Abstract
Chronic obstructive pulmonary disease (COPD) is a common, complex disease associated with substantial morbidity and mortality. COPD is defined by irreversible airflow obstruction; airflow obstruction is typically determined by reductions in quantitative spirometric indices, including forced expiratory volume at 1 s (FEV1) and the ratio of FEV1 to forced vital capacity (FVC). To identify genetic determinants of quantitative spirometric phenotypes, an autosomal 10-cM genomewide scan of short tandem repeat (STR) polymorphic markers was performed in 72 pedigrees (585 individuals) ascertained through probands with severe early-onset COPD. Multipoint variance-component linkage analysis (using SOLAR) was performed for quantitative phenotypes, including FEV1, FVC, and FEV1/FVC. In the initial genomewide scan, significant evidence for linkage to FEV1/FVC was demonstrated on chromosome 2q (LOD score 4.12 at 222 cM). Suggestive evidence was found for linkage to FEV1/FVC on chromosomes 1 (LOD score 1.92 at 120 cM) and 17 (LOD score 2.03 at 67 cM) and to FVC on chromosome 1 (LOD score 2.05 at 13 cM). The highest LOD score for FEV1 in the initial genomewide scan was 1.53, on chromosome 12, at 36 cM. After inclusion of 12 additional STR markers on chromosome 12p, which had been previously genotyped in this population, suggestive evidence for linkage of FEV1 (LOD score 2.43 at 37 cM) to this region was demonstrated. These observations provide both significant evidence for an early-onset COPD-susceptibility locus on chromosome 2 and suggestive evidence for linkage of spirometry-related phenotypes to several other genomic regions. The significant linkage of FEV1/FVC to chromosome 2q could reflect one or more genes influencing the development of airflow obstruction or dysanapsis.
Introduction
Chronic obstructive pulmonary disease (COPD) is a common, complex disease associated with substantial morbidity and mortality (Hoyert et al. 1999). Cigarette smoking is the major environmental determinant of COPD (Davis and Novotny 1989). However, the development of chronic airflow obstruction—the defining characteristic of COPD—is quite variable among smokers (Silverman and Speizer 1996). Airflow obstruction is typically assessed by spirometry. Spirometry involves forced expiratory maneuvers after the subject has inhaled to total lung capacity. Individuals with airflow obstruction have reductions in the forced expiratory volume at 1 s (FEV1) and in the ratio of FEV1 to forced vital capacity (FVC).
Severe alpha 1-antitrypsin (AAT) deficiency is a proven but uncommon genetic determinant of COPD (Larsson 1978; Tobin et al. 1983; Janus et al. 1985). Several types of studies have suggested that genetic factors other than AAT deficiency may be involved in the susceptibility to develop COPD (Lomas and Silverman 2001). Studies of spirometric measurements performed in the general population and in twins have suggested that genetic factors influence variation in pulmonary function (Redline et al. 1989). Studies in relatives of patients with COPD also have supported a role for genetic factors (Cohen et al. 1977; Kueppers et al. 1977; Cohen 1980; Beaty et al. 1987).
To identify genomic regions linked to COPD-related phenotypes, we have enrolled pedigrees ascertained through single probands who had severe early-onset COPD but who did not have severe AAT deficiency. In these families, we have previously demonstrated increased risk, to current or ex-smoking first-degree relatives of probands with early-onset COPD, for airflow obstruction, chronic bronchitis, and increased bronchodilator responsiveness (Silverman et al. 1998; Celedon et al. 1999). In addition, a previous genome-scan linkage analysis of qualitative COPD-related phenotypes in the same data set provided suggestive evidence for linkage of airflow obstruction to chromosome 12p and of chronic bronchitis to chromosome 22q (Silverman et al. 2002). We now report the linkage-analysis results of an autosomal genomewide scan, using quantitative spirometric phenotypes in extended pedigrees of individuals with severe early-onset COPD.
Subjects and Methods
Subjects
The recruitment and assessment of probands with severe early-onset COPD has been described elsewhere (Silverman et al. 1998, 2000). In brief, ascertainment criteria for probands with severe early-onset COPD included (1) FEV1 <40% of predicted value, (2) age <53 years, and (3) absence of severe AAT deficiency (e.g., PI Z, PI null-null). The age threshold of 53 years was chosen at the initiation of study enrollment, to balance our goal of identifying very young probands against our need to ascertain an adequate number of probands. All available first-degree relatives and older second-degree relatives (half sibs, aunts, uncles, and grandparents) of the ascertained probands with COPD were invited to participate. In addition to those enrolled on the basis of this fixed, single-ascertainment scheme, 49 additional relatives were enrolled, including (a) 37 first-degree relatives of pedigree members with moderate airflow obstruction (FEV1 <60% of predicted value and FEV1/FVC <90% of predicted value), (b) 3 other relatives who had been diagnosed with COPD, (c) 2 first-degree relatives of unavailable reportedly affected pedigree members, and (d) 7 other relatives. Seventy-two pedigrees with mean size of 8.1 individuals (range 2–18) were included in the genome-scan linkage analysis.
