.18 (.07) |
| CD19 | .18 (.10) | .57 (.0001) | .44 (.0001) | .07 (.40) | −.31 (.002) | |
| NK | .26 (.01) | .57 (.0001) | −.24 (.02) | .04 (.68) | ||
| CD4 | .47 (.0001) | .40 (.0001) | −.15 (.12) | |||
| CD8 | −.47 (.0001) | −.10 (.30) | ||||
| CD4:CD8 ratio | .08 (.43) | |||||
Heritabilities
Estimates were obtained of familial correlations and their standard deviations, when traits are adjusted by the covariates age and sex (data not shown). These were used to derive heritability estimates (table 4). All six of the immune traits studied are under significant genetic control, with CD8 T cell level being the most heritable trait. These estimates are comparable to estimates derived from twin studies of subjects in the same age range (Hall et al. 2000).
Table 4.
Heritability Estimates (Determined by Use of ASSOC) for Six Immunological Phenotypes in 15 Utah Genetic Reference Project (CEPH) Families
| Trait | Heritability |
| Lymphocyte count | .611 |
| CD19 B cell level | .608 |
| NK cell level | .463 |
| CD4 T cell level | .564 |
| CD8 T cell level | .699 |
| CD4:CD8 ratio | .462 |
Quantitative Linkage Analysis
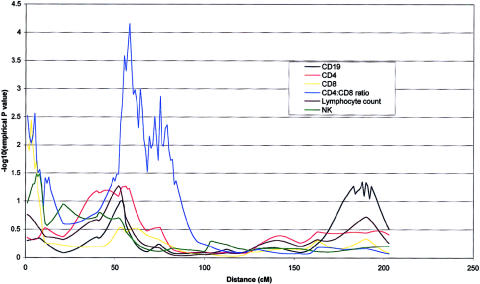
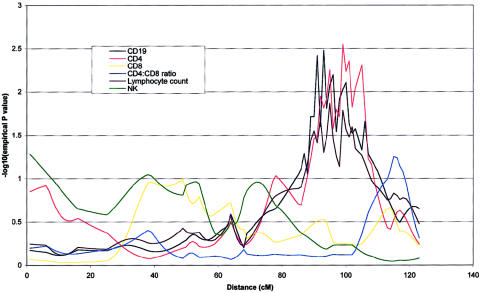
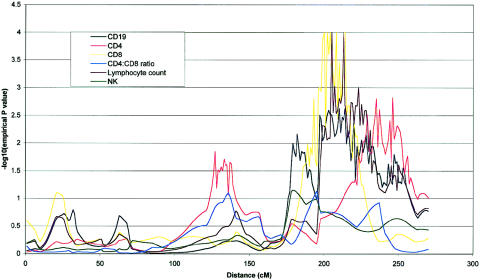
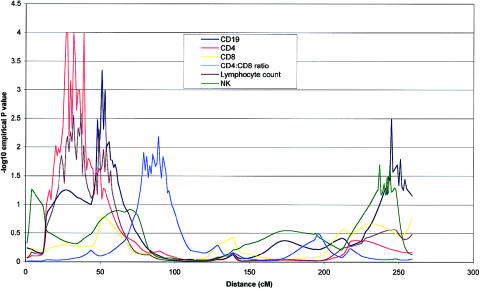
Table 5 gives a summary of the most significant results using the W2 method of a whole-genome scan for the six quantitative immune phenotypes studied. Empirical P values calculated by simulation (P<.01), together with nominal P values are shown for linkages adjusted for the covariates age and sex. Nineteen covariate-adjusted phenotype-region combinations showed nominal evidence of genetic linkage at this significance level. QTLs accounting for significant proportions of the phenotypic variance of lymphocyte subpopulations were detected on chromosomes 1, 2, 3, 4, 8, 9, 11, 12, and 18. The most significant QTL found was for CD4 levels on chromosome 8 at 65–79 cM (empirical P=.00005). Two regions of chromosome 4 showed significant linkage to CD4:CD8 ratio (fig. 1). The centromeric region between 53 and 79 cM showed stronger linkage than the telomeric region between 2 and 5 cM (empirical P=.00007 and P=.003, respectively). A QTL for the highly correlated measure of CD4 and CD19 levels colocalized at 18q21 (each empirical P=.003), as shown in figure 2. This suggests the presence of a QTL that is involved in determining CD4 T cell and B cell levels but does not play a major role in CD8 T cell levels. Similarly, a shared region of chromosome 1 (fig. 3) was linked to CD8 and CD19 levels (empirical P=.0001 and P=.002, respectively) with considerable overlap with a region of linkage to lymphocyte count (empirical P=.001). This region is also adjacent to a region of linkage to CD4 levels (empirical P=.001). Further colocalization was apparent on chromosome 8, with a QTL in an overlapping region showing significant linkage with lymphocyte count and CD19 level (empirical P=.006 and P=.003, respectively). As shown in figure 4, CD4 level and lymphocyte count are linked to a shared region of chromosome 2 (empirical P=.0003 and P=.003, respectively) that overlaps with a region of linkage to CD19 levels (empirical P=.001). QTLs for single lymphocyte subpopulations were found on chromosomes 9 (CD4), 11 (lymphocyte count), and 12 (CD19) (see table 5).
