How important are interactions among mutations for adaptation? Obviously, no gene functions in isolation, but it is possible that assuming that mutations have independent effects could still give a good prediction for how adaptation proceeds. In PNAS, Nahum et al. (1) use an elegant combination of simulations and experiments with Escherichia coli to show that even in adaptation over the course of a few weeks involving only a handful of mutations, interactions among those mutations can have very large effects. The authors detect these effects using a clever indirect method based on the effect of spatial mixing on the evolution of their experimental populations.
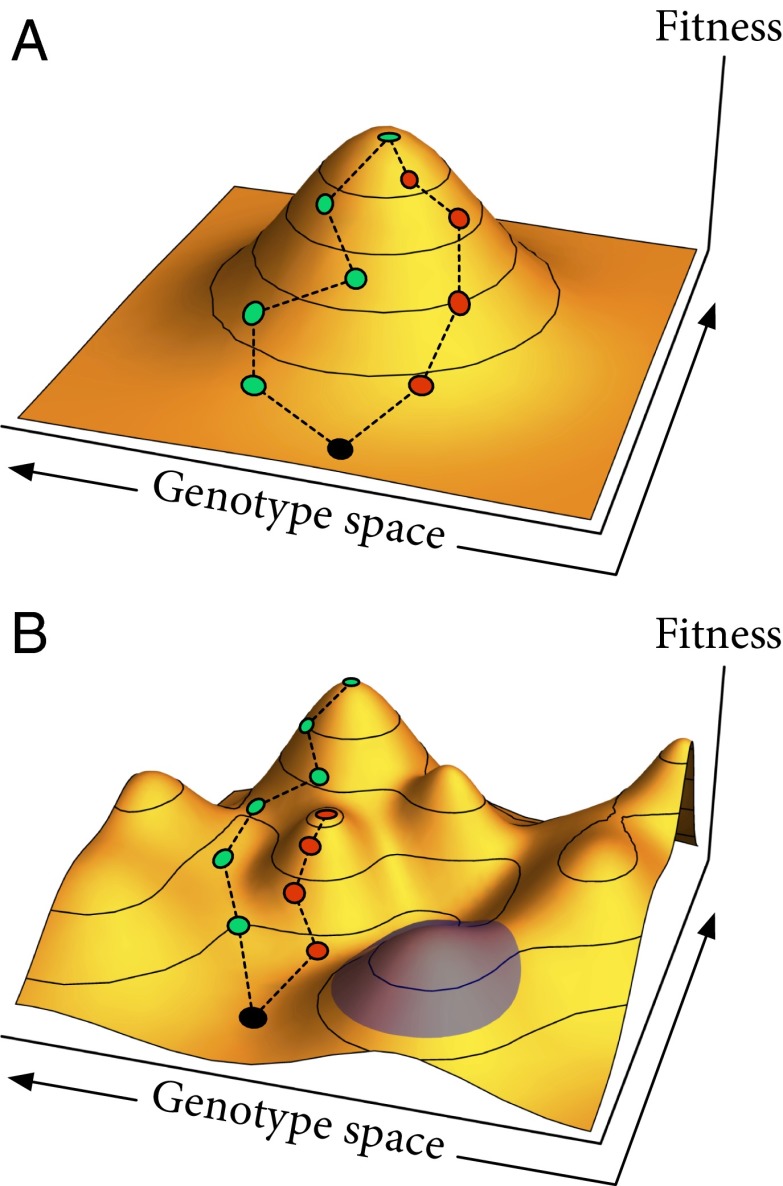
Interactions among mutations are typically visualized using the “fitness landscape” metaphor introduced by Sewall Wright (2), which analogizes contours of organismal fitness with the contours of physical topography. Most commonly, the horizontal axes of the landscape represent the space of possible genotypes with the height of the landscape representing the fitness of the corresponding genotype (3, 4) (Fig. 1). The ruggedness of the fitness landscape is a measure of the prevalence of fitness interactions among genes: in smooth landscapes with a single peak, each mutation has a fixed effect on fitness (Fig. 1A), whereas in rugged landscapes with multiple peaks (Fig. 1B), the effect of each mutation can depend on the other mutations an individual has.
Fig. 1.
