Significance
The origins of novelties pose some of the most difficult experimental challenges in evolutionary biology. Morphological novelties in animals are generally thought to arise through new domains of expression of so-called “toolkit” regulatory genes, but how such changes arise has not been explored in depth. Here, we show that three novel features of wingless gene expression associated with three novel pattern elements are the result of three novel enhancer activities. One of these enhancers is clearly modified from an evolutionarily conserved, pre-existing regulatory sequence. We suggest the modification of extant enhancers is a common path to novelty in gene expression and morphology.
Keywords: enhancers, novelty, gene regulation, development, pigmentation
Abstract
Changes in gene expression during animal development are largely responsible for the evolution of morphological diversity. However, the genetic and molecular mechanisms responsible for the origins of new gene-expression domains have been difficult to elucidate. Here, we sought to identify molecular events underlying the origins of three novel features of wingless (wg) gene expression that are associated with distinct pigmentation patterns in Drosophila guttifera. We compared the activity of cis-regulatory sequences (enhancers) across the wg locus in D. guttifera and Drosophila melanogaster and found strong functional conservation among the enhancers that control similar patterns of wg expression in larval imaginal discs that are essential for appendage development. For pupal tissues, however, we found three novel wg enhancer activities in D. guttifera associated with novel domains of wg expression, including two enhancers located surprisingly far away in an intron of the distant Wnt10 gene. Detailed analysis of one enhancer (the vein-tip enhancer) revealed that it overlapped with a region controlling wg expression in wing crossveins (crossvein enhancer) in D. guttifera and other species. Our results indicate that one novel domain of wg expression in D. guttifera wings evolved by co-opting pre-existing regulatory sequences governing gene activity in the developing wing. We suggest that the modification of existing enhancers is a common path to the evolution of new gene-expression domains and enhancers.
As animals have adapted to diverse habitats they have evolved many new and different kinds of body parts. One of the major outstanding questions in evolutionary biology is: What kinds of mechanisms underlie the origin of morphological novelties? It is well established that the regulatory genes responsible for the formation and patterning of animal bodies and body parts, the so-called “toolkit” genes for animal development, are shared and highly conserved among most animal phyla (1–4). Very different forms are generated by similar sets of developmental genes, and a large body of empirical, comparative studies have led to the general consensus that divergence in the expression and regulation of toolkit genes and the genes they control largely underlies morphological diversity (5–9).
Similarly, several studies have revealed that new features of regulatory gene expression are associated with the evolution of morphological novelties, such as new color-pattern elements on insect wings (10–14). How new patterns of regulatory gene expression evolve, however, has been more difficult to elucidate. In principle, new patterns of gene expression may evolve through: (i) changes in the deployment of upstream trans-acting regulatory factors, (ii) changes in the cis-regulatory sequences of the genes themselves, or (iii) a combination of these mechanisms. For example, the novel, male-specific wing spot in Drosophila biarmipes and a few close relatives evolved through a combination of changes in the spatial expression of the trans-acting Distal-less (Dll) transcription factor and the evolution of Dll and other binding sites in a cis-regulatory element of at least one pigmentation gene (15, 16). In this case, the Dll protein is said to have been co-opted in the evolution of a new morphological trait.
However, the mechanism underlying the co-option of Dll is not known in this case, nor for any other instances of the co-option of regulatory genes. It is not known, for example, whether new features of gene expression evolve via the de novo origin of enhancers or through the transposition or modification of existing enhancers. One distinguishing feature shared by most developmental regulatory gene loci is that, like Dll (17, 18), they often contain vast cis-regulatory regions harboring numerous independent enhancers. To complicate matters, some of these enhancers may be located far away in other genes. The diversity of enhancers belonging to individual regulatory genes is explicit evidence that gene function has expanded in the course of evolution by accumulating additional enhancers, but understanding how this occurs presents significant experimental challenges. To further our understanding of the molecular basis of gene-expression novelties, it is necessary both to identify the novel enhancers in the species of interest and to ascertain their structural and functional relationships to sequences in other species lacking the specific domains of gene expression (19).
The Wingless (Wg) protein is a secreted signaling molecule that acts as a morphogen in the development of numerous structures and pattern elements in Drosophila and other animals (20–23). Here, we have traced the molecular basis of three novel features of wingless (wg) gene expression in Drosophila guttifera that are associated with three distinct features of adult pigmentation. By searching through the wg and adjacent loci of both D. guttifera and Drosophila melanogaster, we found three novel enhancer activities in D. guttifera. We show that one of these enhancers, the novel vein-tip enhancer in D. guttifera, is nestled within a conserved enhancer in other species. We propose that the new enhancer activity evolved through the modification of the pre-existing enhancer.
Results
Novel wg Expression Domains in the D. guttifera Pupal Wing.
Regulatory genes coordinate important developmental events; thus, their expression patterns are constrained and usually conserved, particularly among closely related species. wg expression patterns in larval imaginal discs (wing disc, eye-antennal disc, and leg disc) of D. melanogaster and D. guttifera adhere to this generality and are essentially identical (SI Appendix, Fig. S1). In both species, wg expression was virtually identical in the developing wing pouches and the future nota of wing discs (SI Appendix, Fig. S1 A and D), the anterior-ventral parts of antennae, ventral and dorsal sides of eye discs (SI Appendix, Fig. S1 B and E), and the anterior-ventral parts of leg discs (SI Appendix, Fig. S1 C and F).
