Significance
Pathogens use virulence factors to inhibit key immune cell functions and would be expected to impair immune responses to infection. However, immune responses are still generated against infection, suggesting that the immune system has evolved mechanisms for overcoming pathogenic activity. Here, we demonstrate that cells infected with Legionella pneumophila synthesize IL-1 despite a pathogen-imposed block in host translation, but are unable to produce other critical cytokines. IL-1 signaling allows uninfected bystander cells to produce protective cytokines. Our data thus demonstrate a key role for communication between infected and uninfected bystander cells in overcoming pathogenic activities. This mechanism of immune activation has broad significance for our understanding of how successful immune responses are generated against pathogens.
Keywords: L. pneumophila, IL-1, IL-1R, TNF, type IV secretion system
Abstract
The innate immune system is critical for host defense against microbial pathogens, yet many pathogens express virulence factors that impair immune function. Here, we used the bacterial pathogen Legionella pneumophila to understand how the immune system successfully overcomes pathogen subversion mechanisms. L. pneumophila replicates within macrophages by using a type IV secretion system to translocate bacterial effectors into the host cell cytosol. As a consequence of effector delivery, host protein synthesis is blocked at several steps, including translation initiation and elongation. Despite this translation block, infected cells robustly produce proinflammatory cytokines, but the basis for this is poorly understood. By using a reporter system that specifically discriminates between infected and uninfected cells within a population, we demonstrate here that infected macrophages produced IL-1α and IL-1β, but were poor producers of IL-6, TNF, and IL-12, which are critical mediators of host protection. Uninfected bystander cells robustly produced IL-6, TNF, and IL-12, and this bystander response required IL-1 receptor (IL-1R) signaling during early pulmonary infection. Our data demonstrate functional heterogeneity in production of critical protective cytokines and suggest that collaboration between infected and uninfected cells enables the immune system to bypass pathogen-mediated translation inhibition to generate an effective immune response.
Initiation of innate immune responses to microbial pathogens involves the direct recognition of pathogen-associated molecular patterns (PAMPs) by membrane-bound and cytosolic pattern recognition receptors (PRRs) in infected cells (1, 2). However, virulence factors of many pathogens interfere with essential immune signaling processes, including NF-κB and MAPK signaling and host protein synthesis (3–5). Such virulence factors would be expected to limit cell-intrinsic immune activation of infected cells. The mechanisms that enable the host to successfully overcome pathogen subversion of host cell processes remain poorly understood.
The Gram-negative bacterium Legionella pneumophila encodes a specialized Dot/Icm (for defect in organelle trafficking/intracellular multiplication) type IV secretion system (T4SS) that delivers bacterial effector proteins into host cells to facilitate its intracellular survival and replication (6–8). A subset of effector proteins, Lgt1, Lgt2, Lgt3, SidI, SidL, Pkn5, and Lpg1489, blocks host protein synthesis, in part by disabling elongation factors (9–13). Furthermore, host translational initiation is suppressed during infection due to diminished mTOR signaling (14). These activities result in a greater than 90% decrease in host translation in infected cells (13, 15). Nevertheless, L. pneumophila infection leads to robust production of many key protective proinflammatory cytokines (12, 16–19). Moreover, the presence of the T4SS paradoxically enhances cytokine production, suggesting that much of the host response against L. pneumophila is mediated by cytosolic sensing of bacterial ligands and virulence activities (13, 16, 17, 20).
How the host is able to mount a proinflammatory cytokine response when L. pneumophila potently blocks host translation remains unclear. At the population level, decreased host protein synthesis leads to preferential translation of the most abundant cytokine transcripts (14). At the single cell level, infected cells selectively synthesize IL-1α and IL-1β through a mechanism involving MyD88-dependent translational bypass (21). However, whether mechanisms that enable selective translation of IL-1 also apply to other key cytokines and immune effector proteins has not been determined. Alternatively, as a significant fraction of cells present during infection both in vitro and in vivo remain uninfected bystander cells, we considered the possibility that these uninfected bystander cells might respond to the presence of infection to produce cytokines instead (22). Here, by tracking immune responses in L. pneumophila-infected cells at the single cell level, we demonstrate that although infected cells receiving T4SS effectors synthesize IL-1α and IL-1β, they are poor producers of other key inflammatory proteins. Instead, bystander cells that have not received T4SS effectors are the primary producers of TNF, IL-6, IL-12, and the costimulatory molecule CD86 during both in vitro and in vivo infection. Importantly, lack of IL-1R signaling leads to reduced bystander cytokine production and increased bacterial burden in vivo, suggesting that the IL-1 released from infected cells enables bystander cells to produce proinflammatory cytokines. Our findings indicate that release of IL-1 by infected cells signals the presence of virulent infection, enabling the host to generate a robust innate immune response despite a pathogen-imposed translation block.
Results
Uninjected Cells Produce Key Inflammatory Mediators During in Vitro Infection.
Infection with virulent L. pneumophila expressing a T4SS leads to an enhanced cytokine response despite bacterial inhibition of host translation. How this cytokine response is generated remains unclear. It is possible that directly infected macrophages possess cell-intrinsic mechanisms that enable selective translation of cytokines. Alternatively, cytokines may be produced by bystander cells that are uninfected or have taken up bacteria that failed to translocate effectors (22). To determine whether T4SS-injected cells or uninjected bystander cells produce cytokines, we used a fluorescence-based system that detects the translocated L. pneumophila effector (RalF) fused to β-lactamase (BlaM) (22, 23). In the absence of BlaM activity, 409-nm excitation of the host cell-permeable BlaM fluorescent substrate CCF4-AM results in emission of green fluorescence at 518 nm. However, T4SS-translocated BlaM–RalF results in cleavage of CCF4-AM and a shift in emission to blue fluorescence at 447 nm. This system enables robust discrimination of infected and uninfected cells within tissues in vivo or in cultured cells in vitro (22).
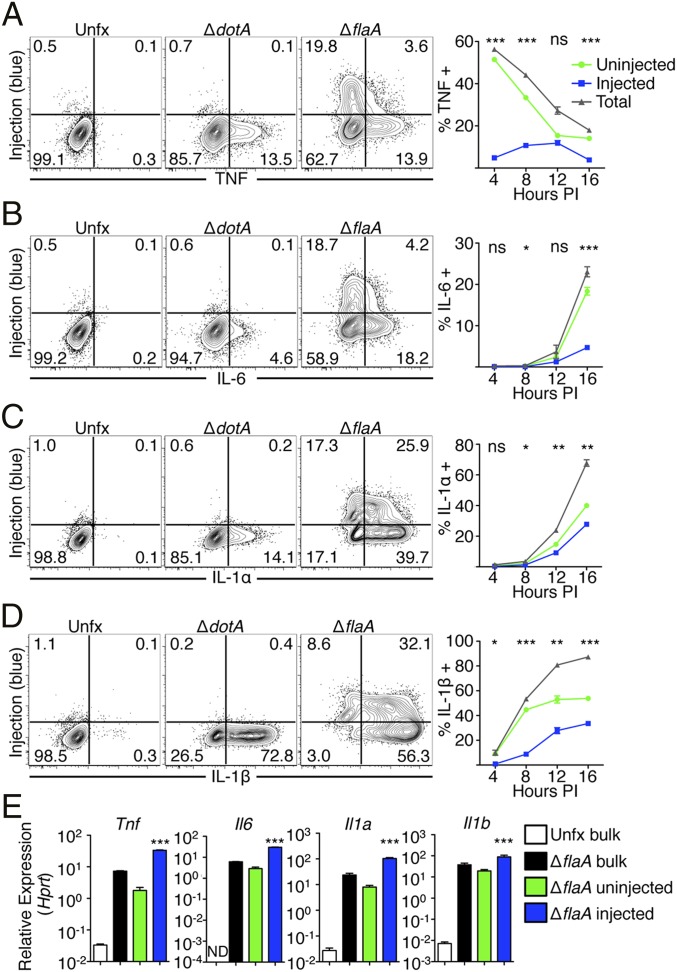
We infected bone marrow-derived macrophages (BMDMs) with L. pneumophila encoding the BlaM–RalF reporter. As flagellin delivered by the T4SS into the host cell cytosol induces rapid cell death via NAIP5 inflammasome activation, we used flagellin-deficient L. pneumophila (ΔflaA) for these studies. ΔflaA L. pneumophila evade NAIP5 detection and replicate in C57BL/6 macrophages and mice (24–26), but still induce NLRP3 and caspase-11 inflammasome activation and IL-1 family cytokine secretion (16, 19, 27). Following infection, BMDMs were loaded with CCF4-AM and intracellularly stained for various cytokines at multiple timepoints postinfection. (Fig. 1 and Fig. S1). BMDMs infected with avirulent L. pneumophila lacking the Dot/Icm T4SS (ΔdotA) demonstrated no BlaM–RalF activity as expected. During ΔflaA infection, we found that TNF is rapidly produced, but T4SS-injected cells poorly produced TNF at all timepoints assayed (Fig. 1A). Instead, uninjected cells were the primary producers of TNF. Likewise, IL-6 was poorly produced by T4SS-injected cells and instead was primarily produced by uninjected cells (Fig. 1B). Similar findings were obtained for IL-12 and the costimulatory protein CD86 (Fig. S1 A and B). In contrast, IL-1α and IL-1β were robustly produced by both T4SS-injected cells and uninjected cells (Fig. 1 C and D), indicating that T4SS-injected cells selectively synthesize IL-1α and IL-1β despite the host translation block, consistent with recent findings (21). To determine whether the decreased cytokine production observed in T4SS-injected cells is due to decreased translation or transcription, we measured cytokine mRNA levels in sorted populations of T4SS-injected and uninjected cells (Fig. 1E). Both injected and uninjected cells displayed marked increases in Il1a, Il1b, Tnf, and Il6 transcript levels relative to uninfected cells, with injected cells exhibiting significantly greater increases in cytokine transcript levels than uninjected cells, consistent with recent findings (21). Thus, the decreased production of immune proteins by injected cells is not due to a lack of transcriptional activation and most likely is due to impaired translation.