Table 5.
Whole-Genome Scan of Quantitative Immune-System Phenotypes[Note]
| Chromosomeand Phenotype | Region(cM) | Empirical Pa | Nominal Pb |
| 1: | |||
| Lymphocyte count | 200–231 | .0013 | .0018 |
| CD19 | 197–228 | .0024 | .0039 |
| CD4 | 227–252 | .0015 | .0057 |
| CD8 | 196–222 | .00013 | .00020 |
| 2: | |||
| Lymphocyte count | 28–54 | .0026 | .019 |
| CD19 | 50–56 | .0010 | .0039 |
| CD4 | 24–40 | .00028 | .00078 |
| CD4:CD8 | 89 | .0013 | .006 |
| 3: | |||
| NK | 192–199 | .0048 | .019 |
| 4: | |||
| CD4:CD8 | 2–5 | .0027 | .012 |
| CD4:CD8 | 53–79 | .00007 | .00012 |
| 8: | |||
| Lymphocyte count | 15–18 | .0064 | .014 |
| CD19 | 16–20 | .0029 | .0040 |
| CD4 | 65–79 | .00005 | .0039 |
| 9: | |||
| CD4 | 74–84 | .00083 | .0022 |
| 11 | |||
| Lymphocyte count | 38–39 | .0052 | .012 |
| 12: | |||
| CD19 | 149–156 | .00082 | .011 |
| 18: | |||
| CD19 | 93–100 | .0033 | .0054 |
| CD4 | 99–105 | .0028 | .0051 |
Note.— Probabilities were calculated using the SIBPAL2 W2 method.
Empirical significance levels (for P<.01) by chromosome regions, calculated using multipoint “new Haseman-Elston” weighted regression method with both age and sex in the model (Haseman and Elston 1972; Xu et al. 2000; SAGE 2001c).
Nominal significance levels, with both age and sex in the model.
Figure 1.
Chromosome 4 multipoint linkage map for six phenotypes under investigation. CD4:CD8 ratio was strongly linked, peaking at 58 cM (P<.0001). A less significant but coincident linkage was seen for CD8 and CD4:CD8 ratio in the p telomeric region (P<.01). QTL analysis was performed with the W2 method in SIBPAL2, using all 15 families (excluding any grandparents who were available). Marker distances were based on the Marshfield Clinic map. P values for linkage were derived empirically, using a Monte Carlo permutation method, and are given as negative logs to base 10 on the abscissa.
Figure 2.
Chromosome 18 multipoint linkage map for six phenotypes under investigation. CD4 and CD19 achieved nominal P values of ⩽.01 for at least one point on the chromosome. CD4, CD19, and lymphocyte counts are distinguished by an apparent region of coincident linkage between 89 and 107 cM. QTL analysis was performed as described in the legend of figure 1.
Figure 3.
Chromosome 1 multipoint map for six phenotypes under investigation. CD8, CD4, CD19, and lymphocyte counts achieved nominal P values of ⩽.01 for at least one point on the chromosome. CD8, CD19, and lymphocyte counts are distinguished by an apparent region of coincident linkage between 193 and 215 cM. QTL analysis was performed as described in the legend of figure 1.
Figure 4.
Chromosome 2 multipoint map for six phenotypes under investigation. CD4, CD19, CD4:CD8 ratio, and lymphocyte count all achieved nominal P values of ⩽.01 for at least one point on the chromosome. CD4 and lymphocyte count are distinguished by an apparent region of coincident linkage around 32 cM. QTL analysis was performed as described in the legend of figure 1.