Participants gave written informed consent and completed a protocol that included a questionnaire, spirometry, and a blood sample. The study protocol was approved by the human research committees of Partners Health Care (Brigham and Women's Hospital and Massachusetts General Hospital) and the Brockton/West Roxbury Veterans Administration Hospital.
Questionnaire
Each participant completed a modified version of the 1978 American Thoracic Society–Division of Lung Diseases Epidemiology Questionnaire, which has been described elsewhere (Ferris 1978; Silverman et al. 1998). Pack-years of cigarette smoking were calculated as the product of the average number of cigarettes smoked per day—which was divided by 20, to convert to packs—and the duration of smoking (in years).
Pulmonary Function Tests
Pulmonary function testing in these pedigrees has been described elsewhere (Silverman et al. 1998, 2000). In brief, spirometry was performed with a Survey Tach Spirometer (Warren E. Collins), in accordance with American Thoracic Society guidelines (1995). Subjects were instructed to withhold, if possible, bronchodilator medications for at least 4 h before spirometry. Although the absolute-volume measurements were the primary phenotypes analyzed, pulmonary function test results also were expressed as percentage of predicted values by use of equations formulated by Crapo et al. (1981) for white adult participants. For white participants of age <18 years, predicted values for FEV1 were determined from equations developed by Hsu et al. (1979), and predicted values for FEV1/FVC were determined from equations developed by Knudson et al. (1983). For African-American participants, predicted spirometric values were determined from equations developed by Hankinson et al. (1999). For seven subjects, previously obtained pulmonary function test results were used, since three of them had undergone lung volume–reduction surgery and four resided at great geographic distance. Spirometry values for three other subjects were unavailable.
Genotyping and Data Management
The Mammalian Genotyping Service of the National Heart, Lung and Blood Institute performed genome-scan genotyping in 607 individuals, using DNA that was extracted from blood samples by Puregene Kits (Gentra Systems). We analyzed 378 autosomal STR markers with an average spacing of 9.1 cM, ranging from 7.4 cM on chromosome 11 to 10.6 cM on chromosome 14. Marker locations were determined on the basis of version 10 of the Marshfield map (see the Center for Medical Genetics, Marshfield Medical Research Foundation, Web site). For markers with exactly the same location on the Marshfield Map, a distance of 0.1 cM was placed between these markers after relative locations on the sequence published by the UCSC Human Genome Project Working Draft (“Golden Path”) and on the Celera Genomics Human Genome Sequence (see the Celera Discovery System Web site) had been determined.
On chromosome 12, an additional 12 STR markers were genotyped, at Brigham and Women’s Hospital. For these additional markers, map locations were also determined on the basis of the Marshfield map, with confirmation from the UCSC Human Genome Project Working Draft and/or the Celera Genomics Human Genome Sequence (see the Celera Discovery System Web site). Fluorescent-labeled and unlabeled primers were obtained from Research Genetics and Applied Biosystems. PCR was performed with Taq Gold Polymerase (Applied Biosystems) in MJ Research PCR machines. Product sizes were assessed on an ABI 3100. GENESCAN and GENOTYPER version 3.7 software were used to assist with genotype determination.
To test for Mendelian inconsistencies in our pedigree data, the RELCHECK program was used to determine pedigree relationships, on the basis of the genome-scan marker data (Broman et al. 1998). Twenty-two subjects failed to match reported familial relationships and were excluded. For the remaining 585 genotyped individuals, inconsistencies at individual markers were resolved by the PEDCHECK program (O'Connell and Weeks 1998). The mean rate of pedigree inconsistencies was less than one inconsistency per marker (0.95). Marker-allele frequencies were estimated by maximum-likelihood estimation by the SOLAR program (Almasy and Blangero 1998).
Linkage Analysis
Phenotypes selected for linkage analysis included FEV1, FVC, and FEV1/FVC. The primary analyses were performed with absolute-volume measurements; confirmatory analyses were performed with percentage of predicted values. The absolute spirometric values appeared to be normally distributed, and tests of model fit (with the FISHER program) suggested that the spirometric variables were acceptably close to normality; therefore, no data transformations were performed (Lange et al. 1988). Covariate adjustments were performed within the linkage analysis, as described below.
Two-point and multipoint linkage analyses of the genome-scan data were performed by a variance-component method, as implemented in the SOLAR program (version 1.7.4, running under Linux Mandrake version 8.0) (Almasy and Blangero 1998). SOLAR partitions observed phenotypic variance into genetic and nongenetic components, by maximum-likelihood methods; additive models, without dominance variance, were used in these analyses. Each model assumed that the distribution of the response phenotype in a pedigree was multivariate normal, with a mean that depended on a particular set of explanatory covariates. Genotypes were imputed for untyped individuals, conditional on all other marker data and on pedigree structure, and marker-specific identical by descent (IBD) matrices among all relative pairs were estimated independently for all autosomal markers. Multipoint IBD matrices were then generated at 1-cM resolution. Expected genetic covariances among relatives in an extended family were specified as a function of the fixed effects of covariates, residual error and random effects reflecting polygenic factors, and an unobserved QTL linked to an observed marker locus.