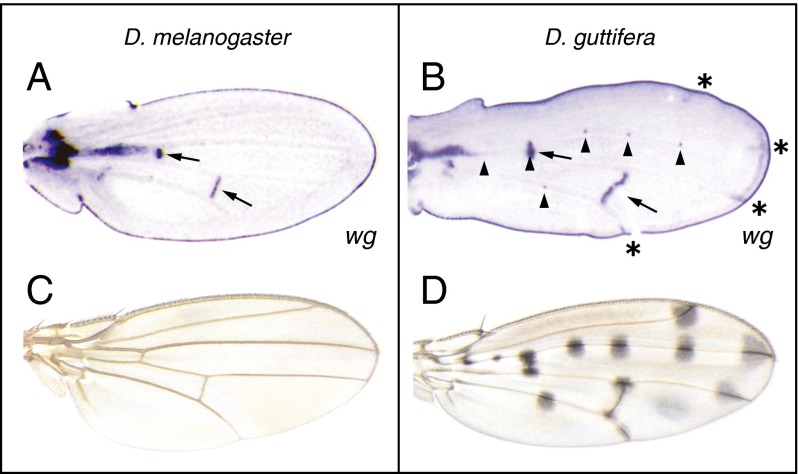
In contrast, in the developing pupal wings of D. guttifera, wg is expressed in two domains that are not present in D. melanogaster pupal wings (14, Fig. 1). Whereas in D. melanogaster, wg is expressed in cells along the developing wing margin (henceforth “margin”) and crossveins (Fig. 1A, arrows), in D. guttifera (Fig. 1B) wg is also expressed at the tips of longitudinal veins (“vein tip”) (Fig. 1B, asterisks) and in precursors of the campaniform sensilla (Fig. 1B, arrowheads). None of the other several species closely related to D. guttifera within the Drosophila quinaria species group (Drosophila deflecta, Drosophila nigromaculata, Drosophila palustris, and D. quinaria) exhibited wg expression in the developing campaniform sensilla or vein tips (SI Appendix, Fig. S2). Both novel wg expression domains correlate with color pattern formation in D. guttifera (Fig. 1D), but not in D. melanogaster (Fig. 1C).
Fig. 1.
Unique wg expression domains in D. guttifera pupal wings correlate with adult pigment spots. (A) wg expression pattern in the pupal wing of D. melanogaster visualized by in situ hybridization. wg is expressed in the developing crossveins and along the wing margin. (B) wg expression pattern in the pupal wing of D. guttifera. wg is expressed in the campaniform sensilla (arrowheads), crossveins (arrows), and longitudinal vein tips (asterisks), and along the entire wing margin. (C) Adult wing of D. melanogaster. (D) Adult wing of D. guttifera.
The wg Enhancers Active in Imaginal Discs Are Conserved Between Species.
Our primary task was to identify the enhancers responsible for these novel features of wg expression in D. guttifera. Because we could not predict where novel enhancers might be located, our approach was to identify functional enhancers across the entire D. guttifera wg region and to compare their activity and structure with the homologous regions of the D. melanogaster wg region. This approach offered the added benefit of enabling a comparison of the overall organization of the cis-regulatory regions of the wg locus of the two species. Because we could not assume that D. guttifera enhancers would have the same activity in D. melanogaster (the usual host for transgenic methods in Drosophila) as in D. guttifera, we constructed reporter genes with DNA from each species and injected them into their species of origin. The D. melanogaster wg locus sequence had been determined previously (20, 22, 24). From a draft assembly of the D. guttifera genome and PCR amplification, we located the D. guttifera wg locus on a 64-kb-long contig. We systematically fused 0.3- to 10-kb (average 4.0 kb) noncoding segments of each species’ wg region to an EGFP/DsRed reporter gene (SI Appendix, Fig. S3).
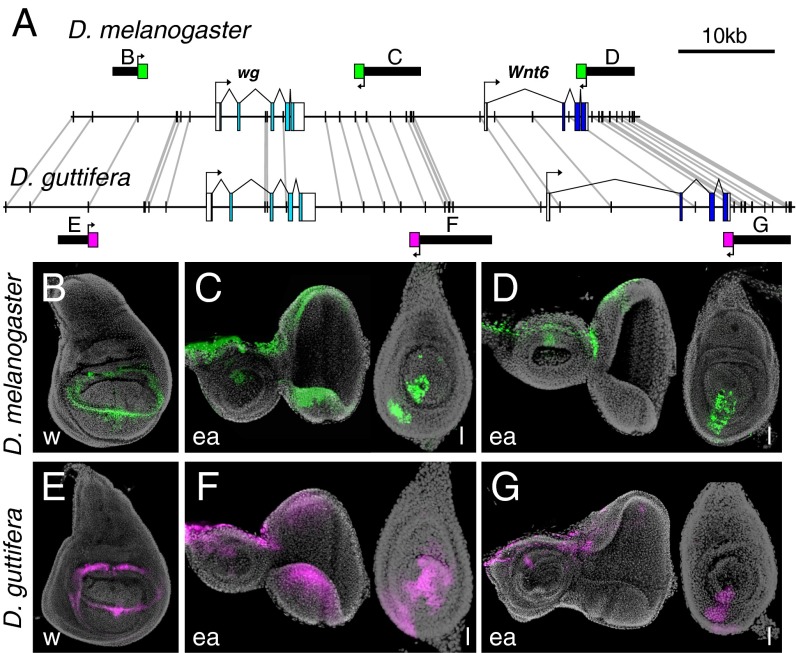
We first monitored the larval imaginal discs for reporter protein activity, and we were able to confirm or identify several orthologous wg enhancers (Fig. 2) including: (i) a previously reported enhancer driving wing pouch expression in the 5′ region in both species (Fig. 2 B and E) [the spadeflag (spdfg) region in D. melanogaster (25)]; (ii) enhancers active in the eye-antennal discs and leg discs that are located in the 3′ region of wg gene (Fig. 2 C and F); and (iii) an enhancer in the 3′ region of the Wnt6 gene (Fig. 2 D and G). These results are consistent with a recent survey describing a large collection of imaginal disc enhancers of D. melanogaster (Flylight) (26). Because the Wnt6 expression pattern is mostly similar to that of wg (27), and the clustering of four Wnt genes is conserved in the Drosophila genus, the loci have been inferred to share regulatory elements (28). We note that the overall position and order of imaginal disc enhancers is largely colinear across the 60- to 74-kb region in both species (Fig. 2A). This result indicates that there have not been any significant inversions or other rearrangements across the region in either lineage since the two species diverged from a common ancestor ∼63 million y ago (the divergence between the subgenus Drosophila and subgenus Sophophora) (29).
