Significance
Knowledge of the origin and evolution of viruses provides important insight into virus emergence involving the acquisition of genes necessary for the infection of new host species or the development of pathogenicity. The family Bunyaviridae contains important arthropod-borne pathogens of humans, animals, and plants. In this study, we provide a comprehensive characterization of two novel lineages of insect-specific bunyaviruses that are in basal phylogenetic relationship to the rodent-borne hantaviruses, the only genus within the Bunyaviridae that is not transmitted by arthropod vectors. These data, together with ancestral state reconstruction of bunyavirus hosts for major virus lineage bifurcations, suggest that the vertebrate-infecting viruses evolved from arthropod-specific progenitors.
Keywords: bunyavirus, arbovirus, insect-specific virus, virus evolution, ancestral reconstruction
Abstract
The evolutionary origins of arboviruses are unknown because their typical dual host tropism is paraphyletic within viral families. Here we studied one of the most diversified and medically relevant RNA virus families, the Bunyaviridae, in which four of five established genera are transmitted by arthropods. We define two cardinally novel bunyavirus groups based on live isolation of 26 viral strains from mosquitoes (Jonchet virus [JONV], eight strains; Ferak virus [FERV], 18 strains). Both viruses were incapable of replicating at vertebrate-typical temperatures but replicated efficiently in insect cells. Replication involved formation of virion-sense RNA (vRNA) and mRNA, including cap-snatching activity. SDS/PAGE, mass spectrometry, and Edman degradation identified translation products corresponding to virion-associated RNA-dependent RNA polymerase protein (RdRp), glycoprotein precursor protein, glycoproteins Gn and Gc, as well as putative nonstructural proteins NSs and NSm. Distinct virion morphologies suggested ancient evolutionary divergence, with bunyavirus-typical morphology for FERV (spheres of 60–120 nm) as opposed to an unusual bimorphology for JONV (tubular virions of 60 × 600 nm and spheres of 80 nm). Both viruses were genetically equidistant from all other bunyaviruses, showing <15% amino acid identity in the RdRp palm domain. Both had different and unique conserved genome termini, as in separate bunyavirus genera. JONV and FERV define two novel sister taxons to the superclade of orthobunyaviruses, tospoviruses, and hantaviruses. Phylogenetic ancestral state reconstruction with probabilistic hypothesis testing suggested ancestral associations with arthropods at deep nodes throughout the bunyavirus tree. Our findings suggest an arthropod origin of bunyaviruses.
Arboviruses are viruses with dual host tropism that are transmitted to their vertebrate hosts during the arthropod host’s blood-feeding. Arboviruses are found in several different RNA virus families as well as in a single DNA virus family, suggesting the dual host tropism has evolved by convergence. However, it is unclear whether arboviruses stem from arthropods or vertebrates, as dual host tropism is a paraphyletic property. All families of interest contain additional taxa with monotropism for either arthropods or vertebrates. Only for the genus Flavivirus within the family Flaviviridae has an evolution from insect-specific viruses been suggested, as new insect-specific flaviviruses have recently been discovered that branch deeper than congeneric arboviruses (1). Here we studied the case of one of the most genetically diversified families of RNA viruses, the family Bunyaviridae (2).
Bunyaviruses contain important pathogens of humans, livestock, and plants. With the exception of the rodent-borne hantaviruses, all bunyaviruses are transmitted by arthropod vectors (3). In addition to the five established genera, we have recently described two novel groups of putatively insect-specific bunyaviruses isolated from mosquitoes (4, 5). One clade, defined by the type species Gouléako virus (GOLV) (4), shares old common ancestors with all members of the genus Phlebovirus, and the second clade, defined by Herbert, Tai, and Kibale viruses (HEBV, TAIV, KIBV) (5), branches from a deep node in sister relationship to the genus Orthobunyavirus. Both virus groups have been proposed to constitute novel bunyavirus genera on the basis of their phylogenetic positions and other criteria such as serological distinction and differences in genome composition, including the absence of NSs and NSm proteins, as well as the lengths and sequences of conserved noncoding elements at genome segment termini. More recently, bona fide bunyavirus sequences distant from all described bunyaviruses were detected in phantom midges as well as in transcriptomes from other insects (6). However, corresponding viruses could not be isolated in cell culture, leaving doubts about whether these virids represent extant viruses.
To further examine the diversity of bunyaviruses, we screened cytopathic Aedes albopictus cell cultures inoculated with mosquitoes from the same region as GOLV and HEBV in Côte d’Ivoire (7). As a new approach, we used sensitivity to temperature to differentiate between insect-specific viruses and arboviruses. Our findings enable a reconciliation of the origin and evolution of the family Bunyaviridae.
Results
Virus Isolation and Morphology.