To adjust for the effects that demographics, body size, and cigarette smoking had on the spirometric measurements, age, sex, race, height, pack-years, age2, height2, and pack-years2 were included as covariates in the linkage analysis. Covariates that were significant at P<.05 were retained in the linkage-analysis model. Heritability values were obtained by SOLAR after adjustment for covariates.
The statistical associations between covariates entered as fixed effects and the response variable were assessed by removal of terms from the mean model and subsequent calculation of a likelihood-ratio χ2-test statistic. The null hypothesis of no linkage at a specific chromosomal location was tested by comparison of a polygenic model to a model with variance components for both a QTL and polygenic factors. Comparing the loge-likelihood values of these two models gives a test statistic that is asymptotically distributed as a mixture of χ2 distributions (Self and Liang 1987). A LOD score for linkage was determined by calculating twice the difference between the log10-likelihood values of these two models (Almasy and Blangero 1998).
Because the pedigrees with early-onset COPD were ascertained through single probands, the ascertainment correction provided in the SOLAR program—which involves conditioning the pedigree likelihood on the probability of the proband’s phenotype for each outcome—was employed (Lange et al. 1988). As noted above, pedigrees were extended to include 49 additional individuals; the impact that inclusion of these individuals had on the linkage results was examined in additional linkage analyses, which used only subjects ascertained through the fixed-ascertainment scheme.
Simulations were performed by SOLAR, to assess the statistical significance of the linkage results. Genotypes for a fully informative unlinked marker were created, and the evidence for linkage was assessed in 100,000 replicates. The number of times that a LOD score exceeded a specified threshold provides an empirical P value for that LOD-score threshold.
Results
Demographics and Spirometry in Families with Early-Onset COPD
A total of 607 individuals were included in the genome-scan data set. Twenty-two subjects were removed because of pedigree inconsistencies, and spirometric data for three subjects were unavailable. The age, smoking history, and spirometry (percentage of predicted values) for the 582 members of 72 pedigrees with spirometric values included in the genome-scan linkage analyses are presented in table 1. The profound degree of airflow obstruction is evident for probands with early-onset COPD, with mean predicted FEV1 values of 17.3%±6.3% and mean predicted FEV1/FVC values of 37.4%±11.4%. In severe COPD, reductions in FVC also are noted; the predicted mean FVC in probands was 47.9%±14.7%. Since the risk of developing COPD increases with age, it is not surprising that mean spirometric values were lower in parents than in siblings and that those in siblings were lower than those in children.
Table 1.
Demographics and Spirometry in Families with Early-Onset COPD
|
Mean ± SD |
|||||
| Group (No. of Individuals) | Age | Pack-Years | FEV1(% of Predicted Value) | FVC(% of Predicted Value) | FEV1/FVC(% of Predicted Value) |
| Probands (72) | 47.7 ± 5.3 | 38.9 ± 21.6 | 17.3 ± 6.3 | 47.9 ± 14.7 | 37.4 ± 11.4 |
| Siblings (148) | 47.0 ± 9.0 | 22.2 ± 20.9 | 80.8 ± 22.8 | 93.9 ± 18.3 | 85.1 ± 15.2 |
| Parents (48) | 72.5 ± 8.1 | 38.0 ± 39.4 | 65.3 ± 25.0 | 81.2 ± 19.1 | 77.6 ± 17.9 |
| Children (124) | 24.7 ± 5.7 | 4.3 ± 6.3 | 90.9 ± 11.6 | 98.0 ± 11.3 | 92.6 ± 7.8 |
| Other relatives (160) | 54.2 ± 18.0 | 23.1 ± 25.7 | 82.1 ± 19.1 | 91.3 ± 14.8 | 89.0 ± 11.9 |
| Spouses (30) | 51.8 ± 6.1 | 37.4 ± 35.0 | 85.5 ± 23.1 | 95.1 ± 18.7 | 89.5 ± 13.3 |
Genomewide Multipoint Variance-Component Linkage Analysis
Genomewide multipoint variance-component linkage results from SOLAR, for the absolute values of FEV1, FVC, and FEV1/FVC, are presented in figure 1 and are summarized in table 2. In these analyses, significant covariates for age, sex, pack-years, height, sex, and quadratic terms were included in the variance-component models, to adjust for these known effects on pulmonary function.
