Abstract
A combined analysis of genome scans for obesity was undertaken using the interim results from the National Heart, Lung, and Blood Institute Family Blood Pressure Program. In this research project, four multicenter networks of investigators conducted eight individual studies. Data were available on 6,849 individuals from four ethnic groups (white, black, Mexican American, and Asian). The sample represents the largest single collection of genomewide scan data that has been analyzed for obesity and provides a test of the reproducibility of linkage analysis for a complex phenotype. Body mass index (BMI) was used as the measure of adiposity. Genomewide linkage analyses were first performed separately in each of the eight ethnic groups in the four networks, through use of the variance-component method. Only one region in the analyses of the individual studies showed significant linkage with BMI: 3q22.1 (LOD 3.45, for the GENOA network black sample). Six additional regions were found with an associated LOD >2, including 3p24.1, 7p15.2, 7q22.3, 14q24.3, 16q12.2, and 17p11.2. Among these findings, the linkage at 7p15.2, 7q22.3, and 17p11.2 has been reported elsewhere. A modified Fisher’s omnibus procedure was then used to combine the P values from each of the eight genome scans. A complimentary approach to the meta-analysis was undertaken, combining the average allele-sharing identity by descent (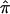 ) for whites, blacks, and Mexican Americans. Using this approach, we found strong linkage evidence for a quantitative-trait locus at 3q27 (marker D3S2427; LOD 3.40, P=.03). The same location has been shown to be linked with obesity-related traits and diabetes in at least two other studies. These results (1) confirm the previously reported obesity-susceptibility locus on chromosomes 3, 7, and 17 and (2) demonstrate that combining samples from different studies can increase the power to detect common genes with a small-to-moderate effect, so long as the same gene has an effect in all samples considered.
) for whites, blacks, and Mexican Americans. Using this approach, we found strong linkage evidence for a quantitative-trait locus at 3q27 (marker D3S2427; LOD 3.40, P=.03). The same location has been shown to be linked with obesity-related traits and diabetes in at least two other studies. These results (1) confirm the previously reported obesity-susceptibility locus on chromosomes 3, 7, and 17 and (2) demonstrate that combining samples from different studies can increase the power to detect common genes with a small-to-moderate effect, so long as the same gene has an effect in all samples considered.
Introduction
The prevalence of obesity has been increasing rapidly in the last several decades, especially in the industrialized countries. Obese individuals are at higher risk of morbidity and mortality from many chronic diseases, such as hypertension, non–insulin-dependent diabetes, lipid abnormalities, and coronary heart disease (Spadano et al. 1999). It is known that obesity-related traits, including BMI, are influenced by both genetic and environmental factors (Bouchard et al. 1998). In the majority of studies, heritability has been estimated to be 40%–90% (Borecki et al. 1998b; Bouchard et al. 1998; Barsh et al. 2000; Luke et al. 2001). Mutations responsible for several rare Mendelian types of obesity have been identified (Chagnon et al. 2000), although the genes that confer susceptibility to the common form of obesity are largely unknown. Segregation analyses have provided evidence of major genes influencing obesity-related traits (Price et al. 1990; Borecki et al. 1998a); however, other studies have inferred that multiple genes contribute, each having small effects individually (Bouchard et al. 1988; Hasstedt et al. 1997; Lecomte et al. 1997; Borecki 1998b). To search for the multiple unknown loci that are hypothesized, linkage analysis has been used with STR markers spread across the genome. Genomewide linkage analysis has been performed in several different populations, including people of European descent (Hager et al. 1998; Lee et al. 1999; Kissebah et al. 2000; Öhman et al. 2000; Van der Kallen et al. 2000; Watanabe et al. 2000; Perola et al. 2001), Mexican Americans (Comuzzie et al. 1997; Mitchell et al. 1999), Pima Indians (Hanson et al. 1998; Norman et al. 1998; Walder et al. 2000), and African Americans (Zhu et al. 2002).
Identifying the susceptibility genes for complex traits such as obesity, diabetes, and hypertension represents a challenging task for human geneticists. Multiple genes interacting with environmental factors usually affect these traits. Because of the moderate effect of each gene on the trait, it is difficult to acquire enough power to detect it with a moderate sample size. One potential solution to this challenge is to combine data from multiple studies (Li and Rao 1996; Gu et al. 1998; Lonjou et al. 1999; Palmer et al. 2001). In the present study, we first performed genome scans for BMI in the eight samples from the National Heart, Lung, and Blood Institute (NHLBI) Family Blood Pressure Program (FBPP). We subsequently conducted linkage analysis in the combined data set by means of a meta-analysis combining P values and also by pooling average allele-sharing identity by descent (IBD),  .
.
