Significance
Tau is an important microtubule-associated protein. Although the structure–function relationship of Tau has been intensively studied for many years primarily by molecular biology and biochemical approaches, little is still known about the molecular mechanisms by which Tau interacts with microtubules and promotes microtubule assembly. Here, we provide detailed insight into the Tau–microtubule association by using NMR spectroscopy and mass spectrometry. We show that Tau binds to microtubules by using small groups of residues, which are important for pathological aggregation of Tau. We further show that Tau stabilizes a straight protofilament conformation by binding to a hydrophobic pocket in between tubulin heterodimers. Our work is thus relevant to normal Tau action development and in Tau-related neurodegenerative diseases.
Keywords: Tau, microtubule, NMR spectroscopy, Alzheimer's disease, chemical cross-linking
Abstract
The structure, dynamic behavior, and spatial organization of microtubules are regulated by microtubule-associated proteins. An important microtubule-associated protein is the protein Tau, because its microtubule interaction is impaired in the course of Alzheimer’s disease and several other neurodegenerative diseases. Here, we show that Tau binds to microtubules by using small groups of evolutionary conserved residues. The binding sites are formed by residues that are essential for the pathological aggregation of Tau, suggesting competition between physiological interaction and pathogenic misfolding. Tau residues in between the microtubule-binding sites remain flexible when Tau is bound to microtubules in agreement with a highly dynamic nature of the Tau–microtubule interaction. By binding at the interface between tubulin heterodimers, Tau uses a conserved mechanism of microtubule polymerization and, thus, regulation of axonal stability and cell morphology.
Microtubules regulate cell division, cell morphology, intracellular transport, and axonal stability and, therefore, play crucial roles in cell function (1). Microtubules are built from tubulin heterodimers that polymerize into protofilaments and associate laterally into microtubules (2). Microtubule dynamics in neurons is modulated by several accessory proteins termed microtubule-associated proteins (3). However, little is known about the mechanism of assembly and stabilization of microtubules by microtubule-associated proteins.
An important microtubule-associated protein is the protein Tau, which promotes formation of axonal microtubules, stabilizes them, and drives neurite outgrowth (4, 5). The adult human brain contains six isoforms of Tau, which are generated from a single gene by alternative splicing. The six isoforms are composed of either three or four repeats, with up to two N-terminal inserts, and range from 37 to 45 kDa (6). The 31- to 32-residue-long imperfect repeats are located in the carboxyl-terminal half of Tau and are highly conserved in several microtubule-associated proteins (7). The repeat domain is flanked by a proline-rich region that enhances binding to microtubules and microtubule assembly (8). Tau isoforms are developmentally regulated and have similar levels in the adult human brain (9).
Impaired interaction of Tau with microtubules plays an important role in the pathology of several neurodegenerative diseases (10, 11). Dysregulation by genetic mutation or hyperphosphorylation affects the Tau–microtubule complex, leads to Tau detachment, causes instability and disassembly of microtubules, and, thus, perturbs axonal transport (12, 13). Microtubule-stabilizing drugs might therefore improve neuronal degeneration (14). When detached from microtubules, Tau can self-aggregate into insoluble aggregates through its hexapeptide motifs in the repeat domain (15). The deposition of aggregated Tau into neurofibrillary tangles and neuritic Tau pathology is one of the hallmarks in Alzheimer’s disease (16).
Biochemical studies have shown that the repeat domain and the neighboring basic proline-rich region contribute strongly to microtubule binding (8, 17). In addition, a variety of binding sites and models of the Tau–microtubule complex were proposed (18–22). The models include binding of Tau to the outer surface of microtubules connecting tubulin subunits either across or along protofilaments (18). Tau might also reach into the interior of the microtubule wall near the binding site of the anticancer drug paclitaxel (19). Often these studies are complicated by the flexibility of the Tau protein, which belongs to the class of intrinsically disordered proteins (23, 24).
Here, we studied the molecular mechanism of the interaction of Tau with microtubules by using a combination of NMR spectroscopy and mass spectrometry. We show that small groups of evolutionary conserved Tau residues bind dynamically at the interface between tubulin heterodimers, thereby promoting microtubule assembly and stabilization.
Results
Tau Binds Microtubules Through Short Sequence Motifs.