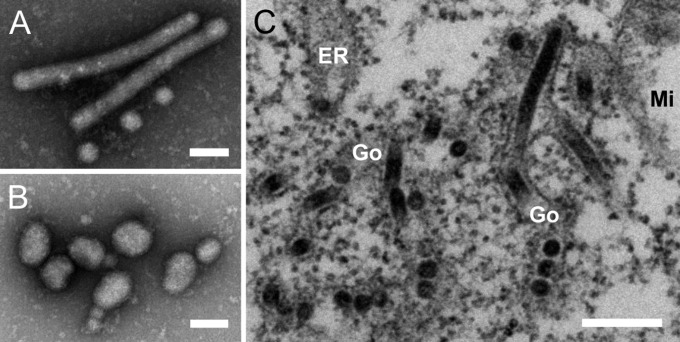
Two viruses, designated Jonchet virus (JONV) and Ferak virus (FERV), were isolated in C6/36 cells from mosquitoes collected in the Taï National Park region, Côte d’Ivoire (sampling described in ref. 7). Both viruses induced strong but distinct cytopathic effects (CPE) 4–5 d postinfection (dpi). JONV induced extensive syncytia formation, with some cells showing stretching and filamentous extensions (Fig. S1A). Cells infected with FERV showed stretching and tapering with slight aggregation and very sporadic formation of syncytia (Fig. S1B). Viral particles were purified by gradient ultracentrifugation from infected cell culture supernatants and examined by electron microscopy. The spherical, enveloped FERV virions were pleomorphic, with a diameter of 60–120 nm (Fig. 1B), morphologically resembling GOLV and HEBV (4, 5). Two types of enveloped virions were associated with JONV. Virions either had a tubular morphology of about 60 nm × up to 600 nm or were spherical with a diameter of about 80 nm (Fig. 1A). Both types of JONV virions co-occurred regularly in ultrathin sections of JONV-infected C6/36 cells in all independent cell culture isolates, but not in noninfected cells and not in cells infected with FERV, suggesting that both forms belong to one virus (Fig. 1C).
Fig. 1.
JONV and FERV morphology. (A and B) Negative-stained virions of JONV (A) and FERV (B) sedimented by ultracentrifugation. (C) Ultrathin sections of JONV-infected C6/36 cells. Mi, mitochondria; Go, Golgi apparatus; ER, endoplasmatic reticulum. (Scale bars, 100 nm in A and B and 250 nm in C.)
Genome Organization and Phylogenetic Analyses.
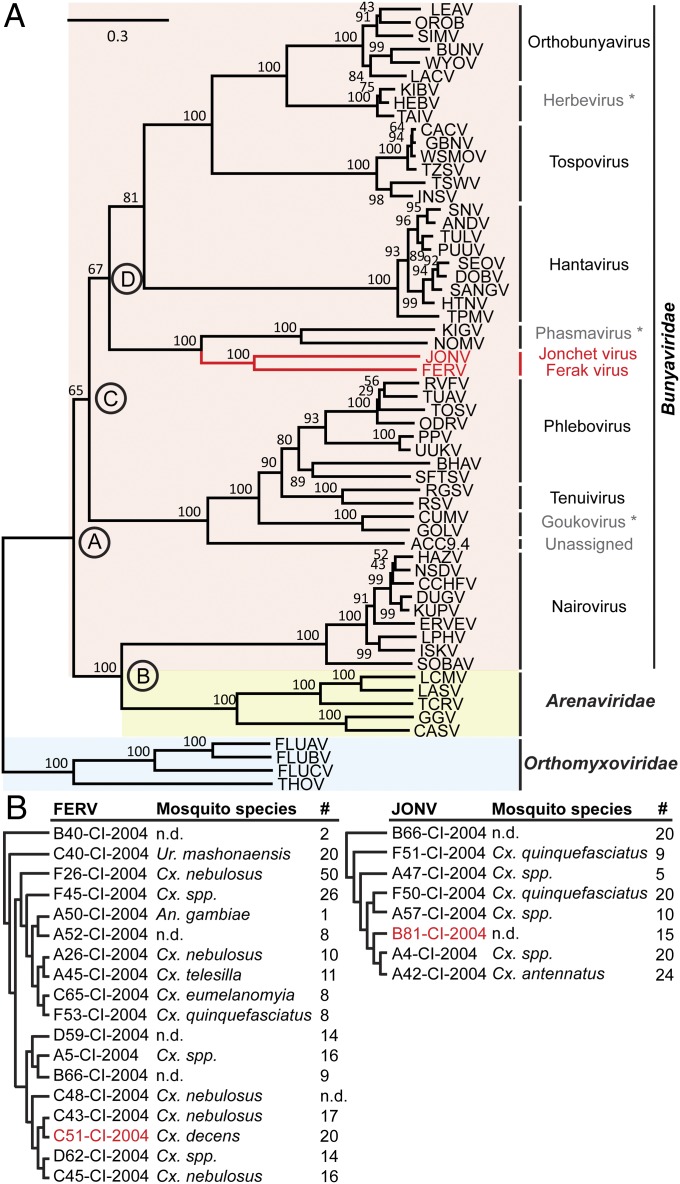
Entire prototype genomes of JONV and FERV were initially determined from infectious cell culture supernatants. RT-PCR on additional CPE-positive cell cultures inoculated with mosquito pools identified a further seven isolates of JONV and 17 isolates of FERV (Fig. 3B). Phylogenetic analysis of sequenced RT-PCR products showed segregation into three clades for FERV and at least two distinct clades for the JONV isolates (Fig. S2 A and B). To determine the purity of viral isolates, the coding regions of three JONV and FERV isolates each were sequenced by next-generation sequencing. Pairwise nt identities among JONV and FERV isolates were higher than 95.9% and 91.6%, respectively. Because of the high similarity in genome organization among isolates, only one prototype genome per virus is described here.
Fig. 3.
Phylogentic relationship of JONV and FERV. (A) ML analyses of polymerase proteins of bunyaviruses, arenaviruses, and orthomyxoviruses. Capital letters in circles indicate tree nodes for which ancestral hypothesis testing was performed (refer to Fig. 5). Bootstrap values are indicated at tree nodes. Refer to Fig. S7 for full virus names and accession numbers. (B) Cladogram and hosts of JONV and FERV isolates.