Fig. 2.
Conserved wg cis-regulatory elements control similar gene-expression patterns in Drosophila imaginal discs. (A) Schematic of enhancers plotted on the wg locus of D. melanogaster and D. guttifera. Solid vertical lines connected by gray lines represent sequences longer than 40 bp with 100% nucleotide conservation between species. (B–D) D. melanogaster third-instar imaginal discs showing reporter expression with D. melanogaster enhancer fragments (EGFP, green). (E–G) D. guttifera third-instar imaginal discs showing very similar reporter expression patterns driven by orthologous D. guttifera enhancer fragments (DsRed, magenta). All discs are oriented with anterior to the left and dorsal on top. ea, eye-antennal disc; l, leg disc; w, wing disc. (Magnification: B–G, 200×.)
The D. guttifera wg Locus Contains a Novel Vein-Tip Enhancer.
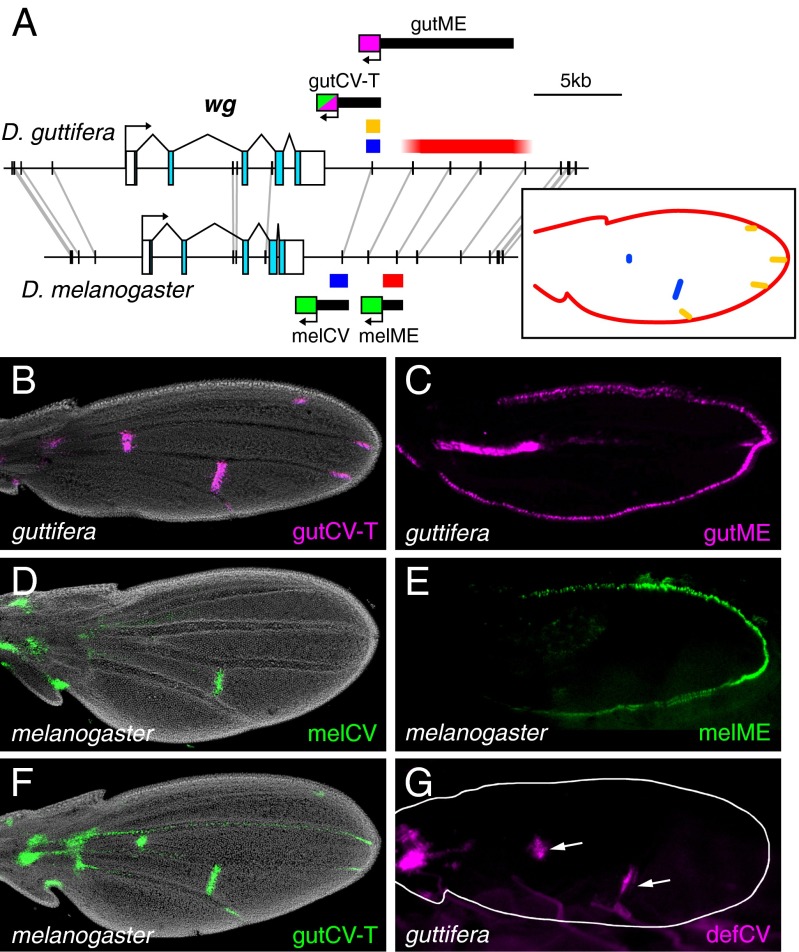
In our search for enhancers that regulate the D. guttifera-specific wg expression domains in pupal wings, we identified two enhancers located 3′ of the D. guttifera wg gene: a crossveins enhancer (gutCV-T) and margin enhancer (gutME), which together account for the conserved wg-expression domains (Fig. 3 A–C; see also Fig. 1B). We also identified the orthologous enhancers, melCV and melME, from D. melanogaster, which drove reporter expression in the crossveins and wing margin, respectively (Fig. 3 D and E). Importantly, in addition to the conserved crossvein expression, gutCV-T also drove reporter expression in the developing wing tips, where the wing veins meet the margin, which is part of the novel wg expression pattern in D. guttifera (Fig. 3B).