Subjects and Methods
Family Ascertainment
Subjects were selected from the four component networks (GenNet, GENOA, HyperGEN, and SAPPHIRe) of the ongoing NHLBI FBPP. In brief, GenNet sampled black and white nuclear families through identification of young-to-middle aged probands with elevated blood pressure (BP). Both GENOA and HyperGEN sampled black and white sibships containing sib pairs with essential hypertension. GENOA also sampled Mexican American sibships containing sib pairs with hypertension, as well as some nonhypertensive sibs in all three ethnic groups. SAPPHIRe recruited three groups of Japanese and Chinese sib pairs: concordant for hypertension, concordant for hypotension, and extremely discordant. Anthropometric measurements were obtained at the time of the clinic visit. The pooled data set analyzed here currently includes genotype data and >120 measured and derived phenotypic variables in 6,849 participants, representing >60% of the total FBPP sample. In the present study, we used BMI as the measure of adiposity, because it is highly correlated with other measures of fat mass (Borecki et al. 1991) and is available for the largest number of individuals. Family relations were verified using the marker data; pedigree errors were corrected, and unrelated individuals were eliminated from the analysis.
Genotyping
DNA was extracted from whole blood by standard methods at each of the four networks and was sent to the Mammalian Genotyping Service in Marshfield, WI for genotyping (Center for Medical Genetics Web site). Screening Set 8 (372 highly polymorphic microsatellite markers) was used for all four networks. This screening set has an average heterozygosity of ∼80% and an average intermarker distance of 10 cM and covers ∼95% of the human genome.
Analytical Methods
The genome scans were performed separately for each of eight sampled populations—that is, for the GenNet white and black samples; the GENOA white, black, and Mexican American samples; the HyperGEN white and black samples, and the SAPPHIRe Asian sample—using the multipoint variance-component (VC) method implemented in the software GENEHUNTER 2 (Kruglyak et al. 1996; Kruglyak Lab Software Programs Web site). The VC method specifies the expected genetic covariances between relatives as a function of the estimated proportion of alleles shared IBD at a marker locus. The IBD probabilities were estimated using a multipoint approach that considers all available genotypes. The likelihood-ratio test was applied to test the null hypothesis of no additive genetic variance due to a quantitative-trait locus (QTL) at a particular location. Outliers for BMI were removed from each population before any analysis. Outliers were chosen as BMI observations that are not only far from the mean (e.g., >4 SD) but also separated from the nearest interior observation by ⩾1 SD. BMI was transformed to approximate normality by taking the inverse normal transformation, since it is known that VC methods can be sensitive to departures from normality in the trait distribution (Allison et al. 1999). We also performed the genome scan using BMI without data transformation, and the results were quite similar (data not shown). Sex, age, and age squared were incorporated as covariates in the analysis. To evaluate significance of linkage, we performed simulations for groups with LOD >3.3, the proposed genomewide significant level (Pratt et al. 2000). We retained phenotypes and pedigree structure in the data sets and randomly generated genotypes under the hypothesis of no linkage. The simulations were repeated 100 times to obtain empiric P values for genomewide significance.
To combine the linkage evidence across the eight data sets, we used Fisher’s omnibus procedure (Fisher 1932). This method is based on the observation that if n independent tests are made of the same hypothesis, resulting in P values P1, P2, …, Pn, then we can calculate a combined P value for all n tests by Σni=1(-2lnPi), which is asymptotically distributed as a χ2 distribution with 2n df. A modification of this method is needed to avoid bias in the pooling of nonparametric LOD scores from GENEHUNTER that are exactly zero (Province 2001). We used .0012 (LOD 2.0) as the suggested significance level for the Fisher test statistic. Since linkage results may differ by ethnic group, we also produced a meta-analysis scan separately for whites and for blacks.
We further combined eight individual studies by pooling average allele-sharing IBD (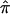 ) for each pair of individuals, estimated separately from each individual study, and performed genome scans using the original Haseman-Elston (HE) regression method (Haseman and Elston 1972) with the pooled
) for each pair of individuals, estimated separately from each individual study, and performed genome scans using the original Haseman-Elston (HE) regression method (Haseman and Elston 1972) with the pooled 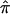 . Because the frequency of individual alleles can vary substantially between different ethnic groups, and the estimation of IBD sharing is sensitive to allele frequencies in the absence of parental genotype data, we first calculated
. Because the frequency of individual alleles can vary substantially between different ethnic groups, and the estimation of IBD sharing is sensitive to allele frequencies in the absence of parental genotype data, we first calculated 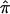 for each pair of individuals from each individual study separately and then pooled
for each pair of individuals from each individual study separately and then pooled 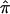 across the studies.
across the studies.