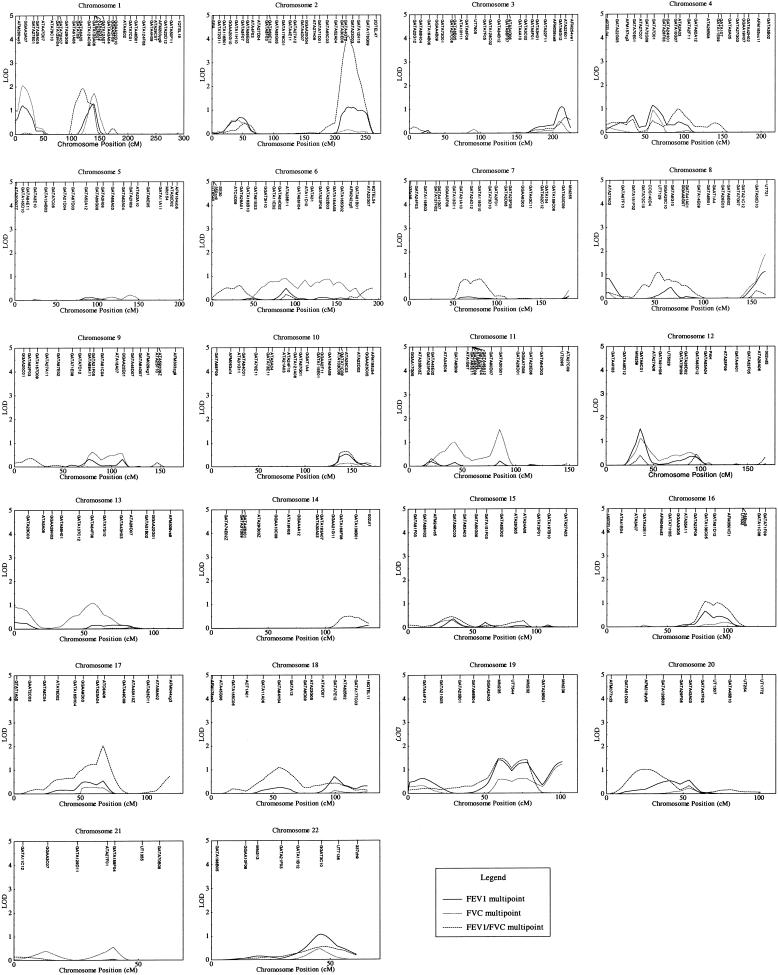
Figure 1.
Multipoint variance-component linkage analysis of all 22 autosomes in pedigrees with early-onset COPD. Results of linkage analysis are presented for FEV1, FEV1/FVC, and FVC, with adjustment for relevant covariates. Genetic distance (in cM), from version 10 of the Marshfield map, is plotted against variance-component LOD score. Significant linkage was demonstrated for FEV1/FVC, on chromosome 2 (LOD score 4.12).
Table 2.
Genome-Scan Multipoint Linkage Analysis with SOLAR: Chromosomal Regions with LOD Scores >1.0
|
Maximum LOD Score (Location)a |
|||
| Chromosome | FEV1 | FVC | FEV1/FVC |
| 1 | 1.28 (139) | 2.05 (13) | 1.92 (120) |
| 2 | 1.12 (222) | .59 (40) | 4.12 (222) |
| 3 | 1.10 (213) | .50 (209) | .64 (219) |
| 4 | 1.15 (60) | .94 (60) | .99 (92) |
| 8 | 1.11 (164) | 1.84 (164) | 1.10 (53) |
| 11 | .21 (85) | 1.53 (85) | .32 (21) |
| 12 | 1.53 (36) | 1.12 (36) | .46 (95) |
| 13 | .29 (0) | 1.08 (56) | .05 (86) |
| 16 | .66 (81) | .18 (98) | 1.07 (81) |
| 17 | .54 (67) | .28 (58) | 2.03 (67) |
| 18 | .70 (99) | .11 (99) | 1.09 (54) |
| 19 | 1.40 (59) | 1.20 (101) | 1.47 (61) |
| 20 | .57 (54) | .24 (54) | 1.04 (24) |
| 22 | 1.07 (46) | .45 (46) | .55 (47) |
Values are highest LOD scores (location in cM), on each chromosome, for each phenotype. Spirometric values were adjusted for known covariates (see text).
For FEV1, the estimated heritability was 35.3% ± 6.3% (P<.0001); significant covariates were those for age (P<.0001), sex (P<.0001), height (P<.0001), pack-years (P<.0001), and pack-years2 (P<.0001). Multipoint LOD scores >1.0 were found on eight chromosomes; the highest LOD score was 1.53, on chromosome 12 (at 36 cM from pter).
For FVC, the estimated heritability was 31.0% ± 6.2% (P<.0001), and significant covariates were those for age (P<.0001), sex (P<.0001), age2 (P=.0002), height (P<.0001), pack-years (P<.0001), pack-years2 (P<.0001), and race (P=.005). Multipoint LOD scores >1.0 were found on six chromosomes; the highest LOD score was 2.05, on chromosome 1 (at 13 cM).
For FEV1/FVC, the estimated heritability was 30.3% ± 5.2% (P<.0001); significant covariates were those for age (P<.0001), pack-years (P<.0001), and pack-years2 (P<.0001). Multipoint LOD scores >1.0 were found on eight chromosomes; the highest LOD score was 4.12, on chromosome 2 (at 222 cM).
Several chromosomal regions demonstrated some evidence for linkage to multiple phenotypes. For example, the chromosome 2 region that demonstrated strong evidence for linkage to FEV1/FVC also had a LOD score of 1.12 for FEV1; the chromosome 12 region with strongest evidence for linkage to FEV1 (LOD score 1.53, at 36 cM) provided a LOD score of 1.12 to FVC, at the same location; the chromosome 1 region at 130–150 cM produced LOD scores >1.0 for FEV1, FVC, and FEV1/FVC; and a broad region of chromosome 19 also demonstrated some evidence for linkage to all three phenotypes.