Fig. 1.
T4SS-injected and uninjected macrophages produce different cytokines in vitro. (A–D) Bone marrow-derived macrophages (BMDMs) were infected with Lp02 strains of L. pneumophila for 4, 8, 12, or 16 h. BMDMs were treated with brefeldin A and monensin, loaded with CCF4-AM, fixed, and stained with antibodies against TNF (A), IL-6 (B), IL-1α (C), and IL-1β (D). Plots show cytokine production at 16 h postinfection (PI). Line graphs show ΔflaA-infected macrophages where total frequency of cytokine-producing cells as well as the contribution of injected and uninjected cells to cytokines are shown. NS, not significant, *P < 0.05, **P < 0.005, ***P < 0.0005 as determined by Student’s t test between uninjected and injected cells at various times PI. (E) BMDMs were infected with Lp02 ΔflaA L. pneumophila for 8 h. Cells were then loaded with CCF4-AM and sorted based upon cleavage of the dye or were collected after going through the sorter without separating the cells based on dye cleavage (bulk). Sorted cells were then lysed and relative abundance of transcripts for proinflammatory cytokines were assayed using RT-PCR. ***P < 0.0005 as determined by Tukey posttest between uninjected and injected samples. All graphs are n = 3 wells per condition per timepoint. All graphs show mean ± SEM and are representative of three independent experiments.
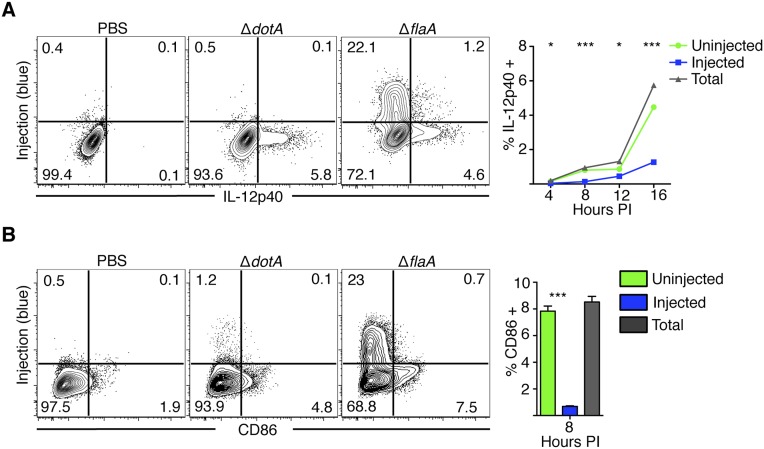
Fig. S1.
IL-12p40 and CD86 are produced by bystander cells during infection. (A) Bone marrow-derived macrophages (BMDMs) were infected with PBS or Lp02 ΔflaA or ΔdotA L. pneumophila at a multiplicity of infection, MOI = 5 for 4, 8, 12, or 16 h. BMDMs were treated with brefeldin A and monensin starting at 3 h before harvest. Cells were then loaded with CCF4-AM, fixed, and intracellularly stained with antibodies against IL-12p40. Plots show cytokine production at 16 h postinfection (PI). Line graph shows ΔflaA-infected macrophages where total = the total frequency of cells positive for the given cytokine. Injected and uninjected = the relative contribution of each subset of cells to the total frequency of cytokine positive cells. n = 3 wells per condition per timepoint. Line graphs show mean ± SEM. (B) Cells infected as in A were harvested 8 h PI, loaded with CCF4-AM, and stained for CD86. Bar graph shows mean ± SEM *P < 0.05,***P < 0.0005 as determined by Student’s t test between uninjected and injected cells at various times PI.
Seven L. pneumophila effectors are known to inhibit host translation (9, 12, 28). To determine if these effectors accounted for impaired cytokine production in injected cells, we infected BMDMs with Δ5 L. pneumophila lacking five of these effectors (Lgt1, Lgt2, Lgt3, SidI, and SidL) or the Δ7ΔflaA strain lacking all seven effectors (Δ5 additionally deleted for Pkn5 and Lpg1489) (12, 13). At 4 h postinfection, cells injected by the Δ5 or Δ7ΔflaA strains produced substantially more TNF than WT- or ΔflaA-injected cells, with an increase both in the frequency of TNF-producing cells and the TNF geometric mean fluorescence intensity (gMFI) (Fig. S2 A and D). IL-1α and IL-1β production was also increased in cells injected with the Δ5 or Δ7ΔflaA strains compared with controls (Fig. S2 G–I). By 16 h postinfection, however, there was no difference in the frequency of TNF-producing injected cells with either the Δ5 or Δ7ΔflaA strains compared with the parental strains, although the TNF gMFI was significantly increased in cells injected by the Δ7ΔflaA strain compared with the ΔflaA strain (Fig. S2 B and E). The frequency of IL-6–producing injected cells was similarly unaffected by the presence or absence of these effectors (Fig. S2 C and F). These data indicate that early in infection, these bacterial effectors suppress cytokine translation, whereas at later timepoints, additional host and/or bacterial mechanisms inhibit cytokine translation in injected cells. These findings would be consistent with previous findings showing that the Δ5 and Δ7ΔflaA strains still partially inhibit host protein synthesis, indicating that yet additional bacterial or host mechanisms suppress host translation. Taken together, these data suggest that although productively infected cells selectively translate IL-1α and IL-1β, they are unable to synthesize TNF, IL-6, IL-12, and CD86. Instead, bystander cells are the primary producers of these key inflammatory proteins.
Fig. S2.
L. pneumophila effectors that interfere with host translation also inhibit early cytokine production, but not late cytokine production, in injected cells. BMDMs were treated with LPS or infected with Lp02 WT, ΔdotA, ΔflaA, Δ5, or Δ7ΔflaA strains of L. pneumophila at an MOI = 5 for the indicated amount of time, loaded with CCF4-AM, and intracellularly stained for the indicated cytokines. Representative flow plots showing TNF production 4 (A) or 16 (B) h PI or IL-6 production at 16 h PI (C). The frequency of TNF-producing injected cells and the geometric MFI (gMFI) of TNF-producing injected cells are quantified for (D) 4 h and (E) 16 h for triplicate samples. The frequency of IL-6–producing injected cells at 16 h is quantified in F. (G) Representative flow plots showing IL-1α production 4 h PI. The frequency of IL-1α–producing cells (H) or IL-1β–producing cells (I) is quantified for 4 and 16 h from flow cytometric data for triplicate samples. Data shown are representative of two independent experiments. Graphs show mean ± SEM. ANOVA was performed and statistics show significance between Δ5 and WT or Δ7ΔflaA and ΔflaA as determined by Tukey posttest. *P < 0.05, **P < 0.005, ***P < 0.0005; NS, not significant.
Bystander Alveolar Macrophages and Neutrophils Produce TNF During L. pneumophila Infection.