The probability of sharing i (i=0, 1, or 2) alleles IBD ( ,
, 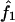 , or
, or  , respectively) for each of the sib pairs was estimated using GENEHUNTER2 (the “dump ibd” subroutine) across the genome in each of the eight studies separately. The proportion of alleles shared IBD for each pair of relatives was calculated as
, respectively) for each of the sib pairs was estimated using GENEHUNTER2 (the “dump ibd” subroutine) across the genome in each of the eight studies separately. The proportion of alleles shared IBD for each pair of relatives was calculated as  . BMI was regressed against sex, age, and age squared within the study to obtain a residual suitable for analysis. This residual was considered as the phenotype in our linkage analysis, and we analyzed only sib pairs with nonmissing phenotype and genotype data. We then pooled samples by race over the different networks (whites and blacks). Because only one Mexican American population was available and because we had already combined the Chinese and Japanese samples for the individual genome scan, there was no pooling for Mexican Americans and Asians. Assuming that the variance of residuals was solely due to the genetic effect and a random environmental effect, we performed an HE regression using the model
. BMI was regressed against sex, age, and age squared within the study to obtain a residual suitable for analysis. This residual was considered as the phenotype in our linkage analysis, and we analyzed only sib pairs with nonmissing phenotype and genotype data. We then pooled samples by race over the different networks (whites and blacks). Because only one Mexican American population was available and because we had already combined the Chinese and Japanese samples for the individual genome scan, there was no pooling for Mexican Americans and Asians. Assuming that the variance of residuals was solely due to the genetic effect and a random environmental effect, we performed an HE regression using the model 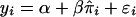 , where yi is the squared difference of the residuals of BMI adjusted for sex, age, and age squared for the ith sib pair, and
, where yi is the squared difference of the residuals of BMI adjusted for sex, age, and age squared for the ith sib pair, and 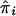 is an estimate of the marker locus IBD proportion. We further pooled whites, blacks, and Mexican Americans together. We did not include Asians in this final pooling exercise, because the mean and variance of the adjusted BMI were quite different from the other three groups and could add substantial heterogeneity to the analysis. To evaluate the significance of the combined IBD analysis, we permuted the age, age squared, and sex-adjusted phenotypes in the HE regression and recalculated the LOD scores through use of these permuted phenotypes. We did the permutation 100 times, to obtain the empiric genomewide P value.
is an estimate of the marker locus IBD proportion. We further pooled whites, blacks, and Mexican Americans together. We did not include Asians in this final pooling exercise, because the mean and variance of the adjusted BMI were quite different from the other three groups and could add substantial heterogeneity to the analysis. To evaluate the significance of the combined IBD analysis, we permuted the age, age squared, and sex-adjusted phenotypes in the HE regression and recalculated the LOD scores through use of these permuted phenotypes. We did the permutation 100 times, to obtain the empiric genomewide P value.
Results
Family Characteristics
The clinical characteristics of the 6,849 family members are presented in table 1 for each study. The mean BMI values for whites, blacks, and Mexican Americans were similar but higher than that for Asians. The proportion of participants with a BMI >30 (a common definition of obesity) was much higher among whites, blacks, and Mexican Americans than among Asians. The lower average BMI in the subjects in the SAPPHIRe network is the result of their primary recruitment in Taiwan, where obesity is less common than in the United Sates. The heritability of BMI was calculated by use of the VC method and was 0.43–0.63 in the eight studies (table 1).
Table 1.
Characteristics of Family Members for BMI Genome Scan from the FBPP Study
