Abstract
Mulibrey nanism is a rare growth disorder of prenatal onset caused by mutations in the TRIM37 gene, which encodes a RING-B-box-coiled-coil protein. The pathogenetic mechanisms of mulibrey nanism are unknown. We have used transiently transfected cells and antibodies raised against the predicted TRIM37 protein to characterize the TRIM37 gene product and to determine its intracellular localization. We show that the human TRIM37 cDNA encodes a peroxisomal protein with an apparent molecular weight of 130 kD. Peroxisomal localization is compromised in mutant protein representing the major Finnish TRIM37 mutation but is retained in the protein representing the minor Finnish mutation. Colocalization of endogenous TRIM37 with peroxisomal markers was observed by double immunofluorescence staining in HepG2 and human intestinal smooth muscle cell lines. In human tissue sections, TRIM37 shows a granular cytoplasmic pattern. Endogenous TRIM37 is not imported into peroxisomes in peroxin 1 (PEX1−/−) and peroxin 5 (PEX5−/−) mutant fibroblasts but is imported normally in peroxin 7 (PEX7−/−) deficient fibroblasts, giving further evidence for a peroxisomal localization of TRIM37. Fibroblasts derived from patients with mulibrey nanism lack C-terminal TRIM37 immunoreactivity but stain normally for both peroxisomal matrix and membrane markers, suggesting apparently normal peroxisome biogenesis in patient fibroblasts. Taken together, this molecular evidence unequivocally indicates that TRIM37 is located in the peroxisomes, and Mulibrey nanism thus can be classified as a new peroxisomal disorder.
Introduction
Mulibrey (muscle-liver-brain-eye) nanism (MUL [MIM 253250]) is an autosomal recessive disorder with severe growth failure of prenatal onset, characteristic dysmorphic features, pericardial constriction, and hepatomegaly. Other common features include J-shaped sella turcica, yellowish dots in the ocular fundi, slight muscular weakness, cutaneous naevi flammei, and enlargement of cerebral ventricles (Perheentupa et al. 1973; Lipsanen-Nyman 1986; Lapunzina et al. 1995). The psychomotor development of patients with mulibrey nanism is within normal limits. Wilms tumor is reported in ∼4% of the patients (Similä et al. 1980; Lipsanen-Nyman 1986; Seemanova and Bartsch 1999). Mulibrey nanism occurs worldwide but is more common in the Finnish population. Of the ∼100 patients that have been diagnosed worldwide, 80 are from Finland.
Mutations in the TRIM37 gene (previously designated MUL) on chromosome 17q22-q23 underlie mulibrey nanism (Avela et al. 2000). The TRIM37 cDNA contains an open reading frame of 2,892 bp and encodes a 964–amino acid protein with a predicted molecular weight of 108 kD. The TRIM37 protein is a new member of the RING-B-box-coiled-coil (RBCC) subfamily of zinc-finger proteins. Four mutations, resulting in a frameshift and predicting a truncated protein, have been described in patients with mulibrey nanism (Avela et al. 2000). As yet, there seems to be no genotype-phenotype correlation (Avela et al. 2000; authors' unpublished data). By RNA in situ hybridization TRIM37 has been found to be expressed in dorsal root and trigeminal ganglia, liver, and epithelia of multiple tissues during early human and mouse embryogenesis (Lehesjoki et al. 2001).
The RING-finger domain identified in the TRIM37 protein is a cysteine-rich, zinc-binding domain found in >200 proteins of diverse eukaryotes (Saurin et al. 1996). RING proteins are localized to the nucleus and cytoplasm, and they mediate diverse cellular processes, such as oncogenesis, apoptosis, viral replication, organelle transport, cell-cycle control, and peroxisomal biogenesis. Recent evidence indicates that RING-mediated protein interactions are critical for transcriptional repression and ubiquitination (Borden and Freemont 1996; Borden 2000; Freemont 2000; Jackson et al. 2000; Joazeiro and Weissman 2000). RING domains are able to mediate protein-protein interactions, particularly the formation of large macromolecular complexes. Thus, their function may be to act both as molecular building blocks and as molecular modifiers that mediate spatial and temporal positioning and specificity in diverse cellular processes (Borden and Freemont 1996; Borden 2000).
In RBCC proteins, the RING domain is associated with another cysteine-rich, zinc-binding domain, the B-box, followed by a coiled-coil domain. The functions of the RBCC subfamily of RING proteins are largely unknown, but they too have been implicated in mediation of protein-protein interactions in diverse cellular processes and compartments (Saurin et al. 1996). For example, midin, the protein defective in X-linked Opitz syndrome, and MURF, which acts in skeletal myoblast differentiation in the mouse, are associated with microtubules (Cainarca et al. 1999; Spencer et al. 2000). The brain-expressed RING-finger protein BERP is another cytoskeleton-associated RBCC protein, which has been shown to bind class V myosins (El-Husseini and Vincent 1999). In contrast, the Caenorhabditis elegans tam-1 and lin-41 are nuclear proteins involved in transcriptional regulation (Hsieh et al. 1999; Slack et al. 2000). PML, RFP, and TIF1 have oncogenic potential when involved in translocations and fusion proteins in man and mouse (Takahashi et al. 1988; de The et al. 1991; Le Douarin et al. 1995). There is growing evidence that, in the RBCC subfamily, the RBCC domain acts as an integral structural unit (Borden 2000; Peng et al. 2000; Reymond et al. 2001). A recent work on a large number of RBCC proteins, also named TRIMs (for tripartite motif), provided evidence that the coiled-coil region is essential for both homo-oligomerization and proper subcellular localization of these proteins, the great majority of which were shown to localize to discrete cytoplasmic or nuclear structures (Reymond et al. 2001).
Recently, TRIM37 was assigned to the family of TNF-receptor-associated factor (TRAF) proteins, because it contains a TRAF domain (Zapata et al. 2001). This domain is responsible for the interaction of TRAF proteins with other proteins, thus acting as scaffold molecules for receptors, kinases, and a variety of regulators in signaling pathways (Aravind et al. 1999; Wajant et al. 2001). The TRAF domain of TRIM37 binds six known TRAF proteins and the cytosolic domains of several TNF-family receptors in vitro, but whether this is physiologically relevant remains unclear (Zapata et al. 2001).
Here, we show that both exogenously expressed and endogenous TRIM37 protein localizes to peroxisomes. This localization is compromised in transiently expressed TRIM37, representing the major Finnish mutation (Finmajor). We conclude that TRIM37 is a peroxisomal protein of as yet unknown function, which allows the classification of MUL as a new peroxisomal disorder.