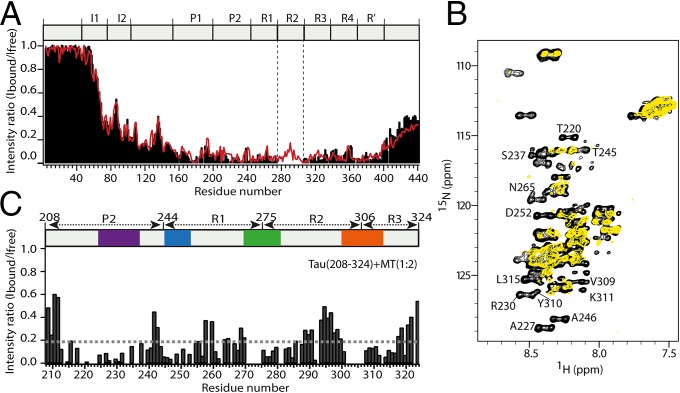
To obtain insight into the Tau–microtubule interaction, we polymerized porcine brain tubulin and validated the formation of microtubules by electron microscopy. Next, we added the microtubules to a solution containing 441-residue Tau. At a twofold excess of microtubules, most Tau residues beyond 70 were strongly attenuated (Fig. 1A). In particular, the resonances of residues 224–398 were just above the noise level in 2D 1H-15N NMR spectra. Decreasing the amount of microtubules resulted in an overall increase in signal intensity (SI Appendix, Fig. S1). In addition, signal intensities now varied strongly in the central region of Tau, consistent with the presence of distinct microtubule-binding motifs (24, 25).
Fig. 1.
Tau binds microtubules through short evolutionary conserved sequence motifs. (A) 1H-15N NMR signal intensities of free Tau, Ifree, are compared with those of Tau bound to microtubules, Ibound (Tau:tubulin heterodimer ratio of 1:3.6). The estimated error in Ibound/Ifree according to the signal-to-noise ratio in the spectra was on average 0.08. Data for three-repeat Tau are shown as red line. The absence of repeat R2 in three-repeat Tau is indicated by vertical dashed lines. Tau’s domain organization is shown on top: I, insert; P, proline-rich region; R, repeat. (B) Superposition of 1H-15N CRINEPT-HMQC-TROSY spectra of Tau(208-324) in the free (black) and microtubule-bound (yellow) state acquired at 30 °C. The Tau(208-324):tubulin heterodimer ratio was 1:2. Selected resonances, which are strongly attenuated when Tau(208-324) is bound to microtubules, are labeled. (C) Signal intensity profile of Tau(208-324) bound to microtubules. NMR intensities were obtained from the spectra shown in B. At a Tau(208-324):tubulin heterodimer ratio of 1:2, Tau(208-324) is fully bound to microtubules (26). The detection of signals from residues in between the microtubule-binding motifs shows that these Tau residues remain flexible in the Tau–microtubule complex.
To obtain further insight into the Tau–microtubule complex, we used a Tau fragment, which is composed of residues 208–324 and binds with higher (submicromolar) affinity to microtubules (26). At a Tau(208-324):tubulin heterodimer ratio of 1:2, resonances of residues 224–237, 245–253, 275–284, and 300–317 were broadened beyond detection (Fig. 1 B and C). The NMR signal broadening is caused by the slow molecular tumbling of microtubules and suggests that the Tau residues, which are not detectable in the presence of microtubules, are tightly bound to microtubules (Fig. 1 B and C). A direct interaction of residues 224–237, 245–253, 275–284, and 300–317 with microtubules is supported by mutational analysis, which showed that Lys224, Lys225, Arg230, Lys274, Lys280, Lys281, and Y310 are important for binding to microtubules (27–29). However, residues in between the microtubule-binding motifs were still observed in the presence of microtubules (Fig. 1 B and C). These Tau regions are thus not rigidly bound to microtubules, demonstrating that Tau does not fold into a single rigid structure upon binding to microtubules.
Different Tau Isoforms Use a Common Mode of Interaction with Tubulin.
The six isoforms of Tau, which are generated from a single tau gene by alternative splicing, differ from each other in the presence or absence of two N-terminal inserts and repeat R2. Biochemical experiments showed that the N-terminal inserts contribute little to the direct binding to microtubules (8, 17). To investigate the influence of the lack of repeat R2 on the microtubule interaction, we analyzed a three-repeat isoform of Tau. The microtubule-induced signal broadening profile closely matched that of the longest isoform with four repeats (Fig. 1A). Thus, three-repeat and four-repeat isoforms use similar mechanisms to bind to microtubules.
Tau has distinct roles at different stages of development. In adult neurons, Tau stabilizes microtubules and suppresses microtubule shortening to retain the structural integrity of axons (29). In addition, Tau drives new microtubule assembly by lowering the critical concentration of tubulin polymerization (5, 23). To dissect the process of Tau-mediated microtubule assembly, we probed the interaction of Tau with unpolymerized α-β-tubulin heterodimers. Experiments were performed at low temperature in the absence of Mg2+ and other polymerization promoting factors to avoid the formation of tubulin rings (SI Appendix, Fig. S2). Addition of α-β-tubulin heterodimers changed the position and intensity of Tau cross-peaks, comparable to the addition of microtubules (SI Appendix, Fig. S2). This finding is in agreement with the ability of Tau to bind to unpolymerized and polymerized tubulin in a similar manner (17).
Tau Competes with Vinblastine for Binding to Tubulin.
