Significance
The lack of effective tools to modulate individual species within a complex microbial community poses a major barrier to studying human microbiome and its associated diseases. We showed targeted removal of human cariogenic Streptococcus mutans within an in vitro oral multispecies community using a high-efficacy antimicrobial peptide—C16G2—as well as drastic reconstruction of the microbial structure following treatment. The findings suggest the potential of using targeted antimicrobials to modulate microbiome and study the ecological function of specific bacterial species.
Keywords: human microbiome, targeted antimicrobial, dental caries, oral microbiota
Abstract
One major challenge to studying human microbiome and its associated diseases is the lack of effective tools to achieve targeted modulation of individual species and study its ecological function within multispecies communities. Here, we show that C16G2, a specifically targeted antimicrobial peptide, was able to selectively kill cariogenic pathogen Streptococcus mutans with high efficacy within a human saliva-derived in vitro oral multispecies community. Importantly, a significant shift in the overall microbial structure of the C16G2-treated community was revealed after a 24-h recovery period: several bacterial species with metabolic dependency or physical interactions with S. mutans suffered drastic reduction in their abundance, whereas S. mutans’ natural competitors, including health-associated Streptococci, became dominant. This study demonstrates the use of targeted antimicrobials to modulate the microbiome structure allowing insights into the key community role of specific bacterial species and also indicates the therapeutic potential of C16G2 to achieve a healthy oral microbiome.
Human microbiome research revealed that every human body contains a variety of microbial communities on various mucosal surfaces that consist of thousands of different microbial species (1–4). Disturbance from host and environmental factors may alter the composition and abundance of these microbial species, leading to various polymicrobial diseases (5–8). With the complexity of these multispecies microbial communities, it is very difficult to determine the functions of individual species contributing to observed physiological and pathological changes, making it one of the most challenging issues in human microbiome research. This study aims to develop and validate a new tool to address this important issue.
The indigenous microbial flora of the human oral cavity consists of over 700 different species of bacteria, with over 100 present in any individual (4, 9, 10). Most bacteria help promote a healthy oral environment by stimulating the immune system and preventing the invasion of pathogenic species (11–13). One pathogenic bacterium, Streptococcus mutans, is predominantly responsible for tooth decay worldwide (14). The carious lesions that S. mutans can cause are generally not considered life-threatening, but they result in an economic burden that leaves many cases in underdeveloped areas untreated, resulting in tooth extraction as the only remedy (15). Currently, there is no effective treatment for S. mutans. Broad-spectrum antibiotics administered to the oral cavity result in destruction of the entire oral bacterial flora, making it prone to reinfection by S. mutans, which reestablishes at pretreatment levels.
To combat S. mutans infection, our group developed C16G2, a synthetic peptide that belongs to a new class of antimicrobials called specifically targeted antimicrobial peptides that can achieve targeted killing of selected pathogens (16). A typical targeted antimicrobial peptide molecule consists of two functionally independent moieties conjoined in a linear peptide sequence: a nonspecific antimicrobial peptide serves as the killing moiety, whereas a species-specific binding peptide comprises the targeting moiety that provides specific binding to a selected pathogen and facilitates the targeted delivery of the attached antimicrobial peptide. Previous studies showed that C16G2 was potent against S. mutans grown in liquid or biofilm states. It displays targeted killing of S. mutans within a three-species biofilm without affecting closely related noncariogenic oral streptococci, including Streptococcus sanguinis and Streptococcus gordonii (16). Additional study showed that C16G2 has a mechanism of action similar to that of traditional antimicrobial peptides and kills S. mutans through disruption of the cell membrane followed by a loss of membrane potential and cell death. Interestingly, this membrane activity is rapid and potent against S. mutans but not against other noncariogenic oral streptococci (17).
In this study, we further investigated the antimicrobial specificity of C16G2 by expanding the panel of testing bacterial species to include more oral streptococci species closely related to S. mutans, nonstreptococcal oral species, and nonoral representatives. To explore its potential clinical application, a saliva-derived in vitro model containing over 100 species approaching the diversity and overall metabolic functionality of the human oral microbiome (18) was used for examining the selective antimicrobial activity of C16G2 against S. mutans within a multispecies community. The main goal of this study is to investigate the impact of targeted removal of S. mutans on the overall structure of a multispecies oral microbial community and provide proof of concept data for using targeted antimicrobials to modulate the microbiome structure and study the ecological role of specific bacterial species.
Results
C16G2 Displayed High Antimicrobial Specificity and Efficacy Against S. mutans in Monospecies Cultures.
