Abstract
Telomeric chromosome rearrangements may cause mental retardation, congenital anomalies, and miscarriages. Automated detection of subtle deletions or duplications involving telomeres is essential for high-throughput diagnosis, but impossible when conventional cytogenetic methods are used. Array-based comparative genomic hybridization (CGH) allows high-resolution screening of copy number abnormalities by hybridizing differentially labeled test and reference genomes to arrays of robotically spotted clones. To assess the applicability of this technique in the diagnosis of (sub)telomeric imbalances, we here describe a blinded study, in which DNA from 20 patients with known cytogenetic abnormalities involving one or more telomeres was hybridized to an array containing a validated set of human-chromosome–specific (sub)telomere probes. Single-copy-number gains and losses were accurately detected on these arrays, and an excellent concordance between the original cytogenetic diagnosis and the array-based CGH diagnosis was obtained by use of a single hybridization. In addition to the previously identified cytogenetic changes, array-based CGH revealed additional telomere rearrangements in 3 of the 20 patients studied. The robustness and simplicity of this array-based telomere copy-number screening make it highly suited for introduction into the clinic as a rapid and sensitive automated diagnostic procedure.
Introduction
Genomic imbalances can cause mental retardation, congenital malformations and miscarriages. The best known example of a genetic imbalance compatible with life is Down syndrome, which is caused by trisomy of chromosome 21 and is present in 1 of 700 newborn children. Relatively small genomic deletions or duplications may also result in clinical phenotypes. Well-known microdeletion syndromes include the Williams (7q11.2), Prader Willi (15q12), Angelman (15q12), Wolf-Hirschhorn (4p16.3), and DiGeorge (22q11.21) syndromes (Battaglia and Carey 1998; Cassidy et al. 2000; Donnai and Karmiloff-Smith 2000; Scambler 2000). In recent years, the development and application of various subtelomeric probes for FISH and microsatellite markers from these regions, led to the awareness that (sub)telomeric regions are often involved in chromosomal rearrangements not visible by routine cytogenetics. These submicroscopic subtelomeric chromosome rearrangements are a significant cause of mental retardation with or without congenital anomalies (Flint et al. 1995; Knight et al. 1999; de Vries et al. 2000; Knight and Flint 2000).
The development of an accurate and sensitive method of screening all subtelomeric regions can facilitate the clinical diagnosis of a considerable group of patients. FISH-based approaches, which are currently employed in the majority of diagnostic centers, are hampered by the need for high-quality metaphase spreads and the limited number of chromosomal loci that can be screened in a single reaction. Array-based comparative genomic hybridization (CGH), the application of CGH to an array of mapped DNA fragments immobilized on glass slides (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Snijders et al. 2001), is rapidly becoming the method of choice for high-resolution screening of genomic copy-number changes (Antonarakis 2001). Array-based CGH builds upon previously well-established CGH procedures (Kallioniemi et al. 1992; Weiss et al. 1999; Lichter et al. 2000) by use of differentially labeled test (patient) and reference (normal) DNAs, to be cohybridized, under in situ suppression hybridization conditions, to cloned genomic fragments in a miniaturized (arrayed) format on a chip. The hybridized DNAs are detected in two different fluorochromes and digitized intensity differences in the hybridization patterns of the DNAs onto these cloned fragments can be interpreted as copy-number differences between the test and reference genomes. This technique, once established and validated, will allow high-throughput “cytogenetic” diagnosis (molecular karyotyping) with a theoretically unlimited resolution. It has already been shown to be useful in rapid amplicon identification (Albertson et al. 2000), differential tumor diagnosis (Wilhelm et al. 2002), and the biological dissection of the role that copy-number changes play in different tumor types (Bruder et al. 2001). The primary purpose of this study was to test the feasibility of array-based CGH for copy-number screening of subtelomeric regions in patients with mental retardation with or without congenital anomalies.
A previously published second-generation set of human-chromosome–specific (sub)telomeric probes (Knight et al. 2000) was used as target for array-based CGH. A blinded study with 20 cytogenetically well-characterized patient samples was performed on these arrays together with a number of normal-versus-normal control and other reproducibility experiments. FISH validation was performed in those patients in which discrepancies between array-based CGH and the conventional cytogenetic analysis were observed. Furthermore, we tested the use of degenerate oligonucleotide-primed PCR (DOP-PCR) (Telenius et al. 1992) protocols for the generation of large amounts of clone DNA and compared the performance of array-based CGH on DOP-PCR products with that of array-based CGH on primary clone DNAs.