Fig. 3.
A novel vein-tip enhancer activity in D. guttifera. (A) Schematic of pupal wing enhancers in D. guttifera and D. melanogaster. Black bars connected by gray lines represent sequences longer than 40 bp with 100% nucleotide conservation between species. (Inset) Schematic of wg expression in the pupal wing that is color-coded for the responsible enhancers. (B) D. guttifera pupal wing showing reporter expression from the gutCV-T enhancer in the crossveins and vein tips (DsRed, magenta). (C) D. guttifera pupal wing showing reporter expression from the gutME enhancer (DsRed, magenta) along the wing margin. (D) D. melanogaster pupal wing showing reporter expression from the melCV enhancer fragment (EGFP, green) in the crossvein. (E) D. melanogaster pupal wing showing reporter expression from the melME enhancer (EGFP, green) along the wing margin. (F) D. melanogaster pupal wing showing reporter expression from the gutCV-T enhancer (EGFP, green) in the crossveins and vein tips. (G) D. guttifera pupal wing showing reporter expression (DsRed, magenta) from the defCV enhancer in the crossveins (asterisks). (Magnification: B–G, 100×.)
The difference in activities between the orthologous melCV and gutCV-T enhancers of D. melanogaster and D. guttifera could be because of differences in trans-acting regulatory factors expressed in each wing, differences in cis-regulatory sequences between the enhancers, or both. To determine which might be the case, we carried out a simple cis-trans test by introducing the gutCV-T enhancer into D. melanogaster. The gutCV-T fragment drove reporter protein expression in both the crossveins and vein tips in pupal D. melanogaster wings (Fig. 3F). This result indicates that the trans-acting factors necessary for the vein tip expression pattern are present in both species. Thus, the differences in activities between the gutCV-T and melCV enhancers must reside in their cis-regulatory sequences.
We also isolated and tested the orthologous cis-regulatory region from D. deflecta, which is one of the most closely related species to D. guttifera but does not have wg expression in the vein tips (SI Appendix, Fig. S2C). This D. deflecta crossveins enhancer (defCV) drove reporter protein expression in the wing crossveins in D. guttifera, but showed no activity in the vein tips (Fig. 3G). This result indicates that the vein-tip enhancer activity is unique to D. guttifera, and that the novel feature of wg expression in D. guttifera wing vein tips arose through the evolution of cis-regulatory sequences in the D. guttifera lineage, after it split off from a common ancestor shared with D. deflecta.
The D. guttifera-Specific Vein-Tip Enhancer Is Nestled Within the Crossvein Enhancer.
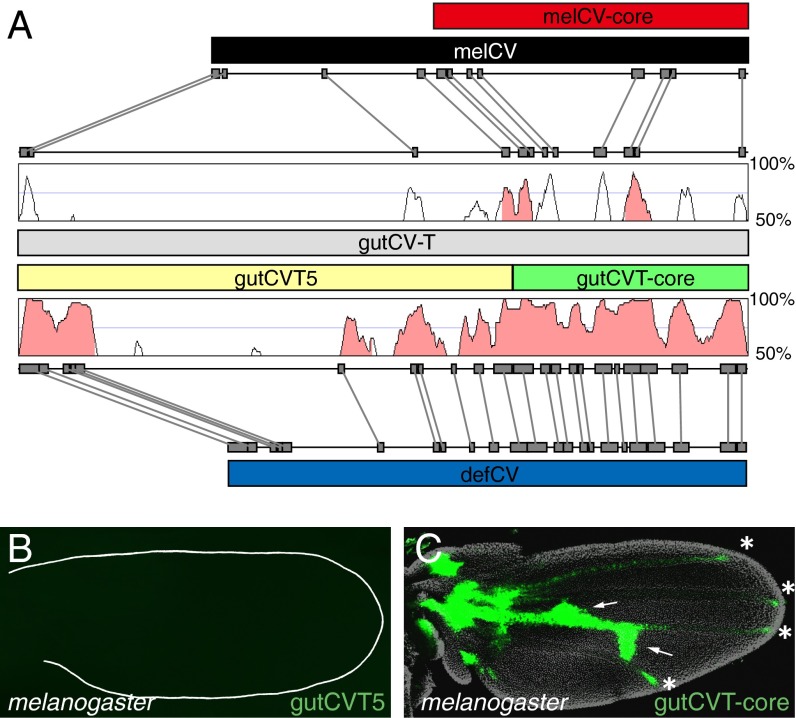
The 2.4-kb gutCV-T enhancer, which drove both crossvein and vein tip expression in D. guttifera (Fig. 3B), shares numerous collinear, highly conserved blocks of sequence with both the 1.8-kb melCV fragment from the D. melanogaster locus and the 1.7-kb defCV fragment from the D. deflecta locus, which both lack vein tip activity (Figs. 3 D and G and 4A). We considered two possibilities to explain how the novel vein tip expression of gutCV-T may have evolved within the domain of the crossvein enhancer: (i) a distinct enhancer element, able to independently drive expression in the vein tips, inserted into the D. guttifera wg locus (by chance next to another pupal wing enhancer); or (ii) a novel activity arose within the crossvein enhancer that used and is dependent upon pre-existing sites in the crossvein enhancer. To attempt to distinguish these possibilities, we compared the D. guttifera, D. deflecta, and D. melanogaster sequences and searched for major insertions or regions unique to D. guttifera. Indeed, we found that the D. guttifera fragment is over 900 bp longer than the orthologous D. melanogaster sequence and 600 bp longer than the orthologous D. deflecta sequence (Fig. 4A). This size difference is largely because of a region in the less conserved 5′ end of the gutCV-T enhancer. This additional sequence did not show any similarity to known transposable elements (when tested by blastn against the National Center for Biotechnology Information nucleotide collection). To test whether this region might contain a distinct enhancer, we divided gutCV-T into two fragments: the insert-containing 5′ 1,653-bp (gutCVT5) and the 3′, highly conserved 756-bp fragment (gutCVT-core) (Fig. 4A). Whereas the gutCVT5 fragment showed no activity, the gutCVT-core fragment drove expression in both the crossveins and vein tips (Fig. 4 B and C). These results reveal that the novel activity in the gutCVT enhancer arose within the 3′ 756-bp region.