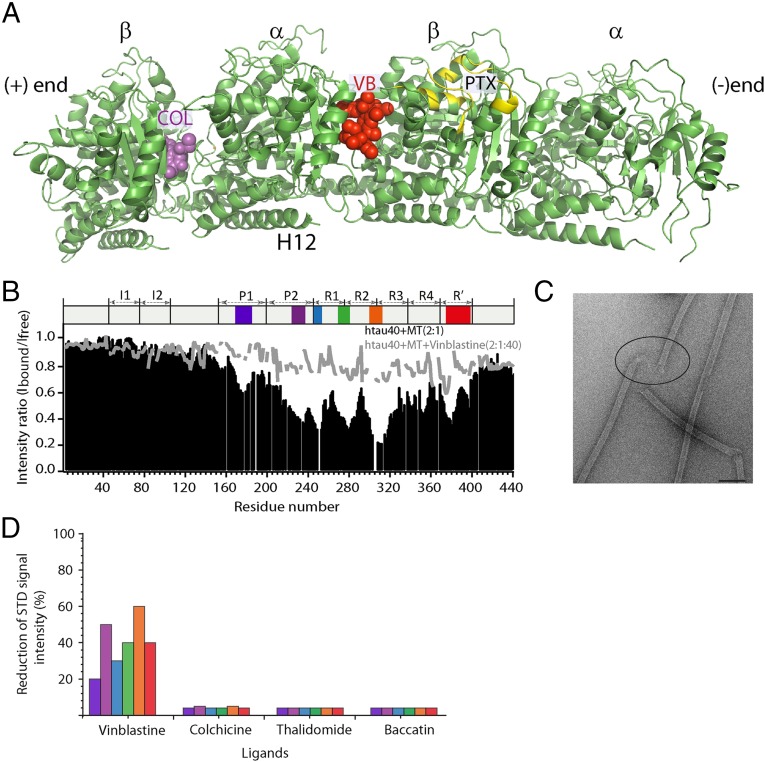
To obtain insight into the binding site of Tau on microtubules, we performed a wide range of competition experiments with different tubulin drugs. The small molecules vinblastine, baccatin, thalidomide, and colchicine were shown to bind to distinct sites on the α-β-tubulin heterodimer (Fig. 2A) (30–32). A 40-fold excess of each small molecule was added to a sample containing Tau and microtubules. When a small molecule binds to a site on tubulin that is involved in binding to Tau, Tau would be released from microtubules and NMR signal intensities would increase. Addition of baccatin, which binds to the paclitaxel pocket in β-tubulin on the microtubule's inner surface, however did not influence the microtubule-interaction of Tau (SI Appendix, Fig. S3). In addition, the depolymerization drug colchicine and the microtubule-stabilizing agent thalidomide did not interfere with the binding of Tau to microtubules (SI Appendix, Fig. S3). In contrast, vinblastine, which binds to a hydrophobic pocket formed by helix H10 and β-strand S9 of α-tubulin (Fig. 2A) (31), resulted in an almost complete recovery of Tau signal intensities (Fig. 2B). Notably, most of the microtubules, which were stabilized by paclitaxel, remained intact during the course of the NMR experiment, despite a vinblastine-induced curling of microtubule ends (Fig. 2C). In addition, vinblastine competed with Tau for binding to unpolymerized tubulin (SI Appendix, Fig. S3D), indicating that the increase in Tau signal intensities upon addition of vinblastine is not simply due to a change in the polymerization state of tubulin/microtubules or is influenced by the paclitaxel-mediated stabilization of microtubules. Competition between Tau and vinblastine, but not baccatin, thalidomide, and colchicine, was further supported by saturation-difference NMR experiments of a variety of Tau peptides with unpolymerized tubulin (Fig. 2D and SI Appendix, Figs. S4 and S5). The combined data suggest that one of the sites where Tau binds to tubulin/microtubules overlaps at least partially with the binding pocket of vinblastine. The same binding pocket is used by a variety of hydrophobic antimitotic peptides (33).
Fig. 2.
Tau competes with vinblastine for binding to tubulin. (A) Location of vinblastine (VB), colchicine (COL), and paclitaxel (PTX) on the 3D structure of the (Tc)2R complex (PDB ID code 1Z2B; ref. 31). Bound colchicine and vinblastine are shown by using the sphere model. Residues involved in paclitaxel binding (54) are highlighted in yellow. (B) Vinblastine competes with four-repeat Tau for binding to microtubules. Black bars represent the NMR line broadening observed in four-repeat Tau upon addition of microtubules (molar ratio of 2:1), whereas the gray line shows the corresponding values after addition of a 20-fold excess (with respect to four-repeat Tau) of vinblastine. (C) Electron micrograph of paclitaxel-stabilized microtubules after addition of vinblastine. The concentrations of Tau, microtubules and vinblastine were 10, 20, and 400 μM, respectively, in 50 mM sodium phosphate buffer, pH 6.8. (D) Competition between Tau fragments and vinblastine for binding to unpolymerized tubulin as probed by saturation-transfer difference NMR (SI Appendix, Figs. S4 and S5). Each of the six Tau fragments Tau(162-188) (blue), Tau(211-242) (purple), Tau(239-267) (light blue), Tau(265-290) (green), Tau(296-321) (orange), and Tau(368-402) (red) overlaps with one of the microtubule-binding motifs (Fig. 1 and SI Appendix, Fig. S1).