Material and Methods
Construction of Expression Plasmids and Site-Directed Mutagenesis
The KIAA0898 clone (GenBank accession number NM_015294; Nagase et al. 1998), containing the full-length 4,111-bp KIAA0898 cDNA, was obtained from the Kazuka Research Institute in Japan. The cDNA insert was excised from the vector and was subcloned at KpnI and NotI sites of pcDNA3.1 (Invitrogen). For N-terminal tagging, an EcoRI site was inserted, by PCR, 6 bp upstream of the ATG codon. The 600-bp PCR fragment was ligated to pCRII (Invitrogen) and was sequenced. The 5′ fragment was subsequently excised with NsiI and XhoI and was ligated to the original KIAA0898 clone in pBluescript (pBS) cut with the same enzymes. The fragment was excised with EcoRI and NotI and was ligated to the pAHC expression vector (kindly provided by T. Mäkelä, University of Helsinki), a derivative of pCIneo (Promega) in frame with the N-terminal hemagglutinin (HA) tag sequence.
Two different mutations were introduced into the TRIM37 coding sequence by use of the QuickChange Site-Directed mutagenesis Kit (Stratagene). Finmajor was reproduced by deletion of base pairs 493–497 of the coding region, and the minor Finnish mutation (Finminor) was reproduced by deletion of a single G at 2212. The template for Finmajor mutagenesis was the 600-bp TRIM37 cDNA 5′ fragment in the pCRII vector from which the mutant fragment was subcloned in frame with the HA tag of pAHC. Finminor was introduced into the pcDNA3.1-TRIM37 expression construct. Final mutant constructs were verified by sequencing the entire coding region of the cDNAs.
In Vitro Translation
In vitro translation was performed with the TnT Quick Coupled Transcription/Translation System (Promega), with the pBS-KIAA0898 plasmid used as the template. 35S-methionine–labeled translation products were analyzed on 7% SDS-PAGE (Laemmli 1970), were transferred onto nitrocellulose membrane, and were visualized by autoradiography.
Cell Culture and Transfections
Cell culture media and supplements were from Sigma, except for fetal bovine serum (Life Technologies) and Ultroser-G (Invitrogen). COS-1 (American Type Culture Collection [ATCC] number CRL-1650), BHK (ATCC number CCL-10), HeLa (ATCC number CCL-2), and HepG2 (ATCC number HB-8065) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), L-glutamine, and antibiotics. Human intestinal smooth muscle cells (HISM cells, from ATCC) were grown in DMEM supplemented with 15% FBS and 2% Ultroser-G. For transfection, the cells were seeded on glass coverslips on 5-cm or 10-cm diameter plates. The next day, the cells were transfected with 2–8 μg of the expression constructs per plate, by use of Fugene reagent (Roche) according to the manufacturer’s protocol. Cells were processed for analysis 16–48 h after transfection. The human skin-fibroblast cell lines (PBD074/PEX7−/−, PBD005/PEX5−/−, and PBD009/PEX1−/−), from patients with peroxisomal disorders, were a kind gift from G. Dodt (Ruhr-Universität Bochum, Germany). They were originally provided by A. B. Moser and H. W. Moser (Kennedy Krieger Institute). All cell lines are referred to by a peroxisome biogenesis disorder (PBD) number given at the Kennedy Krieger Institute and were transformed with the large T-Antigen of SV40 virus, as described elsewhere (Dodt et al. 1995). They were grown in DMEM with 10% FBS and were plated on gelatinized coverslips for immunofluorescence analysis. Fibroblast cultures were established from skin biopsies of three Finnish patients with MUL after informed consent and the approval of the Ethical Review Board of the Helsinki University Hospital were given. The fibroblasts were grown in RPMI-1640 supplemented with 15% FBS and were plated on gelatinized coverslips at passage 2–3 for immunofluorescence analysis.
TRIM37 Antibodies and Antibody Purification
Polyclonal antibodies against human TRIM37 were raised by immunization of rabbits with the synthetic peptides FPDGEQIGPEDLSFNTDENSGR (C-terminal amino acids 942–964) and SVREAKEDEEDEEKIQNEDYHHE (internal amino acids 490–513) coupled to keyhole limpet hemocyanin. The immunizations and antisera preparation were carried out at Sigma Genosys. An aliquot of each antiserum was affinity purified by use of the peptides coupled to CNBr-Sepharose (Amersham Pharmacia Biotech).
Western Blotting and Subcellular Fractionation
Total cell lysates were prepared in SDS lysis buffer (10 mM Tris pH 8, 100 mM NaCl, 2% Triton X-100, 1% SDS, 1 mM EDTA) supplemented with Complete Proteinase Inhibitor Cocktail (Roche). Cells were scraped from the plates and were suspended in lysis buffer. The lysates were passed through a 23G needle to shear DNA and were centrifuged at 13,000g at +4°C for 10 min, and supernatants were recovered. Protein concentration of the lysates was measured by use of the BCA Protein Assay Reagent (Pierce). Per sample, 10–50 μg of total protein were analyzed on SDS-PAGE and were transferred onto Hybond ECL nitrocellulose filter (Amersham Pharmacia Biotech) by use of Bio-Rad blotting apparatus. Filters were blocked in 5% nonfat dry milk in TBS, were rinsed and were incubated with primary antibodies followed by peroxidase-conjugated secondary antibodies (Dako). Detection was performed by use of enhanced chemiluminiscence reagents (Amersham Pharmacia Biotech).
For subcellular fractionation, seven nontransfected 10-cm plates and three 10-cm plates transfected with the HA-tagged TRIM37 construct were washed with PBS and homogenization buffer (25 mM HEPES pH 7.4, 0.25 M sucrose, 1 mM EDTA, 0.1% ethanol, and proteinase inhibitor cocktail), were scraped off the plates, were resuspended in 4.0 ml of ice-cold homogenization buffer, and were homogenized with 20 strokes in a Dounce homogenizer. After serial centrifugations at 1,950g, 23,500g, and 100,000g, the 23,500g pellet (light mitochondrial fraction) was resuspended and subjected to Nycodenz (Axis-Shield) gradient centrifugation according to a published protocol (Graham and Rickwood 1997). The gradient was centrifuged at 35,000g in Sorvall SS-34 rotor for 1 h. Fractions of 0.5 ml were collected from the bottom of the tube by use of a peristaltic pump. The fractions were diluted with an equal volume of homogenization buffer, and an aliquot of each was used for enzyme-activity measurements. The rest of the fractions were pelleted at 16,000g for 20 min. The organelle pellets were resuspended in SDS-lysis buffer and were analyzed by western blotting as described above. The activities of the marker enzymes acid phosphatase (lysosomes) and succinate dehydrogenase (mitochondria) were measured by use of standard spectrophotometric methods according to the recommendation of the manufacturer of the gradient media (Axis-Shield).