| Study and Population (N) | Age(years) | % Male | BMI | % BMI >30 | Heritability |
| GenNet: | |||||
| White (606) | 45.0 ± 14.1 | 47.2 | 29.2 ± 6.1 | 38.5 | .59 |
| Black (621) | 40.5 ± 11.7 | 40.5 | 30.2 ± 8.3 | 42.3 | .57 |
| GENOA: | |||||
| White (753) | 56.3 ± 9.8 | 47.3 | 30.0 ± 6.2 | 41.4 | .43 |
| Black (617) | 56.5 ± 9.4 | 34.5 | 30.4 ± 6.3 | 45.5 | .45 |
| Mexican American (788) | 55.8 ± 11.6 | 38.85 | 30.8 ± 6.1 | 50.4 | .63 |
| HyperGEN: | |||||
| White (1,138) | 60.8 ± 9.4 | 47.31 | 30.4 ± 6.0 | 43.4 | .56 |
| Black (1,256) | 50.7 ± 11.0 | 35.33 | 32.4 ± 7.6 | 56.9 | .62 |
| SAPPHIRe: | |||||
| Asian (1,070) | 54.0 ± 12.2 | 43.6 | 25.6 ± 3.8 | 9.9 | .57 |
Single Genomewide Screen
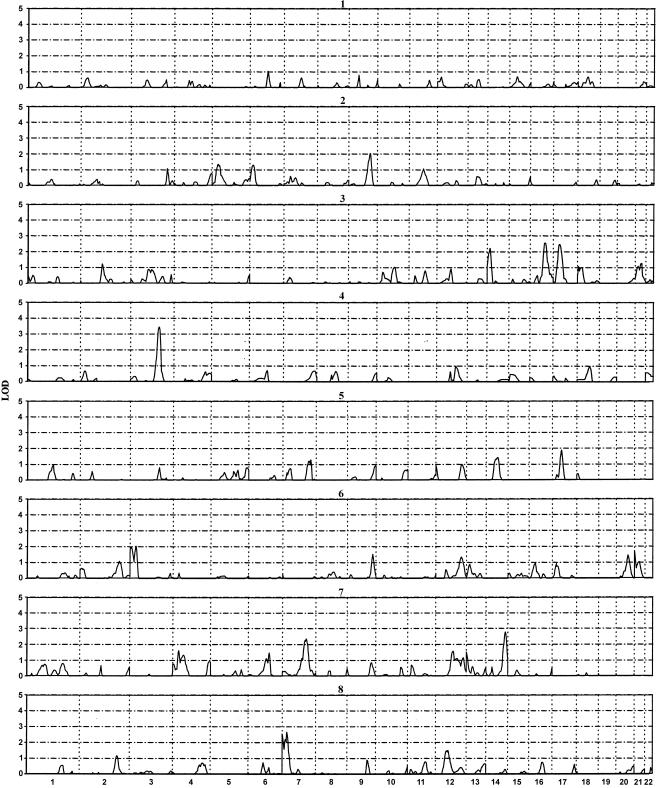
The results of the genome screen in the eight studies considered separately are presented in figure 1. All of the regions with a LOD >2.0 from multipoint VC analysis are listed in table 2. If a nearby region has a LOD >1.0 in other populations, we also listed these regions in table 2 as “replicates.” The analysis revealed one region with multipoint LOD scores >3.3, which is the approximate level of genomewide significance for this method of analysis at α=0.05 (Pratt et al. 2000). This region was centered at 3q22.1 (LOD 3.45 at marker D3S1764 in the GENOA black sample). In 100 simulated genomewide scans, we found a LOD >3.45 only once, which gave an empiric P value of .01. Six more regions were found with a LOD >2 (table 2) in at least one study. The regions are 3p24.1 (LOD 2.03, at marker D3S1259), 7p15.2 (LOD 2.66, at marker D7S3051), 7q22.3 (LOD 2.36, at marker D7S2847), 14q24.3 (LOD 2.15, at marker D14S617), 16q12.2 (LOD 2.55, at marker GATA67G11), and 17p11.2 (LOD 2.47, at marker D17S947).
Figure 1.
Multipoint VC LOD scores for linkage to BMI in each of the eight groups. 1 = GenNet white; 2 = GenNet black; 3 = GENOA white; 4 = GENOA black; 5 = GENOA Mexican American; 6 = HyperGEN white; 7 = HyperGEN black; and 8 = SAPPHIRe Asian. The X-axis is the chromosome location. Each chromosome is scaled according to its length from the genetic map and is separated from the others by vertical dotted lines.
Table 2.
Results of Linkage Analysis from Eight Genome Scans
| Chromosomeand Marker | Location(cM) | Region(cM) | LOD Score | Study (Population) |
| 3: | ||||
| D3S1259 | 31.17 | 0–37 | 2.03 | HyperGEN (white) |
| D3S1764 | 145.45 | 138–155 | 3.45 | GENOA (black) |
| 7: | ||||
| D7S3051 | 24.89 | 0–32 | 2.66 | SAPPHIRe (Asian) |
| D7S2847 | 124.39 | 108–137 | 2.36 | HyperGEN (black) |
| D7S1824 | 146.62 | 124–151 | 1.29a | GENOA (Mexican American) |
| 14: | ||||
| D14S617 | 100.71 | 88–112 | 2.15 | HyperGen (black) |
| 16: | ||||
| GATA67G11 | 78.71 | 66–88 | 2.55 | GENOA (white) |
| 17: | ||||
| D17S947 | 31.33 | 23–48 | 2.47 | GENOA (white) |
| D17S2196 | 47.66 | 36–57 | 1.87a | GENOA (Mexican American) |
Replicate at the same region with LOD >2.
Combined Analyses
The genome scan results pooling the studies with Fisher’s method are presented in figure 2. We first combined the P values for the three white populations and the three black populations separately. Two regions showed suggestive linkage, with LOD >2, in whites: 16q12.2 (LOD 2.15, at marker GATA67G11) and 17 (LOD 2.18, at marker D17S947). Next we combined P values across all eight studies; however, no region showed evidence of linkage with LOD >2. The highest LOD score is around marker D3S1764 (151.3 cM, with LOD 1.58). This is the only region showing strong linkage with BMI in individual studies.
Figure 2.
Results from combining P values by means of the modified Fisher’s method. 1 = combined white; 2 = combined black; and 3 = all groups combined. Scaling and separation of each chromosome are as described in figure 1.
The genome scan result for the combined IBD analysis is presented in figure 3. Table 3 shows the number of sib pairs selected in the analysis. Although no region showed evidence of linkage with LOD >2 in either the black or white samples, both show some evidence of linkage at the same location on chromosome 3 (186.97 cM, at marker D3S2427, with LOD 1.90 in whites vs. LOD 1.88 in blacks). When we combined white, black, and Mexican American populations, we obtained a LOD of 3.40 at the same region (186.97 cM, at marker D3S2427). In 100 permutated genome scans, we found a maximum multipoint LOD >3.40 only three times, which gave a genomewide empiric P value of .03.
