Abstract
The RNASEL gene (2′,5′-oligoisoadenylate-synthetase dependent) encodes a ribonuclease that mediates the antiviral and apoptotic activities of interferons. The RNASEL gene maps to the hereditary-prostate-cancer (HPC)–predisposition locus at 1q24-q25 (HPC1) and was recently shown to harbor truncating mutations in two families with linkage to HPC1. Here, we screened for RNASEL germline mutations in 66 Finnish patients with HPC, and we determined the frequency of the changes in the index patients from 116 families with HPC, in 492 patients with unselected prostate cancer (PRCA), in 223 patients with benign prostatic hyperplasia (BPH), and in 566 controls. A truncating mutation, E265X, was found in 5 (4.3%) of the 116 patients from families with HPC. This was significantly higher (odds ratio [OR]=4.56; P=.04) than the frequency of E265X in controls (1.8%). The highest mutation frequency (9.5%) was found in patients from families with four or more affected members. Possible segregation was detected only in a single family. However, the median age at disease onset for E265X carriers was 11 years less than that for noncarriers in the same families. In addition, of the four missense variants found, R462Q showed an association with HPC (OR=1.96; P=.07). None of the variants showed any differences between controls and either patients with BPH or patients with PRCA. We conclude that, although RNASEL mutations do not explain disease segregation in Finnish families with HPC, the variants are enriched in families with HPC that include more than two affected members and may also be associated with the age at disease onset. This suggests a possible modifying role in cancer predisposition. The impact that the RNASEL sequence variants have on PRCA burden at the population level seems small but deserves further study.
Introduction
Prostate cancer (PRCA) is the most common cancer among men in developed countries (Pentyala et al. 2000). A fraction of patients with PRCA belong to families with hereditary prostate cancer (HPC). Linkage analyses in families with HPC have suggested that multiple genetic loci may harbor PRCA-susceptibility genes, including HPC1 (MIM 601518), at 1q24-q25; ELAC2/HPC2 (MIM 605367), at 17p11; PCAP (MIM 602759), at 1q42.2-q43; HPCX (MIM 300147), at Xq27-q28; CAPB (MIM 603688), at 1p36; and HPC20 (MIM 176807), at 20q13 (Ostrander and Stanford 2000). In spite of the large number of loci reported, only the ELAC2/HPC2 gene, at 17p11, has been implicated as a putative PRCA-susceptibility gene (Tavtigian et al. 2001). Although early studies (Rebbeck et al. 2000) supported the hypothesis that ELAC2/HPC2 mutations played a role in PRCA, recent studies have found little or no evidence that supports this hypothesis (Rökman et al. 2001; Suarez et al. 2001; Vesprini et al. 2001; Wang et al. 2001; Xu et al. 2001).
The first PRCA-susceptibility locus—HPC1, at 1q24-q25 (reported by Smith et al. [1996])—has been proven to be significantly linked to disease in large families with early-onset HPC (Xu 2000). The HPC1 region harbors the 2′,5′-oligoisoadenylate (2-5A)–synthetase–dependent gene RNASEL (MIM 180435), at 1q25 (Zhou et al. 1993), which is involved in the interferon-regulated 2-5A system that mediates antiviral and antiproliferative activities (Hassel et al. 1993; Castelli et al. 1998). RNASEL was directly implicated as a candidate HPC1 gene by the finding of disease-segregating mutations in high-risk U.S. families with HPC (Carpten et al. 2002). A truncating mutation (E265X) and an initiation-codon mutation (M1I) segregated with disease in two families. Furthermore, loss of the wild-type RNASEL allele was found in tumor tissue from an affected patient in a family with the E265X mutation, accompanied by absent protein expression. Here, we investigated the significance of RNASEL germline mutations in 116 Finnish families with HPC, as well as in 492 patients with unselected PRCA, 223 patients with benign prostatic hyperplasia (BPH), and 566 controls.