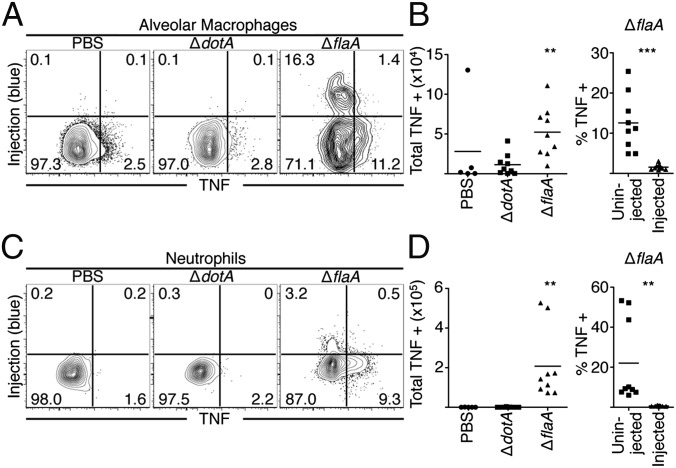
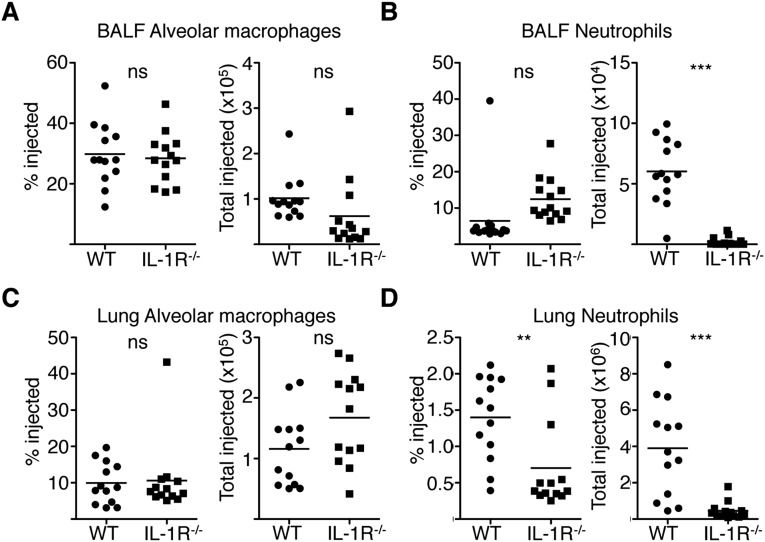
Our in vitro findings indicate that infected cells are unable to produce many key protective cytokines. Instead, these cytokines are primarily generated by bystander macrophages. In vivo, multiple cell populations exist, and distinct immune populations may possess alternative mechanisms for overcoming a pathogen-induced translation block. We therefore examined TNF production during pulmonary infection with ΔflaA or ΔdotA L. pneumophila. At 24 h postinfection, increased levels of bacteria are present during ΔflaA infection, whereas ΔdotA bacteria do not replicate. However, cytokine responses to ΔflaA bacteria do not require increased bacterial load or bacterial replication, suggesting that the cytokine response is primarily driven by cytosolic sensing of T4SS activity (16, 17). We first focused on airway-resident alveolar macrophages and recruited neutrophils, as they are the primary cell types that receive T4SS effectors during pulmonary infection (22). Following intranasal infection with ΔflaA L. pneumophila, there was a significant increase in both the percentage and total numbers of TNF-producing airway alveolar macrophages and neutrophils (Fig. 2), consistent with previous findings showing that cytosolic immune sensing of T4SS activity is required for maximal TNF production (16, 17). However, we observed that T4SS-injected alveolar macrophages and neutrophils did not produce TNF. Instead, TNF was produced almost exclusively by uninjected alveolar macrophages and neutrophils (Fig. 2).
Fig. 2.
Bystander alveolar macrophages and neutrophils produce TNF during L. pneumophila infection. Mice were infected and cells were harvested from the BAL fluid and lung tissue, treated with BFA, loaded with CCF4-AM, stained, and fixed and stained with an antibody against TNF. (A–D) Representative plots from alveolar macrophages (A) and neutrophils (C) collected from the BAL fluid 24 h PI and quantified in B and D. Graphs show individual mice and mean from three pooled independent experiments. Graphs on far right show the relative contribution of uninjected and injected cells to TNF production from ΔflaA-infected mice. n = 2–3 mice per group per experiment. Student’s t tests were performed for total TNF-producing cells between ΔdotA- and ΔflaA-infected mice. **P < 0.005, ***P < 0.0005.
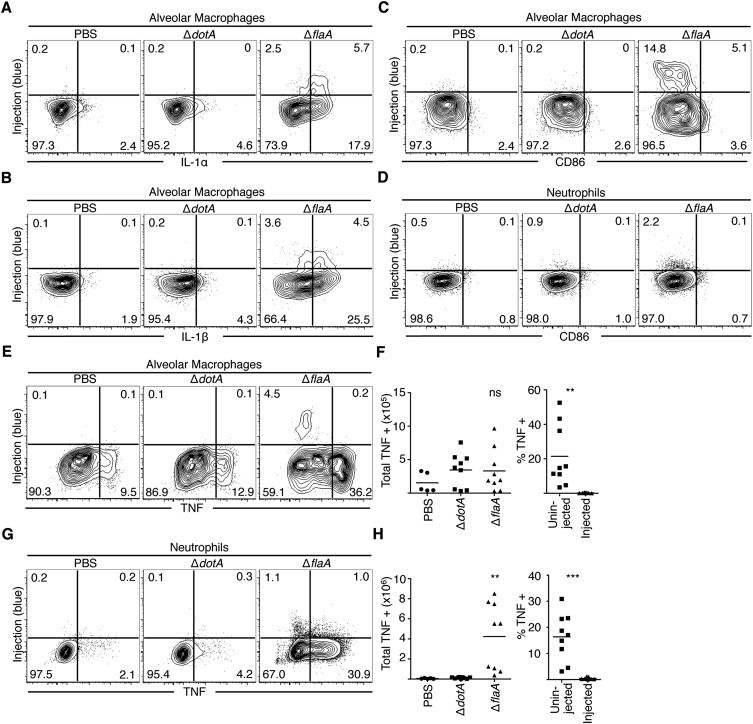
In contrast, both injected and uninjected alveolar macrophages produced IL-1α and IL-1β, in agreement with our in vitro findings (Fig. S3 A and B). PBS vehicle control and avirulent ΔdotA infection yielded no significant increase in TNF-producing alveolar macrophages or neutrophils (Fig. 2), corroborating previous findings that TNF is undetectable in the bronchoalveolar lavage (BAL) fluid or serum from similarly treated mice (16, 17). Similar results were obtained with alveolar macrophages and neutrophils isolated from lung tissue (Fig. S3 E–H). These data indicate that although cytosolic immune sensing of T4SS-translocated bacterial products is critical to elicit TNF production, TNF is produced by bystander alveolar macrophages and neutrophils that have not received T4SS-translocated substrates.
Fig. S3.
T4SS-injected airway alveolar macrophages produce IL-1α and IL-1β, and uninjected lung tissue-isolated alveolar macrophages and neutrophils produce TNF and do not express CD86 during pulmonary infection. (A and B) Mice were intranasally infected with 5 × 106 cfu of JR32 ΔdotA or ΔflaA L. pneumophila or PBS. At 24 h PI, cells were harvested from the lung tissue, treated with BFA, loaded with CCF4-AM, stained for extracellular markers and intracellular IL-1α (A) and IL-1β (B). Representative plots are shown. n = 2–3 mice per group. Data are representative of two independent experiments. (C–H) Mice were intranasally infected with 106 cfu of JR32 ΔdotA or ΔflaA L. pneumophila or PBS. At 24 h PI, cells were harvested from the lung tissue, treated with BFA, loaded with CCF4-AM, and stained with an antibody against CD86. Representative plots for alveolar macrophages (C) and neutrophils (D) are shown. n = 2–3 mice per group. Data are representative of three independent experiments. Alveolar macrophages (E) and neutrophils (G) were harvested from the lung tissue of infected mice and intracellularly stained for TNF production. Representative flow plots are shown, with n = 2–3 mice per group per experiment and are representative of three independent experiments. Graphs show the total numbers and relative percentage of TNF-positive uninjected and injected alveolar macrophages (F) or neutrophils (H) in infected mice from three pooled experiments. Student’s t tests were performed for total TNF-producing cells between ΔdotA- and ΔflaA-infected mice. NS, not significant; **P < 0.005, ***P < 0.0005.
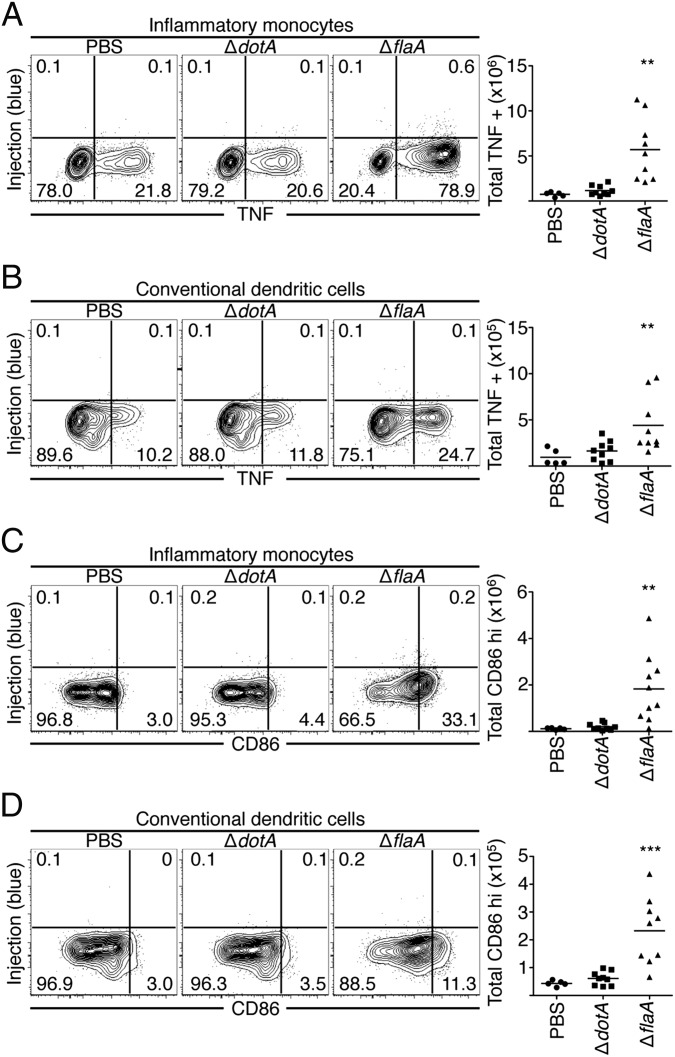
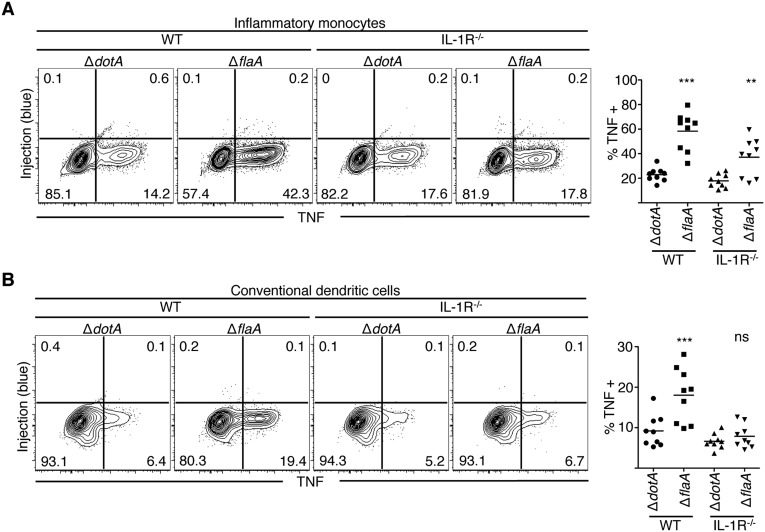
Bystander Inflammatory Monocytes and Dendritic Cells Are the Primary Producers of TNF and CD86 During L. pneumophila Infection.
