Abstract
There is growing concern about elevated blood pressure (BP) in children. The evidence for familial aggregation of childhood BP is substantial. Twin studies have shown that a large part of the familial aggregation of childhood BP is due to genes. The first part of this review provides the latest progress in gene finding for childhood BP, focusing on the combined effects of multiple loci identified from the genome-wide association studies on adult BP. We further review the evidence on the contribution of the genetic components of other family risk factors to the familial aggregation of childhood BP including obesity, birth weight, sleep quality, sodium intake, parental smoking, and socioeconomic status. At the end, we emphasize the promise of using genomic-relatedness-matrix restricted maximum likelihood (GREML) analysis, a method that uses genome-wide data from unrelated individuals, in answering a number of unsolved questions in the familial aggregation of childhood BP.
Keywords: Blood pressure, Childhood, Children, Familial aggregation, Genetic risk scores, Genome-wide association studies, Family risk factors
Introduction
Evidence from epidemiologic studies on blood pressure (BP) in children and adolescents, which have been conducted over the past decade, demonstrates a significant increase in BP level and an increase in the prevalence of hypertension [1]. There is considerable tracking of BP levels from childhood to adulthood, making BP at a young age an important predictor of adult levels [2]. Identification of risk factors contributing to BP elevation and hypertension at childhood will provide opportunities for early prevention of this debilitating disease and its related cardiovascular diseases in adulthood. (Table 1 lists a glossary of genetic terms used in this manuscript.)
Table 1.
A glossary of genetic terms used in this review
| Familial aggregation | The clustering of certain traits, behaviors, or disorders within a given family, which are usually caused by genetic or environmental similarities |
| Genetic architecture | Description of the number of genetic variants, allelic frequencies, and effect sizes of variants that affect a trait and their mode of gene action |
| Heritability h2 | The proportion of observed differences in a trait among individuals of a population that is due to genetic differences or genetic factors. The most commonly used formula is the one for narrow-sense heritability, which means the additive genetic portion of the phenotype variance and is calculated as the variance of additive effect divided by the total phenotypic variance in a population |
| GWAS | Genome-wide association study: a study design in which hundreds of thousands of genetic variants in the genome (usually SNPs) are tested for association with the trait |
| SNP | Single-nucleotide polymorphism, which is a DNA variation occurring within a population in which a single nucleotide (A, T, C, G) differs between members of a biological species |
| Genetic risk score | Multi-locus profiles of genetic risk, which can be used to translate discoveries from genome-wide association studies into tools for population health research |
| Minor allele frequency | The frequency at which the least common allele occurs in a given population |
| Intrapair analysis | The analysis of intrapair differences on measures within monozygotic or dizygotic twin pairs |
| h2SNP | The proportion of the phenotypic variance explained by the common SNPs in human genome |
| Missing heritability | Defined as the fact that individual genes cannot account for much of the heritability of disease, behaviors, and other phenotypes |
| Genetic covariance | The genetic part of the covariance, which can be used to calculate genetic correlation by standardizing the genetic variance-covariance matrix |
| Genetic correlation | The proportion of variance that two traits share due to genetic cause, which indicates how much of the genetic influence on two traits is common to both |
| Pleiotropy | Referring to one gene influencing multiple, seemingly unrelated phenotypes, which means that a mutation in a pleiotropic gene may have an effect on multiple traits simultaneously |
Similar to the studies on adult BP, the strong familial tendency to high (or low) BP has also been found in children. For example, significant sib-sib and mother-child correlations for BP were found in children between 2–14 years of age [3]. Moreover, significant sibling BP correlations with 1-month-old infants [4] and significant parent-offspring BP correlations between mothers and their newborn infants were also observed [5], indicating that familial aggregation of BP is established early in life.
It stands to reason that the potential causes of familial aggregation are genetic factors and/or shared (familial) environmental factors, but it is actually more complex to partition the familial aggregation of BP into these two components because of the intimate connections between them. For example, obesity, the most important risk factor for hypertension and the primary driving force for the elevated BP in children [6, 7], displays strong familial tendency itself. The familial aggregation of BP may therefore to a certain extent be due to the familial aggregation of obesity. Even the other apparently “environmental risk factors” for BP elevation such as sodium intake [8–10] and parental socioeconomic status [11•] have a genetic component. This might also be part of the reasons why the majority of the twin studies found strong evidence for genetic influence but no evidence for influence of shared family environment on childhood BP [12]. To avoid confusion, in this review, we will use “other family risk factors” rather than “family environmental factors” to represent these factors.
In this review, we first give an overview of the latest progress in gene finding for childhood BP, focusing specifically on the combined effects of multiple genetic loci identified from the genome-wide association studies on adult BP. Second, we critically review the evidence on the potential contribution of other family risk factors to familial aggregation of BP. Finally, we highlight the potential use of genomic-relatedness-matrix restricted maximum likelihood (GREML) analysis implemented in the genome-wide complex trait analysis (GCTA) software to better understand the genetic architecture of childhood BP. GREML is a recently developed approach to estimate the proportion of phenotypic variance explained by genome- or chromosome-wide common SNPs for complex traits [13].