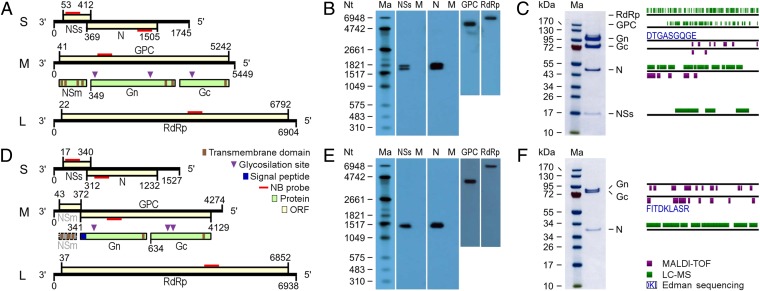
JONV and FERV prototypes were highly distinct from each other and from all other bunyaviruses. The JONV genome consisted of three segments of 1,745 nt (S), 5,449 nt (M), and 6,904 nt (L) (Fig. 2A). The FERV genome was shorter and comprised three segments of 1,527 nt (S), 4,274 nt (M), and 6,938 nt (L) (Fig. 2D). Pairwise aa identities between JONV and FERV ORFs were 22.1%, 10.3%, and 23.8% for S, M, and L segments, respectively. There were seven reverse-complementary terminal nucleotides in JONV genome segments (5′-AGUAGUA), and 11 in FERV genome segments (5′-AGUAGUAAACA). These conserved terminal sequences were different from those of any other bunyaviruses. However, six of seven terminal nucleotides shared between JONV and FERV were also present in the recently described novel clade of bunyaviruses defined by HEBV (5), as well as in all members of the genus Orthobunyavirus (5′-AGUAGU) (2), suggesting a common origin.
Fig. 2.
JONV and FERV genome organization, replication, and expression. (A and D) Schematic diagram of JONV (A) and FERV (D) genomes. (B and E) Northern blot analyses of JONV (B) and FERV (E) RNAs in infected C6/36 cells. RNA from noninfected C6/36 cells was used as mock control (M). (C and F) Major structural proteins of JONV (C) and FERV (F) analyzed by SDS/PAGE, using virions purified by gradient ultracentrifugation.
JONV and FERV L segments comprised ORFs of 2,256 and 2,271 aa in cRNA sense, respectively. Putative 256- and 262-kDa proteins most likely represented the RNA-dependent RNA polymerase (RdRp) proteins. Low similarities (identity <15%) in parts of RdRp protein sequences of bunyaviruses from all genera as well as tenuiviruses were only identified on the protein level. Similarities were only identified for the region 603LVIN-LFGY1287 of JONV and 440SQLH-PKYI1254 of FERV, which correspond to the third conserved region of the bunyavirus RdRp protein (Fig. S3). Within this region, the two novel viruses had low degrees of amino acid identity with representatives of any bunyavirus genus or unclassified bunyaviruses (Fig. S2C). Both viruses’ L proteins contained putative endonuclease domains and conserved palm motives (Fig. S3).
Maximum likelihood (ML) phylogeny using orthomyxoviruses as an outgroup yielded a stable and reconciled topology of the bunyavirus family tree (2, 6) (Fig. 3A; refer to Fig. S4 for the alignment). As proposed earlier, the family Arenaviridae was closely related to bunyaviruses, and in particular to the genus Nairovirus (8). The genus Nairovirus was placed in basal relationship to all other bunyaviruses. The next bifurcation separated the genus Phlebovirus, the clade containing GOLV (tentatively referred to as Goukovirus), and tenuiviruses from all other bunyaviruses. In sister relation to the clade containing the three deep-rooted lineages of JONV, FERV, and phasmaviruses was the common ancestor of the genera Hantavirus, Tospovirus, and Orthobunyavirus as well as the insect-restricted novel clade defined by HEBV [tentatively named Herbevirus (5)].
The M segments of JONV and FERV comprised ORFs of 1,730 and 1,262 aa in cRNA sense that are predicted to encode 193- and 144-kDa glycoprotein precursor (GPC) proteins, respectively (Fig. 2 A and D). No similarities to any known gene were identified on the nucleotide level. The translated JONV M ORF showed low similarities to the glycoprotein gene of yellow head virus (635ISYF-SRQV1541; 23% identity), a crustacean-infecting positive strand RNA virus in the family Roniviridae, order Nidovirales. There was also similarity to a shorter region to the GPC protein of hantaviruses (965IDSM-LNRV1168; 22% identity). No similarities to any viral or cellular protein were identified for the FERV GPC. Putative transmembrane domains, N-linked glycosylation sites, and signal peptide cleavage sites were predicted (Fig. 2 A and D). The FERV M segment encodes a second short ORF in a −1 reading frame that overlaps the N terminus of the GPC ORF by 32 nt (Fig. 2D). For the predicted 12-kDa protein, no similarities to any viral or cellular proteins were identified.