Inflammatory monocytes and conventional dendritic cells (DCs) are also recruited to the lung during L. pneumophila infection. In contrast to alveolar macrophages and neutrophils, these cell populations are not productively infected and do not receive T4SS effectors (18, 22). Even so, inflammatory monocytes and DCs were the primary cell types that produced TNF during ΔflaA L. pneumophila infection (Fig. 3 A and B). In contrast, inflammatory monocytes and DCs from ΔdotA-infected mice did not increase TNF production compared with cells from PBS-treated mice. Inflammatory monocytes and DCs also substantially increased CD86 expression during ΔflaA L. pneumophila infection compared with PBS or ΔdotA infection and were the primary cell types expressing CD86 (Fig. 3 C and D). Alveolar macrophages and neutrophils did not increase CD86 expression during ΔflaA infection (Fig. S3 C and D). These data indicate that although inflammatory monocytes and DCs are not productively infected and do not receive T4SS-translocated products, they are the primary cell types that express TNF and CD86 during virulent L. pneumophila infection.
Fig. 3.
Bystander inflammatory monocytes and conventional dendritic cells produce TNF and express CD86 during infection. Cells were harvested from the lung tissue of mice, treated with BFA, loaded with CCF4-AM, stained, and fixed and stained with an antibody against TNF (A and B) or CD86 (C and D). Representative plots are shown. Graphs show individual mice and mean from three pooled independent experiments. Student’s t tests were performed between ΔdotA-and ΔflaA-infected mice. **P < 0.005, ***P < 0.0005.
IL-1 Signaling Is Required for Bystander Immune Cells to Produce Cytokines and CD86.
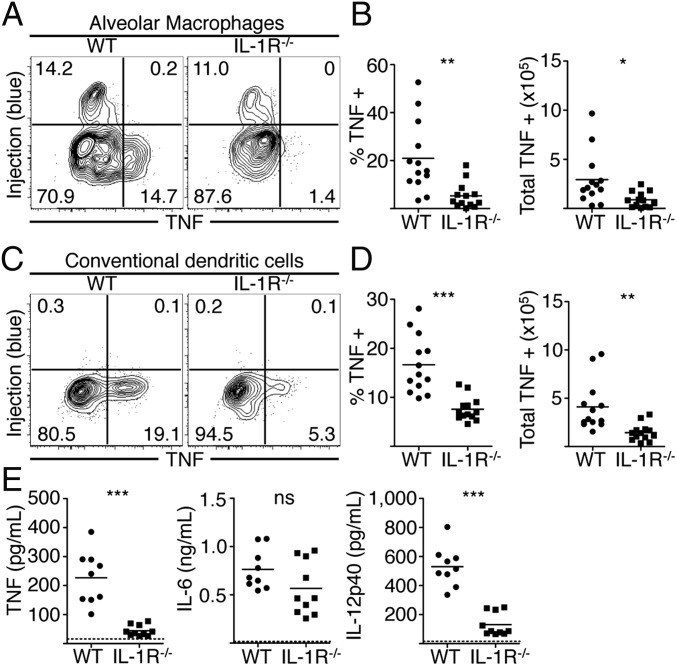
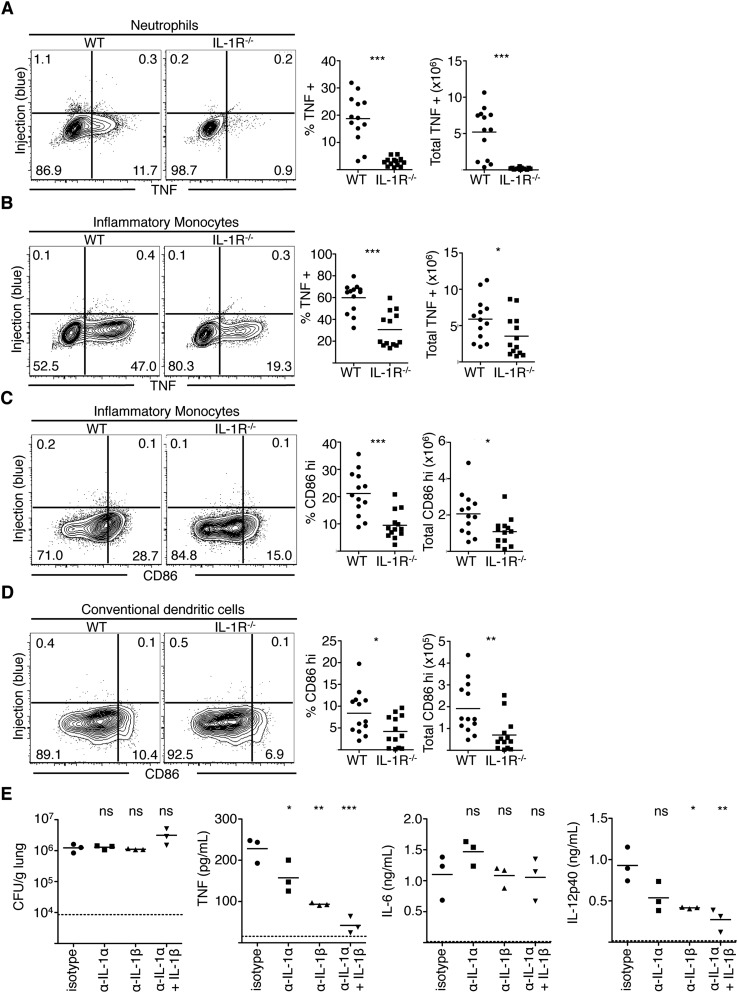
Maximal immune responses to L. pneumophila require cytosolic sensing of T4SS activity (16), yet our data indicate that the majority of cytokine- and CD86-producing cells are bystander innate immune cells that have not encountered T4SS effectors. Given that T4SS-injected cells selectively synthesize IL-1α and IL-1β, we hypothesized that IL-1α and IL-1β released by infected cells would instruct bystander cells to produce inflammatory cytokines, as IL-1α and IL-1β can elicit cytokine expression in other systems (29, 30). Notably, IL-1R signaling is critical for host defense against L. pneumophila, although its contribution has been primarily attributed to eliciting neutrophil-attracting chemokines and recruitment of neutrophils (12, 18, 19, 31). Following intranasal infection of WT or Il1r1−/− (IL-1R−/−) mice with ΔflaA L. pneumophila, equivalent numbers of alveolar macrophages in the lung and airway space were injected by L. pneumophila in WT and IL-1R−/− mice; however, as IL-1R−/− mice fail to recruit neutrophils, the total number of injected neutrophils is decreased in these mice (Fig. S4). Despite carrying an increased bacterial load (19), IL-1R−/− mice exhibited significantly reduced percentages and total numbers of TNF-producing alveolar macrophages, neutrophils, DCs, and inflammatory monocytes compared with WT mice (Fig. 4 and Fig. S5 A and B). Although a percentage of IL-1R−/− DCs and inflammatory monocytes still produced TNF, it was only minimally higher than the basal percentage of TNF-positive cells in WT or IL-1R−/− mice infected with ΔdotA bacteria (Fig. S6). Furthermore, TNF and IL-12p40 levels in the BAL fluid of IL-1R−/− mice were significantly reduced compared with WT mice (Fig. 4E). IL-1R−/− mice also exhibited significantly decreased percentages and total numbers of CD86-expressing inflammatory monocytes and DCs compared with WT controls (Fig. S5 C and D). In contrast, IL-6 levels were unaffected, indicating that IL-1R signaling is not required for all bystander cell responses (Fig. 4E). Finally, antibody-mediated neutralization of IL-1α and/or IL-1β before infection significantly decreased levels of TNF and IL-12p40, but not IL-6, with neutralization of IL-1α and IL-1β together having the greatest effect (Fig. S5E). Overall, these data indicate a crucial role for IL-1R signaling in optimal production of the proinflammatory cytokines TNF and IL-12p40 as well as the costimulatory molecule CD86 by bystander immune cells during pulmonary L. pneumophila infection.