The Genetic Basis of Childhood BP
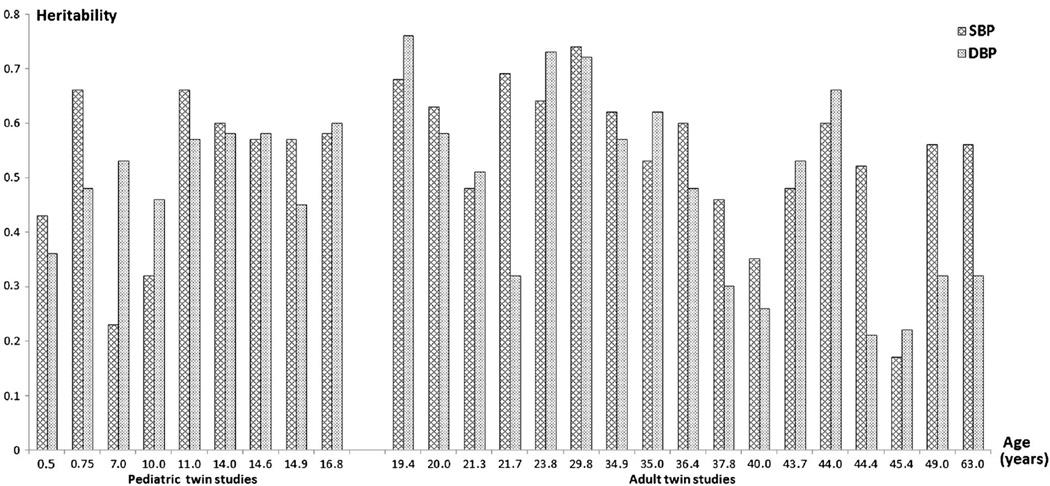
Over the last 30 years, a large number of twin studies have been conducted on BP variation in both children and adults. As shown in Fig. 1, the evidence for a sizable contribution of genetic factors to BP is overwhelming, with most heritability estimates around 50–60 % in both children and adults [12]. However, the gene-finding effort for BP and hypertension turned out to be very challenging, even in the era of genome-wide association studies (GWASs) [14]. It was not until a large number of studies joined forces into the Global BPgen and CHARGE consortia that 13 novel BP loci were identified for BP variation in adulthood [15, 16]. More recently, Global BPgen and CHARGE merged to form the ICBP consortium (International Consortium for BP GWAS), which used a multistage design in 200,000 adults of European ancestry, identified another 16 novel loci, and demonstrated that the 29 loci in total explain ~0.9 % of the variance in both systolic BP (SBP) and diastolic BP (DBP) and a genetic risk score (GRS) based on the 29 loci is significantly associated with hypertension [17]. Detailed results of recent GWASs on adult BP and hypertension can be found in the reviews by Lind et al. [18••] and our group [19].
Fig. 1.
Heritability estimates of SBP and DBP estimated from twin studies of pediatric and adult populations ordered according to the average age of the samples [12]
Several studies have explored whether these genetic variants known to be associated with adult BP are also associated with BP in children and adolescence. Using data from the Cardiovascular Risk in Young Finns cohort in which BP was measured at baseline in 1980 (age 3–18 years) and in follow-ups in 1983, 1986, 2001, and 2007, Oikonen et al. [20••] showed that the GRS of the 13 SNPs identified by the Global BPgen and CHARGE consortia were significantly associated with BP levels both in the longitudinal analyses spanning from childhood to adult age and in the cross-sectional analyses at different age groups. The GRS can explain 0.1 to 0.2 % of the variation in BP, and its significant association with BP can be detected as early as age 9 years for DBP and age 24 years for SBP. Using the same dataset, Juhola et al. [21••] show that the GRS based on all the 29 loci was an independent predictor for the development of adult hypertension at age 24–45 years. Pooling data from two longitudinal birth cohorts with BP measured seven times from age 7 to 17 in the Avon Longitudinal Study of Parents and Children and BP measured five times from age 6 to 17 in the Western Australian Pregnancy Cohort, Howe et al. [22••] examined the associations of the GRS based on the 29 loci with the trajectories of SBP from 6 to 17 years of age. The GRS was significantly associated with SBP at 6 years of age, but showed little or weak evidence in association with SBP changes with age. The GRS explained 0.06 % of the variation in SBP at the first visit (6/7 years) and 0.23 % at the 17-year visit. The highest quintile of the BP genetic risk score had, on average, a higher SBP than the lowest quintile by 1.23 mmHg at the first visit and by 1.37 mmHg at the 17-year visit. In addition to these studies in Caucasians, similar studies have also been conducted in Asians and African-Americans. For example, Xi et al. [23] genotyped six SNPs in 3077 Chinese children aged 6–18 years. These six SNPs were selected from the previous GWAS-identified loci for adult hypertension but have a minor allele frequency >0.30 in Chinese individuals in the HapMap database and a reported effect size of ~1.5 for hypertension. The GRS calculated from these six SNPs was significantly associated with SBP and the risk of hypertension but not with DBP. Bhatnagar et al. [24] conducted a genome-wide association study on SBP in African-American children with sickle cell disease. With the limited sample size (n= 1617), no SNP reached genome-wide significance, but the genetic risk score based on 27 previously reported SBP-associated SNPs was significantly associated with SBP.