Quantitative TDT Analysis
Linkage and association analyses are directed toward identifying candidate genes whose variation accounts for the variation in phenotype. The chromosome 18 region linked to CD4 T cell and CD19 B cell levels contains a number of candidate genes involved in lymphocyte survival and function that are worthy of investigation. One prominent candidate gene in this region is Bcl-2, which is involved in T- and B cell apoptosis and thus in control of the size of these cellular populations. As a first step, we genotyped an STR polymorphism associated with the Bcl-2 gene in the CEPH families. This STR is located 543 bp upstream of the Bcl-2 start codon (Mehrian et al. 1998). Six alleles were observed in the families. QTDT analysis was performed for all six phenotypes under investigation since they were highly correlated. Significant transmission distortion was seen for allele 6, with lymphocyte count and CD4 T cell level indicating association in the presence of linkage (table 6) (Abecasis et al. 2000; Martin et al. 2000). No significant results were seen for the other four phenotypes tested.
Table 6.
Association of Bcl-2 STR Alleles with Lymphocyte Count and CD4 T Cell Levels in UGRP/CEPH Families[Note]
| Bcl-2 STR Alleles | SampleSize | Pa | Corrected Pb |
| Lymphocyte count: | |||
| Allele 1 | 9 | NS | NS |
| Allele 2 | 15 | NS | NS |
| Allele 3 | 37 | NS | NS |
| Allele 4 | 0 | … | … |
| Allele 5 | 17 | .03490 | NS |
| Allele 6 | 7 | <.000001 | <.000006 |
| CD4 T cell count: | |||
| Allele 1 | 9 | NS | NS |
| Allele 2 | 15 | NS | NS |
| Allele 3 | 37 | NS | NS |
| Allele 4 | 0 | … | … |
| Allele 5 | 17 | NS | NS |
| Allele 6 | 7 | .00128 | .00768 |
Note.— QTDT was assessed for Bcl-2 alleles against lymphocyte count and CD4 T cell levels, using the method of George et al. (1999). Significant transmission distortion was seen for allele 6, for lymphocyte count and CD4 T cell level, indicating association in the presence of linkage. P values are only calculated for transmitted alleles. NS = not significant.
Uncorrected significance level.
Significance level with Bonferroni correction.
Discussion
B and T cells are the major populations mediating the adaptive arm of the immune system. In recent years, there has been an explosion in the understanding of the molecular basis of many components of the immune system. Studies of animal models and individuals who have naturally occurring deficiencies of the immune system, along with improvements in molecular biology techniques, have led to the identification of many of the genes that are involved in the development and function of the immune system (DiSanto et al. 1993; Puck 1994; Leonard 1996; Elder 1998; Schuster and Kreth 2000). These include genes responsible for lymphocyte subset differentiation, cell function, immunity to various microorganisms, and lymphocyte activation or regulation. Such studies reveal a complex set of developmental pathways that result in the generation of a large repertoire of B and T lymphocytes, NK cells, monocytes, macrophages, and dendritic cells. Together, these provide the individual with the capability of mounting successful adaptive and innate immune responses. There are a number of genes characterized whose lack of function leads to a total absence of a particular cell type and thus results in a number of immune deficiencies in humans. In contrast, there is little knowledge of how genes may play a role in modulating the actual levels of the individual cell populations while still maintaining a functional balance of the immune system.