Next, we tested binding of Tau to microtubules by using mass spectrometry. Amine-directed cross-linking of Tau to microtubules and tubulin dimers identified cross-links from residues Lys225, Lys240, Lys257, Lys311, and Lys383 in Tau to the side chains of Lys336 and Lys338 of α-tubulin (Fig. 3A and SI Appendix, Table S1). No intermolecular cross-links were found to β-tubulin. Lys225, Lys240, Lys257, Lys311, and Lys383 are within or close to the amino acid patches of Tau that were identified as microtubule binding motifs (Fig. 3B). The two cross-linked residues in α-tubulin, Lys336 and Lys338, are located at the N-terminal end of helix H10 in proximity to the vinblastine binding pocket (Fig. 3C) (31). Chemical cross-links to helix H10 were also detected in complexes of tubulin with the antimitotic agent hemiasterlin (34) and with the N terminus of stathmin (35), which both bind to the vinblastine binding site in α-tubulin.
Fig. 3.
The repeat domain of Tau cross-links to α-tubulin. (A) Separation of cross-linked Tau-tubulin complexes by SDS/PAGE. (B) Map of cross-linked lysine residues. Cross-links are marked by dashed lines. Only cross-links from Tau to Lys336 and Lys338 in α-tubulin were observed. (C) Three-dimensional structure of the (Tc)2R complex (PDB ID code 1Z2B; ref. 31). Vinblastine is shown as red sphere model. Lys336 and Lys338 are highlighted in blue.
Tau Binds at the Interface Between α-β-Tubulin Heterodimers.
The peptide comprising the N-terminal region of a stathmin domain (I19L), which comprises the N-terminal region of stathmin and binds to the vinblastine-binding pocket in α-tubulin (Fig. 4A), impedes tubulin polymerization (36) and keeps tubulin in an unpolymerized state (SI Appendix, Fig. S6 A and B). Consistent with the binding of I19L to the vinblastine-binding domain, vinblastine interfered with the I19L–tubulin interaction (SI Appendix, Fig. S6C). We then asked whether I19L competes with the binding of Tau to tubulin. To address this question, we added an excess of I19L to a sample of Tau bound to unpolymerized tubulin. Addition of I19L released a fraction of Tau from tubulin (Fig. 4B), providing direct evidence that the interface between α-β-tubulin heterodimers is involved in Tau binding.
Fig. 4.
Tau binds at the interface between α-β-tubulin heterodimers. (A) Three-dimensional structure of the N terminus of a stathmin-like domain (green) bound to α-tubulin (55). Lys336 and Lys338 of α-tubulin, which were cross-linked to Tau (Fig. 3), are shown in red. Helix H10 and strand S9 that form the vinblastine binding pocket of α-tubulin are highlighted. (B) Average normalized signal intensity of residues 245–337 (black) in 1H-15N HSQC spectra of four-repeat Tau in the presence of unpolymerized tubulin and after addition of an x-fold molar excess of I19L with respect to Tau. For comparison, averaged normalized intensity values of residues 3–70 are shown in gray. Error bars show the SD over the selected residues.
Discussion
Despite the importance of the regulation of microtubule structure and dynamics by microtubule-associated proteins, little is known about the interaction of microtubule-associated proteins with microtubules. Our study showed that the microtubule-associated protein Tau remains highly dynamic when bound to microtubules. Small groups of Tau residues bind tightly to microtubules, while intervening parts stay flexible. The Tau residues that bind tightly to the microtubule lattice are evolutionarily conserved (37), consistent with a conserved mechanism of microtubule regulation by microtubule-associated proteins. In addition, the microtubule-binding motifs in Tau contain residues that are essential for protein aggregation in support of a mechanism where physiological interaction competes with pathogenic misfolding. The projection domain of Tau up to the proline-rich region remains highly flexible when Tau is bound to microtubules, and also when aggregated into amyloid fibrils (38), and forms an unstructured polypeptide brush covering microtubules and amyloid fibrils (21, 39). Thus, the pathological (fibrillar aggregates) and cellular (microtubule-bound) form of the Tau molecule can show similar properties with different outcome, in agreement with the high structural and functional adaptability of intrinsically disordered proteins.