Fig. S4.
WT and IL-1R−/− mice have similar numbers of injected alveolar macrophages, but IL-1R−/− mice have fewer injected neutrophils during L. pneumophila infection. WT or IL-1R−/− mice were infected with JR32 ΔflaA L. pneumophila intranasally. At 24 h PI, cells from the airway space and lung were isolated and loaded with CCF4-AM. The frequency and total number of injected airway alveolar macrophages (A), neutrophils (B), as well as pulmonary alveolar macrophages (C) and neutrophils (D) were quantified. Graphs show individual mice and mean from four pooled experiments. NS, not significant; **P < 0.005, ***P < 0.0005 as determined by Student’s t test.
Fig. 4.
IL-1 signaling induces cytokine production by bystander cells. Cells were harvested from the lungs of infected WT or Il1r1−/− (IL-1R−/−) mice, treated with BFA, loaded with CCF4-AM, stained, fixed and then stained with an antibody against TNF. Representative plots from alveolar macrophages (A and B) and conventional DCs (C and D) are shown. Graphs show individual mice and mean pooled from four independent experiments. (E) BAL fluid was collected from mice and cytokine levels were measured by ELISA. Graphs show individual mice and mean pooled from three independent experiments. n = 2–4 mice per group. Student’s t tests were performed between pooled groups. NS, not significant, *P < 0.05, **P < 0.005, ***P < 0.0005.
Fig. S5.
IL-1R signaling is required for the expression of CD86 and cytokines by bystander cells. Cells were harvested from the lungs of ΔflaA-infected WT or Il1r1−/− (IL-1R−/−) mice, treated with BFA, loaded with CCF4-AM, and stained for extracellular markers and intracellular TNF (A and B) or CD86 (C and D). Representative flow plots from various cell populations are shown. Graphs show individual mice and mean pooled from four independent experiments. n = 2–4 mice per group. Student’s t tests were performed between pooled groups. *P < 0.05, **P < 0.005, ***P < 0.0005. (E) Mice were injected intraperitoneally with isotype control antibody or neutralizing antibodies specific for IL-1α, IL-1β, or both before intranasal infection with JR32 ΔflaA L. pneumophila. At 24 h PI, BAL fluid and lungs were harvested. The cfus were plated for viable L. pneumophila from lung homogenates and TNF, IL-6, and IL-12p40 were measured in the BAL fluid by ELISA. Graphs show individual mice and mean. Statistics are one-way ANOVA with the stats representing the significance of the Tukey posttest between isotype control-treated mice and the groups of interest. NS, not significant; *P < 0.05, **P < 0.005, ***P < 0.0005.
Fig. S6.
Lack of IL-1R signaling does not alter the production of TNF by avirulent L. pneumophila infection. Cells were harvested from the lungs of ΔdotA- or ΔflaA-infected WT or Il1r1−/− (IL-1R) mice, treated with BFA, loaded with CCF4-AM, and stained for extracellular markers and intracellular TNF. Representative flow plots show TNF production from inflammatory monocytes (A) or conventional dendritic cells (B) from the lung. Bar graphs on Right show individual mice and pooled mean from three independent experiments. NS, not significant; **P < 0.005, ***P < 0.0005.
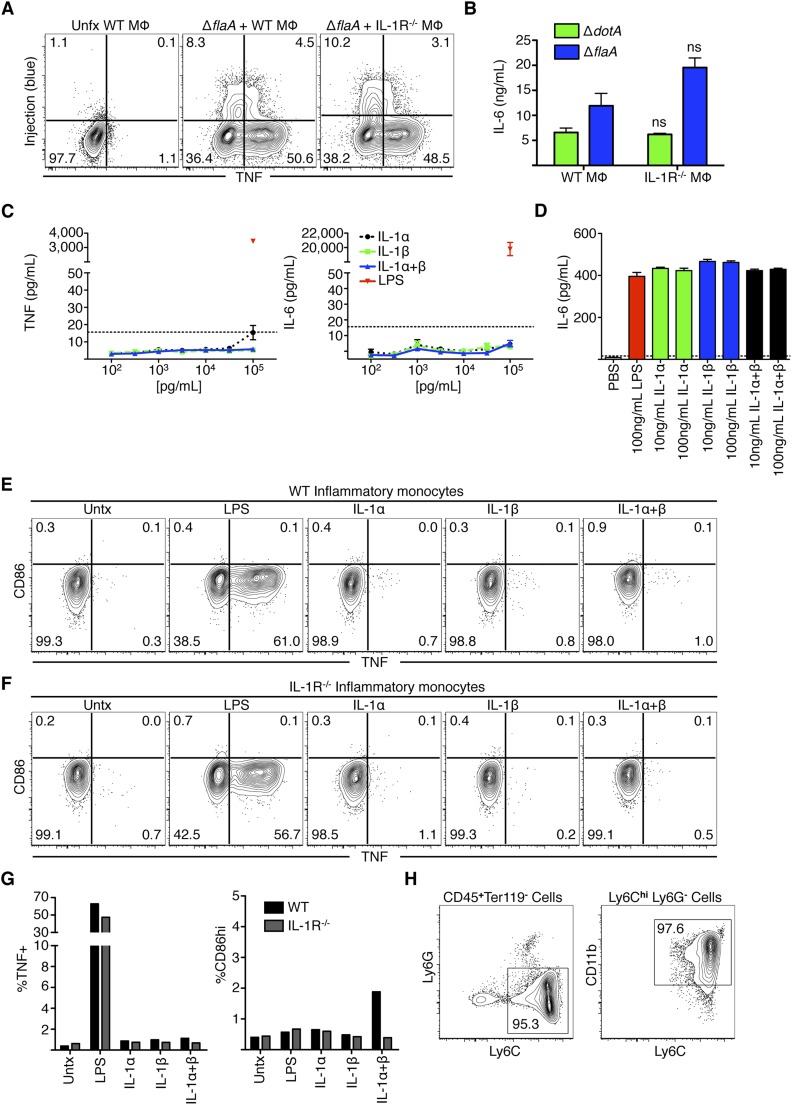
In contrast to our in vivo findings, we observed equivalent levels of TNF and IL-6 in IL-1R−/− and WT BMDMs during in vitro infection, suggesting that IL-1R signaling was not required for bystander responses in these cells (Fig. S7 A and B). Consistently, BMDMs treated with recombinant IL-1α and/or IL-1β did not produce TNF or IL-6, in contrast to LPS treatment, which robustly induced TNF and IL-6 (Fig. S7C). This lack of responsiveness to IL-1 was not due to the inability of IL-1α or IL-1β to induce responses, as treatment of the NIH/3T3 fibroblast line elicited robust IL-6 production (Fig. S7D). These data suggest that signals other than IL-1 are required for induction of bystander cytokine responses in BMDMs during in vitro infection, consistent with the view that BMDMs express very low levels of IL-1R and are poorly responsive to IL-1 (32, 33).
Fig. S7.
Bystander responses in bone marrow-derived macrophages do not require IL-1R, and IL-1 is not sufficient to induce cytokine production in bone marrow-derived macrophages and inflammatory monocytes. (A) BMDMs from WT or IL-1R−/− mice were left untreated or infected with the Lp02 ΔflaA strain of L. pneumophila at an MOI = 5, infected for 4 h, loaded with CCF4-AM, and stained for TNF. (B) BMDMs from WT or IL-1R−/− mice were infected with the Lp02 ΔdotA or ΔflaA strains of L. pneumophila at an MOI = 5 and IL-6 production was measured by ELISA 24 h PI. (C) BMDMs were treated with 100–100,000 pg/mL of recombinant IL-1α, IL-1β, or IL-1α and IL-1β combined, or 100 ng/mL LPS for 24 h. TNF and IL-6 were then measured by ELISA. (D) NIH/3T3 cells were treated with 10–100 ng/mL of recombinant IL-1α, IL-1β, or IL-1α and IL-1β combined, or 100 ng/mL LPS for 24 h and IL-6 was measured by ELISA. Graphs show mean ± SEM; NS, not significant as determined by Student’s t test. (E and F) inflammatory monocytes were isolated from the bone marrow of WT (E) or IL-1R−/− (F) mice. Cells were then treated with 100 ng/mL LPS, IL-1α, IL-1β, or both for 6 h. At 3 h before harvest, BFA was added to the wells. Cells were then stained for TNF and CD86. (G) Quantification of E and F. (H) Gating strategy for isolated inflammatory monocytes. Monocytes are Ly6Chi, CD11bint to CD11bhi and do not express Ly6G. Monocytes were 90–95% pure. Graphs show mean of two independent mice. Data are representative of two independent experiments.