The JONV and FERV S segments comprised ORFs of 372 and 306 aa in cRNA sense that putatively encode 42- and 33-kDa proteins, respectively (Fig. 2 A and D). These correspond on genome position to nucleoprotein ORFs in other bunyaviruses. No primary sequence similarities between JONV and FERV nucleoprotein ORFs and those of other bunyaviruses were identified. Notably, both novel viruses did not encode ORFs for a putative NSs protein, according to coding strategies used for NSs proteins in other bunyaviruses, such as an NSs ORF fully overlapping the N ORF in orthobunyaviruses or an NSs ORF encoded in ambisense in phlebovirus and tospovirus S segments (2). Both viruses comprised ORFs of 119 and 107 aa in a −1 reading frame in cRNA sense upstream of the putative N ORF that were predicted to encode proteins of 13 and 12 kDa, respectively. No conserved protein motifs could be identified for the unassigned ORFs. The mutation frequency in these small ORFs compared with N, GPC, or RdRp ORFs was similar, suggesting the small ORFs are expressed (Fig. S2D; also see following).
Genome Replication, Transcription, and Expression.
Bunyavirus replication involves the transcription of negative-sense viral RNA into cRNA acting as the replicative intermediate, with concomitant transcription of a shorter form of coding-sense RNA acting as the mRNA for protein expression (2). Genomic-length and slightly shorter virus-specific RNAs for the S segment were detected in cells infected with JONV by Northern blot analysis (Fig. 2B). For the JONV M and L segments, as well as for all FERV genome segments, only one RNA species was detected by Northern blot analysis (Fig. 2 B and E). To discriminate vRNA and mRNA, the amounts of positive- and negative-sense viral RNA were quantified by hot-started strain-specific reverse transcription and real-time PCR (Fig. S5). Genome RNA exceeded mRNA initially. Over time, mRNA exceeded genome RNA until both species reached similar levels at 24 h postinfection (hpi), suggesting that both RNA polarities existed in cells and mRNAs were not discriminable in size from vRNAs by Northern blot analysis. Northern blot analysis with S segment-specific probes upstream of the putative NSs ORF yielded no differences in sizes of detected RNA, suggesting no additional mRNA is transcribed for the putative NSs ORFs (Fig. 2 B and E). Bunyavirus mRNAs typically contain 5′-nonvirally templated elements obtained from host cell mRNAs by a cap-snatching mechanism (9, 10). For confirmation, cell lysates were subjected to 5′-rapid amplification of cDNA ends (RACE) with subsequent cloning and analysis of five cDNA clones with appropriate insert sizes per genome segment per virus. Between six and 23 nonvirally templated residues were detected at 5′-ends (Fig. S6), indicating cap-snatching as in other bunyaviruses.
Expressed structural proteins and glycoprotein cleavage sites of JONV and FERV were assessed by SDS/PAGE, followed by limited tryptic digestion and MALDI-TOF, liquid chromatography mass spectrometry analysis, and Edman degradation from gradient purified viral particles. Six proteins of about 250, 200, 100, 80, 50, and 15 kDa were identified for JONV and mapped to the RdRp, GPC, Gn, Gc, and N genes, as well as to the unassigned ORF upstream of the N ORF, respectively (Fig. 2C). According to the nomenclature used for bunyaviruses, the protein was named NSs. The N-terminal sequence of the Gn protein was identified to start at 349DTGA, suggesting that the region upstream of the Gn protein codes for a putative NSm protein of 39 kDa. For FERV, three prominent bands corresponding to 80, 75, and 40 kDa were observed and confirmed to represent the Gn, Gc, and N proteins (Fig. 2F). The observed molecular masses for Gn and Gc were higher than the predicted ones and were compatible with glycosylation. Whereas the weight of JONV glycoproteins was reduced by treatment with peptide-N-glycosidase F, no such evidence for N-linked glycosylation was found for FERV (Fig. S7).
In Vitro Host Range and Sensitivity to Temperature.
To determine growth kinetics of the novel viruses, end-point infectious titers were first determined by parallel serial dilution experiments in C6/36 cell cultures, followed by microscopic inspection for CPE 7 dpi. Peak viral titers were 1.78 × 108 and 1.33 × 108 tissue culture infectious dose 50/mL for JONV and FERV, respectively. To determine growth kinetics, C6/36 cell cultures were inoculated with virus at defined multiplicities of infection (MOIs), and supernatants were sampled daily. Determination of virus RNA concentrations in supernatants by real-time RT-PCR indicated that FERV took 1–2 d longer than JONV to reach peak titers, in particular at lower MOI (Fig. S1 C and D). Peak RNA concentrations in absolute quantitative RT-PCR ranged around 1010 copies/mL for both viruses. Similar growth characteristics were observed for JONV in U4.4 cells, an Aedes albopictus cell line that is competent for the RNAi pathway (Fig. S1C). Growth of FERV was delayed and only detected after 48 hpi in U4.4 cells, reaching RNA concentrations similar to JONV thereafter (Fig. S1D). In contrast to C6/36 cells, both viruses did not induce cytopathic effects in U4.4 cells. The cell line C7/10 derived from Aedes albopictus larvae also supported replication of both viruses (Fig. S1 E and F).