We showed that only vinblastine, but not baccatin, thalidomide, and colchicine, interferes with the Tau–tubulin interaction (Fig. 2 and SI Appendix, Fig. S2). In addition, Tau residues within or next to the microtubule-binding motifs were cross-linked to lysine residues, which are located in α-tubulin in vicinity to the vinblastine-binding pocket (Fig. 3). The combined data suggested that at least one binding site of Tau on tubulin overlaps with the vinblastine-binding pocket at the interface between α-β-tubulin heterodimers. This hypothesis was supported by the ability of a peptide, which comprises the N-terminal region of stathmin and binds to the vinblastine-binding pocket in α-tubulin, to compete with the binding of Tau to tubulin (Fig. 4). The part of stathmin that pairs with β-strand S9 of α-tubulin is homologous to the microtubule-binding motifs in the repeat domain of Tau. Binding at the interface between α-β-tubulin heterodimers is also consistent with the stoichiometry of the Tau:tubulin interaction: Full-length Tau binds to microtubules with a stoichiometry of one Tau molecule for two tubulin dimers (8). In addition, a variety of smaller Tau fragments bind to microtubules with a stoichiometry of up to one Tau molecule for two tubulin dimers, although the affinity of shorter fragments is reduced (8). Binding of Tau at the interdimer interface is also supported by electron microscopy that identified undecagold-labeled Tau in proximity to the tubulin helices H11 and H12 (18). Because of the dynamic nature of the Tau–microtubule interaction, the binding of Tau at the interdimer interface does not exclude additional interactions with other parts of tubulin such as the acidic C-terminal tails (40).
Vinblastine and antimitotic peptides induce a tubulin conformation that promotes curved tubulin rings and impedes microtubule assembly (31). In contrast, Tau stabilizes a straight protofilament conformation that prevents the inside-out curling typical of curved polymorphic forms of tubulin (rings, vinblastine-induced spirals), favors the formation of microtubules, and increases microtubule stability (32, 41). We propose that the two distinct functional consequences of vinblastine and Tau are tightly connected to the flexibility of the structural elements forming the binding pocket at the α-β-tubulin heterodimer interface (33), potentially through an allosteric mechanism. Moreover, flexible regions in between the microtubule-binding motifs in the Tau–microtubule complex (Fig. 1 B and C) support a dynamic mode of binding where microtubule-binding motifs in one Tau molecule can exchange dynamically with the same α-tubulin molecule. Such a multivalent interaction is consistent with the ability of a peptide composed of the Tau residues 274–281—the eight residues that are located at the center of the microtubule-binding motif in repeat 2 (Fig. 1)—to kinetically stabilize steady-state microtubule dynamics in a manner qualitatively similar to full-length Tau (29). In addition, the dynamic structure of Tau makes it possible that different microtubule-binding regions in one Tau molecule bind to distinct tubulin molecules, thereby increasing the local tubulin concentration or connecting adjacent protofilaments, and promoting microtubule assembly.
Together with recent data about the structural transitions in tubulin upon GTP hydrolysis (42), our study supports the importance of remodelling of longitudinal dimer contacts for microtubule assembly and stabilization.
Materials and Methods
Microtubule Assembly.
Porcine brain tubulin was purified as described (8, 43). Tubulin polymerization was performed in microtubule assembly buffer containing 100 mM Na-Pipes, pH 6.9, 1 mM EGTA, 1 mM MgSO4, 1 mM GTP and 1 mM DTT. To induce formation of microtubules, fixed concentrations of tubulin (20–50 μM) were incubated with equal concentrations of paclitaxel at 37 °C for 20–30 min. The suspensions of the samples were fractionated by ultracentrifugation at 40,000 × g for 20 min. For NMR measurements, the microtubule pellet was resuspended in 50 mM phosphate buffer. Analysis of microtubules showed that they remained stable over the duration of the NMR experiments (SI Appendix, Fig. S7).
Protein Expression and Isotope Labeling.
pNG2 plasmids with inserted coding sequences of four-repeat Tau (residues Met1-Leu441), three-repeat Tau(352-residue) and Tau(Met-208-324) were transformed into Escherichia coli strain BL21(DE3) (Merck-Novagen). The bacterial pellet was resuspended in 50 mM Mes, 500 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5 mM DTT, pH 6.8, complemented with a protease inhibitor mixture (Complete Protease Inhibitor; Roche). Cells were disrupted with a French pressure cell and subsequently boiled for 20 min. The soluble extract was isolated by centrifugation, the supernatant was dialyzed twice against 20 mM Mes, 50 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 2 mM DTT, 0.1 mM PMSF, pH 6.8, and loaded onto a FPLC SP-Sepharose column. Proteins were eluted by using a linear gradient of 20 mM Mes, 1 M NaCl, 1 mM EGTA, 1 mM MgCl2, 2 mM DTT, 0.1 mM PMSF, pH 6.8. Tau breakdown products were separated in a second chromatography step by using a Superdex G200 column (GE Healthcare). The buffer was 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, with 1 mM DTT. Protein samples uniformly enriched in 15N were prepared by growing E. coli bacteria in minimal medium containing 1 g/L 15NH4Cl.
Synthetic peptides were purchased from EZBiolab USA or synthesized in house on ABI 433A (Applied Biosystems) and Liberty 1 (CEM) machines. Peptides were synthesized with acetyl and amide protection groups at the N and C termini, respectively. Peptides were purified by reversed-phase HPLC, and the pure product was lyophilized.
NMR Spectroscopy.