Fluorescence Microscopy and Immunohistochemistry
Cells grown on coverslips were rinsed with PBS and were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. They were permeabilized with either 0.2% Triton X-100 or 0.05% saponin in PBS containing 0.5% BSA for 5 min. Selective permeabilization of the plasma membrane was done with 25 μg/ml digitonin (Sigma) for 5 min. The primary polyclonal rabbit antibodies used were anti-TRIM37 (see above), anti-PMP70 (Zymed), anti-SKL (Zymed), and anti-catalase (Calbiochem). The primary mouse monoclonal antibodies were anti-hemagglutinin (12CA5 hybridoma supernatant, kindly provided by M. Bergman, University of Helsinki), anti-human ALDP (MAB2162, Chemicon), anti-human Lamp1 (H4A3, kindly provided by J. T. August and J. E. K. Hildreth, Johns Hopkins University School of Medicine), anti-PDI (StressGen Biotechnologies), anti-CTR433 (kindly provided by M. Bornens, Institut CURIE), anti-EEA1 (Transduction Laboratories), anti-α-tubulin (DM1A, Sigma), and anti-human mitochondrial antigen (MU213-UC, Biogenex). MitoTracker Green (Molecular Probes) was used for the staining of mitochondria in HISM cells. Secondary antibodies included rhodamine, fluorescein, Cy2 or Cy3-conjugated anti-mouse, and anti-rabbit-IgG (Jackson Immunochemicals). Antibody incubations were for 30 min at room temperature. The coverslips were mounted with Mowiol (Calbiochem). For immunohistochemical stainings, antigen affinity-purified fractions of rabbit anti-TRIM37 antisera were used at concentrations of 10 μg/ml for the C-terminal antibody and 5 μg/ml for the internal peptide antibody. Catalase antibody was used as a marker for peroxisomes at a concentration of 2.6 μg/ml. Paraffin sections from normal human ovary, liver, skin, and small intestine were stained using the Vectastain Elite kit (Vector Laboratories) according to the manufacturer’s protocol. For the internal peptide antibody, antigen retrieval was performed by digesting the sections with 0.05% trypsin in PBS for 15 min at +37°C. Diaminobentsidine was used as chromogen, and the sections were counterstained with hematoxylin and mounted under coverslips with Mountex (Medite).
Results
Immunoblot and In Vitro Translation Analysis of the TRIM37 Gene Product
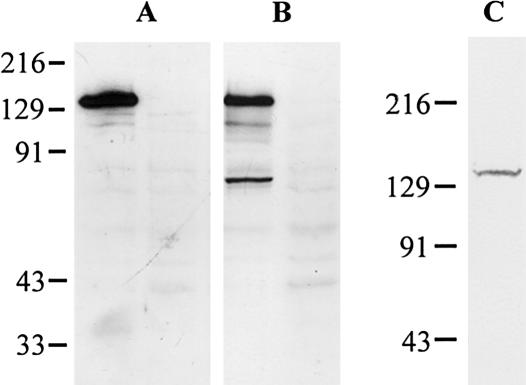
To characterize the TRIM37 gene product, COS-1 cells were transfected with an expression construct containing the full-length TRIM37 cDNA and the expressed polypeptide was analyzed by western blotting. Antibodies raised against a C-terminal peptide (amino acids 942–964) and an internal peptide (amino acids 490–513) of the predicted TRIM37 protein detected a polypeptide of ∼130 kD that was absent in nontransfected COS-1 cells (fig. 1A, 1B). In addition, an antibody against the internal peptide showed weaker bands at ∼110 and ∼70 kD that were not present in nontransfected cells (fig. 1B). The apparent size of the largest TRIM37 polypeptide seen was larger than predicted from the sequence (108 kD). In vitro translation was performed to investigate whether this difference is due to some posttranslational modification occurring in intact cells. The in vitro translation product of TRIM37 was found to migrate at ∼130 kD in the SDS-PAGE analysis (fig. 1C), suggesting that no modification that would markedly affect the molecular weight occurs in the TRIM37 protein.
Figure 1.
Western blot and in vitro translation analysis of the TRIM37 polypeptide. Polyclonal antibodies raised against (A) the C-terminal TRIM37 peptide and (B) the internal TRIM37 peptide detected a strong band at ∼130 kD in transiently transfected COS-1 cells. The internal TRIM37 peptide antibody also detected weaker bands at ∼110 and ∼70 kD. Nontransfected control lanes are shown on the right in panels A and B. C, In vitro transcription-translation of the TRIM37 cDNA in the presence of [35S]methionine produced a polypeptide migrating at ∼130 kD. The molecular weights of the Kaleidoscope marker (Bio-Rad) are shown. For western blot, 15 μg of total protein per lane was analyzed on 10% SDS-PAGE. Preincubation of the antisera with the corresponding antigenic peptides coupled to CNBr-sepharose completely abolished the TRIM37 bands (data not shown). The in vitro translation product was analyzed on 8% SDS-PAGE.
Subcellular Localization of Exogenously Expressed Wild-Type and Mutant TRIM37 Proteins
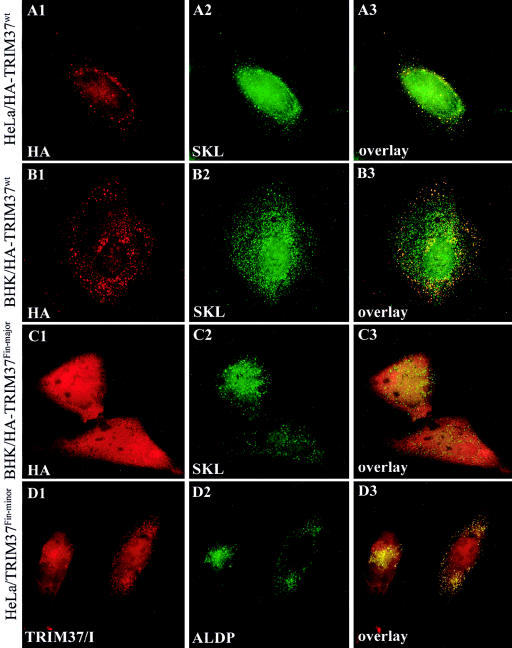
The subcellular localization of the protein encoded by the TRIM37 cDNA was studied by indirect immunofluorescence analysis of transiently transfected cells. BHK and HeLa cells transfected with either nontagged (data not shown) or hemagglutinin (HA)–tagged (fig. 2) TRIM37 expression constructs were first labeled with the two polyclonal anti-TRIM37 peptide antibodies or the monoclonal anti-hemagglutinin marker. Fluorescence microscopy revealed a punctate cytoplasmic staining pattern with numerous small vesicular or granule-like structures (fig. 2A1, 2B1) in most transfected cells. The pattern was identical with both the nontagged and HA-tagged constructs, showing that the N-terminal tag did not interfere with the localization of the protein. This is in line with previously published results (Zapata et al. 2001), where a N-terminally Myc-tagged TRIM37 construct was used. The corresponding preimmune sera produced minimal background staining (data not shown). After the shortest expression time (16 h) a small number of cells showed a few very large vesicular structures or aggregates, instead of the small granules. The number of cells containing these large vesicular structures increased, comprising up to 50% of transfected cells with extended expression time (data not shown).
Figure 2.