Subjects and Methods
Families with HPC
Collection of Finnish families with HPC has been described elsewhere (Schleutker et al. 2000). Samples from the youngest affected patient available in each of 66 families with HPC were initially used for the screening of mutations in the RNASEL gene, by SSCP analysis. These families had either three or more affected members or two affected members with at least the index patient diagnosed with PRCA at <60 years of age. The mean age at diagnosis for the index patients was 62.5 years (range 44–76 years), and the mean number of affected family members was 3.1 (range 2–6). Furthermore, samples from an additional set of 50 families with HPC (from the youngest affected patient available in each family) were also analyzed for specific mutations, bringing the total number of samples from index patients with HPC to 116. This second cohort of families had only two affected members with ages at diagnosis >60 years. The mean age at diagnosis for the index patients was 69.1 years (range 61–86). Specific mutations found in the index patients were then assayed in all other available affected members of the 116 families (n=65) and in all available unaffected members of the five E265X-mutation–positive families (n=6). The PRCA diagnoses and the family histories of the patients were initially obtained from questionnaires and were subsequently confirmed by medical records, the Finnish Cancer Registry, and parish records.
Patients with PRCA, Patients with BPH, and Controls
We analyzed specific RNASEL mutations in 492 patients with unselected PRCA, in 223 patients with BPH, and in 566 healthy male blood donors. The patients with PRCA and the patients with BPH were consecutive patients diagnosed in the Tampere University Hospital in Finland during 1996–99, whereas the controls consisted of DNA samples from blood donors obtained from the Blood Center of the Finnish Red Cross in Tampere. Tampere University Hospital is a regional referral center for all patients in the area who have PRCA, which results in an unselected, population-based collection of patients. The mean age at diagnosis for the patients with unselected PRCA was 68.3 years (range 52–88). Information was available on the tumor grade in 94% (461/492) of patients, on T-stage in 93% (460/492) of patients, and on M-stage (ascertained by bone scan) in 94% (462/492) of patients. Forty-seven (9.6%) of the patients with unselected PRCA reported a positive family history of PRCA. Diagnosis of BPH was based on lower-urinary-tract symptoms, free uroflowmetry, and evidence, by palpation or transrectal ultrasound, of increased prostate size. If prostate-specific antigen (PSA) was elevated, then the patients underwent prostate biopsies to exclude PRCA. Indication for prostate biopsy was total PSA of ⩾4 ng/ml or total PSA of 3.0–3.9 ng/ml with free PSA <16%. Most patients with BPH were followed up for 3–5 years and did not develop PRCA during that time.
Written informed consent was obtained from all living patients and also, for families with HPC, from the unaffected members. The research protocols were approved by the Ethical Committee of the Tampere University Hospital (93175, 95062, and 99228) and the National Human Genome Research Institute (HG-0158). Permission for collection of families in the entirety of Finland was granted by the Ministry of Health and Social Affairs (59/08/95).
Mutation Screening with SSCP Analysis
SSCP analysis of the entire coding sequence of the RNASEL gene was performed using primer sequences that were designed to include all intron-exon boundaries (according to Carpten et al. [2002]). On request, all primers are available from the authors. Genomic DNA was used at 100 ng/15 μl reaction mixture (containing 1.5 mM MgCl2; 20 μM each of dATP, dCTP, dGTP, and dTTP; 0.5 μCi of α[33P]-dCTP [Amersham Pharmacia]; 0.6 μM of each primer; 1.5 U AmpliTaqGold; and the reaction buffer provided by the supplier [PE Biosystems]), with an annealing temperature of 56°C. Radiolabeled PCR products were mixed with 95% formamide dye, were denatured at 95°C for 5 min, and were chilled on ice. The [33P]-labeled PCR products were electrophoresed at 800 V for 13 h at room temperature, in 0.5× (with and without 1% glycerol) and 0.8× MDE (mutation-detection–enhancement) gel (FMC BioProducts) in 0.5× Tris-borate EDTA. After electrophoresis, gels were dried and were exposed to Kodak BioMax MR (maximum-resolution) films for 4 h. All samples in which variant bands were detected, as well as two to three normal bands per exon, were analyzed by sequencing, by use of the same PCR primers. Also, the genotypes of the available family members of the carriers were determined by sequencing, which was performed using an automated ABI Prism 310 Genetic Analyzer (PE Biosystems) according to the instructions of the manufacturer.