Figure 3.
Results from combining IBD-sharing method. 1 = combined white; 2 = combined black; 3 = combined white, black, and Mexican American. Scaling and separation of each chromosome are as described in figure 1.
Table 3.
Number of Sib Pairs Selected in the Combined IBD Analysis
| Population and Study | No. of Sib Pairs |
| White: | |
| GenNet | 384 |
| GENOA | 924 |
| HyperGEN | 1,055 |
| Black: | |
| GenNet | 297 |
| GENOA | 893 |
| HyperGEN | 800 |
| Mexican-American: | |
| GENOA | 468 |
Discussion
In the present study, strong linkage evidence was found for BMI on chromosome 3 (3q27, with LOD 3.40 at marker D3S2427; P=.03), through use of a combined IBD analysis. At least two other analyses support linkage to this region. In a genome scan involving 507 nuclear white families, Kissebah et al. (2000) reported that a QTL on chromosome 3 (3q27) was strongly linked to six traits representing the fundamental phenotypes of metabolic syndrome, including BMI and waist and hip circumferences. The 1-LOD interval for BMI (183–200 cM) from the Kissebah et al. (2000) study is very similar to our result (180–193 cM). Vionnet et al. (2000) also report significant linkage with diabetes or glucose intolerance at age <45 years at the same region in French whites. It is known that obesity is correlated with diabetes, and it is possible that the same variants have an effect on both obesity and diabetes. Several potential candidate genes have been identified around this region. Among these genes, the adiponectin gene (MIM 605441) (at 3q27) seems the most interesting. Adiponectin is the most abundant gene transcript specific to adipose tissue (Maeda et al. 1996). It encodes a secreted protein that circulates in serum of normal individuals. Although the precise function of the adiponectin in energy expenditure and fat partitioning remains unclear, it is known that its circulating level is inversely correlated with BMI (Arita et al. 1999), and its mRNA level is suppressed in the adipose tissue of obese animals and humans (Hu et al. 1996). Kissebah further suggested that the gene for one of glucose transporters (GLUT2 [MIM 138160], at 3q26-q27) might be relevant, at least for the metabolic syndrome (Kissebah et al. 2000). Another candidate gene in this region is apolipoprotein D (Apo-D [MIM 164160]) (at 3q26). Apo-D is a component of high-density lipoprotein and is closely associated with the enzyme lecithin:cholesterol acyltransferase, an enzyme involved in lipoprotein metabolism. In an association study, Vijayaraghavan et al. (1994) found that a marker in this gene was more common in persons with obesity (P=.006).
The only significant linkage in a single genome scan was found at 3q22.1, with a LOD of 3.45 (P=.01) in GENOA blacks. However, this region is ∼30 cM away from the 3q27 region mentioned above, and it is hard to tell whether the two QTLs are the same. Further replication will be required to establish linkage with this region.
Positive evidence for linkage and association with obesity has been reported elsewhere for the leptin gene (MIM 164160) on chromosome 7 (Clement et al. 1996; Duggirala et al. 1996; Reed et al. 1996; Lapsys et al. 1997; Roth et al. 1997). Although we did not show strong evidence for linkage in our pooled analysis, we did find some evidence of linkage in this region in the HyperGEN black (LOD 2.36, at marker D7S2847) and GENOA Mexican American (LOD 1.29, at marker D7S1824) samples.
Kissebah et al. (2000) recently published strong linkage evidence for leptin level, on chromosome 17 (17p11) (LOD 4.97, at marker D17S947; 31–45 cM). Two of our samples show some evidence of linkage for BMI in the same region—namely, the GENOA whites (LOD 2.47, at marker D17S947; 23–48 cM) and GENOA Mexican Americans (LOD 1.87, at marker D17S2196; 36–57 cM). The candidate genes in this region include solute carrier family 4 of the insulin-specific facilitated glucose transporter (GLUT4 [MIM 138190]) and the receptor protein known to bind to globular “heads” of the complement C1q (gC1qR).
Another interesting region emerged at 7p15.2, where “suggestive” evidence for linkage was seen in Asians (LOD 2.66, at marker D7S3051) (table 2). An obvious candidate in this region is neuropeptide Y (NPY [MIM 162640]). NPY is one of the two critical neurochemicals that have been shown to influence feeding behavior. Intracerebroventricular infusion of NPY into the brains of rats results in insatiable eating and weight gain (Morley 1987; Beck et al. 1992). Bray et al. (1999) showed evidence of linkage of NPY with body weight (P=.020) and a composite measure of body mass and size (P=.048) in a Mexican American population.