NMR measurements were performed in 50 mM sodium phosphate buffer, pH 6.8, 10% D2O. Two-dimensional (2D) 1H-15N heteronuclear single quantum coherence (HSQC) experiments (44, 45) of Tau with microtubules or unpolymerized tubulin were recorded at 5 °C on 700, 800, and 900 MHz Bruker spectrometers equipped with cryoprobes. Before the NMR measurements, the samples were incubated at 37 °C for 1 h. Two-dimensional 1H-15N- CRINEPT-HMQC-TROSY (46) and 2D 1H-15N TROSY spectra (47) of 2H, 15N-labeled Tau(208-324) were recorded at 30 °C on a Bruker 900 MHz spectrometer equipped with a TCI cryoprobe (Z-gradient).
Competition experiments between tubulin drugs or Tau fragments and full-length Tau for binding to microtubules, which were stabilized by paclitaxel, were performed by addition of the compounds/peptides to the preincubated Tau-microtubule complex at 37 °C. The trimeric complex was then again incubated at 37 °C for 30 min. The extent of competition of the ligands was monitored by reading out the variation in signal broadening in 2D 1H-15N HSQC NMR spectra of Tau, which were recorded at 5 °C to reduce solvent exchange. The sample used to perform competition experiment with colchicine was incubated for 2 h at 37 °C before the NMR measurements.
To probe the binding of a ligand to nonpolymerized tubulin, saturation-transfer difference NMR spectra (48) of the ligand with and without tubulin (or microtubules) were recorded. The concentration of the Tau peptides/small molecules was 1 mM; that of tubulin 25 μM. The absence of microtubule assembly promoting agents and a concentration of tubulin below the critical concentration for microtubule assembly ensured the absence of large tubulin rings as validated by electron microscopy. In competition experiments, a second ligand was added at equimolar ratio with respect to first ligand, and changes in the saturation-transfer difference signal intensities of the first ligand were measured. Saturation-transfer difference spectra were recorded at 25 °C on a 700-MHz spectrometer equipped with a cryoprobe by using a series of 40 equally spaced 50-ms Gaussian-shaped pulses for saturation of the protein, with a total saturation time of 1.5 s. On- and off-resonance frequencies were set to −0.5 ppm and 60 ppm, respectively.
Electron Microscopy.
Electron microscopy grids were directly prepared after the turbidity assay, and all steps were carried out at 37 °C. Formvar carbon-coated copper grids (200 mesh) were glow-discharged for 30 s in a Pelco easiGlow instrument. Five-microliter samples were mixed with a final concentration of 2% (vol/vol) glycerol and incubated for 3 min on top of the grid. The solution was removed by filter paper, the grid was washed three times with assembly buffer [supplemented with 2% (vol/vol) glycerol], and negatively stained by incubating for 1 min with 2% uranyl acetate, followed by one washing step with ddH2O. The grids were analyzed at 200 kV in a JEOL JEM-2200FS transmission electron microscope.
Chemical Cross-Linking and Mass Spectrometry.
Preincubated Tau–tubulin complexes [Tau–tubulin dimer and Tau microtubules both at a Tau:tubulin heterodimer ratio of 1:2; Tau(208-324)-tubulin ratio of 1:3] were mixed with freshly prepared cross-linker bis(sulfosuccinimidyl) suberate at different cross-linker to protein ratios of 0, 5, 10, 25, 50, 100, and 200. The cross-linking reaction was carried out for 30 min at room temperature and quenched with 2 μL of 2 M Tris pH 7.2. Cross-linked samples were visualized by Coomassie-stained SDS/PAGE and the ratio that exhibited maximum cross-linking yield and minimum formation of higher order aggregates was selected (25 molar excess over the complex). For the identification of cross-linked peptides, the reaction was repeated at the determined optimal cross-linker to tubulin ratio. Preweighted vials of bis(sulfosuccinimidyl) suberate were freshly dissolved in dimethyl sulfoxide and taken to final concentration with 50 mM sodium phosphate buffer, pH 7.2. Reactions were performed in the same buffer. After quenching, samples were analyzed by SDS/PAGE in a 4–12% Bis-Tris gradient gel. Cross-linked complexes derived from four gel lanes were pooled to increase sample amount. In-gel digestion was performed as described (49) on the band at molecular mass ∼150 KDa, which corresponds to the mass of the trimeric complex (tubulin dimer and full-length Tau). After extraction from the gel, samples were immediately analyzed on the mass spectrometer.
For liquid chromatography-MS/MS analyses, samples were dissolved in 30 µL of sample solvent (5% vol/vol acetonitrile, 1% vol/vol formic acid). Five microliters were injected into a nano-liquid chromatography system (Agilent 1100 series; Agilent Technologies) including a ∼2-cm-long, 150-μm inner diameter C18 trapping column in-line with a ∼15 cm long, 75 μm inner diameter C18 analytical column (both packed in-house, C18 AQ 120 Å 5 μm, Dr. Maisch GmbH). Peptides were loaded onto the trapping column at a flow rate of 10 μL/min in buffer A (0.1% formic acid in H2O, vol/vol) and subsequently eluted and separated on the analytical column with a gradient of 7.5–37.5% buffer B (95% acetonitrile, 0.1% formic acid in H2O, vol/vol) with an elution time of 97 min and a flow rate of 300 nL/min.