Fig. 4.
The D. guttifera vein-tip enhancer is nestled within a conserved crossvein enhancer. (A) Schematic comparing crossvein enhancer regions in D. melanogaster, D. guttifera, and D. deflecta. The gutCV-T enhancer (gray bar) aligned with the melCV enhancer (black bar, Top) and the defCV enhancer (blue bar, Bottom) using GenePalette (gray boxes connected with gray lines indicate sequences of 15 bp or longer with 100% conservation between species) and Vista Browser (50-bp sliding window with percent sequence identity indicated, peaks with greater than 80% sequence identity are shaded in pink). Peaks show extent of sequence conservation in a sliding 50-bp window. The gutCV-T enhancer was divided into two fragments, gutCVT5 (yellow bar) and gutCVT-core (green bar). (B) D. melanogaster pupal wing showing absence of reporter expression from gutCVT5 (EGFP, green). (C) D. melanogaster pupal wing showing reporter expression from the gutCVT-core fragment (EGFP, green) in the crossveins (arrows) and vein tips (asterisks). (Magnification: B and C, 100×.)
To examine how this region may have acquired its unique vein-tip activity, we compared it in detail with the orthologous D. deflecta sequence that lacks vein-tip activity. The D. guttifera CVT-core region is 83% similar to the orthologous D. deflecta region, with many large blocks of identical sequence and just a few small (<10 bp) insertions or deletions (SI Appendix, Fig. S4). This pattern of sequence homology indicates that the novel domain of wg expression in the vein tips of D. guttifera is likely caused by a small number of nucleotide changes or small indels nestled within the well-conserved crossvein enhancer.
The Campaniform Sensillum and Thoracic Stripe Enhancers Are in the Distant Wnt10 Region.
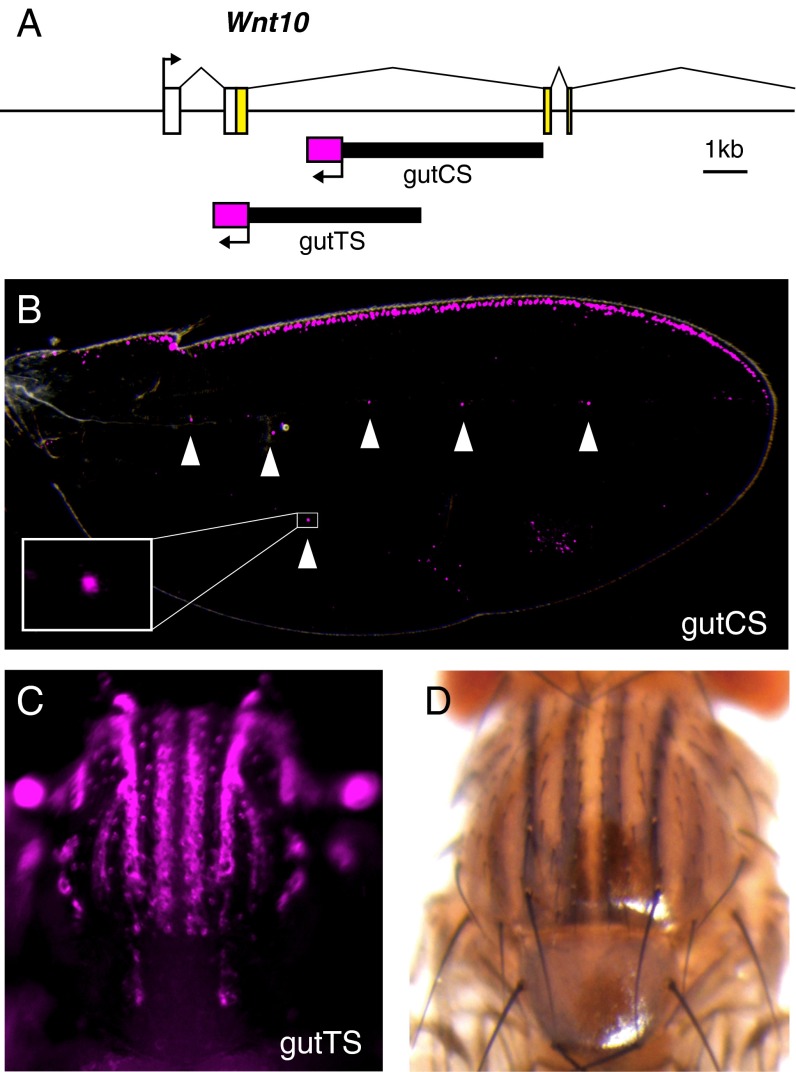
During our initial search for wg enhancer activities in D. guttifera, we were puzzled by our inability to find an enhancer for the novel patterns of wg expression in the developing wing campaniform sensilla, which contributes several spots to the overall polka-dot wing pattern (14). Therefore, we expanded our search into adjacent Wnt loci just in case they might contain enhancers that regulate wg transcription. Using seven additional scaffolds, we extended the region analyzed to include a 174-kb region containing the Wnt4, Wnt6, and Wnt10 genes. We were surprised to find two more distinct enhancer activities in the Wnt10 region, more than 69 kb away from the wg transcription start site and separated from it by the Wnt6 locus (Figs. 2 and 5A, and SI Appendix, Fig. S3). One 5-kb fragment within the second intron of the Wnt10 gene (gutCS) (Fig. 5A) drove reporter expression in the campaniform sensilla and along the anterior margin of the pupal wing (Fig. 5B, arrowheads). Because wg is the only gene in this Wnt cluster that is expressed in campaniform sensilla (SI Appendix, Fig. S5), we conclude that this enhancer controls wg expression.
