Significance
Rnf43 (RING finger protein 43) and Znrf3 (zinc/RING finger protein 3), encoded by stem cell-specific Wnt (wingless-int) target genes, constitute a crucial negative feedback loop in the Wnt signaling pathway. Rnf43 is mutated in subsets of human cancers of the colon, pancreas, stomach, ovary, and liver, while Znrf3 is mutated in adrenocortical carcinoma and osteoblastoma. Indeed, when both genes are mutated simultaneously in small intestinal stem cells in mice, tumors arise within a few weeks. Treatment of mice carrying RZ−/− intestinal neoplasia with a small molecule Wnt secretion inhibitor strongly inhibited growth, while adjacent normal crypts remained intact. These results establish that paracrine Wnt secretion is an essential driver of RZ−/− tumor growth and imply that a therapeutic window exists for the use of porcupine inhibitors for RZ-mutant cancers.
Keywords: RNF43, ZNRF3, LGR5, Wnt, porcupine inhibitor
Abstract
Rnf43 (RING finger protein 43) and Znrf3 (zinc/RING finger protein 3) (RZ) are two closely related transmembrane E3 ligases, encoded by Wnt target genes, that remove surface Wnt (wingless-int) receptors. The two genes are mutated in various human cancers. Such tumors are predicted to be hypersensitive to, yet still depend on, secreted Wnts. We previously showed that mutation of RZ in the intestine yields rapidly growing adenomas containing LGR5+ (leucine-rich repeat-containing G-protein coupled receptor 5) stem cells and Wnt3-producing Paneth cells. We now show that removal of Paneth cells by Math1 mutation inhibits RZ−/− tumor formation. Similarly, deletion of Wnt3 inhibits tumorigenesis. Treatment of mice carrying RZ−/− intestinal neoplasia with a small molecule Wnt secretion inhibitor (porcupine inhibitor C59) strongly inhibited growth, whereas adjacent normal crypts remained intact. These results establish that paracrine Wnt secretion is an essential driver of RZ−/− tumor growth and imply that a therapeutic window exists for the use of porcupine inhibitors for RZ-mutant cancers.
RNF43 (RING finger protein 43) and ZNRF3 (zinc/RING finger protein 3) target Wnt (wingless-int) receptors (the Frizzleds) to the lysosomal degradation pathway by promoting their endocytosis (1, 2). Encoded by stem cell-specific Wnt target genes, they constitute a crucial negative feedback loop in the Wnt signaling pathway (2). RNF43 is mutated in subsets of human cancers of the colon (2, 3), pancreas (4–6), stomach (7), ovary (8), and liver (9), whereas ZNRF3 is mutated in adrenocortical carcinoma (10) and osteoblastoma (11). Indeed, when both genes are mutated simultaneously in small intestinal stem cells in mice, tumors arise that consist almost entirely of LGR5+ (leucine-rich repeat-containing G-protein coupled receptor 5) stem cells and their Wnt3-producing niche cells, the Paneth cells (2). The activity of these E3 ligases is negatively controlled by R-spondins (RSPO) and LGR4/5/6 (LGR) receptors that sequestrate RZ from Frizzled by forming a triple complex (1). The molecular detail of the RSPO/RZ/LGR interaction has been resolved by multiple independent X-ray structures (12–14). In our previous report, we have shown that epithelial organoids (15) derived from RZ-mutant tumors can grow in the absence of the Wnt-amplifier RSPO (2), indicative of their hypersensitivity to Wnt. However, RZ-null mutant organoids die in the presence of porcupine inhibitor IWP1, implying that they require paracrine Wnt (2), which, in organoids, is provided in the form of Wnt3 by Paneth cells (16). Of note, additional Wnts are secreted by nonepithelial cells surrounding crypts, making Wnt3 nonessential in vivo (16–19). This result clearly indicated the Wnt dependency of tumorigenic RZ-null epithelial cells.