Our recent study has shown a more targeted killing of synthetic peptide C16G2 against S. mutans compared with other related oral Streptococcus species, including S. sanguinis and S. gordonii, when tested using monospecies planktonic cultures (16). In this study, we expanded the panel of test bacterial species to include more related oral Streptococcus species, nonstreptococci oral species (such as Fusobacterium nucleatum and Actinomyces naeslundii), and noncommensal oral species, including Escherichia coli and Pseudomonas aeruginosa. To measure the potency of C16G2 against the tested species, the IC50 was determined for each species. The data showed that C16G2 had an IC50 of 5.9 µM against S. mutans and Streptococcus salivarius K12 (Table 1), whereas the IC50 values for the other six oral streptococci tested ranged from 28.7 to 381.0 µM (about 5- to 60-fold higher). Among nonstreptococcal Gram-positive bacteria tested, oral species, including A. naeslundii and Lactobacillus casei, showed IC50 values above 343.2 µM, whereas Micrococcus luteus and Corynebacterium striatum (normal flora of the skin) displayed increased susceptibility to C16G2 compared with S. mutans (both IC50 values <5.9 µM). Meanwhile, all eight Gram-negative bacterial strains tested, including oral species F. nucleatum and Klebsiella pneumoniae as well as nonoral species (such as E. coli and P. aeruginosa), displayed IC50 values above 80 µM, with the exception of Salmonella typhimurium at 23.3 µM.
Table 1.
IC50 of C16G2 against bacteria after a 5-min treatment period
| Bacterium | IC50, µM |
| S. mutans UA140 | 5.9 |
| Streptococcus crista #6 | 257.2 |
| S. gordonii ATCC 10558 | 93.9 |
| S. mitis JM3 | 223.6 |
| Streptococcus pyogenes ATCC 10096 | 28.7 |
| S. salivarius K12 | 5.9 |
| S. sanguinis ATCC 10556 | 59.3 |
| S. oralis ATCC 35057 | 381.0 |
| A. naeslundii ATCC 12104 | 344.7 |
| F. nucleatum ATCC 23726 | 362.3 |
| L. casei ATCC 4646 | >784.5 |
| Bacteroides fragilis 145091 | 533.5 |
| Bacteroides fragilis 90917 | 707.8 |
| C. striatum ATCC 43751 | 3.4 |
| E. coli 49979 | >784.5 |
| K. pneumoniae Kay 2026 | 110.3 |
| S. typhimurium ssp. enterica ATCC 29629 | 23.3 |
| P. aeruginosa PAO1 | 86.8 |
| M. luteus MS2 | 5.1 |
| Yersinia enterolitica ATCC 23715 | >784.5 |
C16G2 Showed Targeted Killing of S. mutans Within a Saliva-Derived in Vitro Oral Microbial Community Spiked with S. mutans.
To further test the selective antimicrobial activity of C16G2 against S. mutans within a multispecies oral microbial community, we first generated an S. mutans-infected multispecies oral microbial community by adding JM11, a spectinomycin-resistant S. mutans strain, to the saliva-derived, SHI medium-cultivated planktonic culture. SHI medium was recently developed in our laboratory and has been shown to be able to support a diversified community with a microbial profile close to that of the original saliva (19). We then treated the planktonic community with either C16G2 or Carrier as a negative control, and the viability of S. mutans within the community after a 30-min treatment was monitored by viability count. PCR-denaturing gradient gel electrophoresis (DGGE) analysis was also performed to monitor the profiles of surviving species within the microbial community right after each treatment.