The conclusion seems warranted that the genetic variants identified for adult BP also influence childhood BP. However, these studies also show that the adult-based genetic risk scores explained less variance in childhood BP than in adult BP and that not all of the individual SNPs (which were also tested in these studies) displayed significant association with childhood BP. This is not surprising and actually consistent with the previous findings from family and twin studies on the age-dependency of genetic effects on BP [12]. BP changing with age and findings of lower parent-offspring correlations compared to those for siblings and dizygotic twins suggest that different genetic and environmental mechanisms have their influence on BP in different periods of life [25]. This was supported by longitudinal studies of adolescent [26] and middle-aged twins [27]. For example, in the longitudinal twin study we conducted on BP for the transition period from childhood to adulthood, the genetic influences were in part the same across the transition time period (58–64 % of the genetic variation at early adulthood was already detected in childhood) and in part age specific (the remaining 36–42 % of the genetic variation at early adulthood was unrelated to that expressed in childhood) [26]. The changes in genetic effects on BP over time were further confirmed by a large-scale investigation on gene-age interactions in BP regulation with the CHARGE, Global BPgen, and ICBP consortia [28•]. In a two-staged design using 99,241 individuals of European ancestry, 20 genome-wide significant loci were identified by using joint tests of the SNP main effect and SNP-age interaction. The genes exhibiting the largest age interactions displayed opposite directions of effect in the young versus the old. A secondary analysis in each age group revealed 22 loci with evidence of age-specific effects.
The age dependency of genetic effects on BP has important implications for gene-finding studies. On the one hand, we should be cautious by pooling data from subjects at different ages to conduct GWAS, especially by pooling data from adolescent and adult subjects. On the other hand, a large genome-wide association consortium specifically focusing on childhood BP is urgently needed. The identification of the genes conferring susceptibility to high BP at early age will provide new avenues for prevention of the development of hypertension in adulthood.
Influence of Other Family Risk Factors on Familial Aggregation of Childhood BP
For hypertension, which is a disease that has clear associations with lifestyle factors, it would seem that familial aggregation of BP may to a large extent reflect the effects of shared family environment, especially in children who usually live in the same family household. However, the majority of the twin studies in both children and adult found no evidence for influence of shared family environment on BP [12, 29]. One reason might be that many twin studies may lack the power to detect moderate size influences of common environment. A few studies that either had large sample size [30, 31] or used a more powerful multivariate approach [32] did find a small contribution of shared environment of around 10–20 %. Another explanation, as we described earlier, might be due to the intimate connections between the genetic and the shared family environment. That is, the so-called environmental risk factors for elevated BP, such as obesity and sodium intake, have a heritable component themselves. In this part, we summarize the evidence on the potential contribution of the genetic components of these family risk factors to the familial aggregation of BP, focusing on known risk factors for childhood BP.
Obesity
In subjects of all ages, body mass index (BMI) is probably the most important correlate of BP. Large population studies showed that the recent increase in BP levels in childhood could largely be explained by the increase in BMI [6, 7], providing strong evidence that increasing rates of overweight and obesity are the major driving forces for the increase in childhood BP levels. Previous twin and family studies have shown that the association between BP and BMI is partly attributed to a common set of genetic factors. For example, Schieken et al. [33] found that the genetic factors influencing BMI account for 8 % of the total variance of SBP in a pediatric population of 11-year-old twins. Similarly, our own study [34] on adult twins found that this figure was 6 % for SBP and 7 % for DBP. This is also supported by the recent GWASs showing that obesity susceptibility loci were associated with BP levels. In Howe et al.’s study [22••] in the two birth cohorts (described earlier), in addition to the GRS calculated from the 29 BP loci, they also examined the association of the GRS calculated from 32 BMI loci on the trajectories of SBP from age 6 to 17. The GRS of the 32 BMI loci was strongly associated with SBP both at age 6 and at age 17. Comparing with the BP genetic risk score, the BMI genetic risk score explains a greater proportion of the variation in SBP at age 6 (0.15 vs. 0.06 %) but a similar proportion of the variation at age 17 (0.16 vs. 0.23 %). The SBP of individuals in the top quintile of the BMI genetic risk score is, on average, 1.51 mmHg higher than the bottom quintile at age 6, with the magnitude of this difference reducing slightly (to 1.21 mmHg) between 7 and 17 years. The role of individual obesity susceptibility loci on BP has also been studied. Take the first GWAS-identified obesity gene, FTO, as an example. In a recent meta-analysis [35•] on 57,464 hypertensive cases and 41,256 controls, the FTO gene variant(s) showed significant association with the risk of hypertension which disappeared on adjustment for BMI, indicating that FTO genotype contributes to obesity-related hypertension. Similar studies have been conducted in children with the majority finding the FTO locus associated with BP levels [36–41].