The genome scans for human complex traits published to date are mostly based on relatively small sample sizes, and it is almost certain that some true linkage will be missed in a single genome scan study in which each gene has a moderate effect. This has recently been shown in a simulation study by Hirschhorn et al. (2001). Thus, replication of some of these findings across multiple studies is critical to identify the true linkage. In obesity genome scan studies, we are beginning to see some of these replications, as was recently reviewed by Comuzzie et al. (2001); this development is particularly encouraging. Four regions (on chromosomes 3, 7, and 17) in the present study have been associated with significant linkage to obesity in publications before, which extends these positive developments.
An obvious step to increase power in linkage analysis of complex traits is to pool data across studies (Lander and Kruglyak 1995; Allison and Heo 1998; Palmer et al. 2001). In a recent review article, Altmuller et al. (2001) compared 31 whole-genome scans for different human complex diseases, with regard to design, methods, and results. They found that the most obvious differences influencing success in finding linkage across studies was sample size. Two methods were used in the present study to combine all eight studies from four networks: combining P value and combining  . Fisher’s method for combining P value is very general and easy to implement; however, highly significant P values from a single study can determine the significance of the Fisher test statistic (Province 2001). Guerra et al. (1999) showed that pooling raw data usually has greater power and results in fewer false-positive results than does combining P values. By combining the original IBD-sharing estimates from all eight studies, we detected a QTL for BMI on chromosome 3 with significant support, although the evidence was weak in any of the individual studies.
. Fisher’s method for combining P value is very general and easy to implement; however, highly significant P values from a single study can determine the significance of the Fisher test statistic (Province 2001). Guerra et al. (1999) showed that pooling raw data usually has greater power and results in fewer false-positive results than does combining P values. By combining the original IBD-sharing estimates from all eight studies, we detected a QTL for BMI on chromosome 3 with significant support, although the evidence was weak in any of the individual studies.
We did not perform ascertainment correction when we applied the VC method, even though the families in the FBPP study were ascertained through hypertension and it is known that there is correlation between hypertension and obesity. This was guided by several considerations. First, on the basis of the simulation study by de Andrade and Amos (2000), the power to detect a major locus and the mean likelihood-ratio test were similar whether the data were corrected for ascertainment or not, although there is some excess of false-positive findings when there is a large genetic background. Their finding is consistent with that of Allison et al. (1999). Second, it is not clear which kind of ascertainment would be the most efficient in our case. When an inefficient ascertainment correction is used, it can decrease power (Comuzzie and Williams 1999).
In summary, by combining the data from eight studies, we found strong evidence for a QTL influencing BMI, in FBPP data, on chromosome 3 (3q27). The same region has been shown to be linked to several traits related to adiposity and diabetes, with highly significant statistical support in at least two other studies. We also replicated the findings on chromosomes 3, 7, and 17 in some of the individual genome scans. Our study shows that by combining different data sets—and, hence, increasing the sample size—we can improve the power to detect genes affecting human complex traits with moderate heritability, such as BMI.
Acknowledgments
We thank Dr. D. C. Rao, of Washington University, for reviewing the manuscript. This work was supported by National Heart, Lung, and Blood Institute grants UO1 HL54485, HL54466, HL65702, HL54481, HL45508, HL47910, HL51021, HL54464, HL54473, HL54496, HL54472, HL54515, HL54495, HL54471, HL54509, 2HHZ598, and HL65702. Genotyping was conducted by the NHLBI Mammalian Genotyping Service, Marshfield, WI.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for genetic maps)
- Kruglyak Lab Software Programs, http://www.fhcrc.org/labs/kruglyak/Downloads/ (for GENEHUNTER)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for adiponectin [MIM 605441], GLUT2 [MIM 138160], Apo-D [MIM 107740], leptin [MIM 164160], GLUT4 [MIM 138190], and NPY [MIM 162640])
References
- Allison DB, Heo M (1998) Meta-analysis of linkage data under worst-case conditions: a demonstration using the human OB region. Genetics 148:859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci–mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmüller J, Palmer LJ, Fischer G, Scherb H, Wjst M (2001) Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 69:936–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y, Kihara S. Ouchi N, Takahashi M, Maeda K, Miyagawa J-I, Hotta K, Shimomura I, Nakamura T, Miyaoka K, Kuriyama H, Nishida M, Yamashita S, Okubo K, Matsubara K, Muraguchi M, Ohmoto Y, Funahashi T, Matsuzawa Y (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun 257:79–83 [DOI] [PubMed] [Google Scholar]