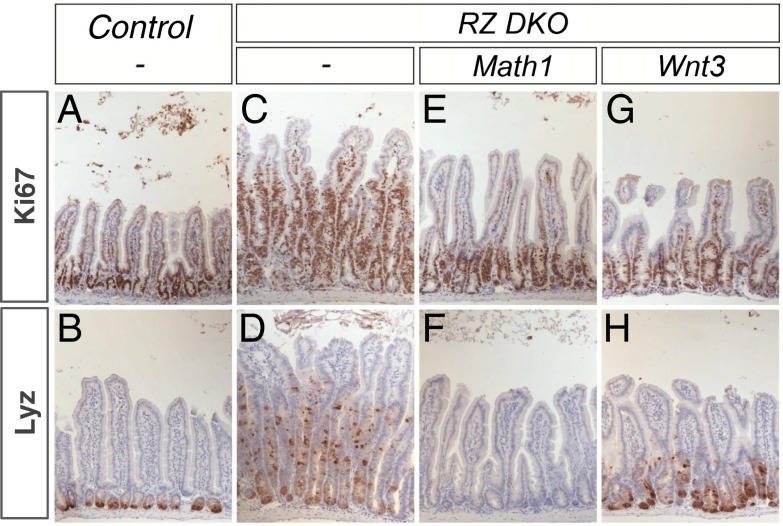
We asked whether Wnt3 produced by Paneth cells is essential for the unrestricted growth of RZ-mutant neoplasia in vivo. We first addressed this Paneth cell dependency by genetically blocking Paneth cell formation by ablating mouse atonal homolog 1 (Math1) (Atoh1, essential for Paneth cell formation; refs. 20 and 21). A floxed Math1 allele was crossed into Rnf43f/f: Znrf3f/f: Villin-CreERT2 conditional knockout mice. Cre-mediated deletion was induced by tamoxifen administration. Rnf43f/f: Znrf3f/f: Villin-CreERT2 mutant intestine showed extensive cellular proliferation with Paneth cell metaplasia, as reported previously (Fig. 1 C and D). Rnf43f/f: Znrf3f/f: Math1f/f: Villin-CreERT2 mutants rapidly lost their Lysozyme+ Paneth cells, whereas the aberrant proliferation significantly attenuated (Fig. 1 E and F). A similar experiment using a conditional Wnt3 allele yielded similar results (Fig. 1 G and H). These observations implied that RZ-null neoplasia depends on Wnt3-secreting Paneth cells.
Fig. 1.
Genetic depletion of Math1 and Wnt3 prevent the formation of tumorigenic neoplasia in the intestine of the Rnf43f/f: Znrf3f/f: Villin-CreERT2 conditional knockout mice. Intestinal sections were analyzed for Ki67 (proliferative cells) (A, C, E, and G) and Lysozyme (Paneth cells) (B, D, F, and H). Rnf43 and Znrf3 (RZ) compound mutant intestine shows dramatic increase in the number of Ki67+ proliferating cells (C) and Lysozyme+ Paneth cells (D) compared with control (A and B). Depletion of Math1 (E) or Wnt3 (G) in the RZ mutant background significantly attenuates the strong proliferative response. Note that Math1 ablation causes the loss of Paneth cells (F).
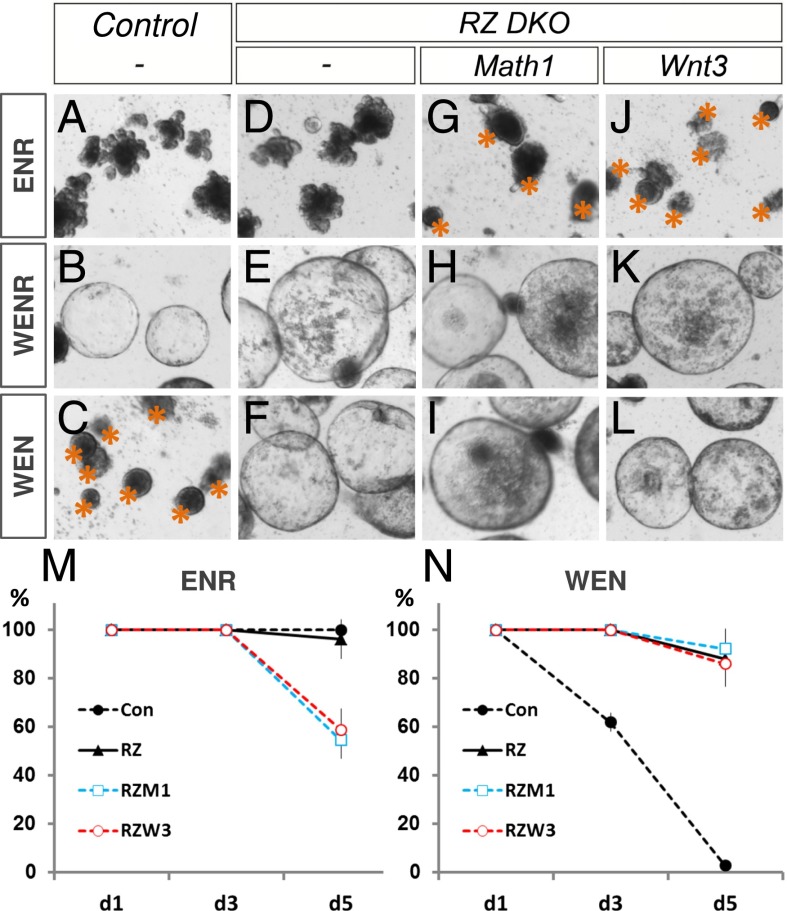
We cultured organoids from Rnf43f/f: Znrf3f/f: Math1f/f: Villin-CreERT2 and from Rnf43f/f: Znrf3f/f: Wnt3f/f: Villin-CreERT2 mutants. Both types of mutant organoids (tumoroids) grow normally when provided with exogenous Wnt3 (Fig. 2 H and K). The mutant organoids were hypersensitive to Wnt, because they did not require RSPO when provided with Wnt3A (Fig. 2 I, L, and N), unlike control organoids (Fig. 2 C and N). This survivability under RSPO withdrawal implied that the triple mutant tumoroids retained the characteristics of the original RZ double mutants. However, triple mutant tumoroids could not be cultured without Wnt ligands (Fig. 2 G, J, and M), unlike control and RZ double mutants (Fig. 2 A, D, and M), which confirms the dependency of RZ mutant tumor cells on Wnt3 secreted by Paneth cells.