However, other chromosomal regions demonstrated linkage to only a single phenotype. For example, chromosome 17 had, at 67 cM, a LOD score of 2.03 for FEV1/FVC but much more modest LOD scores for FEV1 and FVC.
Additional Markers on Chromosome 12p: Variance-Component Linkage Analysis
Additional STR markers on chromosome 12p were genotyped as part of a previous study of qualitative COPD-related phenotypes (Silverman et al. 2002). Therefore, higher-resolution linkage analysis could be performed in this region. Fine mapping was performed with 4 markers (GATA49D12, Mfd259, GATA6C01, and ATA27A06) from the original genomewide scan and 12 additional STR markers located at 18–49 cM on chromosome 12. Because multipoint linkage analysis is sensitive to errors in marker order and in intermarker distances, two-point linkage analysis also was performed in this region, by SOLAR. The maximum observed two-point LOD score for FEV1 was 2.88 (D12S1715, at 36 cM), that for FVC was 1.95 (D12S1581, at 30 cM), and that for FEV1/FVC was 2.68 (D12S1066, at 42 cM).
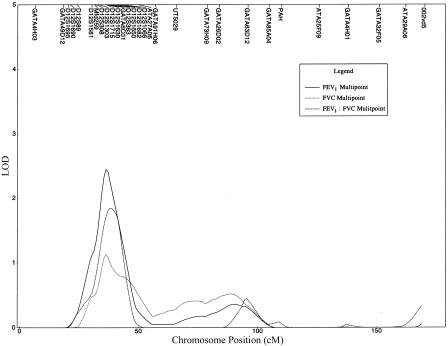
The multipoint variance-component linkage-analysis results for chromosome 12, including the 12 additional flanking markers on 12p, are depicted in figure 2. Compared with the initial genomewide multipoint linkage analysis, increased evidence for linkage was noted for FEV1 and FEV1/FVC. For FEV1, the maximum multipoint LOD score was 2.43, at 37 cM, and, for FEV1/FVC, the maximum multipoint LOD score was 1.84, at 38 cM. With the additional STR markers on 12p, the maximum LOD score for FVC—1.12, at 36 cM—remained unchanged from the initial genomewide scan.
Figure 2.
Multipoint variance-component linkage analysis of chromosome 12, including STR markers from the initial genomewide scan, as well as 12 additional markers. Genetic distance (in cM), from version 10 of the Marshfield map, is plotted against the multipoint variance-component LOD score for FEV1, FEV1/FVC, and FVC. For FEV1, a maximum LOD score of 2.43 was observed at 37 cM.
Confirmatory Analyses
Several additional analyses were performed. Pulmonary function test values are typically expressed as a percentage of predicted values, to adjust for the known effects that age, height, sex, and race have on these measurements. Therefore, we also performed linkage analysis using percentage of predicted values; in these analyses, pack-years (but not age, height, sex, or race) were included as a covariate, to adjust for the effects of cigarette smoking. The genomewide linkage-analysis results when values expressed as percentage of predicted values were used were not substantially different from the results when absolute volumes were used (results not shown).
Additional confirmatory analyses were performed to assess the impact that ascertainment had on the linkage results. Phenotypic values for the 49 subjects enrolled in the study who did not meet the strict ascertainment scheme including all first-degree relatives and all older second-degree relatives (i.e., aunts, uncles, grandparents, and half sibs) were excluded, and the genome-scan linkage analysis was repeated. The linkage-analysis results for these “trimmed” pedigrees were generally quite similar to those for the analyses using all subjects.
To estimate the statistical significance of the FEV1/FVC linkage results, simulations were performed with a fully informative unlinked marker, followed by variance-component linkage analysis of the FEV1/FVC phenotype for each replicate. The highest observed two-point LOD scores in the FEV1/FVC linkage analysis were 2.99, for GATA23D03 (chromosome 2, at 227 cM), and 2.68, for ATC6A06 (chromosome 17, at 67 cM). In 100,000 replicates of an unlinked marker, LOD scores >2.68 were observed 41 times; thus, the empirical P value for the ATC6A06 linkage to FEV1/FVC is .0004. LOD scores >2.99 were observed 20 times, for an empirical P value, for the GATA23D03 linkage, of .0002. Although the simulations were based on two-point linkage analysis, it is of interest that the maximum multipoint LOD score for FEV1/FVC (LOD score 4.12), on chromosome 2, was exceeded in only 2 of 100,000 simulations.
Discussion
Although the development of COPD is clearly influenced by cigarette smoking, genetic determinants are likely to be important as well. To enrich the sample for potential genetic determinants, we have studied extended pedigrees of individuals ascertained through probands with severe early-onset COPD without severe AAT deficiency. Our 10-cM genomewide linkage scan is the first reported investigation of linkage to quantitative spirometric measures in pedigrees with COPD. The highest observed LOD score was 4.12, for FEV1/FVC, on chromosome 2q. On the basis of standard criteria for linkage-analysis results in extended pedigrees (i.e., LOD score 1.9 for suggestive linkage and LOD score 3.3 for significant linkage), this result corresponds to significant linkage (Lander and Kruglyak 1995). The most promising evidence for linkage to FEV1 was found on chromosome 12p, in the initial genomewide scan; with the inclusion of 12 additional markers on 12p, the maximum LOD score for FEV1 was 2.43, which corresponds to suggestive linkage. In addition, suggestive linkage to FEV1/FVC was found on chromosomes 17 (LOD score 2.03, at 67 cM) and 1 (LOD score 1.92, at 120 cM), and suggestive linkage to FVC was found on chromosome 1 (LOD score 2.05, at 13 cM).