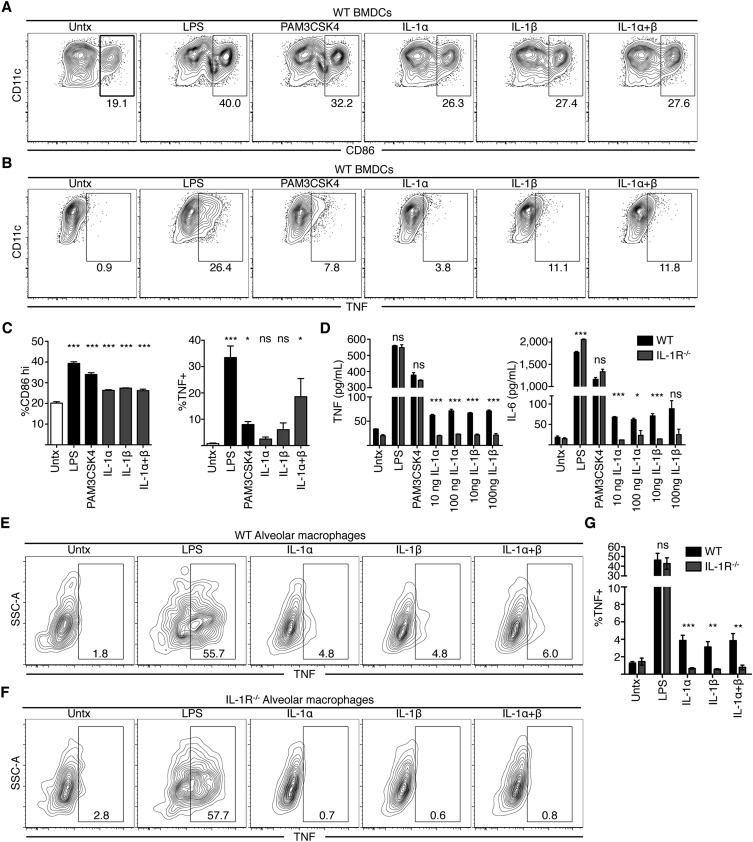
Other key innate immune cell types responded to IL-1 treatment. Indeed, bone marrow-derived DCs up-regulated CD86, TNF, and IL-6 in response to IL-1α and -β (Fig. S8 A–D). Moreover, alveolar macrophages from WT mice also produced TNF upon IL-1α and -β treatment, whereas IL-1R−/− alveolar macrophages did not (Fig. S8 E–G). In contrast, inflammatory monocytes isolated from bone marrow did not produce TNF or CD86 in response to IL-1α and -β (Fig. S7 E–H). Inflammatory monocytes produced TNF but not CD86 in response to LPS, suggesting that additional signals are required for inflammatory monocytes to express CD86 (Fig. S7G). Together, these data indicate that DCs and alveolar macrophages are likely to respond to IL-1 signaling in a cell-intrinsic manner. Other cell types may require either additional signals to produce cytokines or require an alternative signal produced by an intermediate cell in response to IL-1.
Fig. S8.
IL-1 is sufficient to induce cytokine or CD86 production in bone marrow-derived dendritic cells and alveolar macrophages. (A–C) BMDCs were treated with LPS, PAM3CSK4, or 100 ng/mL IL-1α, IL-1β, or both for 6 h. At 3 h before harvest, cells were treated with BFA. Cells were then stained for CD86 (A) or TNF (B). Representative flow plots are shown. (C) Quantification of plots shown in A and B. (D) BMDCs were treated as in A and B for 24 h. Supernatants were then harvested and TNF and IL-6 production were measured by ELISA. Graphs show mean ± SEM. For C, ANOVA was performed and statistics show significance between untreated cells and the appropriate condition as determined by Tukey posttest. For D, Student's t test was performed and statistics show significance between WT and IL-1R−/− cells for each condition. *P < 0.05, **P < 0.005, ***P < 0.0005; NS, not significant. (E–G) Alveolar macrophages from WT (E) or IL-1R−/− (F) mice were isolated and seeded in culture at 10,000 cells per well in 150 μL DMEM + 10% FBS. Cells were then treated with 100 ng/mL LPS, IL-1α, IL-1β, or both for 6 h. At 3 h before harvest, BFA was added to the wells. Cells were fixed and then stained for TNF. (G) Quantification of E and F. Graphs show mean ± SEM of five independent mice from two pooled experiments. Student's t test was performed and statistics show significance between WT and IL-1R−/− cells for each condition. *P < 0.05, **P < 0.005, ***P < 0.0005; NS, not significant.
Discussion
Infection with L. pneumophila leads to a robust proinflammatory cytokine response that requires cytosolic sensing of T4SS activity (13, 16). This findng is paradoxical, as a number of T4SS effectors inhibit host protein synthesis (9–12, 14, 15, 20). We therefore sought to determine how this robust cytokine response is generated. Our data indicate that cells targeted by T4SS effectors selectively synthesize IL-1α and IL-1β, but poorly produce the key immune proteins TNF, IL-6, IL-12, and CD86 (Figs. 1–4). Instead, bystander cells produce the majority of these immune signals in vitro and in vivo. Furthermore, IL-1R signaling is required for bystander cells to produce these immune proteins in vivo. Thus, our data suggest that collaboration between infected cells and bystander cells overcomes the pathogen-imposed translation block by enabling bystander cells to mount an inflammatory cytokine response.
Our findings indicate that uninfected bystander cells play a critical role in host defense by producing immune effector proteins in response to direct or indirect sensing of IL-1 and additional signals released by infected cells. This mechanism provides multiple possible benefits for enabling robust antimicrobial immune defense. First, it allows for initiation of an immune response despite pathogen inhibition of host translation. Second, it may be a strategy for avoiding inappropriate responses to avirulent bacteria, as an inflammatory response against pathogens would be generated only when a second signal, such as IL-1, is specifically released upon sensing of virulence activity (34, 35). Third, bystander cytokine production may provide a means of amplifying early immune signals, allowing for a more rapid and robust response. Bystander activation is likely to be a common strategy used by the immune system for overcoming pathogen subversion, and multiple mechanisms may exist for activating various bystander cell types in the context of different pathogens (36–41). Notably, during influenza A virus infection, IL-1R signaling activates bystander DCs to enable successful priming of naïve CD8+ T cells (42). This study, along with our finding that uninfected bystander cells up-regulate the costimulatory molecule CD86 (Fig. 3 and Fig. S5), has implications for understanding how a T-cell response is generated during L. pneumophila infection. Bystander cells may serve as key antigen presenting cells, perhaps following uptake of dead or dying infected cells (43, 44).
Our findings also elucidate another critical function for IL-1 in early innate immune defense. In addition to the well-established role of IL-1R signaling in eliciting expression of neutrophil-attracting chemokines and subsequent neutrophil recruitment to the site of infection (12, 18, 19, 31), our data indicate that IL-1R signaling mediates cytokine production by bystander immune cells (Fig. 4 and Fig. S5). Immune signals other than IL-1 also likely contribute during in vivo L. pneumophila infection, as IL-1R signaling is required for maximal production of TNF and IL-12, but is dispensable for IL-6 production. Other signals contribute in vitro, as IL-1R signaling is not required for bystander cytokine responses in BMDMs. The nature of these other immune signals and whether they can compensate for the absence of IL-1 in certain infection settings remains to be determined.
It is intriguing that infected cells effectively translate IL-1α and IL-1β, but not other key inflammatory proteins. IL-1α and IL-1β differ from other secreted cytokines, because they are translated by cytosolic ribosomes as precursors that are unconventionally secreted in an inflammasome-dependent manner (45, 46). MyD88 and host modulation of mTOR signaling participate in the selective translation of proinflammatory cytokines during L. pneumophila infection (14, 21) Whether MyD88 and mTOR signaling influence cytokine production in directly infected cells or bystander cells remains to be determined. Previous studies have suggested that mRNA abundance determines which transcripts are selectively translated when translation is suppressed in infected cells (14). Consistent with such a model, our data indicate a slight increase in the total abundance of translated transcripts (Il1a and Il1b) compared with transcripts that are not translated (Il6 and Tnf). It remains to be determined, however, whether mRNA abundance fully explains the ability of injected cells to translate IL-1α and IL-1β but not other cytokines.
We found that the inability of injected cells to produce cytokines is partly due to effector activity, as cells injected by L. pneumophila strains lacking the translation-inhibiting effectors produced increased levels of TNF, IL-1α, and IL-1β early during infection (Fig. S2). However, at later times, cells injected by these mutants produced little TNF and IL-6. Why injected cells are unable to produce TNF and IL-6 later in infection is unclear. Multiple mechanisms may act in concert to impair host translation at later timepoints, including yet-to-be-identified effectors that inhibit host translation or host-driven responses that suppress mTOR signaling (14).
Because IL-1R−/− mice have impaired bystander responses during in vivo infection, we investigated whether IL-1R signaling was sufficient to induce TNF, IL-6, or CD86 expression in various innate immune cell types. Previous studies demonstrated that IL-1 stimulation of both mouse and human immune cells induces TNF production (33, 47), and IL-1 treatment induces CD86 expression on lung DCs (42). We found that alveolar macrophages and bone marrow-derived DCs responded to IL-1 stimulation ex vivo (Fig. S7). The amount of TNF and CD86 elicited by IL-1, however, is moderate compared with LPS treatment or the levels attained during in vivo infection. IL-1 treatment was insufficient for TNF or CD86 expression in inflammatory monocytes ex vivo, although IL-1R signaling is required for these cells to express TNF and CD86 during in vivo infection. We therefore conclude that IL-1R signaling is necessary for robust bystander responses during in vivo infection, but is only partially sufficient. We hypothesize that additional host and/or microbial signals work in concert with IL-1 to drive bystander responses, and some cell types may indirectly respond to IL-1 or other factors via signals produced by an intermediary cell. These other signals may be required for bystander responses in BMDMs and may account for IL-6 production and the remaining TNF made by IL-1R−/− inflammatory monocytes during in vivo infection.