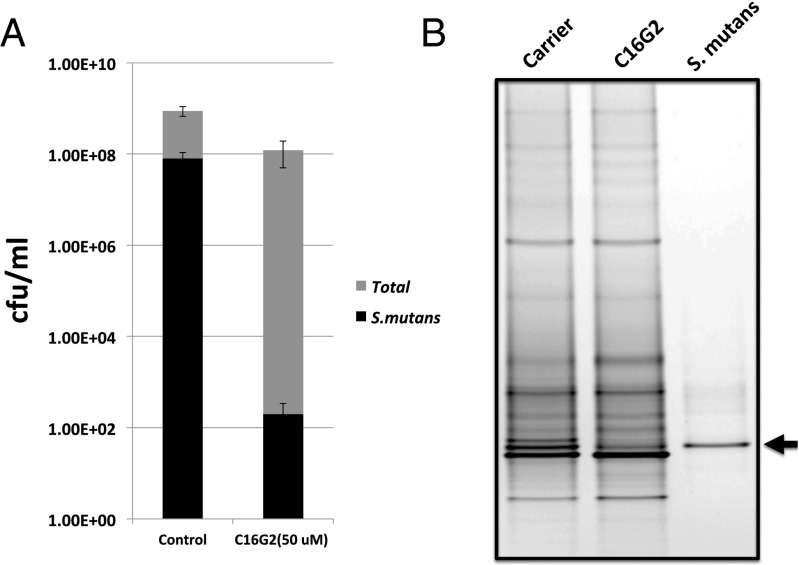
Our data revealed that a 30-min treatment with C16G2 resulted in an approximately 106-fold greater reduction in viable count of S. mutans compared with the Carrier control, whereas the total oral bacterial count was relatively unaffected and only decreased by <10-fold (Fig. 1A). Meanwhile, the specific antimicrobial effect of C16G2 against S. mutans within the multispecies community was further visualized by DGGE. As shown in Fig. 1B, the sample treated with C16G2 for 30 min decreased in intensity of the S. mutans band (black arrow), whereas most of the other bands remained relatively unchanged.
Fig. 1.
C16G2 antimicrobial activity against S. mutans within a multispecies community. The saliva-derived in vitro multispecies community was spiked with S. mutans and exposed to different treatments for 30 min. (A) Viability count. After being washed with PBS, a serial dilution of each sample was spotted onto selective (spectinomycin) and nonselective SHI agar plates, and plates were incubated at 37 °C anaerobically for 3 d followed by colony counting. The data were plotted as cfu per 1 mL and represent averages of the results of at least three independent experiments. (B) PCR-DGGE analysis of the microbial profile of multispecies communities right after treatment. Samples were treated with ethidium monoazide bromide to prevent amplification of DNA from dead bacterial cells and limit the DNA-based PCR-DGGE community analysis to the viable fraction. Two biological replicates were performed, and a representative gel image is shown.
Overall Shift in the Microbial Profiles Was Monitored in the Saliva-Derived Oral Community Recovered After C16G2 Treatment.
To further study the resulting consequence of targeted removal of S. mutans from a complex oral microbial community, we treated the S. mutans-containing in vitro planktonic oral microbial communities with C16G2 or Carrier (negative control) for 30 min followed by extensive washing to remove the residual C16G2 or Carrier. The treated communities were then allowed to recover by being cultured in fresh SHI medium and incubated at 37 °C anaerobically for 24 h. The microbial composition of each recovered community was determined by 454 pyrosequencing analyses to examine how the removal of S. mutans may impact other species within the same community.
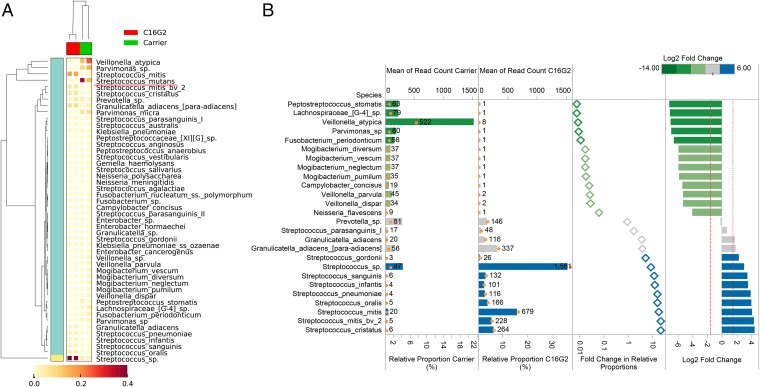
This first assessment of the C16G2 in a mixed microbial community approaching the diversity of the oral microbiome clearly showed that C16G2 treatment resulted in the intended target reduction. Independent experiments showed that the C16G2 treatment prevented S. mutans from regrowing and reduced the average percentage of S. mutans from 24% to 0.1% (Fig. 2). An overall genus-level shift in the composition of the microbial community recovered from C16G2 treatment compared with that of Carrier treatment was also observed (Fig. 2). Data showed that 21 bacterial genera could be detected from regrowth of Carrier-treated samples, with Streptococcus, Veillonella, Parvimonas, Prevotella, and Peptostreptococcus spp. being the most dominant genera. In contrast, only 16 bacterial genera were detected from the regrowth of C16G2-treated samples, with Streptococcus, Granulicatella, and Prevotella being the most dominant ones. Interestingly, although the relative abundance of S. mutans reduced drastically, the overall sequence counts of all Streptococcus spp. increased from 30% to 81% in the culture recovered after C16G2 treatment. Meanwhile, many bacterial genera, most of which are Gram-negative bacteria, including Fusobacteria, Campylobacter, Neisseria, and Parvimonas spp., which were present at less than 5%, could no longer be detected at the depth of sequencing obtained from the regrowth of C16G2-treated samples, whereas genera, such as Veillonella, suffered drastic reduction in relative abundance within the community (from 20% to less than 1%) (Fig. 2).