Mutation Screening with Conformation-Sensitive Gel Electrophoresis (CSGE) Analysis
CSGE analysis (Ganguly et al. 1993) of the coding region of the RNASEL gene was performed using the same primer sequences as in SSCP analysis. 5′-Labeling reaction was performed in a 30-μl volume (containing 45 pmol of the forward primer, 45 μCi of γ[33P]-ATP [Amersham Pharmacia], and 15 U T4 polynucleotide kinase [New England Biolabs]) in 37°C for 60 min. The PCR contained 2.25 mM MgCl2, 200 μM each dNTP, 5.4 pmol of the unlabeled forward primer, 0.6 pmol of the labeled forward primer, 6 pmol of the reverse primer, and 1.25 U of AmpliTaqGold. The amplification conditions were the same as those in the SSCP analysis. The PCR was heated to 95°C for 10 min in a PCR machine, and, then, the machine was switched off to allow the products to slowly form heteroduplexes as they cooled to room temperature. Five microliters of the sample was mixed with 7.5 μl of 30% glycerol containing 0.25% each of xylene cyanol and bromophenol blue. The labeled PCR products were electrophoresed overnight at 5 W (for two gels) in the gel prepared with 10% polyacrylamide (75:1 ratio of acrylamide:bisacrylamide), 10% ethylene glycol, and 15% formamide, in 0.5× TTE buffer (1× TTE is 89 mM Tris, 29 mM taurine buffer, and 0.5 mM EDTA, with pH 9). All samples in which variant bands were detected, as well as two to three normal bands per exon, were analyzed by sequencing, by use of the same PCR primers.
Minisequencing for Large-Scale Population Screening of Identified Variants
The frequencies of the RNASEL variants at the population level were determined by minisequencing (Syvänen 1998). The most interesting variant, E265X, and the G59S variant were screened in the entire sample of patients described above. S406F, R462Q, and D541E missense variants were screened in 176 controls and 167 patients with unselected PRCA. PCR was performed with 100 ng of DNA, 0.2 μM each primer, 0.2 mM each dNTP, 1.5 mM MgCl2, and 1.0 U of AmpliTaqGold (PE Biosystems), in a final volume of 50 μl. Positive results were confirmed by the sequencing, by use of the PCR primers, of two or three positive results from each minisequencing reaction.
Statistical Analyses
Association of the RNASEL genotypes with HPC, PRCA, and BPH was tested by logistic-regression analysis, by use of the SPSS statistical software package (SPSS 1999). Association with demographic, clinical, and pathological features of the disease was tested by the Mann-Whitney test, by use of the SPSS statistical software package and Fisher's exact test with StatXact (Cytel Software 1999; SPSS 1999).
Results
Seven sequence variants were found in the SSCP analysis of the RNASEL gene in 66 index patients from Finnish families with HPC (table 1). These variants included a truncating mutation, E265X, which has also been reported by Carpten et al. (2002). No additional variants were found in the CSGE analysis. E265X was then tested in another, independent set of 50 small families with HPC, bringing the total number of index patients with HPC to 116. The overall E265X-mutation frequency in patients with HPC was 4.3% (table 2). This was significantly higher (odds ratio [OR]=4.56; 95% CI 1.1–19.4; table 3) than the frequency (1.8%) seen in the population sample of 566 blood donors. When stratified by family size, index patients from families with three affected members (OR=7.00; 95% CI 1.1–43.5) and from those with four or more affected members (OR=10.67; 95% CI 1.7–67.7) had significantly elevated E265X-mutation frequencies. Two (9.5%) of 21 patients from families with HPC with four or more affected members were E265X positive. In contrast, in patients from families with HPC with only two affected members, the mutation frequency, 1.6%, was not significantly different from that seen in controls (OR=1.61; 95% CI 0.2–15.7).