In conclusion, we have identified a system used by the innate immune system to ensure generation of proinflammatory cytokines during infection with a bacterial pathogen. Our results demonstrate the existence of heterogeneity in production of critical protective cytokines by innate immune cells during the early response to bacterial infection. These findings define a collaboration between infected and uninfected cells that enables the immune system to bypass pathogen-mediated inhibition of protein synthesis to generate a robust immune response against L. pneumophila. As a variety of pathogens would be expected to disable cytokine production in directly infected cells, uninfected bystander cells may be required to carry out and amplify the early innate signals that eventually confer protective immunity.
Materials and Methods
Ethics Statement.
All experiments performed in this study were done in accordance with the Animal Welfare Act and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (48). The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all procedures (Protocols 803465, 803459, 804714, and 804928).
Bacterial Strains and Plasmids.
All experiments used L. pneumophila serogroup 1 strains. For in vitro studies, macrophages were infected with Lp02 (rpsL, hsdR, and thyA), a thymidine auxotroph derived from strain Lp01, Lp02 ΔdotA, Lp02 ΔflaA, Lp02 Δlgt1Δlgt2Δlgt3ΔsidIΔsidL (WTΔ5), or Lp02 ΔflaAΔlgt1Δlgt2Δlgt3ΔsidIΔsidLΔpkn5Δlpg1489 (Δ7ΔflaA) (12, 13). For in vivo studies, mice were infected with JR32-derived (rpsL and hsdR) ΔdotA or ΔflaA isogenic mutant strains. All strains harbored the pSS128 plasmid encoding an M45-tagged β-lactamase–RalF fusion protein (22). For in vitro and in vivo studies, L. pneumophila was cultured on charcoal yeast extract agar containing 6.25 μg/mL chloramphenicol for 48 h at 37 °C before infection (19, 49–51).
Mice.
C57BL/6 and IL-1R−/− (Il1r1−/−) mice were purchased from The Jackson Laboratories. Mice were maintained in accordance with the guidelines of the University of Pennsylvania Institutional Animal Use and Care Committee. Please refer to SI Materials and Methods on infection of mice, cell culture, isolation of monocytes and alveolar macrophages, flow cytometry and cell sorting, IL-1 stimulation, RT-PCR, and ELISA. Differences were considered significant if the P value was <0.05.
SI Materials and Methods
Mice.
For infections, 8- to 12-wk-old male and female mice were anesthetized by i.p. injection of a ketamine/xylazine/PBS solution at a dose of 100 mg/kg ketamine and 10 mg/kg xylazine. Mice were then infected intranasally with 40 μL of a bacterial suspension containing 1 × 106 cfu L. pneumophila or PBS vehicle control unless otherwise stated. At 24 h postinfection (PI), mice were killed. To isolate lung airway cells and detect cytokines by ELISA, bronchoalveolar lavage (BAL) was performed three times with 1 mL of cold PBS each time. Lungs were then excised and digested for 30 min at 37 °C with occasional shaking in 5 mL of PBS containing 5% (vol/vol) FBS, 250 units/mL of collagenase IV (Worthington Biochemical), and 20 units/mL DNase I (Roche). Lungs were then mechanically homogenized and a single-cell suspension was obtained. To determine bacterial burden, lungs were mechanically homogenized in sterile, distilled H2O and a portion of the lysate was spread onto CYE plates containing chloramphenicol. For antibody neutralizations 16 h before infection, mice were given 200 μg of either isotype antibody (hamster IgG), antibody against IL-1α (α-IL-1α) (ALF-161), IL-1β (α-IL-1β) (B122), or both (BioXcell).
Cell Culture.
For macrophages, C57BL/6 mouse bone marrow cells were differentiated in RPMI containing 30% (vol/vol) L929 cell supernatant and 20% (vol/vol) FBS at 37 °C, 5% CO2 in a humidified incubator. The macrophages were replated in RPMI containing 15% L929 cell supernatant and 10% (vol/vol) FBS (19, 52). For dendritic cells, bone marrow cells were differentiated in RPMI containing 10% FBS, 50 μM β-mercaptoethanol, 2 mM l-glutamine, and 20 ng/mL GM-CSF (Peprotech) (53). Semiadherent dendritic cells were then isolated and replated in medium lacking GM-CSF. NIH/3T3 cells (ATCC) were cultured in DMEM containing 10% (vol/vol) FBS. For infections, cells were treated with 10–100 ng/mL LPS from Escherichia coli strain 055:B5 (Sigma), 0.4 μg/mL PAM3CSK4, 10 μL of bacterial suspension, or 10 μL of PBS vehicle control.
Isolation of Inflammatory Monocytes and Alveolar Macrophages.
Alveolar macrophages were harvested from the airways of mice by bronchoalveolar lavage in 5 × 1 mL cold PBS. For intracellular cytokine staining, cells were centrifuged, counted, and resuspended at 10,000 cells per well in 150 μL DMEM + 10% (vol/vol) FBS. Inflammatory monocytes were isolated from the bone marrow of mice using a magnetic negative selection kit (Miltenyi). Inflammatory monocytes were assayed for purity postselection and were consistently 90–95% CD45+, Ly6Chi, Ly6G−, CD11bint to CD11bhi. For intracellular cytokine staining, inflammatory monocytes were resupsended at 30,000 cells per well in 150 μL DMEM with 10% FBS.
Flow Cytometry and Cell Sorting.
For in vitro experiments, infected cells were treated with 10 μg/mL brefeldin A with or without 2 μM monensin (Sigma) 3 h before harvest. Cells then were lifted and loaded with CCF4-AM (Invitrogen) per the manufacturer’s instructions. Cells were washed and treated with Live/Dead Fixable Dead Cell Stain (Invitrogen). To stain for intracellular cytokines, cells were fixed with BD Cytofix/Cytoperm (BD Biosciences), and then stained with antibodies specific for IL-6 (BioLegend), TNF, IL-1α, and IL-1β (eBioscience). For in vivo studies, lung and airway cells were resuspended in RPMI or DMEM containing 10% (vol/vol) FBS and 10 μg/mL brefeldin A for 3 h. Cells were washed and loaded with CCF4-AM and treated with the Live/Dead Stain. Cells were stained with antibodies specific for the cell surface antigens CD45, CD11c, Ly6G, Ly6C, CD86 (BioLegend), MHCII, (eBioscience), and Siglec F (BD Biosciences). Intracellular staining was performed as described above. Data were collected on a LSR II flow cytometer (BD Biosciences) and postcollection data were analyzed using FlowJo (Treestar). Cells were gated on live singlets that had retained the CCF4-AM dye. Cell sorting experiments were performed on a FACSAria II flow cytometer (BD Biosciences).
IL-1 Stimulation.
For treatment of cells with IL-1, mature recombinant IL-1α, IL-1β, or IL-1α and IL-1β (eBioscience) combined were added to cells at various concentrations. For intracellular staining of BMDCs, inflammatory monocytes, and alveolar macrophages, IL-1 was added at 100 ng/mL for 6 h with BFA added 3 h before harvest. BMDCs were stained for CD11c, MHCII, and CD86. BMDCs were then fixed and stained for TNF. Inflammatory monocytes were stained for CD45, Ly6C, Ly6G, CD11b, Ter119, CD86 and then fixed and stained for TNF. Alveolar macrophages were fixed and then stained for TNF.
RT-PCR.
Sorted bone marrow-derived macrophages were centrifuged and resuspended in RLT lysis buffer and RNA was isolated using an RNeasy kit as per the kit’s protocol (Qiagen). RNA was then reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was then performed using the prepared cDNA with the following primer pair sequences: Il1b (5′-GCA ACT GTT CCT GAA CTA AAC T-3′, 5′-ATC TTT TGG GGT CCG TCA ACT-3′), Il1a (5′-GCA CCT TAC ACC TAC CAG AGT-3′, 5′-TGC AGG TCA TTT AAC CAA GTG G-3′), Il6 (5′-GAC TTC CAT CCA GTT GCC TTC TTG G-3′, 5′-CCA GTT TGG TAG CAT CCA TCA TTT CT-3′), Tnf (5′-GAC GTG GAA CTG GCA GAA GAG-3′, 5′-TTG GTG GTT TGT GAG TGT GAG-3′), Casp4 (5′-ACA AAC ACC CTG ACA AAC CAC-3′, 5′-CAC TGC GTT CAG CAT TGT TAA A-3′), Ifnb (5′-GCA CTG GGT GGA ATG AGA CTA TTG-3′, 5′-TTC TGA GGC ATC AAC TGA CAG GTC-3′), or the housekeeping gene Hprt (5′-GTT GGA TAC AGG CCA GAC TTT GTT G-3′, 5′-GAG GGT AGG CTG GCC TAT AGG CT-3′). Data were analyzed by comparing the CT values obtained for each reaction and then comparing them with the appropriate samples’ housekeeping gene to determine relative expression.