Fig. 2.
Oral microbial taxa detected from regrown cultures after treatment are compared by the overall changes in relative proportions. (A) Hierarchical cluster analyses of oral taxa-weighted abundance profiles obtained from regrowth after treatment with Carrier and C16G2. Relative proportions of the total taxa abundance are indicated in the heat map, which shows how the dominant taxa varied (data filtered for those taxa with >1% relative abundance). (B) In addition to removal of S. mutans from an average abundance of 24% to less than 0.1% after C16G2 treatment, several oral species displayed differential abundance compared with the negative control. Oral taxa after C16G2 treatment that showed a significant fold change relative to the control Carrier treatment are indicated. Relative proportions (horizontal bars) in this plot are corrected for read depth changes given that S. mutans decreased to below 0.1% in the C16G2 treatment. Proportions are, therefore, calculated as a relative percentage of the total population excluding S. mutans counts. Library size normalized read counts are displayed as text and open orange circles. Differences in the relative proportions are expressed as fold changes and log2 fold changes to display the changes between Carrier and C16G2 treatment. All plots are colored according to the log2 fold changes between Carrier and C16G2 treatment.
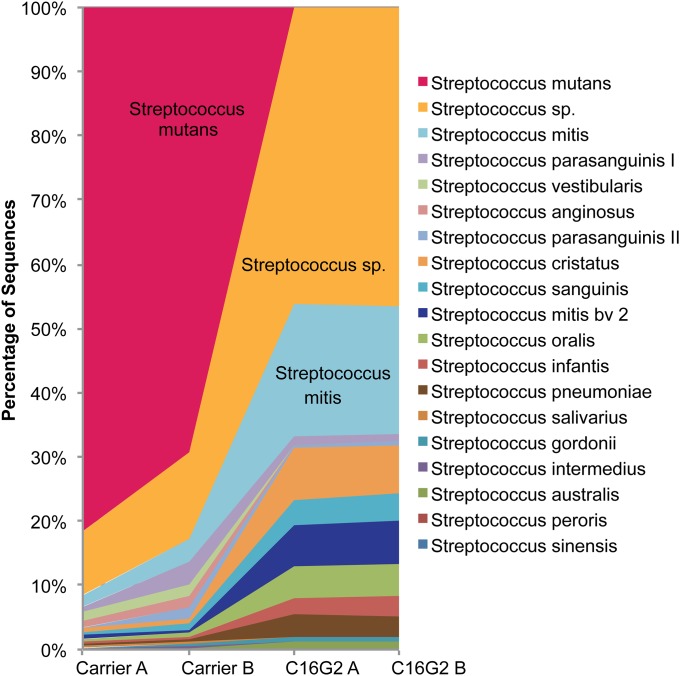
Based on antimicrobial specificity data from monoculture study, C16G2 exhibits far more targeted killing against S. mutans than most of the other tested oral streptococci. To determine the impact of removal of S. mutans on the overall streptococci species profile, we compared the dominant sequences falling within the Streptococcus genus for the communities recovered from C16G2 or Carrier treatment. Our data showed that, after overnight recovery from Carrier (negative control) treatment, S. mutans accounted, on average, for more than 78% of the total Streptococcus spp., whereas this number was reduced to less than 1% in the C16G2-treated cultures (Fig. 3). Furthermore, C16G2 treatment did not significantly affect the diversity of total Streptococcus spp., and the removal of S. mutans by C16G2 within the community was accompanied by a shift in the relative abundance of other Streptococcus spp. (Fig. 3) that were present. Particularly, compared with the Carrier-treated community, an increase in the relative abundance had been observed for unnamed Streptococcus spp. human oral taxa (4) (from 4% to 37%), and members belonging to the mitis group, including Streptococcus mitis (from 2% to 20%), Streptococcus cristatus (from 1% to 8%) and S. sanguinis (from <1% to 4%), within the community recovered after C16G2 treatment (Fig. 3).
Fig. 3.
Comparison of dominant sequences falling within the genus Streptococcus for the microbial community recovered from independent experiments after Carrier treatment (Carrier A and B) and C16G2 treatment (C16G2 A and B).