Patients, Material, and Methods
Patients
A series of 18 patients was selected from our Clinical Genetics patient database on the basis of microscopic chromosomal rearrangements involving at least one subtelomeric region (table 1). In addition, two patients (1 and 19) with an interstitial deletion were included as negative controls (the study was performed in a blinded fashion). In most patients, a direct and accurate cytogenetic diagnosis was possible. In two patients (13 and 14), however, an unambiguous karyotype could not be established because of the limited resolution of the banded chromosomes. Karyotypes from all patients, together with previously performed FISH analyses, were re-evaluated, and the abnormalities were confirmed. All cytogenetic diagnoses were performed on direct cultures of patient material. Peripheral blood lymphocytes from each patient were Epstein-Barr–virus–transformed, and genomic DNAs were isolated from short-term cultures by use of standard procedures. This DNA was used for array-based CGH, as well as FISH and, in one patient, chromosomal CGH. DNAs isolated from blood lymphocytes from cytogenetically normal healthy persons were used as references in the CGH experiments.
Table 1.
Patients, Reasons for Referral, Karyotypes, and Array-Based CGH Results, Including Comparisons with the Original Karyotypes
| Patient | Reason forReferrala | Karyotype | Array-Based CGH | Array-Based CGH vs. Karyotype |
| 2 | MCA | 46,XY,del(18)(q21.2) | Loss 18qterb | Match |
| 3 | MCA/MR | 46,XX,del(18)(p11.2) | Loss 18pter | Match |
| 4 | MCA/MR | 46,XX,del(10)(q26.1) | Loss 10qter | Match |
| 5 | MCA/MR | 46,XX,der(3)t(3;16)(p25;q22) | Loss 3pter, gain 16qter | Match |
| 7 | MCA/MR | 46,XY,del(10)(q26.13) | Loss 10qter | Match |
| 9 | PD | 46,X,del(X)(p22.1) | Loss XpYpter | Match |
| 10 | MCA/MR | 46,XY,del(7)(q36) | Loss 7qter | Match |
| 11 | MR | 46,XY,der(21)t(21;22)(p10;q13.3) | Gain 22qter | Match |
| 12 | MR | 46,XY,der(2)t(2;?)(q37.3;?)c | Loss 2qter | Match |
| 15 | MR | 46,XX,del(2)(q37.3) | Loss 2qter | Match |
| 16 | MCA | 46,XX,der(20)t(20;?)(q13.3;p?)d | Loss 20qter | Match |
| 17 | MR | 46,XX,del(13)(q33) | Loss 13qter | Match |
| 20 | Turner syndrome | 46,XX,del(11)(q23.3) | Loss 11qter | Match |
| 1 | MCA | 46,XY,del(9)(p22p24.2) | Normal | Match (interstitial deletion) |
| 19 | MR | 46,XY,del(3)(q27.3q29) | Normal | Match (interstitial deletion) |
| 6 | MCA | 46,XY,del(13)(q32.2) | Normal | FISH:13qter normal, deletion interstitial |
| 13 | Down syndrome | 46,XX,add(2)(q36.2) | Normal | Chromosome CGH: 2qter normal, gain of 8q24; insertion interstitial |
| 8 | MCA/MR | 45,XY,der(13;14)(q10;q10), dup(17)(qterq24.2) | Gain 17qter and loss 17qter | FISH: gain and loss confirmed, inversion duplication followed by deletion of distal part 17qter |
| 14 | MCA | 46,XX,der(9)t(9;?)(p24;?) | Loss 9pter and gain 7pter | FISH: gain of 7pter confirmed, translocation partner identified |
| 18 | MCA | 46,XY,dup(8)(p23p12) | Loss 8pter | FISH: loss of 8pter confirmed, inversion duplication followed by deletion distal part of 8pter |
MCA = multiple congenital abnormalities; MR = mental retardation; PD = prenatal diagnosis (delXp22.1 present in parent).
The pter and qter clones from the second second-generation set of human-chromosome–specific (sub)telomere probes (Knight et al. 2000).
Satellite at chromosome 2.
Satellite at chromosome 20.