Distribution of exogenously expressed wild-type and mutant TRIM37 proteins in cultured cells, as visualized by indirect immunofluorescence analysis. TRIM37 and HA immunoreactivities (A1, B1, C1, and D1) are shown in red, and the peroxisomal markers SKL (A2, B2, and C2) and ALDP (D2) are shown in green. Yellow color in the overlay images (A3, B3, C3, and D3) indicates overlap of the immunoreactivities. HA-tagged wild-type TRIM37 shows peroxisomal localization in transiently transfected HeLa (A1–A3) and BHK (B1-B3) cells. The full-length wild-type protein is localized in round vesicles throughout the cytoplasm. Consistently in several cell lines analyzed, not all peroxisomes of a transfected cell contained TRIM37 protein. However, all TRIM37-containing vesicles stained positively for the peroxisomal markers. Peroxisomal localization is compromised in the mutant protein representing the Finmajor mutation (C1-C3). The truncated protein is distributed homogenously throughout the cytoplasm and the nucleus (C1) and does not colocalize with the peroxisomal marker (C2 and C3). On the contrary, the mutant protein representing the Finminor mutation retains peroxisomal localization (D1–D3). The Finminor mutant protein was stained with the internal TRIM37 peptide antiserum (“TRIM37/I”), the epitope of which is retained in the truncated protein. Original magnification was 1000×.
To identify the subcellular structures that contained TRIM37 peptide immunoreactivity, double immunofluorescence stainings were performed on the transfected COS-1, BHK, and HeLa cells by use of the TRIM37 peptide and cell organelle–specific antibodies. No colocalization of TRIM37 staining was seen with late endosomes/lysosomes (anti-Lamp1), early endosomes (anti-EEA1), Golgi (anti-CTR433), endoplasmic reticulum (anti-PDI), mitochondria (anti-mitochondrial antigen), or microtubules (anti-α-tubulin) in any of the cell lines (data not shown). The exclusion of TRIM37 from these compartments is in agreement with previously published results (Zapata et al. 2001) where, by double immunofluorescence, the localization of transiently transfected TRIM37 was excluded from mitochondria, Golgi, and lysosomes. In contrast, TRIM37 colocalized with the peroxisomal markers SKL (a Ser-Lys-Leu tripeptide, also called “peroxisomal targeting signal 1” [PTS1]) (fig. 2A1–A3, B1–B3) and PMP70 (peroxisomal membrane protein 70; data not shown). Colocalization of peroxisomal markers and TRIM37 was reproducible but dependent on the expression level. Colocalization was most evident after the shortest expression times (16 h), whereas longer times led to disruption of the small vesicular morphology and the development of very large vesicles with consequent loss of colocalization.
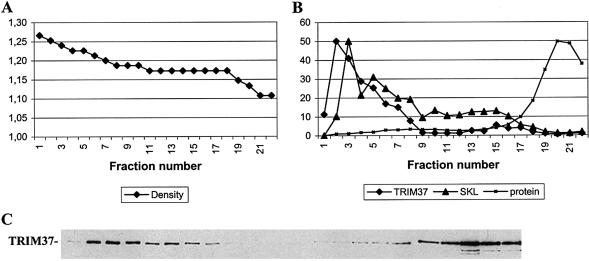
To further assess the subcellular localization of TRIM37 in transfected cells, COS-1 cell lysates were subjected to centrifugal fractionation followed by density gradient centrifugation of the light mitochondrial fraction. TRIM37 was found to be highly concentrated in the heavy (1.22–1.25 g/ml) peroxisomal fractions of the gradient, as determined by western analysis of the fractions with TRIM37 and SKL antibodies (fig. 3).
Figure 3.
Nycodenz density gradient fractionation of the crude peroxisomal fraction of COS-1 cells transfected with HA-tagged TRIM37 construct. Western analysis of TRIM37 protein in the fractions is shown in panel C. An equal volume of each fraction containing 1–50 μg total protein was analyzed on 8% SDS-PAGE. Diagram A shows the density (Y-axis, g/ml) of 22 fractions, as measured by weighing after centrifugal pelleting of the organelles. Diagram B shows the amount of total protein in each fraction (Y-axis, mg/ml) and the relative intensities of TRIM37 and SKL immunoreactive bands per micrograms of total protein in the fraction (Y-axis, arbitrary units). The data for the TRIM37 and SKL immunoreactivity curves were obtained by scanning the western blot films for TRIM37 (C) and SKL (data not shown). The peaks of both TRIM37 and SKL immunoreactivities are within the heavy peroxisomal fractions at densities 1.22–1.25 g/ml (B, fractions 2–6). The presence of TRIM37 in the uppermost light fractions (18–22) likely results from fragmentation of peroxisomes during the fractionation procedure (Graham and Rickwood 1997). A biphasic distribution, such as is seen here, is observed with several peroxisomal enzyme markers, with the activity in the lightest fractions possibly corresponding to a peroxisome subpopulation containing high levels of β-oxidation activity (Baumgart et al. 1996). The majority of acid phosphatase activity, a marker enzyme for lysosomes, was present in the density range 1.11–1.16 g/ml (data not shown), and the activity of the mitochondrial marker succinate dehydrogenase peaked at density ∼1.15 g/ml (data not shown).
The subcellular localization of two mutant TRIM37 proteins was studied by transfection of BHK or HeLa cells with expression constructs carrying the Finmajor (c.493delGAAAG) or the Finminor (c.2212delG) mutations identified in the cDNA of patients with MUL. The predicted protein truncation of these mutated forms of TRIM37 was confirmed by in vitro translation from the corresponding mutant constructs (data not shown). The Finmajor mutant protein, which only retains the 164 N-terminal amino acids of the wild-type 964–amino acid TRIM37 protein showed a homogenous cellular staining without a granular pattern (fig. 2C1). No colocalization with peroxisomal markers was identified (fig. 2C1–C3). In contrast, the truncated protein representing the Finminor mutation identified in compound heterozygotes only and retaining 737 amino acids showed a punctate cytoplasmic localization pattern. The pattern was similar to that of the wild-type protein and colocalized with peroxisomal markers (fig. 2D1–D3).