The extent to which polymorphic genes account for the variation in levels of functional populations of lymphocytes has until recently been an underinvestigated area. Several studies have concentrated on T cell populations, showing that variation in the important clinical parameters of CD4 and CD8 T cell levels and CD4:CD8 ratio are significantly heritable (Amadori et al. 1995; Evans et al. 1999; Hall et al. 2000). It has also been suggested by segregation analysis that CD4:CD8 ratio and CD4 and CD8 T cell levels are under major recessive gene control (Amadori et al. 1996; Clementi et al. 1999). In the present study, we used extended families to provide further heritability estimates of variation among key immune cell populations of lymphocytes, CD4, CD8, CD19 B, and NK cells. We also used a 5-cM genetic map to discover QTLs for variation in these cell populations on a number of different chromosomes. These data open the possibility of fine mapping and identifying positional candidates explaining the observed variation. Nineteen chromosomal regions showed significant levels of linkage over nine different chromosomes. Five of the chromosomes showed linkage to more than one phenotype and, in several instances, a shared chromosomal region showed linkage to more than one correlated phenotype. This suggests the possibility of one or several genes controlling variation in the levels of several correlated lymphocyte subpopulations. Gene-driven correlation between phenotypes may exist, for at least two reasons. First, pleiotropic genes may have direct effects on the variation across multiple cell subpopulations through developmental or homeostatic mechanisms. Second, the immune system is a highly organized and mutually regulated complex. Genes which affect one particular cell population may exert an indirect effect upon a second dependent population. We recently showed that both of these classes of gene control act to shape the covariation in CD4 and CD8 T cell levels (Ahmadi et al. 2001). Examples of shared regions are on chromosome 1 (CD19 and CD8), chromosome 2 (lymphocyte count and CD4) and chromosome 8 (lymphocyte count and CD19). In some instances, these shared chromosomal regions overlap with or are adjacent to a second region of linkage with another correlated variable. Examples of this are the shared region of chromosome 1, which is linked to CD19 B cell and CD8 T cell levels and which overlaps with a region showing linkage to CD4 T cells levels (see table 5). Similarly, the region on chromosome 2 showing linkage to CD4 T cell numbers and lymphocyte count overlaps with a region showing linkage to CD19 B cells numbers. There are also instances where more than one region on the same chromosome shows significant linkage to a phenotype, as is shown by two different regions of chromosome 4 being linked to CD4:CD8 ratio.
Examination of the physical maps within these regions of linkage suggests a number of positional candidate genes which merit further investigation. We examined the chromosome 18 region in some detail. It encompasses the IDDM6, Bcl-2, and RANK loci all of which have been implicated in T cell function and/or autoimmune disease risk (Hawkins and Vaux 1997; Anderson et al. 1997; Bain and Murre 1998; Merriman et al. 2001). In particular, Bcl-2 is recognized as having a profound effect on lymphocyte survival and through associated variation, could be a strong candidate for determining heritable differences in lymphocyte subset levels. To test this idea, we identified a polymorphic STR within the first intron of the Bcl-2 gene and genotyped it in the 15 families. Significant transmission distortion was seen at this locus for allele 6 for the correlated measures of lymphocyte count and CD4 T cell level, indicating association in the presence of linkage. Further work will be necessary to exhaustively identify variation at this QTL and to examine its prediction of lymphocyte and CD4 T cell levels in unrelated individuals. The genetic evidence may also be viewed in light of the fact that Bcl-2 has profound oncogenic potential (Butturini and Gale 1990; Yang and Korsmeyer 1996) and that mouse knockout studies show a key role of the gene in T cell development from hematopoietic stem cells (Nakayama et al. 1994; Matsuzaki et al. 1997).
The results from the QTDT analysis show that, for one phenotype (CD4 T cells), a proportion of the linkage shown to chromosome 18 may be explained by association to Bcl-2, whereas, for CD19 B cells, this was shown not to be the case. Furthermore, there was evidence of association of Bcl-2 allele 6 and lymphocyte count, whereas there had originally not been any evidence of linkage to this region in the data set studied. This would suggest that relying on linkage results to find candidate genes is not sufficient and is consistent with the differences in sensitivity of the two approaches (Risch 2000). Another obvious candidate region for variable immune-system phenotypes is HLA. We found no evidence for linkage between the parameters measured and the HLA region at chromosome 6p21, although this does not preclude effects that could be detected by use of locus-specific markers and powerful association tests.
Until now, studies of the impact of genetic variation on the functioning of the immune system have largely concentrated on qualitative issues of response and nonresponse. Most studies have been conducted in the mouse, and experimental manipulation of the murine genome using knockout and transgenic technologies has enabled insight to be gained in systems that are largely driven to phenotypic extremes. Grupe et al. (2001) have described potential improvements in the mapping of QTLs for complex traits that enable rapid interval mapping between inbred mouse strains. Studies in humans are less advanced. In the present study, we mapped a number of QTLs for variation in multiple immune-system parameters, using the core set of CEPH families previously used for construction of the human genetic map (Dib et al. 1996). The logic of this approach is that a core set of human subjects organized into extended families can be used efficiently for mapping variation that determines multiple human quantitative traits. The approach is suitable for investigating heritable variables across the normal range, and it contrasts with the popular strategy of linkage analysis of disease traits in families with multiple affected individuals. We foresee that our approach may establish a genetic window on the range of variation associated with “normal” human physiology and function, with the eventual aim of understanding the role of selection in shaping the complex network of variation in biological traits. It is unclear to what extent immune-system parameters, such as CD4:CD8 ratio and T cell subset levels, are typical complex traits. Heritabilities of ∼50% fall in the middle of the range for human biological variables, and it is encouraging that major QTLs appear to exist for some of these parameters. Cytofluorographic measurements are objective and very accurate. As a result, calculated heritabilities will have a relatively low error rate, reflecting the low measurement error of the technique. The accurate phenotype measure will increase the power to map QTLs.