Clones
Part of the second-generation set of human-chromosome–specific (sub)telomere probes (Knight et al. 2000) was kindly supplied by Drs. Regina Regan and Jonathan Flint (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, United Kingdom). The remaining clones were commercially obtained (Incyte Genomics and Invitrogen). In addition to the 77 clones from this second-generation set, 3 cosmid clones from the first-generation set were added (Ning et al. 1996). All clones (n=80) were colony purified prior to culturing. Genomic target DNAs were isolated from 200-ml bacterial cultures by use of QIAgen Plasmid Midi Kits (QIAgen), according to the manufacturer's instructions. All clones were PCR verified by use of the STSs developed by Knight et al. (2000), and their mapping positions were confirmed by FISH on normal metaphase spreads by use of routine procedures. DNA from a total of 77 clones passed STS verification and mapped to the correct telomere location. The three clones that did not pass this test were GS-240-G10 (16q), GS-48-O23 (19q), and GS-202-M24 (Xq). DNAs from these clones were subsequently used in the array-based CGH experiments. All of the 41 human subtelomeric chromosome regions were represented, by either one (9 regions), two (29 regions), or three (3 regions) clones.
Array-Based CGH
DNAs from all clones were sonicated to generate fragments 300–3,000 bp in size and were dissolved at a concentration of 1 μg/μl in a 80% dimethyl sulfoxide solution containing 0.3 μg/μl nitrocellulose. The clone DNAs were robotically spotted in triplicate onto glass slides coated with Corning Microarray Technology gamma amino propyl citrate (Corning) by use of an Affymetrix 417 arrayer. The center-to-center spacing of the spots was 250 μm, and the size of the array was 1.44 cm2. Slides were air dried overnight and DNA was ultraviolet crosslinked at 90 mJ by use of a Stratalinker (Stratagene). DNA samples were coded, and an investigator, without knowledge of the cytogenetic diagnosis, performed the array-based CGH analysis. Genomic DNA, from test and reference samples, was also sonicated to generate fragments 300–3,000 bp in size. This DNA was labeled by random priming in a total volume of 80 μl. For this labeling, 500 ng DNA was mixed with 50 mM Tris-HCl (pH 6.8), 5 mM MgCl2, 10 mM 2-mercaptoethanol, and 300 μg/ml random octamers (Bioprime DNA labeling system; 1× Random Primers solution, Invitrogen) and was denatured at 100°C for 10 min. After cooling to 4°C, 64 U Klenow fragment (Bioprime DNA labeling system, Invitrogen), 2 mM dGTP, 2 mM dCTP, 2 mM dATP, 1 mM dTTP (Invitrogen), and 6 nmol fluorolink Cy3-dUTP (test) or Cy5-dUTP (reference) (Amersham Pharmacia Biotech) were added. This solution was incubated for 2 h at 37°C, after which unincorporated fluorescent nucleotides were removed by use of Sephadex G-50 spin columns (Amersham Pharmacia Biotech). Test and reference samples were mixed with 100 μg Cot-1 DNA (Invitrogen), were coprecipitated, and were resuspended in 15 μl of a hybridization solution containing 50% formamide, 10% dextran sulfate, 2× saline sodium citrate (SSC), 4% SDS and 100 μg yeast tRNA (Invitrogen). The hybridization solution was denatured for 10 min at 72°C and subsequently was incubated for 30 min at 37°C, to allow blocking of repetitive sequences. Subsequent hybridization was performed at 37°C under a sealed coverslip for 48 h by use of an Omnislide thermal cycling block (Thermo Hybaid). This was followed by a 15-min posthybridization wash in 50% formamide/2× SSC at 45°C, and a 10-min wash in phosphate buffer at room temperature, both by use of the Omnislide heated wash module (Thermo Hybaid). Slides were dried, after a brief ethanol wash, and were imaged on an Affymetrix 428 scanner (Affymetrix) by use of the Affymetrix 428 scanner software package (version 1.0).
Image Analysis and Processing
The acquired microarray images were analyzed by use of Genepix Pro 3.0 (Axon Instruments). DNA spots were automatically segmented, local background was subtracted, and total intensities—as well as the fluorescence-intensity ratios of the two dyes—were calculated for each spot. For all calculations, we used the median of the pixel-by-pixel ratios of the pixel intensities that have had the median local background intensity subtracted. Spots with poor-quality printing and/or hybridization (indicated in Genepix Pro 3.0 by “Rgn R2 <0.5”), spots with a low signal intensity (indicated in Genepix Pro 3.0 by “%>B635+2SD<30” for the Cy5 channel and “%>B532+2SD<30” for the Cy3 channel), and spots showing autofluorescent particles over the target DNAs were discarded. The mean test-over-reference (T/R) ratio of the triplicate of each clone was calculated and was divided by the median T/R ratio of all targets present on the array, to center the ratios to 1.0. We edited the data files to remove ratios both on clones for which the SD of the triplicates was >20% and on clones for which only one of the triplicates passed the inclusion criteria. For all array hybridizations included in this study, 88% of the clones passed these criteria and were, therefore, included in the final analysis. The average SD for the triplicates was 5%.