Subcellular Localization of Endogenous TRIM37 Immunoreactivity
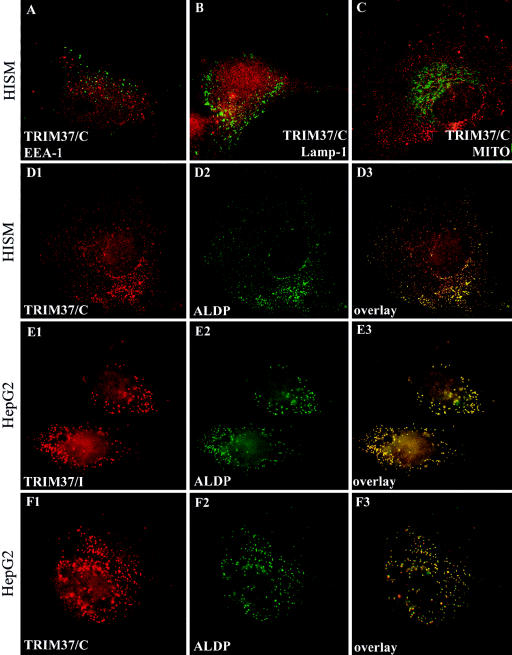
Indirect immunofluorescence staining was performed on a number of cell lines with TRIM37 antisera to search for endogenous immunoreactivity. When either the internal or the C-terminal peptide TRIM37 antiserum was used, a punctate cytoplasmic staining pattern was observed in nontransfected HeLa (data not shown), HISM (fig. 4A, 4B, 4C, 4D1) and HepG2 (fig. 4E1, 4F1) cells, representing staining of endogenous TRIM37. The intensity of the staining was weakest in HeLa cells and strongest in HepG2 cells. No endogenous TRIM37 staining was observed in COS-1 cells, and the preimmune sera produced very low nonspecific staining (data not shown). The staining pattern comprised a large number of vesicles, from <100 to several hundred per cell, round or slightly elongated in shape (fig. 4D1, 4E1, 4F1). In the HepG2 cells, TRIM37 staining was distributed among a smaller number of larger vesicles than those of HISM cells. Double stainings with TRIM37 and organelle-specific antibodies were performed on the cells, as described above. No colocalization of TRIM37 immunoreactivity was observed with late endosomes/lysosomes (anti-Lamp1) (fig. 4B), early endosomes (anti-EEA) (fig. 4A), Golgi (anti-CTR433) (data not shown), endoplasmic reticulum (anti-PDI) (data not shown), or mitochondria (MitoTracker) (fig. 4C). On the contrary, significant colocalization of TRIM37 immunoreactivity was observed with peroxisomal markers, as exemplified by colocalization with ALDP (fig. 4D1–D3, 4E1–E3, 4F1-F3). Unlike the exogenously expressed protein, the endogenous TRIM37 immunoreactivity was consistently present in all peroxisomes of the cell.
Figure 4.
Distribution of endogenous TRIM37 immunoreactivity in HISM and HepG2 cells as visualized by indirect immunofluorescence analysis. Red color represents TRIM37 protein (A, B, C, D1, E1, and F1), the organelle markers are shown in green (A, B, C, D2, E2, and F2) and colocalization of the immunoreactivities results are shown in yellow (D3, E3, and F3). Endogenous TRIM37 immunoreactivity is excluded from organelles other than peroxisomes, as is exemplified by double staining for TRIM37 and endosomes (A), lysosomes (B), or mitochondria (C) in HISM cells. Both the C-terminal peptide (TRIM37/C; D1 and F1) and internal peptide (TRIM37/I; E1) TRIM37 antisera produced a punctate cytoplasmic staining in HISM and HepG2 cells that colocalized with the peroxisomal marker ALDP (D2, D3, E2, E3, F2, and F3). The corresponding preimmune sera showed insignificant background staining (data not shown). Original magnification was 1000×.
Immunohistochemical Localization of TRIM37 in Human Tissue Sections
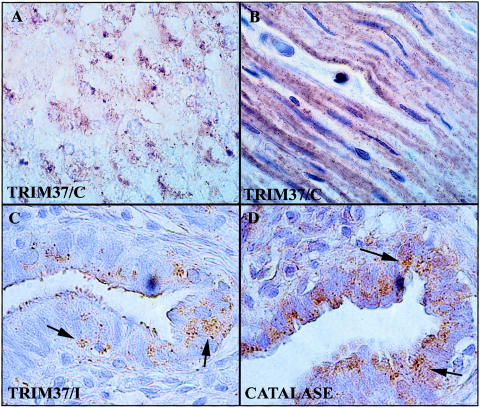
To preliminarily assess the tissue distribution of the TRIM37 protein, immunohistochemical staining for endogenous TRIM37 in paraffin-embedded human tissue material was performed using affinity-purified internal (I) and C-terminal (C) peptide TRIM37 antibodies and a polyclonal anti-human catalase antibody as a marker for peroxisomes. Both the C-terminal peptide and internal peptide TRIM37 antibodies produced a punctate cytoplasmic staining pattern (fig. 5) similar to that of anti-catalase antibody (fig. 5D) and compatible with peroxisomal localization. With the exception of liver (fig. 5A), the tissue distribution of TRIM37 immunoreactivity appeared highly cell-type specific. In the small intestine, the strongest staining was observed in the smooth muscle layer (fig. 5B). Throughout the liver, a rather intense staining of hepatocytes was observed (fig. 5A). It is noteworthy that endogenous TRIM37 was also detected, as shown in fig. 4, in the HepG2 and HISM cell lines derived from liver carcinoma and the intestinal smooth muscle, respectively. The granular pattern (arrows) of both TRIM37 and catalase immunoreactivity is exemplified by sections from an area in the ovary, in which staining was observed in the epithelial lining of the ovarian tube (fig. 5C, D). Control sections where primary antibody was replaced by normal rabbit IgG showed minimal nonspecific staining (data not shown).
Figure 5.
Immunohistochemical detection of endogenous TRIM37 in human liver (A), small intestine (B), and ovary (C) paraffin sections, using antigen-affinity purified TRIM37 peptide antibodies. A fine granular staining pattern was observed in the smooth muscle layer of the small intestine (B), whereas the granularity was more clustered in liver (A). In the ovary (C), TRIM37 staining was observed, among other locations, in the epithelial layer of the ovarian tube. Anticatalase staining of the ovarian tube showed a granular cytoplasmic distribution similar to that of TRIM37 (arrows). Both the C-terminal (C) and the internal peptide (I) antibodies produced similar staining. Nonimmune IgG controls showed minimal nonspecific staining (data not shown). Original magnification was 630× in A and B and 1000× in C and D.