In a previous genomewide linkage scan of these families with early-onset COPD, we studied qualitative phenotypes, including moderate airflow obstruction (FEV1 <60% of predicted value, with FEV1/FVC <90% of predicted value) and mild airflow obstruction (FEV1 <80% of predicted value, with FEV1/FVC <90% of predicted value) (Silverman et al. 2002). For these qualitative phenotypes, the highest observed LOD scores for moderate airflow obstruction were 1.70, on chromosome 12p (at 36 cM), and 1.54, on chromosome 19 (at 42 cM); those regions also provided several of the highest LOD scores for mild airflow obstruction in smokers. When additional STR markers on 12p were included, the maximum LOD score for moderate airflow obstruction, when nonparametric methods were used, was 2.13. Of interest, in the current analysis of quantitative phenotypes, chromosomes 12 and 19 also provide the highest LOD scores for FEV1, suggesting that those loci may be involved in airflow obstruction.
In the qualitative-phenotype linkage analysis, no significant evidence for linkage to chromosome 2q was observed: the maximum LOD score for moderate airflow obstruction was 0.11, at 237 cM, and the maximum LOD score for mild airflow obstruction was 0.14, at 167 cM; however, in the present analysis, significant evidence for linkage of FEV1/FVC (LOD score 4.12, at 222 cM) and nominal evidence for linkage of FEV1 (LOD score 1.12, at 222 cM) were observed. The chromosome 2q locus may represent an airflow-obstruction locus, and quantitative phenotypes may be more powerful at detection of such loci. In particular, the FEV1/FVC ratio may detect earlier airflow obstruction, before significant reductions in FEV1 are observed. Alternatively, the chromosome 2q linkage may represent a locus that influences the development of dysanapsis, which is defined as differences from the normal relationship between airway size and lung size (Mead 1980). Regardless of whether the chromosome 2q linkage to FEV1/FVC represents a gene that influences lung growth and dysanapsis or the early development of smoking-related airflow obstruction, reduction in FEV1/FVC has been shown to be associated with subsequent decline in pulmonary function (Burrows et al. 1987); therefore, genetic determinants of FEV1/FVC likely represent important determinants of COPD.
Subjects with severe COPD may develop reductions in FVC as well as in FEV1. However, compared to FEV1, FVC tends to be relatively preserved in COPD, leading to a reduction in the FEV1/FVC ratio, as a cardinal feature of COPD. Therefore, it is of interest that some regions of nominal linkage to FVC, such as those on chromosomes 8 and 12, showed LOD scores >1.0, for both FEV1 and FVC, whereas other regions, such as that on chromosome 11, only showed evidence for linkage to FVC. Further investigation will be necessary to determine whether regions of linkage to both FEV1 and FVC represent genetic determinants of lung size whereas regions of linkage to FEV1 and FEV1/FVC—but not to FVC—represent genetic determinants of airway obstruction.
In genomewide linkage studies, multiple genomic regions have been linked to asthma-related phenotypes. Evidence for linkage of asthma-related phenotypes to the two most promising regions in this study, chromosome 2q and chromosome 12p, have been reported elsewhere, in some asthma-genetics studies. Nominal evidence for linkage to chromosome 2q (P<.05) was demonstrated for asthma diagnosis in the Collaborative Study of the Genetics of Asthma, for IgE levels in the German Asthma Genetics Study, for airway responsiveness and eosinophil counts in the French Asthma Genetics Study, and for allergy skin tests in the Hutterites (Daniels et al. 1996; Collaborative Study on the Genetics of Asthma 1997; Wjst et al. 1999; Dizier et al. 2000; Ober et al. 2000). On chromosome 12p, several previous asthma-genetics studies have found nominal evidence for linkage to asthma and atopy-related phenotypes in the same general region that, in our study, demonstrates linkage to FEV1 (Collaborative Study on the Genetics of Asthma 1997; Blumenthal et al. 1998). It remains unclear whether asthma and COPD share genetic determinants, and elucidation of the specific genetic determinants of each condition will be necessary to resolve this controversy definitively. However, our results are consistent with the possibility that at least some genetic determinants are shared by asthma and COPD.