- Barsh GS, Farooqi IS, O’Rahilly S (2000) Genetics of body-weight regulation. Nature 404:644–651 [DOI] [PubMed] [Google Scholar]
- Beck B, Stricker-Krongrad A, Nicholas JP, Burlet C (1992) Chronic and continuous intracerebroventricular infusion of neuropeptide Y in Long-Evans rats mimics the feeding of obese Zucker rats. Int J Obes Relat Metab Disord 16:295–302 [PubMed] [Google Scholar]
- Borecki IB, Blangero J, Rice T, Pérusse L, Bouchard C, Rao DC (1998a) Evidence for at least two major loci influencing human fatness. Am J Hum Genet 63:831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borecki IB, Higgins M, Schreiner PJ, Arnett DK, Mayer-Davis E, Hunt SC, Province MA (1998b) Evidence for multiple determinants of the body mass index: the National Heart, Lung, and Blood Institute Family Heart Study. Obes Res 6:107–114 [DOI] [PubMed] [Google Scholar]
- Borecki IB, Rice T, Bouchard C, Rao DC (1991) Commingling analysis of generalized body mass and composition measures: the Quebec Family Study. Int J Obes 15:763–773 [PubMed] [Google Scholar]
- Bouchard C, Perusse L, Leblanc C, Tremblay A, Theriault G (1988) Inheritance of the amount and distribution of human body fat. Int J Obes 12:205–215 [PubMed] [Google Scholar]
- Bouchard C, Perusse L, Rice T, Rao DC (1998) The genetics of human obesity. In: Bray GA, Bouchard C, James WPT (eds) Handbook of obesity. Marcel Dekker, New York, pp 157–190 [Google Scholar]
- Bray MS, Boerwinke E, Hanis CL (1999) Linkage analysis of candidate obesity genes among the Mexican-American population of Starr county, Texas. Genet Epidemiol 16:397–411 [DOI] [PubMed] [Google Scholar]
- Chagnon YC, Perusse L, Weisnagel SJ, Rankinen T, Bouchard C (2000) The human obesity gene map: the 1999 update. Obes Res 8:89–117 [DOI] [PubMed] [Google Scholar]
- Clement K, Garmer C, Hager J, Philippi A, LeDuc C, Carey A, Harris TJ, Jury C, Cardon LR, Basdevant A, Demenais F, Guy-Grand B, North M, Froguel P (1996) Indication for linkage of the human OB gene region with extreme obesity. Diabetes 45:687–690 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitchell BD, Mahaney MC, Dyer TD, Stern MP, MacCluer JW, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet 15:273–276 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Williams JT (1999) Correcting for ascertainment bias in the COGA data set. Genet Epidemiol 17 Suppl 1:S109–S114 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Williams JT, Martin LJ, Blangero J (2001) Searching for genes underlying normal variation in human adiposity. J Mol Med 79:57–70 [DOI] [PubMed] [Google Scholar]
- de Andrade M and Amos CI (2000) Ascertainment issues in variance components models. Genet Epidemiol 19:333–344 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Stern MP, Mitchell BD, Reinhart LJ, Shipman PA, Uresandi OC, Chung WK, Leibel RL, Hales CN, O’Connel P, Blangero J (1996) Quantitative variation in obesity-related traits and insulin precursors linked to the OB gene region on human chromosome 7. Am J Hum Genet 59:694–703 [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1932) Statistical methods for research workers. 4th ed. Okiver & Boyd, London [Google Scholar]
- Gu C, Province M, Todorov A, Rao DC (1998) Meta-analysis methodology for combining non-parametric sibpair linkage results: genetic homogeneity and identical markers. Genet Epidemiol 15:609–626 [DOI] [PubMed] [Google Scholar]
- Guerra R, Etzel CJ, Goldstein DR, Sain SR (1999) Meta-analysis by combining P values: simulated linkage studies. Genet Epidemiol 17 Suppl 1:S605–S609 [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Franck S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nature Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Hasstedt SJ, Hoffman M, Leppert MF, Elbein SC (1997) Recessive inheritance of obesity in familial non-insulin-dependent diabetes mellitus, and lack of linkage to nine candidate genes. Am J Hum Genet 61:668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lindgren CM, Daly MJ, Kirby A, Schaffner SF, Burtt NP, Altschuler D, Parker A, Rioux JD, Platko J, Gaudet D, Hudson TJ, Groop, Lander ES (2001) Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am J Hum Genet 69:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E, Liang P, Spiegelman BM (1996) AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem 271:10697–10703 [DOI] [PubMed] [Google Scholar]
- Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Wber J, Martin L, Blangero J, Comuzzie AG (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA 97:14478–14483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lapsys NM, Furler SM, Moore KR, Nguyen TV, Herzog H, Howard G, Samaras K, Carey DG, Morrison NA, Eisman JA, Chisholm DJ (1997) Relationship of a novel polymorphic marker near the human obese (OB) gene to fat mass in healthy women. Obes Res 5:430–433 [DOI] [PubMed] [Google Scholar]