MS analysis was performed essentially as described in ref. 50. Briefly, online electrospray mass ionization-MS was performed with a LTQ-Orbitrap Velos instrument (Thermo Scientific), operated in data-dependent mode by using A top 8 method. MS fragments were recorded in the m/z range of 300–1,800 at a resolution of 100,000. Only charge states 3 and above were selected. Dynamic exclusion was enabled (30-s repeat duration, repeat count 1). Both precursor ions and fragment ions were scanned in the Orbitrap. Fragment ions were generated by collision dissociation activation (normalized collision energy = 37.5). As precursor ions and fragment ions were scanned in the Orbitrap, the resulting spectra were measured with high accuracy (< 5 ppm) both in the MS and MS/MS level.
Data analysis was performed with MassMatrix as described in their publications and manuals (51–53). Thermo Scientific Raw files were converted to the mzxmL data format with MMConverter and submitted to database search with the following parameters: peptide length between 6 and 40 aa long, 10 ppm MS1 tolerance, 0.02 Da MS2 tolerance, tryptic fragments with a maximum of two missed cleavages. Oxidation in methionine was set as a variable modification, whereas carbamidomethylation of cysteine was set as a fixed modification. Spectra were searched against a FASTA file composed of the sequences of Tau and tubulin and a reversed decoy database. The resulting matches with scores above the decoy scores were manually checked.
Supplementary Material
Acknowledgments
We thank Lukasz Jaremko, Mariusz Jaremko, Christian Griesinger, and Eva Mandelkow for discussions; Sabrina Hübschmann for sample preparations; Kerstin Overkamp for peptide synthesis; and Dietmar Riedel and Gudrun Heim for electron micrographs. E.M. was supported by the Max Planck Gesellschaft consortium Toxic Protein Conformation and the Wellcome Trust. M.Z. was supported by Deutsche Forschungsgemeinschaft Project ZW 71/8-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504081112/-/DCSupplemental.
References
- 1.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6(3):201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 2.Kirschner M, Mitchison T. Beyond self-assembly: From microtubules to morphogenesis. Cell. 1986;45(3):329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 3.Mandelkow E, Mandelkow E-M. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7(1):72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- 4.Drubin DG, Kirschner MW. Tau protein function in living cells. J Cell Biol. 1986;103(6 Pt 2):2739–2746. doi: 10.1083/jcb.103.6.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci USA. 1975;72(5):1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3(4):519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 7.Aizawa H, et al. Molecular cloning of a ubiquitously distributed microtubule-associated protein with Mr 190,000. J Biol Chem. 1990;265(23):13849–13855. [PubMed] [Google Scholar]
- 8.Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33(32):9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- 9.Kosik KS, Orecchio LD, Bakalis S, Neve RL. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2(4):1389–1397. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- 10.Brunden KR, Trojanowski JQ, Lee VM. Advances in tau-focused drug discovery for Alzheimer’s disease and related tauopathies. Nat Rev Drug Discov. 2009;8(10):783–793. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia ML, Cleveland DW. Going new places using an old MAP: Tau, microtubules and human neurodegenerative disease. Curr Opin Cell Biol. 2001;13(1):41–48. doi: 10.1016/s0955-0674(00)00172-1. [DOI] [PubMed] [Google Scholar]
- 12.Hong M, et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science. 1998;282(5395):1914–1917. doi: 10.1126/science.282.5395.1914. [DOI] [PubMed] [Google Scholar]
- 13.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 14.Ballatore C, et al. Microtubule stabilizing agents as potential treatment for Alzheimer’s disease and related neurodegenerative tauopathies. J Med Chem. 2012;55(21):8979–8996. doi: 10.1021/jm301079z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Bergen M, et al. Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming β structure. Proc Natl Acad Sci USA. 2000;97(10):5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: A major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251(4994):675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 17.Butner KA, Kirschner MW. Tau protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol. 1991;115(3):717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol. 2002;157(7):1187–1196. doi: 10.1083/jcb.200201048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kar S, Fan J, Smith MJ, Goedert M, Amos LA. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003;22(1):70–77. doi: 10.1093/emboj/cdg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makrides V, Massie MR, Feinstein SC, Lew J. Evidence for two distinct binding sites for tau on microtubules. Proc Natl Acad Sci USA. 2004;101(17):6746–6751. doi: 10.1073/pnas.0400992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santarella RA, et al. Surface-decoration of microtubules by human tau. J Mol Biol. 2004;339(3):539–553. doi: 10.1016/j.jmb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Gigant B, et al. Mechanism of Tau-promoted microtubule assembly as probed by NMR spectroscopy. J Am Chem Soc. 2014;136(36):12615–12623. doi: 10.1021/ja504864m. [DOI] [PubMed] [Google Scholar]