Immunofluorescence Analysis of Peroxisomes in Fibroblasts of Patients with Peroxisomal Biogenesis Disorder and Patients with MUL
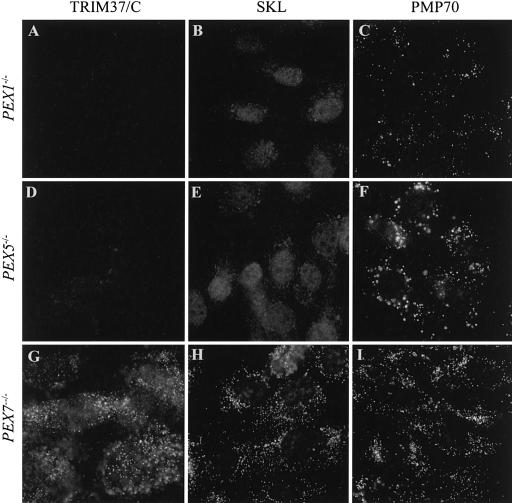
To gain further insight into the peroxisomal localization of TRIM37, immortalized fibroblasts derived from patients with known peroxisomal disorders were stained for endogenous TRIM37. The cell lines included peroxin 1 mutant (PEX1−/−) cells (Zellweger syndrome, complementation group 1; generalized defect in peroxisomal matrix protein import), peroxin 5 mutant (PEX5−/−) cells (autosomal neonatal adrenoleukodystrophy, complementation group 2; an isolated defect in PTS1- and peroxisomal targeting signal 2 [PTS2]-mediated import to peroxisomes) and peroxin 7 mutant (PEX7−/−) cells (rhizomelic chondrodysplasia punctata, complementation group 11; an isolated defect in PTS2-mediated import). Complete loss of peroxisomal TRIM37 immunoreactivity was observed in PEX1−/− and PEX5−/− cells (fig. 6A, 6D) whereas PEX7−/− cells showed normal peroxisomal pattern of TRIM37 staining (fig. 6G), suggesting that TRIM37 is imported to peroxisomes in a PEX5-dependent but PEX7-independent manner. As expected, all three mutant cell lines stained normally for PMP70, indicating the presence of peroxisome membranes (fig. 6C, 6F, 6I). PEX1−/− and PEX5−/− cells showed a diffuse cytosolic staining of SKL (PTS1)–containing matrix proteins (fig. 6B, 6E), whereas SKL staining was granular in PEX7−/− cells (fig. 6H).
Figure 6.
Immunofluorescence analysis of endogenous TRIM37 protein in PEX1, PEX5, and PEX7 mutant fibroblast cell lines. The mutant cell lines stained normally for PMP70, indicating the presence of peroxisome membranes (C, F, and I). PEX1−/− and PEX5−/− cells showed a diffuse cytosolic SKL staining (B and E) reflecting their inability to import PTS1-containing proteins into peroxisomes. On the contrary, SKL staining showed a normal granular pattern in the PEX7−/− cells that are deficient only in PTS2-mediated import (H). Peroxisomal TRIM37 staining is completely lost in PEX1−/− and PEX5−/− cells (A and D) but is present in PEX7−/− cells (G). The peroxisomal localization of TRIM37 in PEX7−/− cells was verified by double staining with ALDP (data not shown). Original magnification was 1000×.
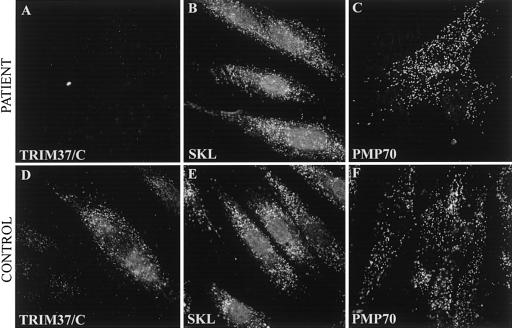
Fibroblasts derived from skin biopsies of three patients with MUL who were homozygous for the Finmajor mutation were analyzed for peroxisomal morphology and TRIM37 protein by immunofluorescence staining, using antibodies against TRIM37, PMP70, and SKL (fig. 7). In control fibroblasts, TRIM37 peptide antibodies produced a granular cytoplasmic staining pattern typical of peroxisomes (fig. 7D) that colocalized with the peroxisomal markers (data not shown). In agreement with the predicted protein truncation of the Finmajor mutant protein, TRIM37 immunoreactivity was absent in patient-derived fibroblasts (fig. 7A). The peroxisomal membrane marker PMP70 and the peroxisomal matrix marker SKL produced a typical peroxisomal staining pattern in both control (fig. 7E, 7F) and patient fibroblasts (fig. 7B, 7C) indicating that peroxisomal morphology and the import of PTS1-containing matrix proteins is not grossly affected in fibroblasts from patients with MUL.
Figure 7.
Immunofluorescence analysis of fibroblasts from patients with MUL, performed by use of TRIM37 and peroxisomal marker antibodies. The upper row shows staining for TRIM37 C-terminus (A), SKL (B), and PMP70 (C) in fibroblasts of a patient with MUL carrying the homozygous Finmajor mutation (c.493-2A→G). The lower row shows the corresponding stainings on normal control fibroblasts (D, E, and F). The patient fibroblasts lacked C-terminal TRIM37 staining (A), whereas a granular staining was seen in control fibroblasts (D). However, SKL (B and E) and PMP70 (C and F) showed a similar staining pattern in patient and control fibroblasts, indicating that both number and morphology of peroxisomes are unaltered in patient fibroblasts. Original magnification was 1000×.
Discussion
TRIM37 is a new member of the RBCC protein family, with a predicted molecular weight of 108 kD (Avela et al. 2000). Western blot analysis of cell lysates transfected with the TRIM37 cDNA showed that TRIM37 protein migrates at ∼130 kD. Migration of in vitro–translated TRIM37 was identical to that of the transiently expressed TRIM37, indicating that no major posttranslational modifications occur in the TRIM37 protein. The smaller fragments detected with the internal peptide antibody most likely represent C-terminally truncated forms of TRIM37, since they were not detected by the C-terminal antibody. Whether they result from nonspecific proteolysis or specific processing of the TRIM37 protein remains to be determined.
In the present study, we used several independent methods to determine the subcellular localization of the TRIM37 protein. Contrary to the sequence-based predictions that indicated a high probability of nuclear localization for TRIM37 (Avela et al. 2000), no sign of nuclear localization was observed. Instead, TRIM37 immunofluorescence in transiently transfected cells showed a cytoplasmic vesicular pattern. A similar pattern was observed in nontransfected HeLa, HISM, and HepG2 cells, as well as by immunohistochemical staining of various tissue samples. These findings are in agreement with recently published results in which Myc-epitope–tagged TRIM37 was shown to be associated with unrecognized cytosolic bodies (Zapata et al. 2001). Using four different peroxisomal marker antibodies, we identified the cellular compartment in which TRIM37 resided as the peroxisome. We could not detect colocalization with any other cell organelle–specific markers. It is noteworthy that normal peroxisomal morphology and colocalization of TRIM37 with peroxisomal markers were rapidly lost by increasing the transfection time, which resulted in the appearance of a few very large vesicles, possibly as a consequence of aggregation of TRIM37 due to high expression level. A similar phenomenon occurs in yeast, in that the number and size of peroxisomes are affected when some of the PEX (peroxin) genes are disrupted or overexpressed. For example, yeast cells lacking Pex11p have only a few large peroxisomes, whereas Pex16p produces enlarged peroxisomes on overexpression (Erdmann and Blobel 1995; Eitzen et al. 1997). We obtained further support for the peroxisomal localization of TRIM37 from subcellular fractionation analysis, in which TRIM37 immunoreactivity was highly enriched in the heavy peroxisomal fractions. We were not able to identify known peroxisomal targeting signals (Titorenko and Rachubinski 2001) in the TRIM37 amino acid sequence. This is not unexpected, since not all of these signals, particularly for peroxisomal membrane proteins, have yet been fully characterized.