Birth Weight
Low birth weight (and catch-up growth after birth) and adverse intrauterine conditions (e.g., preeclampsia) have been well-established etiologies for high BP in childhood [42–45]. Birth weight is a complex multifactorial trait itself with heritability around 20–30 % [46–49]. The importance of genetic factors on birth weight acting independently of the intrauterine environment has also been illustrated by correlations between paternal height or weight and offspring birth weight [50, 51]. Genetic variants or shared environmental factors that are associated both with low birth weight and high BP may account for some of the observed correlation between these two phenotypes. This is supported by several twin studies. For example, Christensen et al.’s study [52] in 1311 pairs of adolescent twins found a decrease in SBP of 1.88 mmHg for every kilogram increase in birth weight in the overall sample, but a reduction of this effect was observed when intrapair analyses were used. This is confirmed by a recent meta-analysis [53] in 3901 twin pairs in which the decrease in SBP for every kilogram increase in birth weight was −2.0 mmHg in the unpaired analysis but only −0.4 mmHg in the paired analysis. Further support comes from the recent GWAS on birth weight in 69,308 individuals of European descent [54•]. Out of the seven loci identified for birth weight, one locus, the ADRB1 rs1801253 (Arg389Gly), is known to be associated with adult BP. The associations between birth weight and the 29 BP loci identified by the ICBP consortium were also tested. While no strong evidence of deviation from the null was observed, associations between the SBP-raising allele and lower birth weight achieved P<0.01 at GUCY1A3/GUCY1B3-rs13139571 and CYP17A1/NT5C2-rs11191548.
Sleep Behavior
Several aspects of sleep behavior such as poor sleep quality and short sleep time as well as sleep apnea syndrome have been linked to hypertension in children and adolescents [55–57]. Twin studies have suggested that genetic influences partially underline the variation seen for many sleep-related traits across life span as reviewed in detail by Barclay et al. [58]. To what extent shared genetic and environmental factors may contribute to the association between sleep behavior and BP has not been reported. In our recent two studies on ambulatory blood pressure in youth [59] and young adult [60], we observed that nighttime BP not only shared genes with daytime BP but a sizeable part was explained by its own specific genetic determinants. This indicates that the underlying mechanisms for BP regulation partly change with the day-night shift, either via switching on or off of relevant genes or via a change in the amplitude of their expression. Since day-night shift is related to sleep behavior and sleep apnea has been associated with changes in nighttime BP levels [61–63], it is possible that genes responsible for sleep apnea also contribute to nighttime BP regulation. This needs to be confirmed by, e.g., multivariate modeling in twin and family studies.
Sodium Intake, Parental Smoking, and Parental Socioeconomic Status
These are also recognized risk factors for hypertension and elevated BP in children. A recent study using twins (age≥30) and their first-degree family members observed a strong genetic component for both sodium intake (indexed by half-day urine sodium excretion) and salt habit (evaluated by questionnaire) with heritability ranging from 0.31 to 0.34 [8]. This is consistent with a previous twin study [9] on adult female twins which observed a heritability of 0.43 for 24 h urine sodium excretion and our own twin study [10] of youth and young adults on overnight urine sodium excretion, which observed a heritability of 0.55 in European Americans and a heritability of 0.29 in African-Americans. Consistent evidence has also implicated genetic factors in smoking behavior, and the recent GWASs have identified several loci for smoking amount, smoking initiation, or smoking cessation [64]. Although socioeconomic status (SES) is typically viewed as an environmental variable, recently, Marioni et al. [65] observed a heritability of 71 % for SES in the Scottish Family Health Study. Due to the fact that children’s environment such as parental smoking and parental SES are the same for children growing up together in a family, the extent to which the genetic components of parental smoking and SES might also contribute to the familial aggregation of childhood BP has not been explored in twin and family studies. With recent methodological developments prompted by GWAS [66••], it is now possible to use DNA itself to estimate genetic influence in any sample of unrelated individuals rather than relying on comparisons between monozygotic twins and dizygotic twins. Using this method, Trzaskowski et al. [11•] examined the genetic influence on family SES at ages 2 and 7 as well as on its association with children’s IQ at ages 7 and 12 in 3000 unrelated children. They found that 19 and 20 % of the family SES variance can be explained by common SNPs at age 2 and age 7, respectively. Significant genetic correlation was also observed between family SES and children’s IQ. Similar studies are warranted for childhood BP.