Table 1.
Summary of RNASEL Sequence Variants Found in 66 Patients with HPC in the SSCP Analysis
| Exon | Codon | Positiona; Nucleotide Change (Codon/Amino Acid Change) |
| 2 | 59b | 175; G→A (GGC→AGC/Gly→Ser) |
| 2 | 265b | 793; G→T (GAG→TAG/Glu→Stop) |
| 2 | 393 | 1179; G→A |
| 2 | 406 | 1217; C→T (TCT→TTT/Ser→Phe) |
| 2 | 462b | 1385; G→A (CGA→CAA/Arg→Gln) |
| 4 | 541b | 1623; T→G (GAT→GAG/Asp→Glu) |
| 7 | 724 | 2172; G→A |
Numbering is according to the cDNA starting at the A in the start codon.
Also reported by Carpten et al. (2002).
Table 2.
Genotype Frequencies of RNASEL Sequence Variants in Controls, Patients with Unselected PRCA, and Patients with HPC
|
No./Total (Frequency) in |
|||
| Patients with |
|||
| Variant and Genotype | Controls | PRCA | HPC |
| E265Xa: | |||
| G | 556/566 (98.2%) | 482/492 (98.0%) | 111/116 (95.7%) |
| GT | 10/566 (1.8%) | 10/492 (2.0%) | 5/116 (4.3%) |
| S406F: | |||
| C | 173/176 (98.3%) | 160/167 (95.8%) | 63/66 (95.5%) |
| CT | 3/176 (1.7%) | 7/167 (4.2%) | 3/66 (4.5%) |
| R462Q: | |||
| G | 69/176 (39.2%) | 60/167 (35.9%) | 28/66 (42.4%) |
| GA | 84/176 (47.7%) | 83/167 (49.7%) | 23/66 (34.8%) |
| A | 23/176 (13.1%) | 24/167 (14.4%) | 15/66 (22.7%) |
| D541E: | |||
| T | 29/176 (16.5%) | 21/167 (12.6%) | 8/66 (12.1%) |
| TG | 91/176 (51.7%) | 94/167 (56.3%) | 32/66 (48.5%) |
| G | 56/176 (31.8%) | 52/167 (31.1%) | 26/66 (39.4%) |
Because of the complete linkage disequilibrium, frequencies for G59S are the same as those for E265X.
Table 3.
Association of the Truncating Mutation E265X and Missense Variant R462Q of the RNASEL Gene with Patients with BPH, Unselected PRCA, or HPC
| Patient or Family Sampleand Mutation | No. of Carriers/Total (Frequency) | OR | 95% CI | P |
| E265X: | ||||
| Controls | 10/566 (1.8%) | 1.00 | … | … |
| Patients with BPH | 7/223 (3.1%) | 1.80 | .68–4.79 | .75 |
| Patients with unselected PRCA | 10/492 (2.0%) | 1.15 | .48–2.80 | .24 |
| All patients with HPC | 5/116 (4.3%) | 4.56 | 1.07–19.42 | .04a |
| Two affecteds | 1/64 (1.6%) | 1.61 | .17–15.71 | .68 |
| Three affecteds | 2/31 (6.5%) | 7.00 | 1.12–43.53 | .04a |
| Four or more affecteds | 2/21 (9.5%) | 10.67 | 1.68–67.72 | .01a |
| R462Q homozygotes: | ||||
| Controls | 23/176 (13.1%) | 1.00 | … | … |
| Patients with unselected PRCA | 24/167 (14.4%) | 1.12 | .60–2.07 | .73 |
| All patients with HPC | 15/66 (22.7%) | 1.96 | .95–4.03 | .07 |
| Two affecteds | 2/19 (10.5%) | 0.78 | .17–3.61 | .75 |
| Three affecteds | 7/26 (26.9%) | 2.45 | .93–6.47 | .07 |
| Four or more affecteds | 6/21 (28.6%) | 2.66 | .94–7.55 | .07 |
Statistically significant.