All four reported mutations underlying MUL result in a frameshift and predict protein truncation. We determined the subcellular localization in transfected cells of two mutant proteins representing disease mutations. The Finmajor mutant TRIM37 protein, retaining 164 amino acids of the wild-type protein followed by 10 nonsense amino acids, was found to lose peroxisomal targeting and displayed diffuse cellular staining. However, in agreement with previous results (Zapata et al. 2001), cytoplasmic vesicular localization was retained in a deletion mutant containing the full RBCC domain (data not shown). The results are concordant with the data of Reymond et al. (2001), who showed that the coiled-coil region of RBCC proteins is critical for their subcellular targeting. Alternatively, the sequence of 10 nonsense amino acids, 3 of which are cysteines, could interfere with the targeting of the mutant polypeptide. In contrast to the Finmajor mutant protein, the Finminor (c.2212delG) mutant protein, retaining 737 amino acids of the wild-type TRIM37 protein, was found to localize to peroxisomes. When these localization data are taken together, the primary defect in the Finmajor mutant protein seems to be disturbed targeting to peroxisomes, whereas the Finminor mutant is correctly targeted, thus indicating a defective function of TRIM37 in the peroxisome. As yet, we have not measured TRIM37 transcript levels in patient cells. It is possible that the mutant mRNAs are rapidly degraded by nonsense-mediated mRNA decay. If, however, the mRNAs are stable and translated in significant amounts, the stability and distribution of mutant proteins in patient tissues remain to be determined.
Peroxisomes are single-membrane–bound organelles, <100 to >1,000 copies of which are present in nearly all eukaryotic cells (Sacksteder and Gould 2000; Titorenko and Rachubinski 2001). The peroxisome matrix is involved in numerous metabolic processes, such as the β-oxidation of long- and very-long-chain fatty acids; the biosynthesis of plasmalogens, cholesterol, and bile acid; and the degradation of amino acids and purine. Several inherited disorders are known to be due to mutations in genes encoding peroxisomal proteins. These can be broadly categorized into two groups: disorders resulting from a single enzyme defect and those resulting from defective biogenesis of the peroxisome due to so-called peroxin gene mutations (Moser 2000; Sacksteder and Gould 2000). The latter are called “peroxisome biogenesis disorders” (PBDs), and they result in defective import of peroxisomal matrix proteins, the majority of which are metabolic enzymes. Consequently, multiple metabolic functions of the peroxisome are affected. More than 20 PEX genes have been suggested to be involved in human peroxisome biogenesis, on the basis of complementation studies using either yeast or Chinese hamster ovary cells defective in peroxisome biogenesis (Sacksteder and Gould 2000; Titorenko and Rachubinski 2001). Eleven PEX genes have been shown to be mutated in PBDs, and three of them, PEX2, PEX10, and PEX12, encode proteins with a C-terminally located RING domain (Slawecki et al. 1995; Moser 2000; Sacksteder and Gould 2000). PEX2, PEX10, and PEX12 encode integral membrane proteins implicated to be involved in the translocation of proteins across the peroxisome membrane, possibly through RING finger–mediated protein-protein interactions (Shimozawa et al. 1992; Okumoto et al. 1998, 2000; Sacksteder and Gould 2000). We studied the localization of endogenous TRIM37 in three PEX mutant cell lines, including PEX1−/− cells, which have a generalized defect in the import of peroxisomal matrix proteins, PEX5−/− cells, which have a defect in both PTS1- and PTS2-mediated import to peroxisomes, and PEX7−/− cells, which have an isolated defect in PTS2-mediated import. Interestingly, we found that TRIM37 was not localized to peroxisomes in PEX1−/− and PEX5−/− cells, suggesting that its peroxisomal import is PTS1 dependent. However, TRIM37 apparently lacks PTS1, a C-terminal SKL, or a conservative variant of it. Further, the fact that the Finminor mutant protein that lacks the C-terminal 227 amino acids (see above) is targeted to peroxisomes indicates that the C-terminus of TRIM37 is not required for correct subcellular localization. Moreover, the full-length RBCC domain (Zapata et al. 2001; authors' unpublished data) also shows granular cytoplasmic localization. It is therefore possible that the import of TRIM37 into peroxisomes is dependent on the interaction with an as-yet-unidentified protein that uses PTS1 to enter the peroxisomes. For example, in yeast, the peroxisomal enzyme Eci1p may be imported into peroxisomes through dimerization with DCi1p, another peroxisomal enzyme that has a PTS1 signal (Yang et al. 2001). Interestingly, TRIM37 staining was absent after selective permeabilization of the plasma membrane with digitonin, supporting the view that TRIM37 protein resides within a membrane-bound organelle (data not shown).
Although MUL shares features with several syndromes that result from single peroxisomal enzyme defects, the combination of features in MUL most closely resemble those found in PBDs. Notably, prenatal-onset growth failure, facial dysmorphism, hepatomegaly, pigmentary changes in retina and muscular weakness, which are characteristic for patients with MUL (Perheentupa et al. 1973; Lipsanen-Nyman 1986), are also typical of PDB disorders like Zellweger syndrome (MIM 214100), neonatal adrenoleukodystrophy (MIM 202370), infantile Refsum disease (MIM 266510) or rhizomelic chondrodysplasia punctata type 1 (MIM 215100) (Moser 2000). The fact that some patients with MUL develop fatty and cirrhotic liver and hypoplasia of various endocrine glands (Lipsanen-Nyman 1986; M. Lipsanen-Nyman, unpublished data) is in agreement with a peroxisomal disease. Also, sensorineural hearing loss has been observed in patients with MUL (M. Lipsanen-Nyman, unpublished data). However, patients with MUL are neurologically intact, and their life span is mainly affected by the cardiopathy (M. Lipsanen-Nyman, unpublished data). Because of the overlap of clinical features, it has been speculated previously that MUL may be a peroxisomal disorder. Plasma concentrations of several fatty acids, the profile of very-long-chain fatty acids, and de novo plasmalogen biosynthesis in cultured fibroblasts, as well as the activity in plasma of a peroxisomal enzyme involved in fatty acid metabolism, were found to be in the normal range in two patients studied (Schutgens et al. 1994). We found that fibroblasts of three patients with MUL lacked TRIM37 immunoreactivity but contained peroxisomes that were apparently normal in morphology and number. The fibroblasts also stained normally for PTS1, suggesting apparently unaffected import of PTS1-containing subset of peroxisomal matrix proteins. Given the relatively mild course of MUL, the absence of striking peroxisomal defects, like those seen in some of the PBD patient fibroblasts, is not surprising.
Further study of the distribution of TRIM37 in adult tissues is under way. Whether a tissue-specific peroxisomal defect is involved in pathogenesis of MUL remains to be investigated. Moreover, the peroxisomal function in patients with MUL needs to be characterized in more detail to determine which subset of metabolic functions are affected in this disorder. Improved knowledge on TRIM37 function will not only advance our understanding of the molecular pathogenesis of MUL but will also give new insights into peroxisomal function and cell biology.