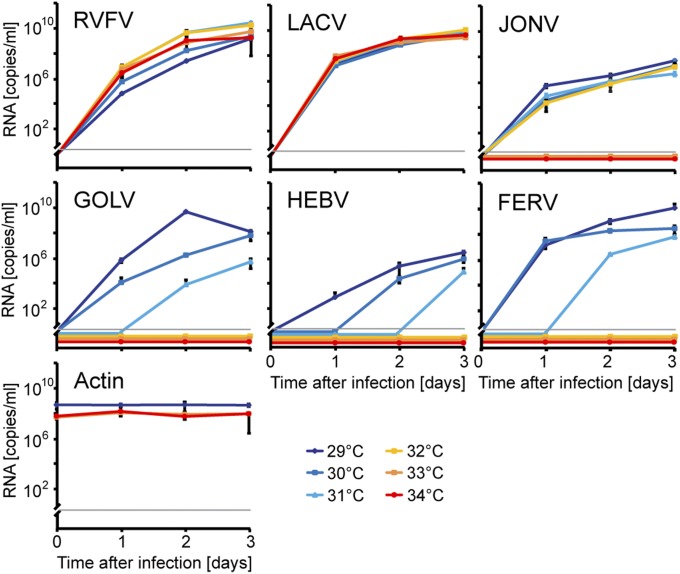
To determine the potential to grow in vertebrate cells, primate, rodent, bat, goat, and frog cells were inoculated with both viruses and cultivated for 4 wk. No growth, as evident by real-time RT-PCR, was seen in any of the vertebrate cell cultures with any virus (Fig. S1 E and F). Temperature permissiveness was used as an additional criterion for potential vertebrate tropism. To compare JONV’s and FERV’s permissiveness at vertebrate body temperatures, arboviruses were selected from the two pathogenic bunyavirus genera Phlebovirus and Orthobunyavirus, as well as their insect-restricted sister taxa, Goukovirus and Herbevirus, and cultured under temperature gradients. This involved Rift Valley fever virus (RVFV) as an arbovirus representative for the genus Phlebovirus and GOLV as a bona fide insect-restricted virus of the Goukovirus clade (4). For the genus Orthobunyavirus, the arbovirus La Crosse virus as well as the prototypic insect-restricted virus HEBV were chosen (5). Both tested arboviruses replicated efficiently across the whole temperature range of 29–34 °C (Fig. 4). All four bona fide insect-restricted viruses were impaired in replication above 32 °C and completely blocked above 33 °C. Some viruses were impaired already, starting from 29 °C. FERV and JONV were completely blocked above 30 and 32 °C, respectively. To exclude that JONV’s and FERV’s inability to replicate in vertebrate cells might only be determined by temperature conditions, JONV, FERV, GOLV, and HEBV were infected in Vero cells at MOI of 10 and incubated at the permissive temperature of 30 °C. No viral replication was evident by real-time RT-PCR for any of the viruses, whereas RVFV and La Crosse virus (LACV) replicated as previously observed.
Fig. 4.
Temperature sensitivity of bunyaviruses. C6/36 cells were infected with the indicated viruses, and cells were incubated at temperatures from 29 to 34 °C. Viral genome copy numbers were measured by real-time RT-PCR. Actin copy numbers in mock-infected cells were determined for 29, 33, and 34 °C. Limit of detection is indicated by a gray line.
Ancestral Reconstruction.
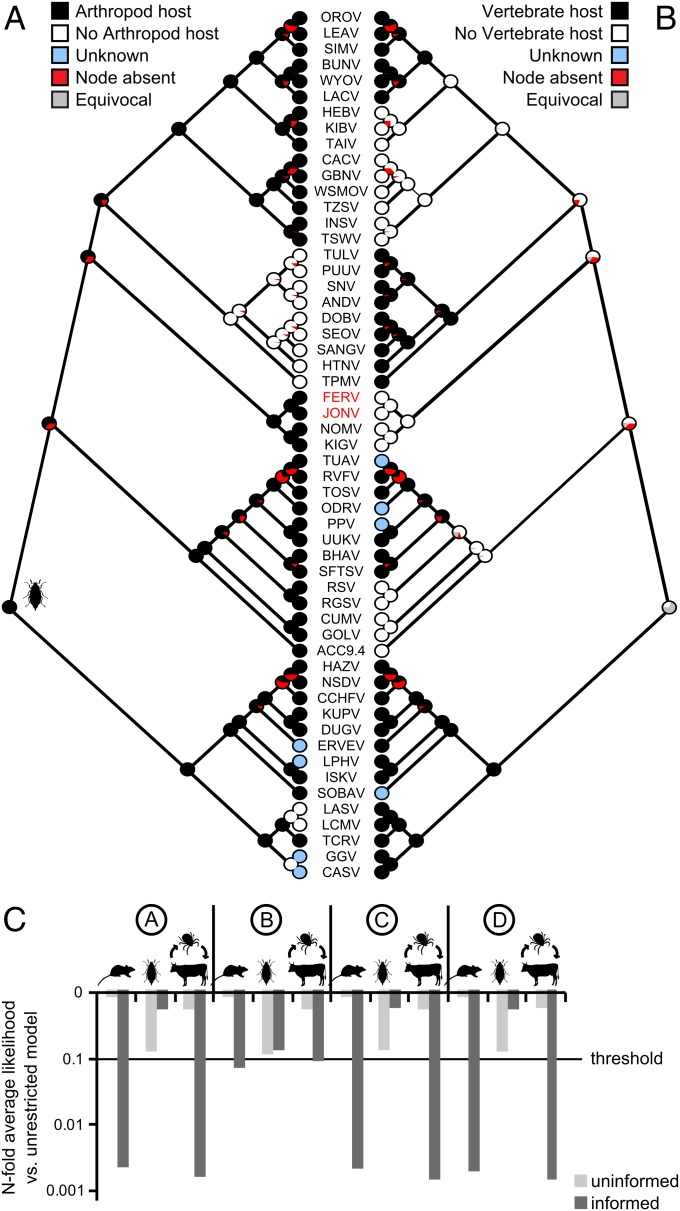
To determine whether the novel viruses might have evolved independently to be restricted to arthropods or inherited their arthropod restriction as a property from common ancestors, phylogeny-based reconstruction of ancestral traits was attempted. We used a parsimony-based algorithm that reconstructs ancestral traits at all internal tree nodes by calculating the minimum number of trait changes along the tree that is necessary to explain the present state of host associations at tree tips (11). The model used two binary host traits that identify whether a virus has an arthropod host (yes/no) or vertebrate host (yes/no). Using this approach, an arthropod host was reconstructed at the bunyavirus root in 100% of 1,000 bootstrap tree replicates used for the analysis (Fig. 5A). Association with a vertebrate host could not be deduced (Fig. 5B). However, these analyses did not account for the uncertainty about associations with vertebrate hosts for a number of novel viral taxa included in the tree (JONV, FERV, Hebeviruses, Goukoviruses, Phasmaviruses). Moreover, parsimony-based models cannot take branch lengths into account, which vary considerably in the given tree. To incorporate branch length information and optionally include new knowledge about host associations based on cell culture studies, we conducted probabilistic hypotheses testing in a maximum likelihood framework (12, 13). This approach determines the most likely trait change matrix along the bunyavirus phylogeny as well as the loss of likelihood that occurs when restricting traits at given tree nodes (12, 13). Host properties were ascribed to tree taxa in the form of combined binary traits as summarized in Table S1. In one dataset version termed the uninformed dataset, known host traits were ascribed to all bunyaviruses except the novel viruses JONV, FERV, GOLV, HEBV, and KIBV, for which host traits were left open. In an alternative, informed dataset, the host trait information obtained from cell culture infection experiments