Fitness landscapes. The horizontal axes represent the space of different combinations of genotypes, and the vertical axis is individual fitness as a function of genotype. (A) Smooth fitness landscape with a single peak and no fitness interactions among genes. Different evolutionary trajectories lead to the same peak. (B) Rugged fitness landscape with multiple peaks and pervasive interactions among genes. Different evolutionary trajectories can reach different peaks even from the same initial genotype. The blue highlighted region shows that each peak in the rugged landscape still looks like a smooth, single-peaked landscape locally.
On any fitness landscape, adaptation starts with beneficial genotypes arising and proceeds as these types increase in frequency and take over the population. The first step, generating beneficial types, requires genetic diversity, but the second step tends to reduce that diversity as the population converges on the fittest existing type. There is thus a potential trade-off between exploration and exploitation, with populations actually adapting slower if the best immediate mutations spread too quickly and wipe out others that lead in more productive directions. On smooth landscapes, however, there is no trade-off because the best single mutations lead in the best directions (Fig. 1A). In this case, because the best new genotype is always near the best current genotype, the population should focus its exploration in this region and there is no point in maintaining diversity elsewhere. Such smooth landscapes may seem unlikely, but precisely this pattern has been found in some experimental yeast populations (5).
In contrast, there is accumulating evidence from experiments in E. coli (6–8) and other microorganisms (reviewed in ref. 9) that genetic interactions and rugged fitness landscapes are common. Wright was an early proponent of the importance of rugged fitness landscapes and proposed in his “shifting balance theory” (SBT) (2, 10) that spatial population structure is crucial for populations evolving in rugged landscapes from low peaks to higher peaks. Although the SBT was influential among biologists involved in the “modern evolutionary synthesis” in the first half of the 20th century (11), it later was criticized in an influential review (12) for having important conceptual problems.
A crucial observation of Nahum et al. (1) and others (5) is that the advantage of population structure for adaptation on rugged landscapes is both simpler and more general than Wright (2) had originally proposed in the SBT. Population structure has the more general effect of slowing down the spread of beneficial mutations, temporarily shielding genetic variation. This extra variation allows for a broader search of genotype space, which may be important for adaptation on rugged fitness landscapes. Crucially, this general effect is largely free from the issues that plague the SBT.
Nahum et al. (1) confirm that population structure can allow a broader search of rugged landscapes by using the classic NK model of Kauffman and Levin (3). The NK model uses a parameter K to measure landscape ruggedness, where larger values of K imply more ruggedness. Nahum et al. (1) evolve populations with either unrestricted migration (weak population structure) or restricted migration (strong population structure). When fitness landscapes are smooth, they find that both the weakly and strongly structured populations evolve the same final fitness, but the weakly structured populations adapt faster. When landscapes are rugged, the weakly structured populations initially adapt faster but are overtaken by the strongly structured populations, which reach a higher final fitness (figure 2 of ref. 1). This pattern holds for increasing levels of fitness landscape ruggedness K. The simulations also showed that the strongly structured populations accumulated more mutations when the fitness landscape was rugged.
To test whether the theoretical results might be relevant to real organisms, Nahum et al. (1) conducted evolution experiments with E. coli. They created replicate populations starting from the same ancestral strain and let them adapt to the same environment with weak and strong population structure treatments that paralleled the simulations. The experimental results matched the pattern seen in the simulations of rugged landscapes. At first, the weakly structured populations adapted faster, as beneficial mutations rapidly swept through. As time went on however, the strongly structured populations caught up and then overtook the weakly structured populations as their increased diversity allowed them to find the best combinations of mutations (which apparently involved mutations that were less beneficial individually). Whole-genome sequencing of clones from the final populations showed that the strongly structured populations had indeed acquired more mutations than the weakly structured ones.
These results stand in clear contrast to those of a similar experiment done in yeast by Kryazhimskiy et al. (5), which found that spatial structure always slowed adaptation, suggesting a smooth fitness landscape. That these two experiments gave opposite results raises the depressing prospect that each experiment’s results may be specific to the particular species and phenotype studied, with only a limited ability to extrapolate. However, Nahum et al. (1) suggest a more encouraging possibility: they note that near a fitness peak, all landscapes look smooth regardless of their overall roughness (Fig. 1B), so we might expect that experiments starting from better-adapted organisms would tend to find smoother landscapes. And indeed, although Kryazhimskiy et al. (5) started from a fairly well-adapted ancestor whose fitness increased by no more than 10% over 500 generations, Nahum et al. (1) deliberately created an ancestor carrying costly mutations and saw fitness gains of more than 70% in fewer than 200 generations, suggesting that they started much farther from a fitness peak. Thus, we might generally expect that the roughness of the fitness landscape will be important for large but not small increases in fitness.