Normal clone-to-clone variation in fluorescence-intensity ratios was measured in five normal-versus-normal experiments, by averaging the ratios obtained for each clone and calculating the SD from this average. These control experiments represent an important quality measurement of possible label-related problems and allow a good estimate of slide-to-slide variation. The average ratio of each clone in the five experiments varied between 0.94 and 1.17 (mean 1.0, as a result of the centering step described above). This intrinsic clone-to-clone variability was corrected in the analysis of the patients' DNAs by division of each intensity ratio by the mean intensity ratio of that particular clone in the normal-versus-normal hybridizations. The SD in these five control experiments ranged from 0.01 to 0.14, with a mean of 0.05. One clone (GS-963K6, located on 4qter) showed a SD >0.1, and this clone was excluded from further analyses. On basis of these experiments, we decided that a safe normal range would be 0.8–1.2, a range that is broader than calculated on basis of two times the SD for each clone calculated in the normal-versus-normal hybridizations.
DOP-PCR
From each clone, DNA was amplified using routine DOP-PCR protocols (Telenius et al. 1992). In brief, PCRs were performed in a total volume of 100 μl 1× PCR buffer, containing 50 ng template DNA, 5 U Taq DNA polymerase, 200 μM dNTPs, 3 mM MgCl2, and 1.5 μM of DOP primer (5′-CCG ACT CGA GNN NNN NAT GTG G-3′) (Invitrogen). Samples were processed in a 96-well reaction plate at 94°C for 3 min, followed by 30 cycles of 95°C for 30 s, 37°C for 30 s, a linear ramp from 37°C to 72°C over 10 min, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. All reactions were performed in a PCR Express machine (Thermo Hybaid). DOP-PCR products were purified using the QIAquick filter system (QIAgen) and were dissolved in 70 μl water. These PCR products were processed in exactly the same manner as described above for primary clone DNAs. Arrays were constructed containing both primary DNAs and DOP-PCR products from the entire subtelomeric clone set.
FISH and Chromosomal CGH Validation Experiments
FISH validation experiments were performed on metaphase spreads prepared from patient-derived cell lines by use of routine procedures. Probe labeling, slide preparation, and hybridization were performed essentially as described elsewhere (de Bruijn et al. 2001). CGH analysis was performed for validation of one patient, as described by Simons et al. (1999). A Zeiss epifluorescence microscope equipped with appropriate filters was used for visual examination of the slides. Digital images were captured by use of a high-performance cooled CCD camera (Photometrics) coupled to a Macintosh Quadra 950 computer. The Image FISH software package (Intergen) was used for analysis of the FISH images. Inverted images of slides stained with 4′,6-diamino-2-phenylindole were used for chromosome identification. Examination of the CGH images was performed by use of the Quips CGH analysis software (Vysis).
Results
Our subtelomeric array contained whole clone DNAs isolated from a second-generation set of 77 subtelomeric genomic fragments cloned into PACs, P1s or BACs, confirmed to be located within 500 kb of all human telomeres (Knight et al. 2000). The applicability of array-based CGH in detecting subtelomeric copy-number changes was tested in a blinded fashion by hybridizing total genomic DNAs from 20 cytogenetically preselected patients to this array in the presence of normal reference DNA. The subtelomeric copy-number profile of these patients was analyzed through fluorescence ratios indicating copy-number gains or losses, and the results were compared with the original diagnoses based on routine karyotyping (fig. 1 and table 1). Thresholds for copy-number gain (1.2) and loss (0.8) were set on the basis of five normal-versus-normal control experiments (see the “Material and Methods” section).
Figure 1.