Several biologically plausible candidate genes are located within the two chromosomal regions that are most promising in our analysis (the candidate-gene locations were obtained from the April 2001 freeze at the UCSC Human Genome Project Working Draft [“Golden Path”] Web site). The interleukin-8 receptor alpha gene (IL8RA) is located within the region of linkage to FEV1/FVC, on chromosome 2q. Interleukin-8 is a cytokine involved in neutrophil recruitment to the lung, which could lead to increased delivery of neutrophil proteases and to increased risk for lung-elastin destruction (Baggiolini et al. 1989). Microsomal glutathione S-transferase 1 (MGST1), an enzyme involved in the detoxification of oxygen radicals such as the reactive species produced by cigarette smoke, and matrix Gla protein (MGP), a lung extracellular-matrix component that is mutated in a syndrome (i.e., Keutel syndrome) including diffuse pulmonary calcification, are located near the region of strongest evidence for linkage to FEV1, on chromosome 12p (Munroe et al. 1999; Kelner et al. 2000).
This study has several significant limitations. The generalizability of linkage results in the families with severe early-onset COPD to COPD at later ages is undetermined. We have included the primary environmental determinant of COPD, cigarette smoking, as a covariate in our analyses, but we have not formally tested for genotype × environment interactions. Our ascertainment scheme did include single probands, but additional extended relatives were included in some families; however, the linkage results were not significantly altered when these additional relatives were excluded. Finally, we have not adjusted for the multiple phenotypes analyzed, because these spirometric phenotypes are significantly correlated.
There are likely to be multiple genetic determinants of COPD, and these linkage analyses of quantitative spirometric phenotypes have identified both one region of significant linkage and several regions of suggestive linkage, on which to focus fine-mapping efforts. Both the use of additional STR markers to evaluate the regions of linkage that have been observed in our study and replication of our findings in other samples will be necessary. If novel genetic determinants of COPD can be identified, important new insights into COPD pathophysiology—and, ultimately, treatment—could result.
Acknowledgments
We appreciate assistance, from a variety of physicians, in the recruitment of subjects for this study. We are grateful for the high-quality genotyping performed by the National Heart, Lung and Blood Institute Mammalian Genotyping Service. We are especially thankful for the enthusiastic support that the members of the families with early-onset COPD have given to this study. We would like to acknowledge helpful discussions with Dr. Steven Shapiro, as well as computer-programming support from Soma Datta. This study was supported by National Institutes of Health grants HL 61575 to Brigham and Women's Hospital (support to E.K.S.); P50 HL56383 to Brigham and Women's Hospital (support to J.M.D.); Program in Genomic Applications grant U01 HL 66795 to Brigham and Women's Hospital (support to S.T.W.); HL 67204 to the University of California, San Francisco (support to H.A.C.); HL 46440 to the University of Utah Health Sciences Center (support to E.J.C.); an American Lung Association Research Grant to the Channing Laboratory, Brigham and Women's Hospital (support to E.K.S.); and a gift from the Overholt Thoracic Foundation to the Division of Pulmonary and Critical Care Medicine, Brigham and Women's Hospital.
Electronic-Database Information
URLs for data in this article are as follows:
- Celera Discovery System, http://cds.celera.com/cds/login.cfm
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- UCSC Human Genome Project Working Draft (“Golden Path”), http://genome.cse.ucsc.edu/
References
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Thoracic Society (1995) Standardization of spirometry: 1994 update. Am J Respir Crit Care Med 152:1107–1136 [DOI] [PubMed] [Google Scholar]
- Baggiolini M, Walz A, Kunkel SL (1989) Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Invest 84:1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Liang KY, Seerey S, Cohen BH (1987) Robust inference for variance components models in families ascertained through probands. II. Analysis of spirometric measures. Genet Epidemiol 4:211–221 [DOI] [PubMed] [Google Scholar]
- Blumenthal MN, Rich SS, King R, Weber J (1998) Approaches and issues in defining asthma and associated phenotypes map to chromosome susceptibility areas in large Minnesota families: The Collaborative Study for the Genetics of Asthma (CSGA). Clin Exp Allergy 28 Suppl 1:51–55; discussion 65–66 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows B, Knudson RJ, Camilli AE, Lyle SK, Lebowitz MD (1987) The “horse-racing effect” and predicting decline in forced expiratory volume in one second from screening spirometry. Am Rev Respir Dis 135:788–793 [DOI] [PubMed] [Google Scholar]
- Celedon JC, Speizer FE, Drazen JM, Weiss ST, Campbell EJ, Carey VJ, Reilly JJ, Ginns L, Silverman EK (1999) Bronchodilator responsiveness and serum total IgE levels in families of probands with severe early-onset COPD. Eur Respir J 14:1009–1014 [DOI] [PubMed] [Google Scholar]