in the present study was added. Using the program Bayestraits (12), a hypothesis-free reconstruction run was performed for reference, recording the median likelihood of trait change reconstructions over 1,000 bootstrap tree replicates. Fossil host assumptions were then defined at deep tree nodes, restricting the optimization space for the ML algorithm. This analysis was conducted on three different alternative tree topologies, as shown in Fig. S8. Analysis of the uninformed dataset failed to reject any host hypotheses on a high significant level (Fig. 5C and Fig. S8). Only the analysis of the informed dataset convincingly rejected vertebrate hosts and dual hosts at all deep nodes, including the root. This was unanimously the case for all alternative tree topologies, including a topology that assumed arenaviruses to belong to the bunyavirus family, as proposed earlier (8) (Fig. S8). Vertebrate hosts at all analyzed deep nodes were 158–794-fold less likely than arthropod hosts. Dual hosts were 63–398-fold less likely. Hypotheses of exclusive arthropod association at all deep nodes including the root left the overall likelihood unaffected (Fig. 5C and Fig. S8).
Fig. 5.
Ancestral reconstruction of bunyavirus hosts. (A and B) Parsimony-based ancestral reconstruction of arthropod (A) and vertebrate (B) host associations. “Node absent” refers to the fraction of 1,000 bootstrap tree replicates in which the node of interest was not supported (i.e., loss of monophyly of dependent clade). (C) Hypotheses testing based on ML inferences of trait change models. Hypothesized (fossilized) ancestral host assumptions at deep tree nodes A–D (refer to Fig. 3) are symbolized by vertebrate and arthropod silhouettes. Bars represent the resulting loss of likelihood of trait change models conferred by fossilization (averaged results over 1,000 bootstrap tree replicates). The significance threshold was 10-fold loss of likelihood.
Discussion
In this study we characterized prototype strains and numerous additional viral isolates representing two novel groups of viruses within the family Bunyaviridae. The type viruses, JONV and FERV, branch from an old common ancestor in sister relationship to unclassified phasmavirid sequences (6). The novel viruses and Phasmaviruses branch deep in the bunyavirus family tree and in sister relationship to a viral superclade comprising the three established genera Orthobunyavirus, Tospovirus, and Hantavirus, as well as the novel unclassified clade of Herbeviruses (5). The lengths of the phylogenetic stem lineages of JONV and FERV, respectively, are similar to stem lineage lengths for other accepted genera including Orthobunyavirus, Tospovirus, Phlebovirus, and Nairovirus. The deep phylogenetic separation between JONV and FERV and the low level of amino acid identity suggest these viruses define two independent novel genera. Kigluaik phantom virus (KIGV) and Nome phantom virus (NOMV) might as well define two additional independent genera, pending further study of live viral isolates (6).
Unfortunately, the classification of bunyavirus genera does not obey any strictly defined criteria and would thus, in addition to phylogenetic analyses, have to rely on genetic, structural, or functional features that typically vary between and are conserved within accepted genera. As bunyaviruses are not known to show morphological heterogeneities within genera, the striking morphological differences between JONV and FERV could be considered as evidence for different genera. A genetic auxiliary criterion is the composition of genome segment termini. Seven nucleotides (AGUAGUA) are conserved between JONV and FERV, which is the same number of nucleotides conserved between the genus Orthobunyavirus and its insect-restricted sister taxon Herbevirus. It is remarkable that eight additional nucleotides are shared between JONV L and S segments, but not the M segment, suggesting the M segment might have been acquired from an unknown source by reassortment after the separation of the FERV-specific stem lineage. However, reassortment within bunyavirus genera has not been investigated sufficiently to use it as a formal criterion for taxonomic classification.