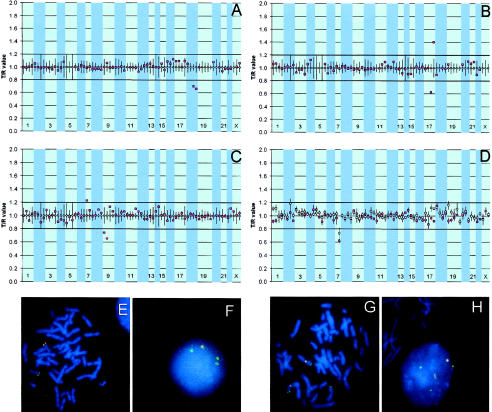
Telomere screening by array-based CGH. A–C, Telomere profiles of three patients. The arrays were composed of 77 cloned subtelomeric genomic DNA targets, ordered within each chromosome from pter to qter. Results for clone GS-963-K6, located on 4qter, are not included because of the high variability observed in the normal-versus-normal hybridizations (see the “Material and Methods” section). The unblackened squares represent the T/R values of the control hybridizations, individually normalized to a value of 1. The vertical lines represent twice the SD for each target clone in the control hybridizations. Next to that, the dark horizontal lines indicate the thresholds for copy-number loss (0.8) and gain (1.2). The blackened squares represent the normalized T/R ratios for patient 2 (46,XY,del[18][q21.2]) (A), patient 8 (46,XY,der[13;14][q10;q10],dup[17][qterq24.2]) (fig. 1B) and patient 14 (46,XX,der[9]t [9;?)[p24;?]) (fig. 1C) versus reference hybridization. A shows a clear loss of both telomeric targets mapping to the subtelomeric region of the long arm of chromosome 18. In B, gain of one 17qter clone and deletion of the other 17qter clone is observed in patient 8. In C, gain of the 7pter clone and loss of both 9pter clones is observed. D, Quadruplicate array-based CGH experiment of patient 10 (46,XY,del[7][q36]). The unblackened squares represent the mean T/R values obtained with the DOP-PCR products, and the blackened squares represent the mean T/R values obtained with the whole clone DNAs as targets. The vertical lines represent the SD for each target clone in the four hybridizations. The deletion of the distal part of the long arm of chromosome 7 is clearly identified in the whole clone DNAs and, to a lesser extent, in the DOP-PCR products (in one experiment, the T/R value for this clone was 0.87). In addition, the SDs of the DOP-PCR products of two clones, GS-1011-O17 on 2qter and GS-546-C11 on 19pter, crossed the threshold for copy-number gain because of a false-positive result in one of the four repeat experiments. E and F, FISH validation experiments of patient 8. The duplicated 17qter clone is labeled in green; the deleted 17qter clone is labeled in red. The duplication is visible in interphase (F), and the deletion is visible in both interphase (F) and metaphase (E). G and H, FISH validation experiments of patient 14 (G metaphase, H interphase). The 7pter clone is labeled in green, and chromosome 7 centromere is labeled in red for chromosome identification. The 7pter clone is present on both chromosome 7 homologs, as well as on the short arm of chromosome 9, thus confirming the unbalanced translocation as identified by array-based CGH.
In 16 patients, specific subtelomeric regions (represented by either one, two, or three clones) showing copy-number gains (mean 1.28) or losses (mean 0.67) could be identified, whereas, in four patients, no fluorescence ratios crossed the thresholds for copy-number gain or loss. As a representative example, figure 1A shows the array-based CGH profile of patient 2 (karyotype: 46,XY,del[18][q21.2]). Both clones mapping to the subtelomere of 18q exhibit intensity ratios <0.8, the threshold for copy-number loss. Equally important is the observation that the fluorescence ratios of the other subtelomeric clones are nicely distributed within the normal range. Comparison of the overall array-based CGH diagnoses with the original cytogenetic diagnoses showed a perfect match in 15 patients and disparities in 5 patients. These latter patients are individually described below (see also fig. 1 and table 1).
The array-based CGH profile of patient 6 (karyotype: 46,XY,del[13][q32.2]) did not confirm the cytogenetic finding in two independent hybridizations to the telomeric array, although this aberration (i.e., loss of the distal part of the long arm of chromosome 13) had been interpreted as a terminal deletion, on the basis of high-resolution chromosome studies. FISH on metaphase chromosomes was performed with both 13qter clones present on the array and with clone RP11-235O20 located at 13q32.2. The deletion at 13q32.2 could indeed be identified by use of this latter clone as a probe. In contrast, both 13qter clones were present on the normal chromosome 13, as well as on the chromosome 13 containing the 13q32.2 deletion, in all metaphases examined. Therefore, the observed deletion should be reclassified as an interstitial deletion that leaves the subtelomeric part of the chromosome intact.
Similarly, no abnormalities at the telomeres of chromosome 2 were identified by array-based CGH in patient 13 (46,XX,add[2][q36.2]). Unfortunately, no patient material was available for additional FISH validation. Chromosomal CGH could, however, be performed with DNA from the same batch as was used for array-based CGH. The results of this experiment showed a gain at 8q24 (not shown), suggesting an insertion of chromosome 8 material into the long arm of chromosome 2, leaving the subtelomeric regions of the derivative chromosome 2 unaffected.