- Cohen BH (1980) Chronic obstructive pulmonary disease: a challenge in genetic epidemiology. Am J Epidemiol 112:274–288 [DOI] [PubMed] [Google Scholar]
- Cohen BH, Ball WC Jr, Brashears S, Diamond EL, Kreiss P, Levy DA, Menkes HA, Permutt S, Tockman MS (1977) Risk factors in chronic obstructive pulmonary disease (COPD). Am J Epidemiol 105:223–232 [DOI] [PubMed] [Google Scholar]
- Collaborative Study on the Genetics of Asthma (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–392 [DOI] [PubMed] [Google Scholar]
- Crapo RO, Morris AH, Gardner RM (1981) Reference spirometric values using techniques and equipment that meet ATS recommendations. Am Rev Respir Dis 123:659–664 [DOI] [PubMed] [Google Scholar]
- Daniels S, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, le Souef PN, Lathrop GM, Musk AW, Cookson WOCM (1996) A genome-wide search for quantitative trait loci underlying asthma. Nature 383:247–250 [DOI] [PubMed] [Google Scholar]
- Davis RM, Novotny TE (1989) Changes in risk factors: the epidemiology of cigarette smoking and its impact on chronic obstructive pulmonary disease. Am Rev Respir Dis 140:S82–S84 [DOI] [PubMed] [Google Scholar]
- Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, Charpin D, et al (2000) Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med 162:1812–1818 [DOI] [PubMed] [Google Scholar]
- Ferris BG (1978) Epidemiology Standardization Project. Am Rev Respir Dis Suppl 118(part 2): 1–120 [PubMed] [Google Scholar]
- Hankinson JL, Odencrantz JR, Fedan KB (1999) Spirometric reference values from a sample of the general US population. Am J Respir Crit Care Med 159:179–187 [DOI] [PubMed] [Google Scholar]
- Hoyert DL, Kochanek KD, Murphy SL (1999) Deaths: final data for 1997. Natl Vital Stat Rep 47:1–104 [PubMed] [Google Scholar]
- Hsu KHK, Jenkins DE, Hsi BP, Bourhofer E, Thompson V, Tanakawa N, Hsieh GSJ (1979) Ventilatory functions of normal children and young adults—Mexican-American, white, and black. I. Spirometry. J Pediatr 95:14–23 [DOI] [PubMed] [Google Scholar]
- Janus ED, Phillips NT, Carrell RW (1985) Smoking, lung function, and alpha 1-antitrypsin deficiency. Lancet 1:152–154 [DOI] [PubMed] [Google Scholar]
- Kelner MJ, Bagnell RD, Montoya MA, Estes LA, Forsberg L, Morgenstern R (2000) Structural organization of the microsomal glutathione S-transferase gene (MGST1) on chromosome 12p13.1-13.2: identification of the correct promoter region and demonstration of transcriptional regulation in response to oxidative stress. J Biol Chem 275:13000–13006 [DOI] [PubMed] [Google Scholar]
- Knudson RJ, Lebowitz MD, Holberg CJ, Burrows B (1983) Changes in the normal maximal expiratory flow-volume curve with growth and aging. Am Rev Respir Dis 127:725–734 [DOI] [PubMed] [Google Scholar]
- Kueppers F, Miller RD, Gordon H, Hepper NG, Offord K (1977) Familial prevalence of chronic obstructive pulmonary disease in a matched pair study. Am J Med 63:336–342 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed] [Google Scholar]
- Larsson C (1978) Natural history and life expectancy in severe alpha 1-antitrypsin deficiency, Pi Z. Acta Med Scand 204:345–351 [DOI] [PubMed] [Google Scholar]
- Lomas DA, Silverman EK (2001) The genetics of chronic obstructive pulmonary disease. Respir Res 2:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead J (1980) Dysanapsis in normal lungs assessed by the relationship between maximal flow, static recoil, and vital capacity. Am Rev Respir Dis 121:339–342 [DOI] [PubMed] [Google Scholar]
- Munroe PB, Olgunturk RO, Fryns JP, Van Maldergem L, Ziereisen F, Yuksel B, Gardiner RM, Chung E (1999) Mutations in the gene encoding the human matrix Gla protein cause Keutel syndrome. Nat Genet 21:142–144 [DOI] [PubMed] [Google Scholar]
- Ober C, Tsalenko A, Parry R, Cox NJ (2000) A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 67:1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redline S, Tishler PV, Rosner B, Lewitter FI, Vandenburgh M, Weiss ST, Speizer FE (1989) Genotypic and phenotypic similarities in pulmonary function among family members of adult monozygotic and dizygotic twins. Am J Epidemiol 129:827–836 [DOI] [PubMed] [Google Scholar]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, Wain J, Speizer FE (1998) Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 157:1770–1778 [DOI] [PubMed] [Google Scholar]
- Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, Province MA, Rao DC, Reilly JJ, Ginns LC, Speizer FE, Weiss ST (2002) Genomewide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet (in press) [DOI] [PubMed] [Google Scholar]
- Silverman EK, Speizer FE (1996) Risk factors for the development of chronic obstructive pulmonary disease. Med Clin North Am 80:501–522 [DOI] [PubMed] [Google Scholar]
- Silverman EK, Weiss ST, Drazen JM, Chapman HA, Carey V, Campbell EJ, Denish P, Silverman RA, Celedon JC, Reilly JJ, Ginns LC, Speizer FE (2000) Gender-related differences in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med 162:2152–2158 [DOI] [PubMed] [Google Scholar]
- Tobin MJ, Cook PJL, Hutchison DCS (1983) Alpha 1-antitrypsin deficiency: the clinical and physiological features of pulmonary emphysema in subjects homozygous for Pi type Z. Br J Dis Chest 77:14–27 [DOI] [PubMed] [Google Scholar]
- Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A, et al (1999) A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics 58:1–8 [DOI] [PubMed] [Google Scholar]