Fig. 5.
The distant Wnt10 region contains two novel and distinct wg enhancers in D. guttifera. (A) Schematic showing the location of two enhancer fragments in the second intron of Wnt10. (B) D. guttifera pupal wing showing reporter expression driven by the gutCS enhancer (DsRed, magenta) in the campaniform sensilla (arrowheads). (C) D. guttifera pupal thorax showing a striped reporter expression pattern driven by the gutTS enhancer (DsRed, magenta). (D) Stripes of black pigmentation on the thorax of an adult D. guttifera. (Magnification: B, 80×; C, 50×; D, 32×.)
A second, partially overlapping 4.3-kb fragment (gutTS) (Fig. 5A) drove reporter expression in a series of thoracic stripes (Fig. 5C) that correspond well with the adult thoracic striped pigmentation pattern (Fig. 5D). We were not able to confirm by in situ hybridization that this reflects a native wg expression domain because gene probes did not yield reliable signals in pupal thoracic body wall tissues. However, we performed RT-PCR on thoracic body wall total RNA to ascertain which Wnt genes were active in this tissue. Only wg showed strong expression, whereas the other, adjacent Wnt genes (Wnt4, Wnt6, and Wnt10) exhibited weak or no expression (SI Appendix, Fig. S6). These results, and the strong correlation with thoracic pigmentation, indicate that wg is expressed in the thorax and regulated by the gutTS enhancer.
Cis-Regulatory Sequence Evolution Is Partly Responsible for Novel Distant Enhancer Activities.
We next sought to identify the relative contribution of cis-acting and trans-acting regulatory factors in the evolution of the D. guttifera gutCS and gutTS enhancer activities. We conducted reciprocal tests of the D. guttifera and homologous D. melanogaster sequence in the other species’ genetic background. In contrast to the gutCV-T enhancer, the D. guttifera CS enhancer was not active in D. melanogaster wings, indicating a role for trans-acting factors in enhancer activity in D. guttifera (SI Appendix, Fig. S7C). In addition, the homologous D. melanogaster fragment (45.7% similarity) was not active in either D. guttifera or D. melanogaster, indicating an additional contribution of cis-regulatory changes in the gutCS enhancer (SI Appendix, Fig. S7 A and B). Taken together, these results indicate that both cis-regulatory and trans-regulatory changes were responsible for the evolution of the novel wg expression domain in campaniform sensilla.
We performed a similar set of reciprocal experiments with the gutTS enhancer and homologous D. melanogaster sequence (SI Appendix, Fig. S8). The homologous fragment from D. melanogaster (46.3% similarity) was inactive in both D. melanogaster (SI Appendix, Fig. S8A) and D. guttifera (SI Appendix, Fig. S8B), whereas the D. guttifera TS enhancer was weakly active in stripes in the D. melanogaster thorax (compare SI Appendix, Fig. S8 C and D). These results indicate that cis-regulatory changes are largely responsible for the novel activity of the gutTS enhancer and that some, but perhaps not all, of the trans-acting factors involved in regulating the enhancer are deployed in D. melanogaster.
Discussion
A large body of comparative studies has shown that changes in the spatiotemporal expression of toolkit genes and the target genes they regulate correlate with the evolution of morphological traits. In a considerable number of instances, these spatiotemporal changes in gene expression have been demonstrated to involve the modification of enhancers (6, 7, 30–36). However, there are relatively few cases in which the origins of new enhancers have been elucidated, and none involving regulatory genes themselves.