The karyotype of patient 18 indicated an inversion and duplication of part of the short arm of chromosome 8 (46,XY,dup[8][p23p12]). Array-based CGH results of both 8pter clones, however, revealed a deletion of the subtelomeric region of 8p. These results were confirmed by FISH with both clones present on the array and indicated that the inversion duplication of the distal part of 8p indeed coincided with a deletion of the very distal (subtelomeric) region of 8p, a previously reported cytogenetic anomaly (de Die-Smulders et al. 1995; Guo et al. 1995).
An inversion and duplication was also indicated at the distal part of the long arm of chromosome 17 in patient 8 (45,XY,der[13;14][q10;q10],dup[17][qterq24.2]). DNA copy-number analysis by array-based CGH resulted in a remarkable profile (fig. 1B). One 17qter clone (GS-50C4, with an estimated maximum physical distance of 100–300 kb from the telomere [Knight et al. 2000]) revealed a copy-number gain, completely in line with the karyotype. However, the other clone (GS-362K4, with a maximum physical distance of 90 kb from the telomere [Knight et al. 2000]) clearly revealed a copy-number loss. These results were confirmed by FISH and show that the inversion duplication of the distal part of 17q coincided with a deletion of the last 100 kb of the subtelomeric region. These results are reminiscent of those obtained for patient 18.
Finally, the potential of array-based CGH for the detection of cryptic cytogenetic abnormalities was shown most clearly in patient 14 (46,XX,der[9]t[9;?][p24;?]). In this patient, the translocation partner of chromosome 9p could not be identified by routine cytogenetic analysis. Array-based CGH clearly showed loss of both 9p subtelomeric clones and, next to that, a copy-number gain of the 7pter clone (fig. 1C). FISH validation experiments confirmed translocation of the corresponding 7p region to 9pter, thereby refining the cytogenetic diagnosis as an unbalanced translocation between chromosomes 7 and 9.
Next to the correct identification of the affected telomeric regions in all patients, six clone hybridizations showed mean intensity ratios (merely) crossing the thresholds of 0.8 and 1.2 in these 20 experiments. Although no FISH experiments were performed to validate these observations, we assume that these were false-positive results, on the basis of the presence of unaltered profiles for the other clone(s) located in the same subtelomeric regions.
To further determine the accuracy and reproducibility of array-based CGH for the detection of subtelomeric copy-number gains and losses, we hybridized genomic DNA from one patient with a deletion of the distal part of the long arm of chromosome 7 in four different experiments to the telomeric array (patient 10; fig. 1D). These experiments were performed onto a new batch of arrays containing whole clone DNAs as well as DOP-PCR products from all clones, to compare the performance of array-based CGH on DOP-PCR products with that of array-based CGH on primary clone DNAs. The intensity ratios observed for the 7q clone GS-3K23 varied, for total clone DNA, between 0.58 and 0.67 (mean 0.62) and, for the DOP-PCR product, between 0.69 and 0.85 (mean 0.73), which, except for one of the four DOP-PCR products, is well below the threshold for copy-number loss of 0.8. Clones mapping to other subtelomeric regions showed mean ratios between 0.87 and 1.18 (mean 1 and mean SD 0.05 for the clone DNAs, as well as for the DOP-PCR products), none of which was above or below the threshold for copy-number gain and loss, respectively. The SDs of the DOP-PCR products of two clones, GS-1011-O17 on 2qter and GS-546-C11 on 19pter, however, crossed the threshold for copy-number gain due to a false-positive result in one of the four repeat experiments.
Discussion
We here demonstrate the application of a new array-based subtelomeric assay, capable of screening all human subtelomeric regions in a single hybridization reaction. Telomeric abnormalities were correctly detected in all patients with previously identified telomeric deletions and duplications. In addition a refined cytogenetic diagnosis was established by array-based CGH in 5 patients. The mean T/R ratios obtained for single copy alterations are comparable to measurements performed by Pinkel et al. (1998) by use of cell populations containing one to five copies of the X chromosome and calculations recently reported by Snijders et al. (2001). Importantly, no false-negative results were obtained in this study, and the number of presumed false-positive clone signals was very low (0.4%). With further optimization of this technology, it is to be expected that the false-positive rate will decrease even further, requiring only minimal FISH-based validation. At this moment, however, we strongly suggest for clinical diagnosis to confirm all positive results by FISH.