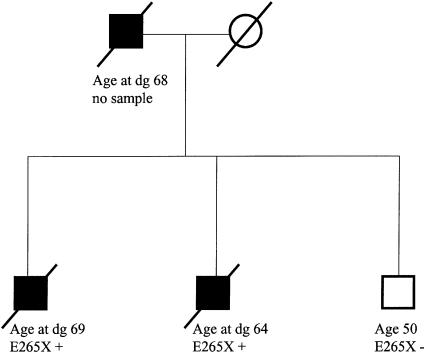
We then tested additional samples from the other affected members of the 116 families with HPC, as well as all available samples from unaffected members of the E265X-mutation–positive families, for the E265X mutation. Suggestive evidence of segregation between the E265X mutation and PRCA was seen only in a single family with three affected members. In this family, two affected brothers were mutation carriers, whereas their unaffected, 50-year-old brother did not carry the mutation (fig. 1). No sample was available from their affected father. Of the remaining four E265X-positive families with HPC, two had samples available only from a single affected sib pair, one had samples available from three affected members, and one had samples available from four affected members. However, in each of these families, only one affected person carried the E265X mutation. None of the unaffected family members tested were mutation positive. In the five families in which the E265X mutation was found, the median age at diagnosis for the six mutation carriers was 11 years younger than that for the seven affected members who were not carriers (P=.07; Mann-Whitney test).
Figure 1.
Pedigree structure of an E265X-positive family with HPC. E265X-mutation carriers are denoted by a plus sign (+), and noncarriers are denoted by a minus sign (−). Age or age at diagnosis (dg) (in years) is indicated below the symbol for each family member.
The frequencies of the E265X (table 2) and G59S variants were then compared, at the population level, in families with HPC (n=116), in controls (n=566), in patients with unselected PRCA (n=492), and in patients with BPH (n=223). All E265X carriers found were heterozygotes. E265X was found to be in complete linkage disequilibrium with G59S, but neither variant showed a significant association with either PRCA or BPH. S406F, R462Q, and D541E missense variants were analyzed in the index patients from 66 families with HPC, 176 controls, and 167 patients with PRCA (table 2). All gene frequencies were found to be in Hardy-Weinberg equilibrium. R462Q homozygotes were found to be more common among patients with HPC (22.7%) than among the controls (13.1%), indicating a possible association with HPC risk (OR=1.96; 95% CI 0.9–4.0; P=.07; table 3). The frequency was highest (28.6%) in families with four or more affected members. The other missense variants did not show any significant associations with either HPC or PRCA. We also analyzed the association between these variants and disease phenotype, including T-stage, M-stage, and tumor grade, among patients with unselected PRCA (Fisher's exact test). No significant associations emerged from these analyses (data not shown).
Discussion
RNASEL was recently suggested as a candidate HPC1 gene, on the basis of the finding that a truncating mutation (E265X) and an initiation-codon mutation (M1I) segregate in two families with HPC with linkage to HPC1 (Carpten et al. 2002). The E265X variant has been proven to cause a dramatic decrease in the enzymatic activity of RNASEL (Dong and Silverman 1999). Loss of the wild-type allele and loss of the protein expression in tumor tissues from a patient in an E265X-mutation–positive family with linkage to HPC1 were also reported (Carpten et al. 2002).