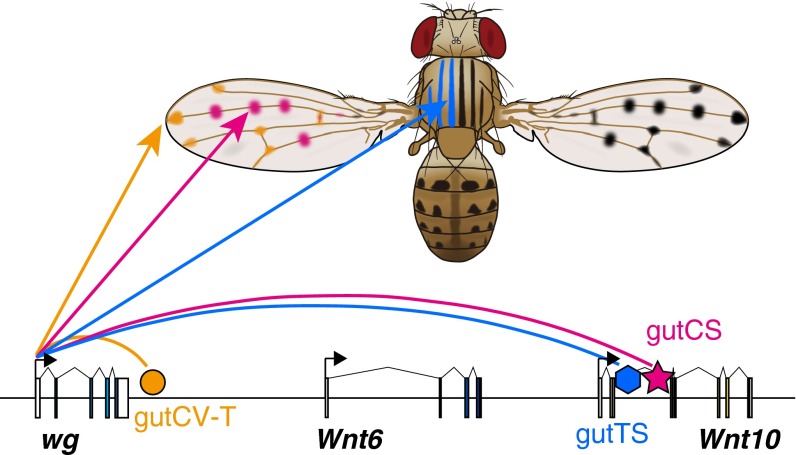
Here, we have shown that three novel domains of wg expression in D. guttifera are governed by three novel enhancers, respectively (Fig. 6). We found that the evolution of wg cis-regulatory sequences within the D. guttifera lineage played a role in the gain of each enhancer activity, and that the evolution of trans-acting regulatory factors was also necessary for the activity of two elements (gutCS and gutTS). Detailed analysis of the D. guttifera vein-tip enhancer revealed that it evolved within another conserved enhancer, whereas two other enhancers (the campaniform sensilla and thoracic stripe enhancers) arose within in an intron of the distant Wnt10 locus. These results bear on our understanding of the mechanisms underlying the evolution of new enhancers and domains of gene expression.
Fig. 6.
Three novel wg enhancers drive D. guttifera-specific pigmentation patterns. The genomic organization of the D. guttifera Wnt region is shown with colored shapes corresponding to enhancers from this study. The pupal expression domains of each enhancer are mapped by their respective color onto the pigmentation patterns of the adult animal.
The Origin of the Vein-Tip Enhancer via Co-option of an Existing Enhancer.
The D. guttifera vein-tip enhancer activity was localized within a 756-bp DNA segment that was also active in the developing pupal crossveins. This DNA segment is orthologous to segments of DNA in D. melanogaster and D. deflecta that were only active in the crossveins. The segments are all collinear, and contain numerous blocks of identical sequence, which suggests that the vein-tip enhancer activity evolved within the pre-existing crossvein enhancer.
One explanation for the presence of two activities in this one fragment is that they share functional sites: that is, binding sites for common transcription factors. Because both activities appear in the pupal wing, it is likely that they use common tissue-specific (wing) and temporal (pupal) inputs. The evolution of a new activity in the vein tips could have arisen through the addition of DNA-binding sites for transcription factors that were already present active in cells at vein tips. In this scenario, the novel enhancer activity would have resulted from the evolutionary co-option of an existing enhancer.
There is precedent for multifunctional enhancers and for this mechanism of co-option. For example, one enhancer of the D. melanogaster even-skipped gene governs two domains of gene expression that are controlled by shared inputs (37). In addition, Rebeiz et al. (19) demonstrated that a novel optic lobe enhancer of the Drosophila santomea Neprilysin-1 gene arose via co-option of an existing enhancer. Moreover, it was shown that co-option had occurred in just a few mutational steps. The co-option of existing elements is an attractive explanation for the evolution of novel enhancers because it requires a relatively short mutational path.
The Evolution of Distant cis-Regulatory Elements.
One surprising property of enhancers is their ability to control gene transcription at promoters located at considerable linear distances away in the genome (38–40). For example, the enhancer that drives Sonic hedgehog (Shh) expression in the developing amniote limb bud is located in the intron of another gene ∼1 Mb from the Shh locus (41, 42). A growing body of evidence indicates that long segments of DNA are looped out in accommodating long-range enhancer–promoter interactions (43, 44). The ability of enhancers to act over such long ranges suggests that new enhancers could evolve at considerable distances from the promoters that they regulate.
Here, we identified two enhancers in an intron of the D. guttifera Wnt10 gene that control transcription of the wg gene from a distance of ∼70 kb, and separated by the Wnt6 locus. Our data suggest that the gutTS enhancer preferentially regulates wg transcription and not Wnt10 or Wnt6 transcription, although we cannot offer any explanation at present for this preference. The origins of the gutCS and gutTS enhancers are not as clear as the vein-tip enhancer. We did not detect any pupal enhancer activity in the orthologous DNA segments of D. melanogaster, so we do not have any evidence of enhancer co-option. Nor did we find any obvious insertions in these DNA segments such as a transposon. Nevertheless, the discovery of these novel, distant elements reflects the functional flexibility of cis-regulatory elements and their contribution to the evolution of gene regulation and morphological diversity.
Materials and Methods
Fly Strains and Genomic DNA.
D. melanogaster Canton-S (wild-type) was used for genomic DNA preparation and expression analysis of Wnt genes. We obtained D. guttifera (stock no.15130–1971.10), D. deflecta (15130-2018.00), D. quinaria (15130-2011.00), and D. palustris (15130-2001.00) from the Drosophila Species Stock Center at University of California, San Diego, and D. nigromaculata (strain no. E-14201) from EHIME-Fly, Ehime University, Japan. Genomic DNA was extracted and purified using a squish method (45) and Genomic tip-20/G columns (Qiagen).
In Situ Hybridization.