Genome-Wide Complex Trait Analysis
In addition to the main approach of GWAS which tests each SNP individually for an association with the trait using a very stringent P value, the GWAS SNP data can also be used to estimate the genetic relationship between unrelated individuals. The approach calculates to what extent phenotypic similarities between pairs of unrelated individuals can be attributed to their SNP similarity allowing an estimate of the extent to which phenotypic variance can be explained by genetic variance. The method is called genomic-relatedness-matrix restricted maximum likelihood (GREML) and is implemented in the genome-wide complex trait analysis (GCTA) software [13]. It was first introduced by Yang et al. in 2010 [66••] and has now been widely applied to many traits and diseases. Different from the heritability estimated from twin and family data which captures the entire genome, the heritability estimated from the genetic relationships of unrelated individuals only reflects the part explained by common SNPs (i.e., h2SNP=common SNP heritability).
The GREML-GCTA approach can help to elucidate the genetic architecture of common complex traits. For example, despite the fact that GWAS has identified many loci for BP and hypertension in adults, these loci only explain ~1 % of the variance of BP. There has not been any consensus on the explanation of the “missing heritability.” Possible explanations include a large number of common variants with small effects, rare variants with large effects, and DNA structural variation. Recently, using the GCTA approach, Vattikuti et al. [67] observed that the h2SNP was 24 % for SBP, which is about 50 % of the heritability of SBP, indicating that a large part of the heritability for SBP is hiding rather than missing because of many SNPs with small effects.
A bivariate extension of GREML-GCTA can estimate the genetic covariance and hence genetic correlation between different traits and disorders to provide estimates of genome-wide pleiotropy [68••]. These traits or disorders can be collected from the same or from different individuals. For example, Vattikuti [67] explored the genetic correlation between metabolic traits (measured in the same individuals) using bivariate GCTA and observed large genetic correlations between BMI and waist-hip ratio as well as between triglyceride and high-density lipoprotein. Using GWAS data for different diseases from the Wellcome Trust Case Control Consortium, Lee et al. [68••] observed that the estimated genetic correlation between hypertension and type 2 diabetes was 0.31, indicating shared genetic etiology between these two diseases.
For familial aggregation of childhood BP, the GCTA approach has the potential to shed light on many unsolved questions. For example, univariate GREML can estimate the extent to which the heritability of childhood BP is explained by common genetic variants. Bivariate GREML-GCTA tests can determine the contribution of the genetic components of other family risk factors to the familial aggregation of BP, even for the risk factors usually shared within the family such as parental smoking and parental SES. Bivariate GCTA test can also determine to what extent the same or different common genetic variants contribute to the stability and change in BP from childhood to adulthood. However, the prerequisite is the availability of GWAS data in children with BP measured. This further emphasizes the importance of large genome-wide association studies in cohorts of children to study childhood BP.
Conclusion
Familial aggregation of BP is established early in life. Twin and family studies have shown that the familial aggregation of BP is largely due to genes. The gene finding efforts showed that the genetic loci identified from GWAS on BP and hypertension in adults also influence childhood BP. However, the age dependency of genetic effects on BP requires large GWAS studies on childhood BP. We also highlight the evidence on the contribution of the genetic components of other family risk factors to the familial aggregation of childhood BP including obesity, birth weight, sleep quality, sodium intake, parental smoking, and parental SES. At the end, we emphasize the promise of using GREML-GCTA, a newly developed quantitative genetic method that uses GWAS data from unrelated individuals, in answering a number of unsolved questions in the familial aggregation of childhood BP.
Acknowledgments
This work was supported by NIH grants R01HL104125 and R01HL105689.
Footnotes
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest Xiaoling Wang, Xiaojing Xu, Shaoyong Su, and Harold Snieder declare that they have no conflicts of interest.
Contributor Information
Xiaoling Wang, Email: xwang@gru.edu, Georgia Prevention Center, Medical College of Georgia, Georgia Regents University, HS-1640, Augusta, GA 30912, USA.
Xiaojing Xu, Georgia Prevention Center, Medical College of Georgia, Georgia Regents University, HS-1640, Augusta, GA 30912, USA.
Shaoyong Su, Georgia Prevention Center, Medical College of Georgia, Georgia Regents University, HS-1640, Augusta, GA 30912, USA.