Telomere screening by array-based CGH has many advantages over other methods available for telomere screening, including microsatellite analysis (Flint et al. 1995; Slavotinek et al. 1999), the Multiprobe T assay (Knight et al. 1997), primed in situ labeling (Bonifacio et al. 2001), and the very recently developed M-TEL assay (Brown et al. 2001) and MAPH-technique (Armour et al. 2000; Sismani et al. 2001). A total of 77 subtelomeric probes were screened in a single reaction and required only 500 ng genomic DNA from the patient. This clone set can easily be extended, to further increase the sensitivity for detecting genomic imbalances. The use of DNA instead of metaphase spreads used in FISH-based approaches greatly enhances applicability and (semi)automation of the assay. In addition, extensive labeling procedures as performed for the M-TEL assay (Brown et al. 2001) are not required, since array-based CGH builds upon heavily optimized and simplified CGH protocols that use only two fluorochromes (Weiss et al. 1999; Lichter et al. 2000). Until recently, the major drawback of array-based CGH was the laborious isolation of suitable quantities of DNA from the target large-insert clones to be spotted on the arrays. This procedure can now be replaced by a robotic miniprep procedure in combination with DOP-PCR amplification, as we have shown in this study. In the small test experiment, array-based CGH on DOP-PCR products performed slightly less well than array-based CGH on whole BAC DNAs, and this will have to be optimized. In addition, alternative PCR-based amplification procedures have also been developed and will be of help in preparing high-quality target DNA (Snijders et al. 2001).
Array-based subtelomeric screening is expected to have a profound impact on the diagnosis and genetic counseling of patients with mental retardation. Recent reports in the literature (Knight et al. 1999; Slavotinek et al. 1999; de Vries et al. 2001) suggest that cryptic subtelomeric rearrangements may account for 6%–8% of unexplained mental retardation and congenital anomalies. Automated detection of subtle deletions and/or duplications by array-based CGH brings telomere screening within reach of most diagnostic cytogenetic laboratories at a reasonable cost.
As a next step, molecular karyotyping by array-based CGH may be extended to thousands of randomly selected clones, thereby improving the resolution of copy-number screening from ∼5 Mb (routine karyotyping) to ultimately as little as 100 kb (30,000 clones evenly spread over the genome). Currently, 7,600 large-insert clones have been integrated into the draft sequence of the human genome (Cheung et al. 2001). Collectively, they represent ideal targets to be used for array-based CGH. With these tools in hand, copy-number alterations, as detected by array-based CGH, can be translated directly to the gene level by use of clone-specific sequence information present in the public domain. This will soon allow a whole-genome copy-number screening at unprecedented resolution in a single reaction and concurrent disease-gene identification.
Acknowledgments
We thank Drs. Jonathan Flint and Regina Regan (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, United Kingdom) for providing part of the second-generation set of human-chromosome–specific (sub)telomere probes. We acknowledge Dr. Graeme Hodgson (Cancer Genetics, UCSF Cancer Center, San Francisco), for the DOP-PCR protocol; Anthony van Kampen, Jaqueline de Ruijter, and Iris Zandman, for expert technical assistance; and Quinten Ruhé, for help in selection of samples. This work was sponsored by an investment from the PPS-Mibiton foundation.