S. mutans Displayed Physical Interaction with Particular Oral Species with Elimination from the C16G2-Treated Culture That Is Concurrent with a Significantly Reduced S. mutans Population.
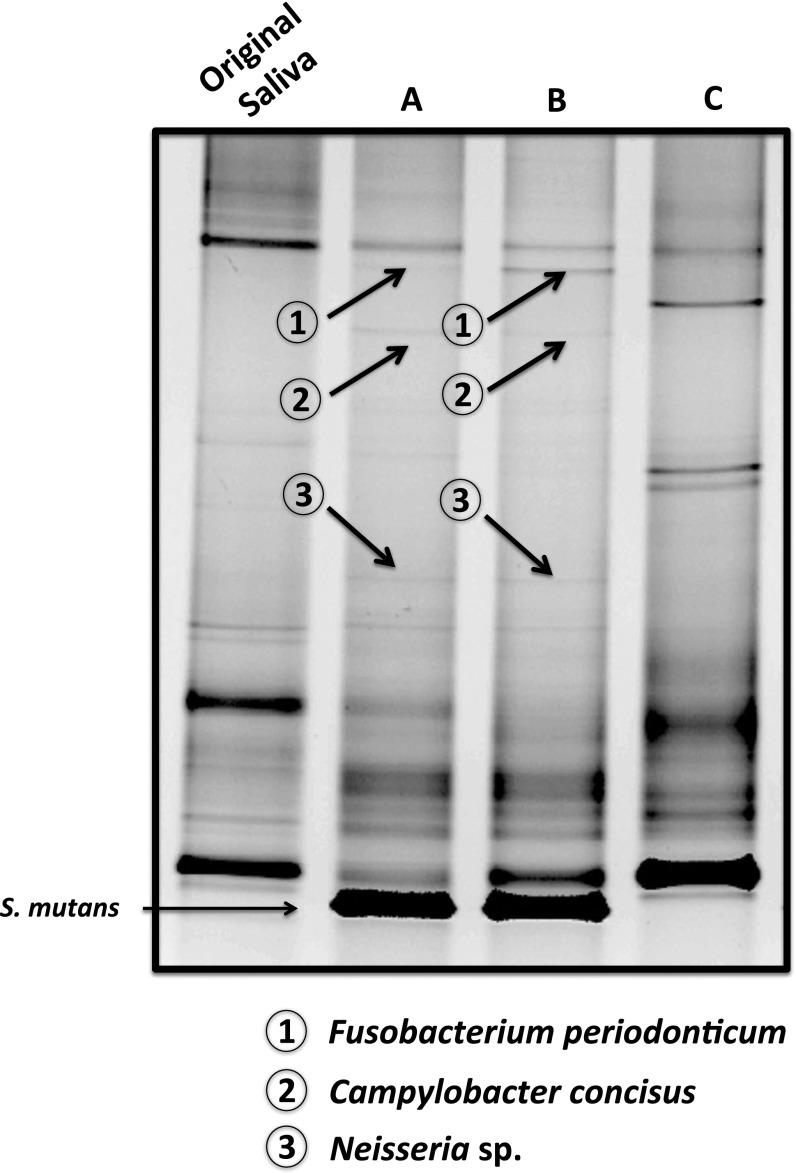
The sequencing data revealed that, within the community recovered from C16G2 treatment, the reduction in the S. mutans population was concurrent with reduced abundance or elimination of other oral bacterial species. This observation could result from either nonspecific killing by C16G2 or a complex interplay of several factors, such as direct or indirect metabolic- as well as direct physical contact-dependent optimal growth of certain bacterial species on the targeted species, particularly within a multispecies community. To address this question, we performed a pull-down assay to identify oral species that could display direct physical interaction with S. mutans. Our result showed that S. mutans biofilm was able to recruit and form tight physical association with specific oral bacterial species, including Fusobacterium periodonticum, Campylobacter concisus, and one of the Neisseria spp., which has a presence that cannot be detected within the community recovered from C16G2 treatment (Fig. 4). Meanwhile, the oral strain representing F. periodonticum showed strong coaggregation with S. mutans and displayed an IC50 value of C16G2 significantly higher than the treatment concentration.
Fig. 4.
The pull-down assay. Saliva was added to overnight S. mutans biofilm; after 3 h of incubation, unattached cells were washed, and the attached biofilm was collected either (A) immediately or (B) after 24 h of incubation in fresh Brain-Heart Infusion medium. (C) Twenty-four–hour saliva biofilm served as the control to exclude any salivary species that could potentially attach to the surface not covered by S. mutans. Samples were subjected to PCR-DGGE analysis. Bands shown in A or B but not C were cut out, and corresponding bacterial species were identified by sequencing as described in SI Materials and Methods.