Fig. 2.
(A–L) Tumorigenic Rnf43 and Znrf3 mutant intestinal organoids depend on the paracrine Wnt source. Various Intestinal organoids (control, RZ, RZ:Math1, and RZ:Wnt3) were isolated and induced by tamoxifen before being cultured under indicated conditions (ENR, WENR, and WEN medium; E, EGF; N, Noggin; R, R-spondin; W, Wnt3a). All RZ mutant organoid lines (RZ, RZ:Math1, and RZ:Wnt3) show unaffected growth in the absence of R-spondin (F, I, and L), in contrast to control organoids (C, asterisks), whereas the removal of exogenous Wnt3a suppresses the growth of RZ:Math1 (G, asterisks) and RZ:Wnt3 (J, asterisks) organoids. (M, N) The percentage of surviving organoids were scored at three time points [day 1 (d1), day 3 (d3), and day 5 (d5)] after seeding in indicated conditions [ENR (M) and WEN (N)].
These observations suggested that blocking the interaction between the tumorigenic intestinal stem and niche cell mediated by Wnt ligand can be an effective therapeutic intervention of tumors with mutations in Rnf43 and/or Znrf3. Small molecule porcupine inhibitors block palmitoylation of Wnt proteins, an essential step for secretion of Wnts (22). It has been tested in the Wnt1-driven mouse mammary tumor model (MMTV-Wnt1; refs. 23 and 24).
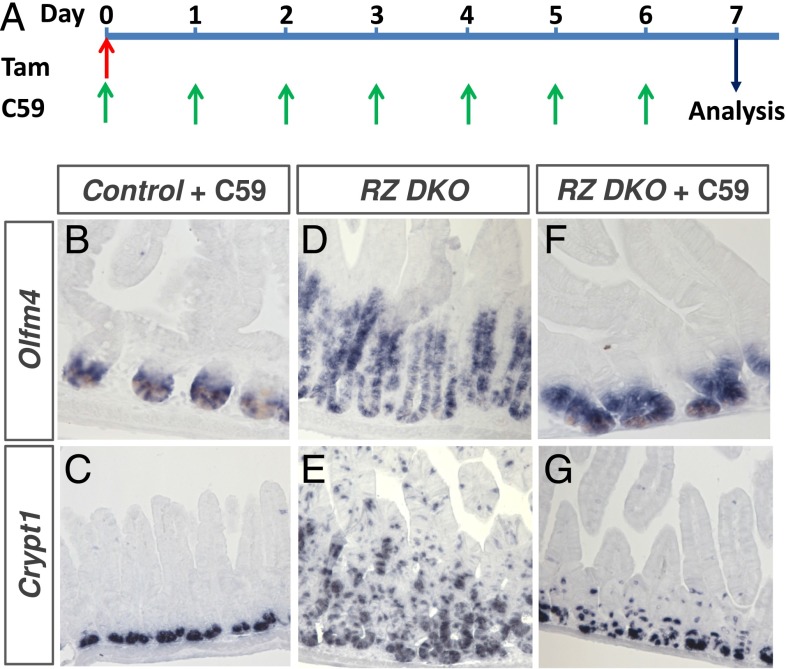
We treated 8-wk-old Rnf43f/f: Znrf3f/f: Villin-CreERT2 conditional knockout mice with C59 porcupine inhibitor (50 mg/kg) directly after inducing RZ-null neoplasia by tamoxifen administration (Fig. 3A). At this dosing, we noted no effects on functioning of normal crypts as shown by the presence of olfactomedin-4 (Olfm4+) intestinal stem cells (Fig. 3B) and Cryptdin1+ Paneth cells (Fig. 3C), although the presence of intestinal stem cells and Paneth cells crucially depend on Wnt (16–19). However, the C59 porcupine inhibitor strongly suppressed the formation of RZ-null tumorigenic epithelium when provided from the neoplasia induction date onward (Fig. 3 F and G versus Fig. 3 D and E, respectively). Additional analysis for stem cell (Lgr5) and Paneth cell (Lysozyme) markers showed similar results (Fig. S1).