Acknowledgments
We thank Anne Vikman and Maarit Takatalo for technical assistance, Dr. Petra Eskelin and Dr. Paula Salmikangas for helpful advice, Dr. Ralf Bützow for help with tissue section analysis, and Dr. Kalervo Hiltunen for critical reading of the manuscript. We are thankful for Dr. Gabriele Dodt for the PEX mutant cell lines and helpful comments in finalizing the manuscript. This study was supported by the Ulla Hjelt Foundation and the Academy of Finland (project 50011). This study was performed in the Centre of Excellence in Disease Genetics of the Academy of Finland (project 44870, Finnish Centre of Excellence Programme 2000–2005).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Axis-Shield, http://www.axis-shield-poc.com/density/dtech.htm
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for TRIM37 [accession number NM_015294])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MUL [MIM 253250], Zellweger syndrome [MIM 214100], neonatal adrenoleukodystrophy [MIM 202370], infantile Refsum disease [MIM 266510], and rhizomelic chondrodysplasia punctata type 1 [MIM 215100])
References
- Aravind L, Dixit VM, Koonin EV (1999) The domains of death: evolution of the apoptosis machinery. Trends Biochem Sci 24:47–53 [DOI] [PubMed] [Google Scholar]
- Avela K, Lipsanen-Nyman M, Idänheimo N, Seemanova E, Rosengren S, Mäkelä TP, Perheentupa J, de la Chapelle A, Lehesjoki A-E (2000) Gene encoding a new RING-B-box-coiled-coil protein is mutated in mulibrey nanism. Nat Genet 25:298–301 [DOI] [PubMed] [Google Scholar]
- Baumgart E, Fahimi HD, Stich A, Völkl A (1996) L-Lactate dehydrogenase A4- and A3B isoforms are bona fide peroxisomal enzymes in rat liver. J Biol Chem 271:3846–3855 [DOI] [PubMed] [Google Scholar]
- Borden KLB (2000) RING domains: master builders of molecular scaffolds? J Mol Biol 295:1103–112 [DOI] [PubMed] [Google Scholar]
- Borden KLB, Freemont PS (1996) The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol 6:395–401 [DOI] [PubMed] [Google Scholar]
- Cainarca S, Messali S, Ballabio A, Meroni G (1999) Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet 8:1387–1396 [DOI] [PubMed] [Google Scholar]
- de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A (1991) The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66:675–684 [DOI] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet 9:115–125 [DOI] [PubMed] [Google Scholar]
- Eitzen GA, Szilard R, Rachubinski RA (1997) Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane protein. J Cell Biol 137:1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Vincent SR (1999) Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J Biol Chem 274:19771–19777 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol 128:509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS (2000) RING for destruction? Curr Biol 10:R84–R87 [DOI] [PubMed] [Google Scholar]
- Graham JM, Rickwood D (1997) Subcellular fractionation—a practical approach. Oxford University Press, Oxford, pp 143–167 [Google Scholar]
- Hsieh J, Liu J, Kostas SA, Chang C, Sternberg PW, Fire A (1999) The RING finger/B-Box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev 13:2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10:429–439 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 1:549–552 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Lapunzina P, Rodriguez JI, de Matteo E, Gracia R, Moreno F (1995) Mulibrey nanism: three additional patients and a review of 39 patients. Am J Med Genet 55:349–355 [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R (1995) The N-terminal part of TIF1 a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors is fused to B-raf in the oncogenic protein T18. EMBO J 14:2020–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehesjoki A-E, Reed VA, Gardiner RM, Greene NDE (2001) Expression of MUL, a gene encoding a novel RBCC family ring-finger protein, in human and mouse embryogenesis. Mech Develop 108:221–225 [DOI] [PubMed] [Google Scholar]
- Lipsanen-Nyman M (1986) Mulibrey-nanismi. PhD thesis, University of Helsinki, Helsinki [Google Scholar]
- Moser HW (2000) Molecular genetics of peroxisomal disorders. Front Biosci 5:D298–D306 [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1998) Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 5:355–364 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Abe I, Fujiki Y (2000) Molecular anatomy of the peroxin Pex12p. J Biol Chem 33:25700–25710 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y (1998) Mutations in PEX10 is the cause of Zellweger peroxisome deficiency syndrome of complementation group B. Hum Mol Gen 7:1399–1405 [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ (2000) Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol 5:1139–1162 [DOI] [PubMed] [Google Scholar]
- Perheentupa J, Autio S, Leisti S, Raitta C, Tuuteri L (1973) Mulibrey nanism, an autosomal recessive syndrome with pericardial constriction. Lancet 2:351–355 [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A (2001) The tripartite motif family identifies cell compartments. EMBO J 20:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ (2000) The genetics of peroxisome biogenesis. Ann Rev Genet 34:623–652 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS (1996) Does this have a familiar RING? Trends Biochem Sci 21:208–214 [PubMed] [Google Scholar]
- Schutgens RBH, Ryynänen M, Wanders RJA (1994) Peroxisomal functions in mulibrey nanism. J Inher Metab Dis 17:626 [DOI] [PubMed] [Google Scholar]
- Seemanova E, Bartsch O (1999) Mulibrey nanism and Wilms tumor. Am J Med Genet 85:76–78 [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y (1992) A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science 255:1132–1134 [DOI] [PubMed] [Google Scholar]
- Simila S, Timonen M, Heikkinen E (1980) A case of Mulibrey nanism with associated Wilms' tumor. Clin Genet 17:29–30 [DOI] [PubMed] [Google Scholar]
- Slack FJ, Bosson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5:659–669 [DOI] [PubMed] [Google Scholar]
- Slawecki ML, Dodt G, Steinberg S, Moser AB, Moser HW, Gould SJ (1995) Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci 108:1817–1829 [DOI] [PubMed] [Google Scholar]
- Spencer JA, Eliazir S, Ilaria RL Jr, Richardson JA, Olson EN (2000) Regulation of microtubule dynamics and myogenic differentiation by MURF a striated muscle RING-finger protein. J Cell Biol 150:771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Inaguma Y, Hiai H, Hirose F (1988) Developmentally regulated expression of a human “finger”-containing gene encoded by the 5′ half of the ret transforming gene. Mol Cell Biol 8:1853–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001) The life cycle of the peroxisome. Nat Rev Mol Cell Biol 2:357–368 [DOI] [PubMed] [Google Scholar]
- Wajant H, Henkler F, Scheurich P (2001) The TNF-receptor-associated factor family. Scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal 13:389–400 [DOI] [PubMed] [Google Scholar]
- Yang X, Purdue PE, Lazarow PB (2001) Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur J Cell Biol 80:126–138 [DOI] [PubMed] [Google Scholar]
- Zapata JM, Pawlowski K, Haas EC, Ware F, Godzik A, Reed JC (2001) A diverse family of proteins containing TRAF domains. J Biol Chem 276:24242–24252 [DOI] [PubMed] [Google Scholar]