Further criteria that are different between but shared within bunyavirus genera include the existence and coding strategy for noncoding elements as well as overall genome segment length variation. Another important difference exists in growth kinetics, particularly in RNAi-competent insect cells. Whereas the presence of an NSs ORF upstream of the N ORF is a unique feature of JONV and FERV, it represents a commonality rather than a contrast between the two taxa. NS proteins are typically involved in viral replication (14, 15) or host cell interference (16–18). ORFs compatible with NSs were also detected in KIGV and NOMV that share a most recent common ancestor (MRCA) with JONV and FERV (6). The striking differences in CPE, virion morphology, genome organization, and sizes of expressed proteins, as well as phylogenetic separation, provide support for the definition of JONV and FERV as independent taxonomic entities at the rank of genera. If we add these viruses to our previous findings of Herbeviruses and Goukoviruses, four highly diverged clades that might form novel genera are now known in mosquitoes originating from one tropical rainforest region (4, 5). These data nearly double the number of major bunyavirus taxa and suggest that more diverse bunyaviruses exist in other regions and arthropod species. Phasmavirids may not provide the only further example (6).
We are currently lacking consensus experimental conditions to evaluate whether previously unknown pathogens discovered in hematophagus arthropods are arboviruses or arthropod-specific viruses. Classification of arthropod-specific viruses has so far relied on multiple negative in vitro and in vivo experiments, such as the absence of viral replication in cell lines derived from different vertebrate hosts (4, 5, 19–24) or replication studies in intracerebrally inoculated newborn mice (20, 23, 25, 26). In the case of flaviviruses, in vitro replication studies of mosquito-borne, tick-borne, and insect-specific viruses, as well as vertebrate flaviviruses of the “no known-vector” group, were in agreement with the natural host range and phylogenetic grouping of these viruses, providing evidence that in vitro replication studies can be useful for host range evaluation (19). Additional in vivo infection experiments, particularly in neonatal mice, have been used, but to our knowledge they yielded no examples of arboviruses replicating in neonatal mice if they cannot replicate in vertebrate cell culture. To provide an ecologically more relevant scenario and avoid animal experiments, we chose temperature gradient kinetics in cell culture. Arboviruses have to be capable of replicating at high temperatures according to the body temperature of mammals and birds (36.5–42 °C). In contrast, arthropod-restricted viruses should be adapted to ambient temperatures, which range around 28 °C in the studied tropical rainforest habitat. Temperature gradient experiments followed the rationale that departure from ambient temperature will only be tolerated if the virus has adapted the capability to replicate at higher temperature by selection in dual host replication cycles (27). Prototype arbo-bunyaviruses from the genera Phlebovirus and Orthobunyavirus indeed were not affected by temperature, as expected because of their vertebrate tropism. As expected, all tentative arthropod-specific viruses were highly sensitive to temperature. Viral replication was completely blocked at temperatures above 31–33 °C, depending on the virus. In summary, these findings suggest that JONV and FERV, as well as the clades containing GOLV and HEBV, are insect-specific viruses without any vertebrate tropism. Vertical transmission (transovarial and transveneral) plays a major role for the transmission of insect-specific flaviviruses (28, 29). Other routes of transmission may also be important for the insect-specific bunyaviruses that have diverse insect host associations.
Our studies of ancestral trait reconstruction have taken these new data on insect-specific viruses into account. Whereas the parsimony-based approach could not provide any conclusive reconstruction of ancestral traits, particularly for the vertebrate association of ancestral bunyaviruses, the more comprehensive hypothesis testing studies led to a clear rejection of vertebrate hosts at deep bunyavirus nodes, including the common bunyavirus ancestor. In addition, a dual host tropism was rejected if informing the model with the novel experimental insight from temperature gradient cultures. In summary, these data suggest arbo-bunyaviruses have evolved from arthropod-specific ancestors. Rejection of the vertebrate host hypothesis for several deep sister nodes in the bunyavirus tree implies that vertebrate or dual host tropism must have evolved several times convergently. The existence of different arbo-bunyaviruses in humans or mammalian livestock demonstrates that no barriers such as population immunity exist against convergent conquest of host. For the genus Hantavirus, our analysis infers that arthropod tropism has been lost in favor of vertebrate monotropism, rather than preserving vertebrate tropism from ancestral viruses. This may have happened with or without a transitory stage via dual host tropism (30). It is tempting to speculate that the loss of dual or arthropod tropism must have taken place in mammals ancestral to bats and insect eaters, whose extant relatives seem to contain the largest diversity of hantaviruses (31, 32), and come in contact with arthropods intensively via diet. Similar scenarios may be applicable to explain the origin and evolution of other viral families that contain arboviruses and vertebrate viruses at the same time, such as the flavivirus family, wherein at least three extant genera (Hepacivirus, Pegivirus, Pestivirus) are thought to have exclusive vertebrate tropism.
Methods
Virus Characterization.
Virus isolation was performed on C6/36 cells, and entire genomes were sequenced by next-generation sequencing. Details on growth analyses, Northern blotting, and protein sequencing are provided in SI Methods.
Phylogenetic Analyses.