Discussion
C16G2 is a synthetic peptide designed to achieve targeted killing of the cariogenic oral pathogen S. mutans (16). In this study, we tested C16G2 against a panel of 20 different bacterial species, including both oral and nonoral Gram-positive as well as Gram-negative species in monoculture. Our data revealed an overall low potency of C16G2 against Gram-negative species, and among all of the oral Gram-positive bacteria tested, C16G2 is most potent in killing S. mutans and S. salivarus. Interestingly, C16G2 also exhibited high killing efficacy against two of the Gram-positive species from human skin flora. Recent studies indicated that C16G2 kills S. mutans through membrane disruption, a mechanism of action similar to that of traditional antimicrobial peptides (17). Although the exact mechanism of selective membrane disruption by C16G2 remains unclear, it may involve early membrane binding or partition steps governed by the targeting moiety of C16G2 (17). It is possible that Gram-positive membrane components, such as lipids, exopolysaccharides, or teichoic acids, play a role in the mechanism of action of C16G2. These components may be very similar in S. mutans and other species, such as S. salivarus and M. luteus, which may explain the killing efficacy achieved with C16G2 in these species (Table 1). Our data confirmed data from an earlier study, which showed the specificity and efficacy of C16G2 against S. mutans compared with a group of diverse oral bacterial species.
To further test the selective antimicrobial activity of C16G2 against S. mutans in a multispecies community of biological relevance, we first generated an S. mutans-infected multispecies oral microbial community by adding JM11, a spectinomycin-resistant S. mutans strain, to a saliva-derived, SHI medium-cultivated planktonic culture. We then treated the statically grown community with either C16G2 or Carrier control as control, and therefore, the selectivity of C16G2 could be shown in a highly diverse oral flora background. Because current protocols used for preparing DNA for metagenomic analysis also include DNA from dead or nonviable bacteria, we performed a modified PCR-DGGE analysis instead for monitoring the surviving bacterial species right after C16G2 treatment. In this assay, samples were treated with ethidium monoazide bromide to prevent amplification of DNA from dead bacterial cells and limit DNA-based PCR-DGGE community analysis to the viable fraction (20). Our viability and PCR-DGGE data strongly suggested that C16G2 displayed superior selectivity in killing S. mutans within a multispecies community (Fig. 1). However, because of the semiquantification nature and limited resolution of the DGGE technique (21), we cannot rule out the potential nonspecific killing of C16G2 against certain oral bacterial species other than S. mutans within the in vitro salivary community. Nevertheless, our data strongly indicate that C16G2 is selectively killing S. mutans within a multispecies environment to the potential benefit of the other species present.
The main purpose of the study is to develop a proof of concept that we could use to potentially examine the community role and function of individual species within a complex microbial community by knocking out or knocking down one particular species with a targeted antimicrobial (in this case, C16G2) and then tracking the impact on the rest of the species within the same community. Intriguingly, a significant shift in the community diversity and relative abundance of various Streptococcus spp. was revealed by metagenomic analysis (Figs. 2 and 3). Bacteria within the oral community display extensive and complex interactions, such as competition between bacteria for nutrients, synergistic/mutualistic interactions, which may stimulate the growth or survival of one or more residents, and production of an antagonist by one resident, which inhibits the growth of another (10, 22–24). Because of these factors, the elimination of one or a few community members could potentially affect the growth of many bacterial species and result in an overall shift of the microbial composition within the community.
In this study, C16G2 treatment induced a shift in the relative abundance of various Streptococcus spp. The reduction in S. mutans population was accompanied by an increase in the abundance of several streptococci from the mitis group, including S. mitis, S. cristatus, Streptococcus oralis, and S. sanguinis (Fig. 4), which are among the most prevalent bacterial species detected within the oral cavity of healthy human subjects (4, 9, 25). The antagonism between S. mutans and streptococci of the mitis group, particularly S. sanguins and S. gordonii, at the ecological level has been well-documented (26). Epidemiological studies revealed that high levels of S. mutans are always concurrent with low levels of S. sanguinis (27), whereas high levels of S. sanguinis in the oral cavity correlate with delayed S. mutans colonization (28). Recent work by Kreth et al. (29) showed sophisticated interspecies interactions between these two species that might play an essential role in balancing competition and coexistence within the oral community. The targeted removal of S. mutans could shift the balance and provide a competitive growth advantage to the mitis group.