Harold Snieder, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Din-Dzietham R, et al. High blood pressure trends in children and adolescents in national surveys, 1963 to 2002. Circulation. 2007;116(13):1488–1496. doi: 10.1161/CIRCULATIONAHA.106.683243. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Wang Y. Tracking of blood pressure from childhood to adulthood: a systematic review and meta-regression analysis. Circulation. 2008;117(25):3171–3180. doi: 10.1161/CIRCULATIONAHA.107.730366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinner SH, Levy PS, Kass EH. Familial aggregation of blood pressure in childhood. N Engl J Med. 1971;284(8):401–404. doi: 10.1056/NEJM197102252840801. [DOI] [PubMed] [Google Scholar]
- 4.Hennekens CH, et al. Aggregation of blood pressure in infants and their siblings. Am J Epidemiol. 1976;103(5):457–463. doi: 10.1093/oxfordjournals.aje.a112247. [DOI] [PubMed] [Google Scholar]
- 5.Lee YH, et al. Familial aggregation of blood pressures of newborn infants and their mother. Pediatrics. 1976;58(5):722–729. [PubMed] [Google Scholar]
- 6.Dong B, et al. Trends in blood pressure and body mass index among Chinese children and adolescents from 2005 to 2010. Am J Hypertens. 2013;26(8):997–1004. doi: 10.1093/ajh/hpt050. [DOI] [PubMed] [Google Scholar]
- 7.Muntner P, et al. Trends in blood pressure among children and adolescents. JAMA. 2004;291(17):2107–2113. doi: 10.1001/jama.291.17.2107. [DOI] [PubMed] [Google Scholar]
- 8.Kho M, et al. Genetic and environmental influences on sodium intake determined by using half-day urine samples: the healthy twin study. Am J Clin Nutr. 2013;98(6):1410–1416. doi: 10.3945/ajcn.113.067967. [DOI] [PubMed] [Google Scholar]
- 9.Hunter DJ, et al. Genetic contribution to renal function and electrolyte balance: a twin study. Clin Sci (Lond) 2002;103(3):259–265. doi: 10.1042/cs1030259. [DOI] [PubMed] [Google Scholar]
- 10.Ge D, et al. Stress-induced sodium excretion: a new intermediate phenotype to study the early genetic etiology of hypertension? Hypertension. 2009;53(2):262–269. doi: 10.1161/HYPERTENSIONAHA.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trzaskowski M, et al. Genetic influence on family socioeconomic status and children’s intelligence. Intelligence. 2014;42(100):83–88. doi: 10.1016/j.intell.2013.11.002. Using GCTA approach, this study for the first time evaluated the contribution of genetic factors to family SES.
- 12.Wang X, Snieder H. In: Familial aggregation of blood pressure, in pediatric hypertension. Flynn JT, Ingelfinger JR, Porman RJ, editors. New York: Springer Science+Business Media New York; 2013. pp. 195–209. [Google Scholar]
- 13.Yang J, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12(1):17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41(6):677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41(6):666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lind JM, Chiu CL. Genetic discoveries in hypertension: steps on the road to therapeutic translation. Heart. 2013;99(22):1645–1651. doi: 10.1136/heartjnl-2012-302883. This paper provides the most recent review on GWAS results in adult BP and hypertension.
- 19.Wang X, et al. Beyond genome-wide association studies: new strategies for identifying genetic determinants of hypertension. Curr Hypertens Rep. 2011;13(6):442–451. doi: 10.1007/s11906-011-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oikonen M, et al. Genetic variants and blood pressure in a population-based cohort: the Cardiovascular Risk in Young Finns study. Hypertension. 2011;58(6):1079–1085. doi: 10.1161/HYPERTENSIONAHA.111.179291. This is the first study exploring the effect of GWAS loci identified for adult BP on childhood BP in a longitudinal youth cohort.
- 21. Juhola J, et al. Childhood physical, environmental, and genetic predictors of adult hypertension: the Cardiovascular Risk in Young Finns study. Circulation. 2012;126(4):402–409. doi: 10.1161/CIRCULATIONAHA.111.085977. This paper evaluated whether the 29 GWAS loci identified for adult BP can increase the prediction of hypertension in early adulthood in a longitudinal youth cohort.
- 22. Howe LD, et al. Genetic influences on trajectories of systolic blood pressure across childhood and adolescence. Circ Cardiovasc Genet. 2013;6(6):608–614. doi: 10.1161/CIRCGENETICS.113.000197. This paper evaluates the associations of (1) a GRS of 29 BP GWAS loci, (2) a GRS of 180 height GWAS loci, and (3) a GRS of 32 BMI loci with SBP trajectories from 5–17 years of age in two longitudinal birth cohorts.