ML phylogenies were inferred in PhyML, and ancestral host reconstructions were performed using Mesquite and Bayestraits. Details are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank F. Leendertz for assistance with fieldwork design and F. Weber for providing RVFV and LACV, as well as P. Trippner for technical assistance. We thank the Ivorian Ministry of Environment and Forest, the Ministry of Research, and the directorship of the Taï National Park to permit mosquito sampling. The project was supported by the Deutsche Forschungsgemeinschaft (Grant Agreement JU 2857/3-1 to S.J. and DR722/10-1 to C.D.) and by the Deutsches Zentrum für Infektionsforschung (infrastructural support to C.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.G. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP710232, KP710238–KP710245, KP710246, KP710262–KP710269, KP710233–KP710237, and KP710247–KP710261).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502036112/-/DCSupplemental.
References
- 1.Cook S, et al. Molecular evolution of the insect-specific flaviviruses. J Gen Virol. 2012;93(Pt 2):223–234. doi: 10.1099/vir.0.036525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plyusnin A, et al. Family Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; London: 2012. [Google Scholar]
- 3.Schmaljohn C, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 2. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1741–1789. [Google Scholar]
- 4.Marklewitz M, et al. Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J Virol. 2011;85(17):9227–9234. doi: 10.1128/JVI.00230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marklewitz M, et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J Virol. 2013;87(23):12850–12865. doi: 10.1128/JVI.01862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballinger MJ, Bruenn JA, Hay J, Czechowski D, Taylor DJ. Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes. J Virol. 2014;88(16):8783–8794. doi: 10.1128/JVI.00531-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junglen S, et al. Examining landscape factors influencing relative distribution of mosquito genera and frequency of virus infection. EcoHealth. 2009;6(2):239–249. doi: 10.1007/s10393-009-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieth S, Torda AE, Asper M, Schmitz H, Günther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318(1):153–168. doi: 10.1016/j.virol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Bishop DH, Gay ME, Matsuoko Y. Nonviral heterogeneous sequences are present at the 5′ ends of one species of snowshoe hare bunyavirus S complementary RNA. Nucleic Acids Res. 1983;11(18):6409–6418. doi: 10.1093/nar/11.18.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons JF, Pettersson RF. Host-derived 5′ ends and overlapping complementary 3′ ends of the two mRNAs transcribed from the ambisense S segment of Uukuniemi virus. J Virol. 1991;65(9):4741–4748. doi: 10.1128/jvi.65.9.4741-4748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddison WP, Maddison DR. 2014. Mesquite: A modular system for evolutionary analysis. Available at mesquiteproject.org. Accessed April 14, 2015.
- 12.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53(5):673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 13.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabtree MB, et al. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: Potential role for NSm in mosquito infection. PLoS Negl Trop Dis 2012;6(5):e1639.
- 15.Szemiel AM, Failloux AB, Elliott RM. 2012. Role of Bunyamwera Orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl Trop Dis 6(9):e1823. [DOI] [PMC free article] [PubMed]
- 16.Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci USA. 2001;98(2):664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda A, et al. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002;532(1-2):75–79. doi: 10.1016/s0014-5793(02)03632-3. [DOI] [PubMed] [Google Scholar]
- 18.Won S, Ikegami T, Peters CJ, Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol. 2007;81(24):13335–13345. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuno G. Host range specificity of flaviviruses: Correlation with in vitro replication. J Med Entomol. 2007;44(1):93–101. doi: 10.1603/0022-2585(2007)44[93:hrsofc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Evangelista J, et al. Characterization of a novel flavivirus isolated from Culex (Melanoconion) ocossa mosquitoes from Iquitos, Peru. J Gen Virol. 2013;94(Pt 6):1266–1272. doi: 10.1099/vir.0.050575-0. [DOI] [PubMed] [Google Scholar]
- 21.Junglen S, et al. A new flavivirus and a new vector: Characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J Virol. 2009;83(9):4462–4468. doi: 10.1128/JVI.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huhtamo E, et al. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J Virol. 2009;83(18):9532–9540. doi: 10.1128/JVI.00529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huhtamo E, et al. Novel flaviviruses from mosquitoes: Mosquito-specific evolutionary lineages within the phylogenetic group of mosquito-borne flaviviruses. Virology. 2014;464-465:320–329. doi: 10.1016/j.virol.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasar F, et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci USA. 2012;109(36):14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auguste AJ, et al. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J Gen Virol. 2014;95(Pt 2):481–485. doi: 10.1099/vir.0.058412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Attoui H, et al. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: Isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus) Virology. 2005;343(2):212–223. doi: 10.1016/j.virol.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 27. Aliota MT, Kramer LD (2012) Replication of West Nile virus, Rabensburg lineage in mammalian cells is restricted by temperature. Parasit Vectors 5:293. [DOI] [PMC free article] [PubMed]
- 28.Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg. 2011;85(1):169–177. doi: 10.4269/ajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saiyasombat R, Bolling BG, Brault AC, Bartholomay LC, Blitvich BJ. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae) J Med Entomol. 2011;48(5):1031–1038. doi: 10.1603/me11043. [DOI] [PubMed] [Google Scholar]
- 30.Yu XJ, Tesh RB. The role of mites in the transmission and maintenance of Hantaan virus (Hantavirus: Bunyaviridae) J Infect Dis. 2014;210(11):1693–1699. doi: 10.1093/infdis/jiu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo WP, et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9(2):e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plyusnin A, Sironen T. Evolution of hantaviruses: Co-speciation with reservoir hosts for more than 100 MYR. Virus Res. 2014;187:22–26. doi: 10.1016/j.virusres.2014.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.