Nahum et al.’s (1) results emphasize how understanding and predicting adaptation requires understanding something about the structure of fitness landscapes. Currently, however, we know very little about the shape
Nahum et al.'s results emphasize how understanding and predicting adaptation requires understanding something about the structure of fitness landscapes.
of these landscapes. A number of studies have addressed this problem by exhaustively measuring small regions of landscapes (e.g., refs. 6 and 13–15). This approach has yielded important insights, but it faces a fundamental limitation: because the full fitness landscape is exponentially large in the size of the genome, we can never hope to measure more than a tiny fraction of it. Nahum et al.’s (1) approach, inferring some aspects of the landscape from adaptive trajectories under different conditions, offers a possible solution to this problem by focusing on just the limited set of features that are relevant to adaptation. Encouragingly, an increasing number of experiments are being done along these lines (e.g., ref. 16), and we can hope that general patterns will begin to emerge.
Crucially, this work will need to be extended to natural populations, whose fitness landscapes may be quite different from those in the laboratory. Pathogens may be the best natural populations to consider first. There is a desperate need to predict how they will evolve, and we are beginning to have the data and theoretical tools to make such predictions (17, 18). In fact, one of the most important adaptations, drug resistance, often involves multiple interacting mutations (6, 19), so understanding natural fitness landscapes is essential.
Footnotes
The authors declare no conflict of interest.
See companion article on page 7530.
References
- 1.Nahum JR, et al. A tortoise–hare pattern seen in adapting structured and unstructured populations suggests a rugged fitness landscape in bacteria. Proc Natl Acad Sci USA. 2015;112:7530–7535. doi: 10.1073/pnas.1410631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. In: Jones DF, editor. Proceedings of the Sixth International Congress on Genetics. Vol 1. Brooklyn Botanic Garden; Brooklyn, NY: 1932. pp. 356–366. [Google Scholar]
- 3.Kauffman S, Levin S. Towards a general theory of adaptive walks on rugged landscapes. J Theor Biol. 1987;128(1):11–45. doi: 10.1016/s0022-5193(87)80029-2. [DOI] [PubMed] [Google Scholar]
- 4.Gavrilets S. Evolution and speciation on holey adaptive landscapes. Trends Ecol Evol. 1997;12(8):307–312. doi: 10.1016/S0169-5347(97)01098-7. [DOI] [PubMed] [Google Scholar]
- 5.Kryazhimskiy S, Rice DP, Desai MM. Population subdivision and adaptation in asexual populations of Saccharomyces cerevisiae. Evolution. 2012;66(6):1931–1941. doi: 10.1111/j.1558-5646.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 6.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 7.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332(6034):1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 8.Woods RJ, et al. Second-order selection for evolvability in a large Escherichia coli population. Science. 2011;331(6023):1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szendro IG, Schenk MF, Franke J, Krug J, de Visser JAGM. Quantitative analyses of empirical fitness landscapes. J Stat Mech Theory Exp. 2013;2013(01):P01005. [Google Scholar]
- 10.Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobzhansky T. Genetics and the Origin of Species. Columbia Univ Press; New York: 1937. [Google Scholar]
- 12.Coyne JA, Barton NH, Turelli M. Perspective: A critique of Sewall Wright’s shifting balance theory of evolution. Evolution. 1997;51(3):643–671. doi: 10.1111/j.1558-5646.1997.tb03650.x. [DOI] [PubMed] [Google Scholar]
- 13.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acevedo A, Brodsky L, Andino R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature. 2014;505(7485):686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bank C, Ewing GB, Ferrer-Admettla A, Foll M, Jensen JD. Thinking too positive? Revisiting current methods of population genetic selection inference. Trends Genet. 2014;30(12):540–546. doi: 10.1016/j.tig.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Levy SF, et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519(7542):181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507(7490):57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 18.Neher RA, Russell CA, Shraiman BI. Predicting evolution from the shape of genealogical trees. eLife. 2014;3:e03568. doi: 10.7554/eLife.03568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328(5983):1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]