ELISA.
Harvested supernatants from BAL were assayed using capture and detection antibodies specific for TNF, IL-6 (BioLegend), and IL-12p40 (BD Biosciences).
Statistical Analysis.
Plotting of data and statistical analysis were performed using GraphPad Prism software. Statistical significance was determined using the unpaired, two-tailed Student’s t test or one-way ANOVA with Tukey posttest. Differences were considered significant if the P value was <0.05.
Supplementary Material
Acknowledgments
We thank Russell Vance for providing the WTΔ5 and Δ7ΔflaA L. pneumophila strains, Mike Askenase in Yasmine Belkaid’s laboratory for providing protocols on inflammatory monocyte isolation and ex vivo stimulation, members of the Brodsky laboratory for helpful discussions, and Igor Brodsky for critical reading of the manuscript. This work was supported in part by National Institutes of Health Grants K99/R00AI087963 (to S.S.) and T32GM007229 (to A.M.C.), American Lung Association Grant RG-268528-N (to S.S.), University of Pennsylvania University Research Foundation (S.S.), an Institute for Immunology Pilot Award (to S.S.), American Heart Association Grant 13BGIA14780070 (to S.S.), and the Roy and Diana Vagelos Scholars Program in Molecular Life Sciences (M.M.D.). This material is based upon work supported by the National Science Foundation under Grant DGE-0822 (to C.N.C., a graduate research fellowship).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501289112/-/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krachler AM, Woolery AR, Orth K. Manipulation of kinase signaling by bacterial pathogens. J Cell Biol. 2011;195(7):1083–1092. doi: 10.1083/jcb.201107132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8(11):1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 5.Viboud GI, Bliska JB. Yersinia outer proteins: Role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 6.Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89(20):9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 8.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 9.Belyi Y, Tabakova I, Stahl M, Aktories K. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol. 2008;190(8):3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen X, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11(6):911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belyi Y, et al. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc Natl Acad Sci USA. 2006;103(45):16953–16958. doi: 10.1073/pnas.0601562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry KC, Fontana MF, Portman JL, Dugan AS, Vance RE. IL-1α signaling initiates the inflammatory response to virulent Legionella pneumophila in vivo. J Immunol. 2013;190(12):6329–6339. doi: 10.4049/jimmunol.1300100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana MF, et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7(2):e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov SS, Roy CR. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol. 2013;14(12):1219–1228. doi: 10.1038/ni.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCusker KT, Braaten BA, Cho MW, Low DA. Legionella pneumophila inhibits protein synthesis in Chinese hamster ovary cells. Infect Immun. 1991;59(1):240–246. doi: 10.1128/iai.59.1.240-246.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin S, et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4(11):e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spörri R, Joller N, Albers U, Hilbi H, Oxenius A. MyD88-dependent IFN-gamma production by NK cells is key for control of Legionella pneumophila infection. J Immunol. 2006;176(10):6162–6171. doi: 10.4049/jimmunol.176.10.6162. [DOI] [PubMed] [Google Scholar]
- 18.LeibundGut-Landmann S, Weidner K, Hilbi H, Oxenius A. Nonhematopoietic cells are key players in innate control of bacterial airway infection. J Immunol. 2011;186(5):3130–3137. doi: 10.4049/jimmunol.1003565. [DOI] [PubMed] [Google Scholar]
- 19.Casson CN, et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9(6):e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontana MF, Shin S, Vance RE. Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect Immun. 2012;80(10):3570–3575. doi: 10.1128/IAI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asrat S, Dugan AS, Isberg RR. The frustrated host response to Legionella pneumophila is bypassed by MyD88-dependent translation of pro-inflammatory cytokines. PLoS Pathog. 2014;10(7):e1004229. doi: 10.1371/journal.ppat.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copenhaver AM, et al. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infect Immun. 2014;82(10):4325–4336. doi: 10.1128/IAI.01891-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knapp T, Hare E, Feng L, Zlokarnik G, Negulescu P. Detection of beta-lactamase reporter gene expression by flow cytometry. Cytometry A. 2003;51(2):68–78. doi: 10.1002/cyto.a.10018. [DOI] [PubMed] [Google Scholar]
- 24.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2(3):e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7(3):318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 26.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203(4):1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case CL, et al. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci USA. 2013;110(5):1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belyi Y, Jank T, Aktories K. Cytotoxic glucosyltransferases of Legionella pneumophila. Curr Top Microbiol Immunol. 2013;376:211–226. doi: 10.1007/82_2013_338. [DOI] [PubMed] [Google Scholar]
- 29.Kohase M, May LT, Tamm I, Vilcek J, Sehgal PB. A cytokine network in human diploid fibroblasts: Interactions of beta-interferons, tumor necrosis factor, platelet-derived growth factor, and interleukin-1. Mol Cell Biol. 1987;7(1):273–280. doi: 10.1128/mcb.7.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87(6):2095–2147. [PubMed] [Google Scholar]
- 31.Mascarenhas DPA, Pereira MSF, Manin GZ, Hori JI, Zamboni DS. Interleukin 1 receptor-driven neutrophil recruitment accounts to MyD88-dependent pulmonary clearance of legionella pneumophila infection in vivo. J Infect Dis. 2015;211(2):322–330. doi: 10.1093/infdis/jiu430. [DOI] [PubMed] [Google Scholar]
- 32.Demuth A, Goebel W, Beuscher HU, Kuhn M. Differential regulation of cytokine and cytokine receptor mRNA expression upon infection of bone marrow-derived macrophages with Listeria monocytogenes. Infect Immun. 1996;64(9):3475–3483. doi: 10.1128/iai.64.9.3475-3483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu K, et al. IL-1 receptor type 2 suppresses collagen-induced arthritis by inhibiting IL-1 signal on macrophages. J Immunol. 2015;194(7):3156–3168. doi: 10.4049/jimmunol.1402155. [DOI] [PubMed] [Google Scholar]
- 34.Fontana MF, Vance RE. Two signal models in innate immunity. Immunol Rev. 2011;243(1):26–39. doi: 10.1111/j.1600-065X.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- 35.Franchi L, et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13(5):449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreux M, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12(4):558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolowschiak T, et al. Potentiation of epithelial innate host responses by intercellular communication. PLoS Pathog. 2010;6(11):e1001194. doi: 10.1371/journal.ppat.1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel SJ, King KR, Casali M, Yarmush ML. DNA-triggered innate immune responses are propagated by gap junction communication. Proc Natl Acad Sci USA. 2009;106(31):12867–12872. doi: 10.1073/pnas.0809292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasper CA, et al. Cell-cell propagation of NF-κB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity. 2010;33(5):804–816. doi: 10.1016/j.immuni.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503(7477):530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christian DA, et al. Use of transgenic parasites and host reporters to dissect events that promote interleukin-12 production during toxoplasmosis. Infect Immun. 2014;82(10):4056–4067. doi: 10.1128/IAI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang IK, Ichinohe T, Iwasaki A. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8⁺ T cell responses to influenza A virus. Nat Immunol. 2013;14(3):246–253. doi: 10.1038/ni.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yrlid U, Wick MJ. Salmonella-induced apoptosis of infected macrophages results in presentation of a bacteria-encoded antigen after uptake by bystander dendritic cells. J Exp Med. 2000;191(4):613–624. doi: 10.1084/jem.191.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trunk G, Oxenius A. Innate instruction of CD4+ T cell immunity in respiratory bacterial infection. J Immunol. 2012;189(2):616–628. doi: 10.4049/jimmunol.1200924. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson FT, Torrano F, Locksley RM, Lovett DH. Interleukin 1: The patterns of translation and intracellular distribution support alternative secretory mechanisms. J Cell Physiol. 1992;152(2):223–231. doi: 10.1002/jcp.1041520202. [DOI] [PubMed] [Google Scholar]
- 46.Rathinam VAK, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13(4):333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikejima T, Okusawa S, Ghezzi P, van der Meer JW, Dinarello CA. Interleukin-1 induces tumor necrosis factor (TNF) in human peripheral blood mononuclear cells in vitro and a circulating TNF-like activity in rabbits. J Infect Dis. 1990;162(1):215–223. doi: 10.1093/infdis/162.1.215. [DOI] [PubMed] [Google Scholar]
- 48. National Research Council (2011) Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), 8th Ed.
- 49.Feeley JC, et al. Charcoal-yeast extract agar: Primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10(4):437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nogueira CV, et al. Rapid pathogen-induced apoptosis: A mechanism used by dendritic cells to limit intracellular replication of Legionella pneumophila. PLoS Pathog. 2009;5(6):e1000478. doi: 10.1371/journal.ppat.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neild A, Murata T, Roy CR. Processing and major histocompatibility complex class II presentation of Legionella pneumophila antigens by infected macrophages. Infect Immun. 2005;73(4):2336–2343. doi: 10.1128/IAI.73.4.2336-2343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engel P, et al. The B7-2 (B20) costimulatory molecule expressed by monocytes and activated B lymphocytes is the CD86 differentiation antigen. Blood. 1994;81:1402–1407. [PubMed] [Google Scholar]
- 53.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]