The degree to which the extremes of normal variation may represent risk factors for common diseases remains to be established more generally. Crude measures, such as white cell count, were shown to be significantly influenced by genetic factors and to predict all-cause mortality and specific mortality from coronary artery disease and cancer (Grimm et al. 1985; Whitfield and Martin 1985; Hansen et al. 1990; Ensrud and Grimm 1992). It is likely that heritable variation in immune-system parameters has had adaptive value in the past. This is most likely to have been the case in environments where chronic and intense pathogen immune stimulation was common, rather than in a contemporary Western environment, such as the United States, from which the families were sampled. However, rapid changes in environmental conditions, such as the introduction of new pathogens like HIV, may allow expression of the adaptive value of the residual genetic and phenotypic variation. The QTL which predicts CD4:CD8 ratio on chromosome 4, for example, would have a profound effect on the rate of progression of HIV disease in an untreated subject (Fahey et al. 1990; Burcham et al. 1991). This and other QTLs that determine levels of lymphocyte subsets may impact upon human health in a variety of ways. However, the initial benefits of these findings will be in furthering our understanding of the development and homeostasis of the immune repertoire in humans. We have applied conventional techniques to the novel area of immune-system architecture. In a general sense, the use of an integrated human gene-mapping approach for heritable molecular phenotypes in large pedigrees should be considered a powerful tool for unraveling the effects of polymorphic genes in human health and disease.
Acknowledgments
M.A.H. is an Arthritis Research Campaign Postdoctoral Research Fellow. We would like to thank the family members for their generous participation in this study and the rest of the Utah Genetics Reference Project team for their contribution. The core support for this project was provided by the W. M. Keck Foundation. Some of the results of the present article were obtained by using the program package SAGE, which is supported by U.S. Public Health Service Resource Grant 1 P41 RR03655 from the National Center for Research Resources.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CEPH Genotype Database, http://www.cephb.fr/cephdb/ [Google Scholar]
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CD4 antigen [MIM 186940], CD8 antigen [MIM 186910], CD19 antigen [MIM 107265], and CD4/CD8 T cell ratio [MIM 601083])
- SAGE documentation, http://darwin.cwru.edu/sage40/sage40.html
References
- Abecasis GR, Cardon LR, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi KR, Hall MA, Norman P, Vaughan RW, Snieder H, Spector TD, Lanchbury JS (2001) Genetic determinism in the relationship between human CD4 + and CD8+ T lymphocyte populations? Genes Immun 2:381–387 [DOI] [PubMed] [Google Scholar]
- Amadori A, Zamarchi R, Chieco-Bianchi L (1996) CD4: CD8 ratio and HIV infection: the “tap-and-drain” hypothesis. Immunol Today 17:414–417 [DOI] [PubMed] [Google Scholar]
- Amadori A, Zamarchi R, De Silvestro G, Forza G, Cavatton G, Danieli GA, Clementi M, Chieco-Bianchi L (1995) Genetic control of the CD4/CD8 T-cell ratio in humans. Nat Med 1:1279–1283 [DOI] [PubMed] [Google Scholar]
- Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, Roux ER, Teepe MC, DuBose RF, Cosman D, Galibert L (1997) A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390:175–179 [DOI] [PubMed] [Google Scholar]
- Bain G, Murre C (1998) The role of E-proteins in B- and T-lymphocyte development. Semin Immunol 10:143–153 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham J, Marmor M, Dubin N, Tindall B, Cooper DA, Berry G, Penny R (1991) CD4% is the best predictor of development of AIDS in a cohort of HIV-infected homosexual men. AIDS 5:365–372 [DOI] [PubMed] [Google Scholar]