References
- Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D (2000) Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet 25:144–146 [DOI] [PubMed] [Google Scholar]
- Antonarakis SE (2001) BACking up the promises. Nat Genet 27:230–232 [DOI] [PubMed] [Google Scholar]
- Armour JA, Sismani C, Patsalis PC, Cross G (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 28:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Carey JC (1998) Wolf-Hirschhorn syndrome and Pitt-Rogers-Danks syndrome. Am J Med Genet 75:541 [DOI] [PubMed] [Google Scholar]
- Bonifacio S, Centrone C, Da Prato L, Scordo MR, Estienne M, Torricelli F (2001) Use of primed in situ labeling (PRINS) for the detection of telomeric deletions associated with mental retardation. Cytogenet Cell Genet 93:16–18 [DOI] [PubMed] [Google Scholar]
- Brown J, Saracoglu K, Uhrig S, Speicher MR, Eils R, Kearney L (2001) Subtelomeric chromosome rearrangements are detected using an innovative 12-color FISH assay (M-TEL). Nat Med 7:497–501 [DOI] [PubMed] [Google Scholar]
- Bruder CE, Hirvela C, Tapia-Paez I, Fransson I, Segraves R, Hamilton G, Zhang XX, et al (2001) High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet 10:271–282 [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Dykens E, Williams CA (2000) Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet 97:136–146 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, et al (2001) The BAC Resource Consortium: integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, Geurts Van Kessel A (2001) Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet 92:310–331 [DOI] [PubMed] [Google Scholar]
- de Die-Smulders CE, Engelen JJ, Schrander-Stumpel CT, Govaerts LC, de Vries B, Vles JS, Wagemans A, Schijns-Fleuren S, Gillessen-Kaesbach G, Fryns JP (1995) Inversion duplication of the short arm of chromosome 8: clinical data on seven patients and review of the literature. Am J Med Genet 59:369–374 [DOI] [PubMed] [Google Scholar]
- de Vries BB, White SM, Knight SJ, Regan R, Homfray T, Young ID, Super M, McKeown C, Splitt M, Quarrell OW, Trainer AH, Niermeijer MF, Malcolm S, Flint J, Hurst JA, Winter RM (2001) Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet 38:145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnai D, Karmiloff-Smith A (2000) Williams syndrome: from genotype through to the cognitive phenotype. Am J Med Genet 97:164–171 [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Callif-Daley F, Zapata MC, Miller ME (1995) Clinical and cytogenetic findings in seven cases of inverted duplication of 8p with evidence of a telomeric deletion using fluorescence in situ hybridization. Am J Med Genet 58:230–236 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D (1992) Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258:818–821 [DOI] [PubMed] [Google Scholar]
- Knight SJ, Flint J (2000) Perfect endings: a review of subtelomeric probes and their use in clinical diagnosis. J Med Genet 37:401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Horsley SW, Regan R, Lawrie NM, Maher EJ, Cardy DL, Flint J, Kearney L (1997) Development and clinical application of an innovative fluorescence in situ hybridization technique which detects submicroscopic rearrangements involving telomeres. Eur J Hum Genet 5:1–8 [PubMed] [Google Scholar]
- Knight SJ, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DL, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J (2000) An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 67:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, Horsley SW, Kearney L, Homfray T, Winter RM, Bolton P, Flint J (1999) Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet 354:1676–1681 [DOI] [PubMed] [Google Scholar]
- Lichter P, Joos S, Bentz M, Lampel S (2000) Comparative genomic hybridization: uses and limitations. Semin Hematol 37:348–357 [DOI] [PubMed] [Google Scholar]
- Ning Y, Roschke A, Smith ACM, Macha M, Precht K, Riethman H, Ledbetter DH, Flint J, Horsley S,Regan R, Kearney L, Knight S, Kvaloy K, Brown WRA (1996) A complete set of human telomeric probes and their clinical application. Nat Genet 14:86–89 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 20:207–211 [DOI] [PubMed] [Google Scholar]
- Scambler PJ (2000) The 22q11 deletion syndromes. Hum Mol Genet 9:2421–2426 [DOI] [PubMed] [Google Scholar]
- Simons A, Schepens M, Forus A, Godager L, van Asseldonk M, Myklebost O, van Kessel AG (1999) A novel chromosomal region of allelic loss, 4q32-q34, in human osteosarcomas revealed by representational difference analysis. Genes Chromosomes Cancer 26:115–124 [PubMed] [Google Scholar]
- Sismani C, Armour JA, Flint J, Girgalli C, Regan R, Patsalis PC (2001) Screening for subtelomeric chromosome abnormalities in children with idiopathic mental retardation using multiprobe telomeric FISH and the new MAPH telomeric assay. Eur J Hum Genet 9:527–532 [DOI] [PubMed] [Google Scholar]
- Slavotinek A, Rosenberg M, Knight S, Gaunt L, Fergusson W, Killoran C, Clayton-Smith J, Kingston H, Campbell RH, Flint J, Donnai D, Biesecker L (1999) Screening for submicroscopic chromosome rearrangements in children with idiopathic mental retardation using microsatellite markers for the chromosome telomeres. J Med Genet 36:405–411 [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264 [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer 20:399–407 [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725 [DOI] [PubMed] [Google Scholar]
- Weiss MM, Hermsen MA, Meijer GA, van Grieken NC, Baak JP, Kuipers EJ, van Diest PJ (1999) Comparative genomic hybridisation. Mol Pathol 52:243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Veltman JA, Olshen A, Jain A, Moore DH, Kovacs G, Presti JC, Waldman FM (2002) Array based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res 62:957–960 [PubMed] [Google Scholar]