Furthermore, the treatment of C16G2 also induced a significant change in the overall microbial profiles within the oral community (Fig. 2). Our data showed that, after overnight regrowth, the C16G2-treated community showed decreased microbial diversity compared with the negative control. Many Gram-negative species, such as Veillonella, experienced drastic reduction in the abundance, whereas F. periodonticum, Campylobacter, Gemella, and Neisseria could not be detected by pyrosequencing from communities recovered from the C16G2 treatment, although they were only present at abundances of less than 5%. The results could be caused by nonspecific killing of the peptide; however, the data show that some of these species, including F. periodonticum, displayed high levels of resistance against C16G2 treatment, suggesting that the reduction or elimination of certain species could be directly or indirectly related to the removal of S. mutans. For example, it has been shown that lactic acid, one of the metabolic products of S. mutans, is required for the growth of Veillonella spp. (30). The reduction in S. mutans population as a result of C16G2 treatment may, therefore, have a negative effect on the growth of Veillonella spp., such as was seen in our metagenomic data (Fig. 2).
Meanwhile, the elimination of certain oral species, including Fusobacterium spp., Campylobacter, and Neisseria, could also result from a shifted community structure caused by the targeted removal of S. mutans. For example, the increased abundance of mitis group streptococci could potentially modify and generate a different microenvironment by producing hydrogen peroxide (31), which could exert inhibitory effects on these specific Gram-negative species. Furthermore, direct physical contact between different species has recently been implicated in coordinating interacting partner members, which achieves synergistic growth and better survival within a multispecies community (32, 33). In an effort to further investigate this intriguing finding, we performed pull-down assays. Our data clearly revealed several oral species, including F. periodonticum, C. concisus, and Neisseria spp., that could form direct physical associations with S. mutans (Fig. 4). Interestingly, all of these species recovered to less than 5% relative abundance in the Carrier treatment but could not be detected from the C16G2-treated multispecies culture at the depth of sequencing in this study, although F. periodonticum showed high levels of resistance to C16G2 when tested in monoculture. Our data suggested the possibility that the diminished population of S. mutans within the C16G2-treated community could negatively affect growth of species, such as F. periodonticum, with persistence and optimal growth within the multispecies that might require directly physical interaction with S. mutans. Our follow-up study has confirmed the strong physical binding between S. mutans and F. periodonticum. We are in the process of identifying the molecular components involved in interspecies coadherence and investigating the suggested physically dependent optimal growth of these species on S. mutans, particularly within a multispecies community.
Treatment of infectious diseases using conventional antimicrobials often results in indiscriminate killing of microbes including commensal indigenous microbiota. The past decade has witnessed increasing effort to develop more targeted approaches to combat infections (34, 35). Our study suggested a strong potential for C16G2 to be developed into therapeutics that could selectively eliminate cariogenic pathogens while achieving a more benign oral microbial community. The potential clinical application of C16G2 is supported by the fact that the community that recovered from C16G2 treatment contained increased populations of S. mitis and S. sanguinis, signature bacterial species identified from the oral microbial community of healthy subjects (9), as well as a decrease in many Gram-negative species, including fusobacteria, implicated in the pathogenesis of periodontal disease (36).
The most intriguing finding of our study was that the targeted removal of specific bacterium has a community-level impact on the species composition and abundance within the same community. From the evolutionary and ecological point of view, the development of environmental or host-associated microbial communities often is the result of complex intraspecies, interspecies, and microbe–host interactions. Increasing lines of evidence indicate that established microbial communities often exist and act as an entity instead of the random mixture of different bacterial species (37, 38). These unique entities behave in an analogous manner to a cell, where each member can be regarded as a functional gene unit and the interaction among different residential species eventually determines the microflora’s community-level behavior, such as community invasion resistance or polymicrobial pathogenesis. Meanwhile, any change in the abundance of particular species within the community could have drastic effects on its interacting members, eventually resulting in a change of community profile as well as community-level functions, similar to the effect of single-gene knockout/knockdown within a cell on the overall gene expression and behavior of the cell. In this regard, the ability of targeted antimicrobial peptide technology to target specific bacterial species within the multispecies community could be used for investigating the potential roles of specific bacterial species in maintaining the stability of the community as well as contributing to community-associated physiology and pathogenicity.