- Lecomte E, Herbeth B, Nicaud V, Rakotovao R, Artur Y, Tiret L (1997) Segregation analysis of fat mass and fat-free mass with age- and sex-dependent effects: the Stanislas Family Study. Genet Epidemiol 14:51–62 [DOI] [PubMed] [Google Scholar]
- Lee JH, Reed DR, Li W-D, Xu W, Joo E-J, Kilker RL, Nanthakumar E, North M, Sakul H, Bell C, Price RA (1999) Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet 64:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Rao DC (1996) Random effects model for meta-analysis of multiple quantitative sibpair linkage studies. Genet Epidemiol 13:377–383 [DOI] [PubMed] [Google Scholar]
- Lonjou C, Collins A, Ennis S, Tapper W and Morton NE (1999) Meta-analysis and retrospective collaboration: two methods to map oligo-genes for atopy and asthma. Clin Exp Allergy 29 Suppl 4:57–59 [PubMed] [Google Scholar]
- Luke A, Guo X, Zhu X, Rotimi C, Adeyemo A, Wilks R, Forrester T, Lowe W, Comuzzie A, Martin LJ, Cooper R (2001) Heritability of obesity-related traits among Nigerians, Jamaicans and US black people. Int J Obes Relat Metab Disord 25:1034–1041 [DOI] [PubMed] [Google Scholar]
- Maeda K, Okubo K, Shimomura I, Funahsahi T, Matsuzawa Y, Matsubara K (1996) cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1). Biochem Biophys Res Commun 221:286–289 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Comuzzie AG, Laura A, Blangero J, MacCluer JW, Hixson JE (1999) A quantitative trait locus influencing BMI maps to the region of the β-3 adrenergic receptor. Diabetes 48:1863–1867 [DOI] [PubMed] [Google Scholar]
- Morley JE (1987) Neuropeptide regulation of appetite and weight. Endocr Rev 8:256–287 [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhman M, Oksanen L, Kaprio J, Koskenvuo M, Mustajoki P, Rissanen A, Salmi J, Kontula K, Peltonen L (2000) Genome-wide scan of obesity in Finnish sibpairs reveals linkage to chromosome Xq24. J Clin Endocrinol Metab 85:3183–3190 [DOI] [PubMed] [Google Scholar]
- Palmer LJ, Barnes KC, Burton PR, Chen H, Cookson WOCM, Collaborative Study on the Genetics of Asthma, Deichmann KA, Elston RC, Holloway JW, Jacobs KB, Laitinen T, Wjst M (2001) Meta-analysis for linkage to asthma and atopy in the chromosome 5q31-33 candidate region. Hum Mol Genet 10:891–899 [DOI] [PubMed] [Google Scholar]
- Perola M, Öhman M, Hiekkalinna T, Leppävuori J, Pajukanta P, Wessman M, Koskenvuo M, Palotie A, Lange K, Kaprio J, Peltonen L (2001) Quantitative-trait-locus analysis of body-mass index and of stature, by combined analysis of genome scans of five Finnish study groups. Am J Hum Genet 69:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RA, Ness R, Laskarzewski P (1990) Common major gene inheritance of extreme overweight. Hum Biol 62:747–765 [PubMed] [Google Scholar]
- Province MA (2001) The significance of not finding a gene. Am J Hum Genet 69:660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Ding Y, Xu W, Cather C, Green ED, Price RA (1996) Extreme obesity may be linked to markers flanking the human OB gene. Diabetes 45:691–694 [DOI] [PubMed] [Google Scholar]
- Roth H, Hinney A, Ziegler A, Barth N, Gerber G, Stein K, Bromel T, Mayer H, Siegfried W, Schafer H, Remschmidt H, Grzeschik KH, Hebebrand J (1997) Further support for linkage of extreme obesity to the obese gene in a study group of obese children and adolescents. Exp Clin Endocrinol Diabetes 105:341–344 [DOI] [PubMed] [Google Scholar]
- Spadano MA, Coakley EH Field AE, Colditz G, Dietz WH (1999) The disease burden associated with overweight and obesity. JAMA 282:1523–1529 [DOI] [PubMed] [Google Scholar]
- Van der Kallen CJH, Cantor RM, van Greevenbroek MMJ, Geurts JMW, Bouwman FG, Aouixerat BE, Allayee H, Buurman WA, Lusis AJ, Rotter JI, de Bruin TWA (2000) Genome scan for adiposity in Dutch dyslipidemic families reveals novel quantitative trait loci for leptin, body mass index and soluble tumor necrosis factor receptor super family 1A. Int J Obes Relat Metab Disord 24:1381–1391 [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan S, Hitman GA, Kopelman PG (1994) Apolipoprotein D polymorphism: a genetic marker of obesity and hyperinsulinemia. J Clin Endocrinol Metab 79:568–570 [DOI] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Leprêtre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P (2000) Genomewide search for type 2 diabetes–susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2–diabetes locus on chromosome 1q21-q24. Am J Hum Genet 67:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder K, Hanson RL, Kobes S, Knowler WC, Ravussin E (2000) An autosomal genomic scan for loci linked to plasma leptin concentration in Pima Indians. Int J Obes Relat Metab Disord 24:559–565 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL, et al (2000) The Finland-United States investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. II. An autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–200 [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Cooper RS, Luke A, Chen G, Wu X, Chakravati A, Weder AB (2002) A genome-wide scan for obesity in African Americans. Diabetes 51:541–544 [DOI] [PubMed] [Google Scholar]