- Butturini A, Gale RP (1990) Oncogenes and leukemia. Leukemia 4:138–160 [PubMed] [Google Scholar]
- Clementi M, Forabosco P, Amadori A, Zamarchi R, De Silvestro G, Di Gianantonio E, Chieco-Bianchi L, Tenconi R (1999) CD4 and CD8 T lymphocyte inheritance: evidence for major autosomal recessive genes. Hum Genet 105:337–342 [DOI] [PubMed] [Google Scholar]
- Coates RA, Farewell VT, Raboud J, Read SE, Klein M, MacFadden DK, Calzavara LM, Johnson JK, Fanning MM, Shepherd FA (1992) Using serial observations to identify predictors of progression to AIDS in the Toronto Sexual Contact Study. J Clin Epidemiol 45:245–253 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- DiSanto JP, Bonnefoy JY, Gauchat JF, Fischer A, de Saint Basile G (1993) CD40 ligand mutations in X-linked immunodeficiency with hyper-IgM. Nature 361:541–543 [DOI] [PubMed] [Google Scholar]
- Elder ME (1998) ZAP-70 and defects of T-cell receptor signaling. Semin Hematol 35:310–320 [PubMed] [Google Scholar]
- Elston RC, Buxbaum S, Jacobs KB, Olson JM (2000) Haseman and Elston revisited. Genet Epidemiol 19:1–17 [DOI] [PubMed] [Google Scholar]
- Ensrud K, Grimm RH Jr (1992) The white blood cell count and risk for coronary heart disease. Am Heart J 124:207–213 [DOI] [PubMed] [Google Scholar]
- Evans DM, Frazer IH, Martin NG (1999) Genetic and environmental causes of variation in basal levels of blood cells. Twin Res 2:250–257 [DOI] [PubMed] [Google Scholar]
- Fahey JL, Taylor JM, Detels R, Hofmann B, Melmed R, Nishanian P, Giorgi JV (1990) The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N Engl J Med 322:166–172 [DOI] [PubMed] [Google Scholar]
- George V, Tiwari HK, Zhu X, Elston RC (1999) A test of transmission/disequilibrium for quantitative traits in pedigree data, by multiple regression. Am J Hum Genet 65:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm RH Jr, Neaton JD, Ludwig W (1985) Prognostic importance of the white blood cell count for coronary, cancer, and all-cause mortality. JAMA 254:1932–1937 [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G (2001) In silico mapping of complex disease-related traits in mice. Science 292:1915–1918 [DOI] [PubMed] [Google Scholar]
- Hall MA, Ahmadi KA, Norman P, Snieder H, Macgregor A, Vaughan RW, Spector TD, Lanchbury JS (2000) Genetic influence on peripheral blood T lymphocyte levels. Genes Immun 1:423–427 [DOI] [PubMed] [Google Scholar]
- Hansen LK, Grimm RH Jr, Neaton JD (1990) The relationship of white blood cell count to other cardiovascular risk factors. Int J Epidemiol 19:881–888 [DOI] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Vaux DL (1997) The role of the Bcl-2 family of apoptosis regulatory proteins in the immune system. Semin Immunol 9:25–33 [DOI] [PubMed] [Google Scholar]
- Hersh EM, Reuben JM, Mansell PW, Rios A, Gutterman JU, Munn G, Murray JL, Spector SP, Goldstein AL, Newell GR (1984) Immunologic studies of the acquired immune deficiency syndrome: relationship of immunodeficiency to extent of disease. Ann NY Acad Sci 437:364–372 [DOI] [PubMed] [Google Scholar]
- Janeway CA, Travers P (1996) Immunobiology: the immune system in health and disease. Churchill Livingstone, London [Google Scholar]
- Leonard WJ (1996) The molecular basis of X-linked severe combined immunodeficiency: defective cytokine receptor signaling. Annu Rev Med 47:229–239 [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Nakayama K, Tomita T, Isoda M, Loh DY, Nakauchi H (1997) Role of bcl-2 in the development of lymphoid cells from the hematopoietic stem cell. Blood 89:853–862 [PubMed] [Google Scholar]
- Mehrian R, Quismorio FP Jr, Strassmann G, Stimmler MM, Horwitz DA, Kitridou RC, Gauderman WJ, Morrison J, Brautbar C, Jacob CO (1998) Synergistic effect between IL-10 and bcl-2 genotypes in determining susceptibility to systemic lupus erythematosus. Arthritis Rheum 41:596–602 [DOI] [PubMed] [Google Scholar]