Fig. 3.
Porcupine inhibitor C59 prevents neoplastic outgrowth of Rnf43 and Znrf3 mutant Olfm4+ intestinal stem cells. Schematic drawing of drug treatments (A). Mice were treated with C59 porcupine inhibitor for seven consecutive days upon tamoxifen (Tam) administration to control (B and C) and RZ (F and G) mutant mice. C59 untreated, but tamoxifen treated RZ mutant mouse was used as tumorigenic-positive control (D and E). Sections were analyzed by Olfm4, a robust marker for intestinal stem cells (B, D, and F), and Cryptin1, a marker for Paneth cells (C, E, and G) via in situ hybridization.
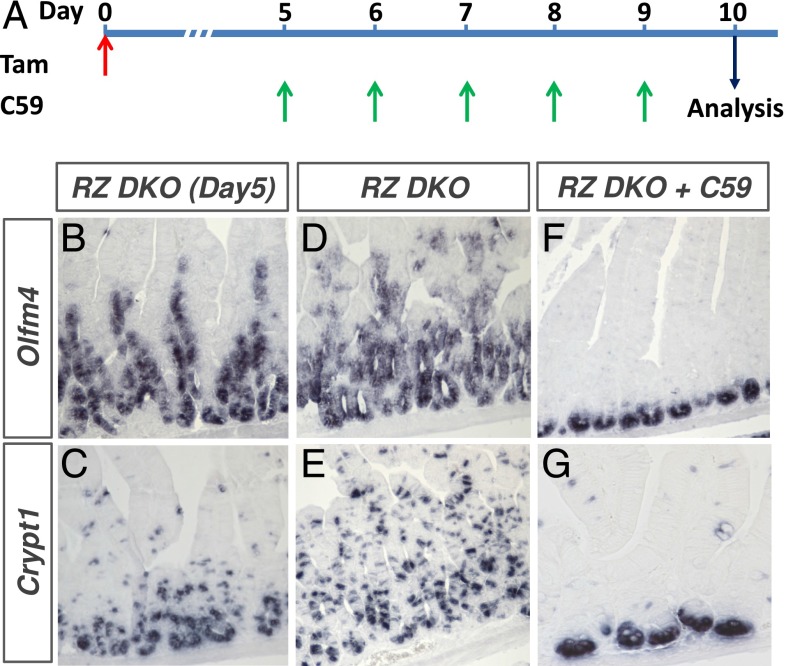
Next, we tested whether C59 porcupine inhibitor affected established RZ-null intestinal neoplasia. RZ mutations were induced first by tamoxifen administration to Rnf43f/f: Znrf3f/f: Villin-CreERT2 conditional knockout mice. C59 porcupine inhibitor treatment was initiated 5 d after tamoxifen induction. At this time point, the mutant intestine is filled with Olfm4+ stem cells (Fig. 4B) and Cryptdin1+ Paneth cells (Fig. 4C) throughout the crypt epithelium. Again, C59 porcupine treatment for 5 d suppressed the outgrowth of RZ-mutant intestinal epithelium (Fig. 4 F and G versus Fig. 4 D and E, respectively). Additional stainings for stem cell (Lgr5) and Paneth cell (Lysozyme) markers showed similar results (Fig. S2).
Fig. 4.
Porcupine inhibitor C59 eradicates preestablished neoplasia induced by Rnf43 and Znrf3 loss. Schematic drawing of drug treatments (A). Mice were treated with C59 porcupine inhibitor 5 d after initial tamoxifen (Tam) administration to RZ (F and G) mutant mice. C59 untreated, but tamoxifen-treated RZ mutant mice were used as control (D and E). RZ mutant mice were analyzed on day 5 to examine the status of hyperplasia at the time point when C59 treatment was started (B and C). Sections were analyzed for Olfm4 (intestinal stem cells) (B, D, and F) and Cryptdin 1 (Paneth cells) expression (C, E, and G) via in situ hybridization.