- 23.Cleveland DW, Hwo SY, Kirschner MW. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol. 1977;116(2):227–247. doi: 10.1016/0022-2836(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 24.Mukrasch MD, et al. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7(2):e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukrasch MD, et al. The “jaws” of the tau-microtubule interaction. J Biol Chem. 2007;282(16):12230–12239. doi: 10.1074/jbc.M607159200. [DOI] [PubMed] [Google Scholar]
- 26.Fauquant C, et al. Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J Biol Chem. 2011;286(38):33358–33368. doi: 10.1074/jbc.M111.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goode BL, et al. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol Biol Cell. 1997;8(2):353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goode BL, Chau M, Denis PE, Feinstein SC. Structural and functional differences between 3-repeat and 4-repeat tau isoforms. Implications for normal tau function and the onset of neurodegenetative disease. J Biol Chem. 2000;275(49):38182–38189. doi: 10.1074/jbc.M007489200. [DOI] [PubMed] [Google Scholar]
- 29.Panda D, Goode BL, Feinstein SC, Wilson L. Kinetic stabilization of microtubule dynamics at steady state by tau and microtubule-binding domains of tau. Biochemistry. 1995;34(35):11117–11127. doi: 10.1021/bi00035a017. [DOI] [PubMed] [Google Scholar]
- 30.Amos LA. What tubulin drugs tell us about microtubule structure and dynamics. Semin Cell Dev Biol. 2011;22(9):916–926. doi: 10.1016/j.semcdb.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Gigant B, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435(7041):519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 32.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391(6663):199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 33.Cormier A, Marchand M, Ravelli RB, Knossow M, Gigant B. Structural insight into the inhibition of tubulin by vinca domain peptide ligands. EMBO Rep. 2008;9(11):1101–1106. doi: 10.1038/embor.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunes M, et al. Two photoaffinity analogues of the tripeptide, hemiasterlin, exclusively label alpha-tubulin. Biochemistry. 2005;44(18):6844–6857. doi: 10.1021/bi0474766. [DOI] [PubMed] [Google Scholar]
- 35.Müller DR, et al. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73(9):1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 36.Clément M-J, et al. N-terminal stathmin-like peptides bind tubulin and impede microtubule assembly. Biochemistry. 2005;44(44):14616–14625. doi: 10.1021/bi0512492. [DOI] [PubMed] [Google Scholar]
- 37.Cleveland DW, Spiegelman BM, Kirschner MW. Conservation of microtubule associated proteins. Isolation and characterization of tau and the high molecular weight microtubule associated protein from chicken brain and from mouse fibroblasts and comparison to the corresponding mammalian brain proteins. J Biol Chem. 1979;254(24):12670–12678. [PubMed] [Google Scholar]
- 38.Bibow S, et al. The dynamic structure of filamentous tau. Angew Chem Int Ed Engl. 2011;50(48):11520–11524. doi: 10.1002/anie.201105493. [DOI] [PubMed] [Google Scholar]
- 39.Wischik CM, et al. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USA. 1988;85(13):4884–4888. doi: 10.1073/pnas.85.13.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano L, Montejo de Garcini E, Hernández MA, Avila J. Localization of the tubulin binding site for tau protein. Eur J Biochem. 1985;153(3):595–600. doi: 10.1111/j.1432-1033.1985.tb09342.x. [DOI] [PubMed] [Google Scholar]
- 41.Dye RB, Fink SP, Williams RC., Jr Taxol-induced flexibility of microtubules and its reversal by MAP-2 and Tau. J Biol Chem. 1993;268(10):6847–6850. [PubMed] [Google Scholar]
- 42.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157(5):1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandelkow EM, Herrmann M, Rühl U. Tubulin domains probed by limited proteolysis and subunit-specific antibodies. J Mol Biol. 1985;185(2):311–327. doi: 10.1016/0022-2836(85)90406-1. [DOI] [PubMed] [Google Scholar]
- 44.Bax A, Ikura M, Kay LE, Torchia DA, Tschudin R. Comparison of different modes of 2-dimensional reverse-correlation Nmr for the study of proteins. J Magn Reson. 1990;86(2):304–318. [Google Scholar]
- 45.Bodenhausen G, Ruben DJ. Natural abundance N-15 Nmr by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69(1):185–189. [Google Scholar]
- 46.Riek R, Wider G, Pervushin K, Wüthrich K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA. 1999;96(9):4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed. 1999;38(12):1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 50.Christian H, Hofele RV, Urlaub H, Ficner R. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014;42(2):1162–1179. doi: 10.1093/nar/gkt985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9(6):1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Hsu PH, Zhang L, Tsai MD, Freitas MA. Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J Proteome Res. 2010;9(7):3384–3393. doi: 10.1021/pr100369y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7(1):138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313(5):1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 55.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.