Species specific, partial sequences of Wnt genes (Wnt4, wg, Wnt6, and Wnt10) were amplified by PCR from genomic DNA and cloned into the pGEM-TEasy vector (Promega). PCR products reamplified from the plasmid clones were in vitro transcribed to produce DIG-RNA probes (35). Imaginal discs of late third-instar larvae and wings of P6 stage pupae (46) were subjected to in situ hybridization, as described previously (14, 47). Specimens were mounted and imaged under a stereomicroscope SZX-16 (Olympus).
Genomic Sequence of the Wnt Locus.
The genome sequence reads of D. guttifera were obtained with a Genome Analyzer IIx (Illumina), and assembled with CLC workbench (CLC Bio). The Wnt locus of D. guttifera was reconstructed with seven genomic scaffolds and genomic PCR products (Accession no. KP966547) (SI Appendix, Fig. S3). For the comparison of sequences from multiple species, we used GenePalette software (48). All primers are listed in SI Appendix, Table S1.
EGFP/DsRed Reporter Assay for Enhancer Activity Using Transgenic Drosophila.
For the site-specific integration of transgenes into D. melanogaster, the plasmid vector S3aG (36) and fly strains VK00006 (cytogenetic location 19E7), VK00018 (cytogenetic location 53B2) (49), and ZH-attP-51D (cytogenetic location 51D) (50) were used. D. guttifera transgenics were made according to the previously described method (14), using the cloning shuttle vector pSLfa1180fa harboring DsRed2 or DsRed.T4, the piggyBac transposon vector pBac {3xP3-EGFPafm} (51, 52), and the piggyBac helper plasmid phspBac (53). Fluorescent reporter expression was observed under a stereomicroscope SZX-16 and a confocal laser-scanning microscope FV1000 (Olympus).
Supplementary Material
Acknowledgments
We thank Jane Selegue and Steve Paddock for their technical help; Henry Chung, Héloïse D. Dufour, Cédric Finet, Noah Dowell, David W. Loehlin, Thomas M. Williams, Troy Shirangi, and Mark Rebeiz for fruitful discussions and comments; and Kiyokazu Agata and Naoyuki Fuse for sharing experimental facilities. This work was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (to S.K.) and the Howard Hughes Medical Institute (S.B.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. 1805591 and KP966547).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509022112/-/DCSupplemental.
References
- 1.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd Ed Blackwell Publishing; Oxford: 2004. [Google Scholar]
- 2.McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 3.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265(5173):785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 4.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3(7):e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 7.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittkopp PJ. Evolution of cis-regulatory sequence and function in Diptera. Heredity (Edinb) 2006;97(3):139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 9.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 10.Brakefield PM, et al. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384(6606):236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- 11.Carroll SB, et al. Pattern formation and eyespot determination in butterfly wings. Science. 1994;265(5168):109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- 12.Martin A, et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA. 2012;109(31):12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed RD, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333(6046):1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 14.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464(7292):1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 15.Arnoult L, et al. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science. 2013;339(6126):1423–1426. doi: 10.1126/science.1233749. [DOI] [PubMed] [Google Scholar]
- 16.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433(7025):481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 17.McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136(1):61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vachon G, et al. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell. 1992;71(3):437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- 19.Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci USA. 2011;108(25):10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: The spatial distribution of a transcript in embryos. EMBO J. 1987;6(6):1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- 22.Rijsewijk F, et al. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987;50(4):649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 23.Swarup S, Verheyen EM. Wnt/Wingless signaling in Drosophila. Cold Spring Harb Perspect Biol. 2012;4(6):a007930. doi: 10.1101/cshperspect.a007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 25.Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development. 1996;122(6):1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- 26.Jory A, et al. A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Reports. 2012;2(4):1014–1024. doi: 10.1016/j.celrep.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janson K, Cohen ED, Wilder EL. Expression of DWnt6, DWnt10, and DFz4 during Drosophila development. Mech Dev. 2001;103(1-2):117–120. doi: 10.1016/s0925-4773(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 28.Gieseler K, et al. DWnt4 and wingless elicit similar cellular responses during imaginal development. Dev Biol. 2001;232(2):339–350. doi: 10.1006/dbio.2001.0184. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21(1):36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 30.Belting HG, Shashikant CS, Ruddle FH. Modification of expression and cis-regulation of Hoxc8 in the evolution of diverged axial morphology. Proc Natl Acad Sci USA. 1998;95(5):2355–2360. doi: 10.1073/pnas.95.5.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong S, et al. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132(5):783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 33.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448(7153):587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 34.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440(7087):1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 35.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326(5960):1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134(4):610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175(2):314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 38.Forrester WC, et al. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4(10):1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 39.Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113(3):735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- 40.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113(3):405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 41.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 42.Sagai T, et al. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh) Mamm Genome. 2004;15(1):23–34. doi: 10.1007/s00335-033-2317-5. [DOI] [PubMed] [Google Scholar]
- 43.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32(4):623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 44.Su W, Jackson S, Tjian R, Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991;5(5):820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 45.Gloor GB, et al. Type I repressors of P element mobility. Genetics. 1993;135(1):81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- 47.Sturtevant MA, Roark M, Bier E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev. 1993;7(6):961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- 48.Rebeiz M, Posakony JW. GenePalette: A universal software tool for genome sequence visualization and analysis. Dev Biol. 2004;271(2):431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314(5806):1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 50.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line–specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104(9):3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horn C, Jaunich B, Wimmer EA. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000;210(12):623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- 52.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210(12):630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 53.Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8(4):449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.