Our data imply that cancer cells, carrying mutations in RNF43 and/or ZNRF3 as drivers of tumor growth, require a Wnt-secreting niche. We demonstrate that blocking this Wnt niche function by porcupine inhibition attenuates aberrant RZ-null hyperplasia at doses that do not affect normal crypts. The existence of this therapeutic dose of porcupine inhibitor is particularly interesting with a high incidence of RNF43 mutations in various types of human cancer (2–9), including colorectal cancer (18%) (3). These observations indicate that the growing numbers of small molecule Wnt inhibitors may be tested in clinical trials of patients carrying malignant tumors with demonstrated mutations in RNF43 or ZNRF3.
Methods
Mice.
The conditional Rnf43, Znrf3, Wnt3, and Math1 alleles and the transgenic Villin-CreERT2 line have been described (2, 25–27). The different mice were intercrossed to generate the Rnf43f/f: Znrf3f/f: Villin-CreERT2, Rnf43f/f: Znrf3f/f: Math1f/f: Villin-CreERT2, and Rnf43f/f: Znrf3f/f: Wnt3f/f: Villin-CreERT2 mice and various genotypic controls. The Cre enzyme was induced by a single i.p. injection of 200 μL of Tamoxifen (5 mg/200 μL; Sigma Aldrich) dissolved in sunflower oil. The porcupine inhibitor C59 (50 mg/kg; Cellagen Technology) was mixed with 0.5% methylcellulose and 0.1% Tween 80 and then administrated by oral gavage (19). The mice were killed on the indicated days and the intestines isolated. All procedures were performed in compliance with local animal welfare laws, guidelines, and policies.
Histology.
The isolated mouse intestine was immediately fixed in formalin and embedded in paraffin by using standard procedures. The immunohistochemistry and in situ hybridization was carried out as described (2). For immunohistochemistry, the primary antibodies were as follows: mouse anti-Ki67 (1:250, Monosan) and rabbit anti-Lysozyme (1:1,500; DAKO) and for in situ hybridization, the (DIG)-labeled RNA probes were as follows: Lgr5 (a kind gift of Fred de Sauvage, Genentech Inc., South San Francisco), Olfm4 (image clone 1078130), and Cryptdin1 (image clone 1096215).
Intestinal Organoid Culture.
Crypt isolation, cell dissociation, and culture have been described (15). For the analysis of organoid’s growth under various conditions, established organoid cultures were dissociated into small pieces and seeded in matrigel (BD Biosciences) under the appropriate growth condition. Growth morphology of organoids was imaged, and viability of each organoid was scored.
Supplementary Material
Acknowledgments
B.-K.K. is supported by Wellcome Trust with the Henry Dale Fellowship. J.H.v.E. and H.C. are supported by Grant KWF/PF-HUBR 2007-3956 and NIH/MIT Subaward 5710002735.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508113112/-/DCSupplemental.
References
- 1.Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 2.Koo BK, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 3.Giannakis M, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa T, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA. 2011;108(52):21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2013;110(31):12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 8.Ryland GL, et al. Australian Ovarian Cancer Study Group RNF43 is a tumour suppressor gene mutated in mucinous tumours of the ovary. J Pathol. 2013;229(3):469–476. doi: 10.1002/path.4134. [DOI] [PubMed] [Google Scholar]
- 9.Ong CK, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44(6):690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 10.Assié G, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46(6):607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- 11.Nord KH, et al. Recurrent chromosome 22 deletions in osteoblastoma affect inhibitors of the Wnt/beta-catenin signaling pathway. PLoS ONE. 2013;8(11):e80725. doi: 10.1371/journal.pone.0080725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen PH, Chen X, Lin Z, Fang D, He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27(12):1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng WC, et al. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS ONE. 2013;8(12):e83110. doi: 10.1371/journal.pone.0083110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zebisch M, et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4:2787. doi: 10.1038/ncomms3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 16.Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143(6):1518–1529.e7. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 17.Durand A, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci USA. 2012;109(23):8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TH, Escudero S, Shivdasani RA. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci USA. 2012;109(10):3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabiri Z, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141(11):2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294(5549):2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 21.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takada R, et al. Monounsaturated fatty acid modification of Wnt protein: Its role in Wnt secretion. Dev Cell. 2006;11(6):791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci USA. 2013;110(50):20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proffitt KD, et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 25.Barrow JR, et al. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 2003;17(3):394–409. doi: 10.1101/gad.1044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 27.Shroyer NF, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132(7):2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.