- 23.Xi B, et al. Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children. J Hum Hypertens. 2014;28(1):32–36. doi: 10.1038/jhh.2013.50. [DOI] [PubMed] [Google Scholar]
- 24.Bhatnagar P, et al. Genome-wide meta-analysis of systolic blood pressure in children with sickle cell disease. PLoS One. 2013;8(9):e74193. doi: 10.1371/journal.pone.0074193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iselius L, Morton NE, Rao DC. Family resemblance for blood pressure. Hum Hered. 1983;33(5):277–286. doi: 10.1159/000153391. [DOI] [PubMed] [Google Scholar]
- 26.Kupper N, et al. Emergence of novel genetic effects on blood pressure and hemodynamics in adolescence: the Georgia cardiovascular twin study. Hypertension. 2006;47(5):948–954. doi: 10.1161/01.HYP.0000217521.79447.9a. [DOI] [PubMed] [Google Scholar]
- 27.Colletto GM, Cardon LR, Fulker DW. A genetic and environmental time series analysis of blood pressure in male twins. Genet Epidemiol. 1993;10(6):533–538. doi: 10.1002/gepi.1370100634. [DOI] [PubMed] [Google Scholar]
- 28. Simino J, et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am J Hum Genet. 2014;95(1):24–38. doi: 10.1016/j.ajhg.2014.05.010. This is the first study assessing the gene-age interactions on BP using common variants from GWAS data.
- 29.Evans A, et al. The genetics of coronary heart disease: the contribution of twin studies. Twin Res Hum Genet. 2003;6(5):432–441. doi: 10.1375/136905203770326439. [DOI] [PubMed] [Google Scholar]
- 30.Snieder H, et al. Heritability of central systolic pressure augmentation: a twin study. Hypertension. 2000;35(2):574–579. doi: 10.1161/01.hyp.35.2.574. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y, et al. Genetic and environmental influences on blood pressure in elderly twins. Hypertension. 1994;24(6):663–670. doi: 10.1161/01.hyp.24.6.663. [DOI] [PubMed] [Google Scholar]
- 32.Boomsma DI, et al. Heritability of blood pressure increases during mental stress. Twin Res. 1998;1(1):15–24. doi: 10.1375/136905298320566447. [DOI] [PubMed] [Google Scholar]
- 33.Schieken RM, et al. Multivariate genetic analysis of blood pressure and body size. The Medical College of Virginia Twin Study. Circulation. 1992;86(6):1780–1788. doi: 10.1161/01.cir.86.6.1780. [DOI] [PubMed] [Google Scholar]
- 34.Wu T, et al. Genetic and environmental influences on blood pressure and body mass index in Han Chinese: a twin study. Hypertens Res. 2011;34(2):173–179. doi: 10.1038/hr.2010.194. [DOI] [PubMed] [Google Scholar]
- 35. He D, et al. FTO gene variant and risk of hypertension: a meta-analysis of 57,464 hypertensive cases and 41,256 controls. Metabolism. 2014;63(5):633–639. doi: 10.1016/j.metabol.2014.02.008. This study presents a meta-analysis confirming that FTO genotype contributes to obesity-mediated hypertension.
- 36.Luczynski W, Zalewski G, Bossowski A. The association of the FTO rs9939609 polymorphism with obesity and metabolic risk factors for cardiovascular diseases in Polish children. J Physiol Pharmacol. 2012;63(3):241–248. [PubMed] [Google Scholar]
- 37.Yang M, et al. The effects of genetic variation in FTO rs9939609 on obesity and dietary preferences in Chinese Han children and adolescents. PLoS One. 2014;9(8):e104574. doi: 10.1371/journal.pone.0104574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xi B, et al. Associations of obesity susceptibility loci with hypertension in Chinese children. Int J Obes (Lond) 2013;37(7):926–930. doi: 10.1038/ijo.2013.37. [DOI] [PubMed] [Google Scholar]
- 39.Melka MG, et al. Genome-wide scan for loci of adolescent obesity and their relationship with blood pressure. J Clin Endocrinol Metab. 2012;97(1):E145–E150. doi: 10.1210/jc.2011-1801. [DOI] [PubMed] [Google Scholar]
- 40.Pausova Z, et al. A common variant of the FTO gene is associated with not only increased adiposity but also elevated blood pressure in French Canadians. Circ Cardiovasc Genet. 2009;2(3):260–269. doi: 10.1161/CIRCGENETICS.109.857359. [DOI] [PubMed] [Google Scholar]
- 41.Olza J, et al. Influence of FTO variants on obesity, inflammation and cardiovascular disease risk biomarkers in Spanish children: a case-control multicentre study. BMC Med Genet. 2013;14:123. doi: 10.1186/1471-2350-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National High Blood Pressure Education Program Working Group on High Blood Pressure in Children and Adolescents. The fourth report on the diagnosis, evaluation, and treatment of high blood pressure in children and adolescents. Pediatrics. 2004;114(2 Suppl):555–576. 4th Report. [PubMed] [Google Scholar]
- 43.Lurbe E, et al. Management of high blood pressure in children and adolescents: recommendations of the European Society of Hypertension. J Hypertens. 2009;27(9):1719–1742. doi: 10.1097/HJH.0b013e32832f4f6b. [DOI] [PubMed] [Google Scholar]
- 44.Bonamy AK, Kallen K, Norman M. High blood pressure in 2.5-year-old children born extremely preterm. Pediatrics. 2012;129(5):e1199–e1204. doi: 10.1542/peds.2011-3177. [DOI] [PubMed] [Google Scholar]