Materials and Methods
Detailed experimental procedures are provided in SI Materials and Methods. Monocultures of 20 bacterial species, including multiple oral streptococci, nonstrep oral species, and nonoral human-associated species as well as environmental species, were used for testing the specificity and efficacy of C16G2 in targeted killing of S. mutans. The S. mutans-infected multispecies oral microbial community was generated by adding S. mutans to the SHI medium-cultivated, human saliva-derived microbial community. The resultant communities were treated with C16G2 or Carrier as a negative control. Viability assay and DGGE analysis were performed to monitor the specific killing of S. mutans within the multispecies community right after treatment. Meanwhile, cultures with different treatment were allowed to recover in fresh medium, and the microbial profiles were obtained by pyrosequencing. A pull-down assay was performed to identify oral species that exhibit direct physical interaction with S. mutans. DNA extraction and processing of pyrosequencing data as well as identification and phylogenetic analyses of 16S genes were performed as previously described (18), and the detailed procedures are provided in SI Materials and Methods; 16S rRNA gene sequence data are available at depts.washington.edu/jsmlab/downloads/.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants 1-R01-DE020102 and 1-R01-DE023810-01 and by C3 Jian, Inc.
Footnotes
Conflict of interest statement: The authors declare that R.E., C.W.K., P.K., B.V., and W.S. are employees of C3 Jian, Inc., which has licensed technologies from University of California Regents that could be indirectly related to this research project.
This article is a PNAS Direct Submission.
Data deposition: The 16S rRNA gene sequence data are available at depts.washington.edu/jsmlab/downloads/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506207112/-/DCSupplemental.
References
- 1.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice EA, et al. NISC Comparative Sequencing Program A diversity profile of the human skin microbiota. Genome Res. 2008;18(7):1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8(4):435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontol 2000. 2004;36:14–26. doi: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- 6.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 7.Hugot JP. Inflammatory bowel disease: A complex group of genetic disorders. Best Pract Res Clin Gastroenterol. 2004;18(3):451–462. doi: 10.1016/j.bpg.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71(4):653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy S, et al. Bacteria in oral health-probiotics and prebiotics a review. Int J Biol Med Res. 2011;2(4):1226–1233. [Google Scholar]
- 12.He X, et al. Community-based interference against integration of Pseudomonas aeruginosa into human salivary microbial biofilm. Mol Oral Microbiol. 2011;26(6):337–352. doi: 10.1111/j.2041-1014.2011.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X, McLean JS, Guo L, Lux R, Shi W. The social structure of microbial community involved in colonization resistance. ISME J. 2014;8(3):564–574. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44(2):331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dye BA, et al. Trends in oral health status: United States, 1988-1994 and 1999-2004. Vital Health Stat 11. 2007;248(2007):1–92. [PubMed] [Google Scholar]
- 16.Eckert R, et al. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother. 2006;50(11):3651–3657. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan CW, et al. Selective membrane disruption: Mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob Agents Chemother. 2011;55(7):3446–3452. doi: 10.1128/AAC.00342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edlund A, et al. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 2013;1(1):25. doi: 10.1186/2049-2618-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian Y, et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol. 2010;25(5):357–367. doi: 10.1111/j.2041-1014.2010.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol. 2006;72(3):1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolenbrander PE. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander PE, Andersen RN, Kazmerzak K, Wu R, Palmer RJ., Jr Spatial organization of oral bacteria in biofilms. Methods Enzymol. 1999;310:322–332. doi: 10.1016/s0076-6879(99)10026-0. [DOI] [PubMed] [Google Scholar]
- 24.Kolenbrander PE, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 25.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111(28):E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190(13):4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loesche WJ, Rowan J, Straffon LH, Loos PJ. Association of Streptococcus mutants with human dental decay. Infect Immun. 1975;11(6):1252–1260. doi: 10.1128/iai.11.6.1252-1260.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caufield PW, et al. Natural history of Streptococcus sanguinis in the oral cavity of infants: Evidence for a discrete window of infectivity. Infect Immun. 2000;68(7):4018–4023. doi: 10.1128/iai.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187(21):7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190(24):8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125(2):237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 34.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310(5748):670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 35.Aoki W, Kuroda K, Ueda M. Next generation of antimicrobial peptides as molecular targeted medicines. J Biosci Bioeng. 2012;114(4):365–370. doi: 10.1016/j.jbiosc.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13(2):25–36. [PubMed] [Google Scholar]
- 37.Delhaes L, et al. The airway microbiota in cystic fibrosis: A complex fungal and bacterial community—implications for therapeutic management. PLoS ONE. 2012;7(4):e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wade W. 2009. The normal oral microbiota. Periodontal Medicine and Systems Biology, eds Henderson B, Curtis M, Seymour R, Donos N (Wiley-Blackwell, Oxford), pp 139–148.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.