- Merriman TR, Cordell HJ, Eaves IA, Danoy PA, Coraddu F, Barber R, Cucca F, et al (2001) Suggestive evidence for association of human chromosome 18q12-q21 and its orthologue on rat and mouse chromosome 18 with several autoimmune diseases. Diabetes 50:184–194 [DOI] [PubMed] [Google Scholar]
- Nakayama K, Negishi I, Kuida K, Sawa H, Loh DY (1994) Targeted disruption of Bcl-2 alpha beta in mice: occurrence of gray hair, polycystic kidney disease, and lymphocytopenia. Proc Natl Acad Sci USA 91:3700–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayi GS, Lanchbury JS, Kingsley GH (1992) The importance of the T cell in initiating and maintaining the chronic synovitis of rheumatoid arthritis. Arthritis Rheum 35:729–735 [DOI] [PubMed] [Google Scholar]
- Pardoll D (2001) T cells and tumours. Nature 411:1010–1012 [DOI] [PubMed] [Google Scholar]
- Puck JM (1994) Molecular basis for three X-linked immune disorders. Hum Mol Genet 3:1457–1461 [DOI] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Saah AJ, Munoz A, Kuo V, Fox R, Kaslow RA, Phair JP, Rinaldo CR Jr, Detels R, Polk BF (1992) Predictors of the risk of development of acquired immunodeficiency syndrome within 24 months among gay men seropositive for human immunodeficiency virus type 1: a report from the Multicenter AIDS Cohort Study. Am J Epidemiol 135:1147–1155 [DOI] [PubMed] [Google Scholar]
- SAGE (2001a) Statistical analysis for genetic epidemiology, 4.0. Computer program package available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland. Section 5, pp 37–47 [Google Scholar]
- ——— (2001b) Statistical analysis for genetic epidemiology, 4.0. Computer program package available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland. Section 15, pp 126–143 [Google Scholar]
- ——— (2001c) Statistical analysis for genetic epidemiology, 4.0. Computer program package available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland. Section 15.3.5, p 137 [Google Scholar]
- ——— (2001d) Statistical analysis for genetic epidemiology, 4.0. Computer program package available from the Department of Epidemiology and Biostatistics, Rammelkamp Center for Education and Research, MetroHealth Campus, Case Western Reserve University, Cleveland. Section 15.2.4.6, p 132 [Google Scholar]
- Schuster V, Kreth HW (2000) X-linked lymphoproliferative disease is caused by deficiency of a novel SH2 domain-containing signal transduction adaptor protein. Immunol Rev 178:21–28 [DOI] [PubMed] [Google Scholar]
- Selwyn PA, Alcabes P, Hartel D, Buono D, Schoenbaum EE, Klein RS, Davenny K, Friedland GH (1992) Clinical manifestations and predictors of disease progression in drug users with human immunodeficiency virus infection. N Engl J Med 327:1697–1703 (erratum 328:671 [1993]) [DOI] [PubMed] [Google Scholar]
- Shete S, Jacob KJ, Elston RC (1999) Adding further power to the Haseman and Elston Method for detecting linkage. Genet Epidemiol 17:194 [DOI] [PubMed] [Google Scholar]
- Taylor JM, Fahey JL, Detels R, Giorgi JV (1989) CD4 percentage, CD4 number, and CD4:CD8 ratio in HIV infection: which to choose and how to use. J Acquir Immune Defic Syndr 2:114–124 [PubMed] [Google Scholar]
- White R, Leppert M, Bishop DT, Barker D, Berkowitz J, Brown C, Callahan P, Holm T, Jerominski L (1985) Construction of linkage maps with DNA markers for human chromosomes. Nature 313:101–105 [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Martin NG (1985) Genetic and environmental influences on the size and number of cells in the blood. Genet Epidemiol 2:133–144 [DOI] [PubMed] [Google Scholar]
- Xu X, Weiss S, Xu X, Wei LJ (2000) A unified Haseman-Elston method for testing linkage with quantitative traits. Am J Hum Genet 67:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Korsmeyer SJ (1996) Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood 88:386–401 [PubMed] [Google Scholar]