- 45.Wolfenstetter A, et al. Altered cardiovascular rhythmicity in children born small for gestational age. Hypertension. 2012;60(3):865–870. doi: 10.1161/HYPERTENSIONAHA.112.196949. [DOI] [PubMed] [Google Scholar]
- 46.Mook-Kanamori DO, et al. Heritability estimates of body size in fetal life and early childhood. PLoS One. 2012;7(7):e39901. doi: 10.1371/journal.pone.0039901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson L, et al. Genetic and environmental influences on infant growth: prospective analysis of the Gemini twin birth cohort. PLoS One. 2011;6(5):e19918. doi: 10.1371/journal.pone.0019918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubois L, et al. Genetic and environmental influences on body size in early childhood: a twin birth-cohort study. Twin Res Hum Genet. 2007;10(3):479–485. doi: 10.1375/twin.10.3.479. [DOI] [PubMed] [Google Scholar]
- 49.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG. 2000;107(3):375–381. doi: 10.1111/j.1471-0528.2000.tb13234.x. [DOI] [PubMed] [Google Scholar]
- 50.Knight B, et al. Evidence of genetic regulation of fetal longitudinal growth. Early Hum Dev. 2005;81(10):823–831. doi: 10.1016/j.earlhumdev.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Klebanoff MA, et al. Father’s effect on infant birth weight. Am J Obstet Gynecol. 1998;178(5):1022–1026. doi: 10.1016/s0002-9378(98)70542-3. [DOI] [PubMed] [Google Scholar]
- 52.Christensen K, Stovring H, McGue M. Do genetic factors contribute to the association between birth weight and blood pressure? J Epidemiol Community Health. 2001;55(8):583–587. doi: 10.1136/jech.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNeill G, Tuya C, Smith WC. The role of genetic and environmental factors in the association between birthweight and blood pressure: evidence from meta-analysis of twin studies. Int J Epidemiol. 2004;33(5):995–1001. doi: 10.1093/ije/dyh260. [DOI] [PubMed] [Google Scholar]
- 54. Horikoshi M, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45(1):76–82. doi: 10.1038/ng.2477. This is a GWAS on birth weight. The findings highlight genetic links between fetal growth and postnatal growth and metabolism.
- 55.Archbold KH, et al. Effects of sleep patterns and obesity on increases in blood pressure in a 5-year period: report from the Tucson Children’s assessment of sleep apnea study. J Pediatr. 2012;161(1):26–30. doi: 10.1016/j.jpeds.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Javaheri S, et al. Sleep quality and elevated blood pressure in adolescents. Circulation. 2008;118(10):1034–1040. doi: 10.1161/CIRCULATIONAHA.108.766410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bixler EO, et al. Blood pressure associated with sleep-disordered breathing in a population sample of children. Hypertension. 2008;52(5):841–846. doi: 10.1161/HYPERTENSIONAHA.108.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barclay NL, Gregory AM. Quantitative genetic research on sleep: a review of normal sleep, sleep disturbances and associated emotional, behavioural, and health-related difficulties. Sleep Med Rev. 2013;17(1):29–40. doi: 10.1016/j.smrv.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, et al. Genetic influences on daytime and night-time blood pressure: similarities and differences. J Hypertens. 2009;27(12):2358–2364. doi: 10.1097/HJH.0b013e328330e84d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, et al. Specific genetic influences on nighttime blood pressure. Am J Hypertens. 2014 doi: 10.1093/ajh/hpu162. pii:hpu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li AM, et al. Blood pressure is elevated in children with primary snoring. J Pediatr. 2009;155(3):362.e1–368.e1. doi: 10.1016/j.jpeds.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 62.Weber SA, et al. Ambulatory blood pressure monitoring in children with obstructive sleep apnea and primary snoring. Int J Pediatr Otorhinolaryngol. 2012;76(6):787–790. doi: 10.1016/j.ijporl.2012.02.041. [DOI] [PubMed] [Google Scholar]
- 63.Xu Z, Li B, Shen K. Ambulatory blood pressure monitoring in Chinese children with obstructive sleep apnea/hypopnea syndrome. Pediatr Pulmonol. 2013;48(3):274–279. doi: 10.1002/ppul.22595. [DOI] [PubMed] [Google Scholar]
- 64.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marioni RE, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence. 2014;44(100):26–32. doi: 10.1016/j.intell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. This is the first study using the GCTA approach. It showed that 45 % of the height variance can be explained by the common SNPs in the human genome.
- 67.Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8(3):e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee SH, et al. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. This is the first study using the bivariate GCTA approach. A significant positive genetic correlation between risk of Type 2 diabetes and hypertension was observed.