Our most significant finding—which suggests that the RNASEL gene is involved in PRCA predisposition—came from the higher frequency of the E265X truncating mutation in patients from Finnish families with HPC, as compared to its frequency in controls. An OR of 4.56 was obtained for E265X when all patients with familial disease were considered, and an OR of 10.67 was obtained when families with HPC with four or more affected members were considered.
We found that the prevalence (5/116 [4.3%]) of E265X mutations in the Finnish index patients with HPC was somewhat higher than that which had been found in the U.S. study reported by Carpten et al. (2002), in which the E265X mutation was found in only 1 (3.8%) of the 26 patients with HPC whom they studied. This is despite the fact that Carpten et al. (2002) specifically studied families with linkage to HPC1, whereas the families that we studied were not selected on the basis of prior linkage results. In fact, families with linkage to HPC1 are rare in Finland (Schleutker et al. 2000), and, of the seven candidate families with HPC1 linkage (defined as LOD score >0.25) that have been studied here, none carried E265X mutations of the RNASEL gene.
Consistent with this view, possible evidence of segregation of the E265X mutation was seen only in a single Finnish family, in which two affected brothers were carriers and in which an unaffected brother was a noncarrier. Their father was also diagnosed with PRCA, but he was unavailable for study (fig. 1). In four other E265X-positive families, no segregation was found. In the E265X-positive families, the diagnosis of PRCA was based on symptomatic disease, not on PSA testing. Similarly, Carpten et al. (2002) reported segregation of the E265X mutation with disease in a single family. Therefore, our results indicate a significant disease association when E265X-mutation frequencies are compared between index patients with HPC and controls, but they indicate a lack of evidence of segregation with disease in these families. This observation could be explained by the fact that the HPC1 mutations are not sufficient to cause disease but perhaps act as modifiers of disease onset. In support of this hypothesis, we found that, in all families with E265X mutations, there was a tendency for PRCA to be diagnosed ∼11 years earlier in mutation carriers than in noncarriers (P=.07).
Besides E265X, we found four missense variants—G59S, S406F, R462Q, and D541E—and two silent base substitutions. Of these, S406F was not reported in the study by Carpten et al. (2002). R462Q and D541E are also found in an SNP database maintained by the National Center for Biotechnology Information (see the dbSNP Home Page). To evaluate the impact that the RNASEL missense variants have at the population level, we determined their frequencies in patients with HPC, in patients with unselected PRCA, and in a group of male blood donors. G59S frequencies were significantly different in the patients with HPC than in the controls, because of the complete linkage disequilibrium with E265X. Also, R462Q homozygotes were more common in families with HPC than in the controls. Therefore, two RNASEL variants showed an association with HPC, but no effect that the RNASEL variants had at the population level could be distinguished.
In conclusion, our results suggest that the E265X and R462Q variants of the RNASEL gene may influence disease onset in large families with HPC. The variants are most often seen in patients from families with HPC with three or more affected members. However, neither variant alone seems to explain the familial clustering of prostate cancer. Furthermore, in the analysis of >1,300 individuals, we were not able to demonstrate a significant impact that any of the RNASEL variants had in the causation of PRCA at the population level. The present results warrant further studies of the role that RNASEL variants have as risk factors for HPC and unselected PRCA in other populations.
Acknowledgments
We thank Minna Sjöblom and Kati Rouhento for excellent technical assistance and Eeva Rauhala and Riitta Vaalavuo for assistance. In addition, we thank all patients with PRCA and their families for participation. The present study was supported by grants from the Medical Research Fund of Tampere University Hospital, the Reino Lahtikari Foundation, the Academy of Finland, and the Sigrid Juselius Foundation. A.R. received support from the Pirkanmaa Cancer Society, the Finnish Cancer Society, the Foundation for the Development of Laboratory Medicine, and the University of Tampere; N.N. received support from the Finnish Cultural Foundation. Ascertainment of families with HPC in Finland was supported by grant NO1-HG-55389, from the National Human Genome Research Institute, National Institutes of Health.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for SNP database)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HPC1 [MIM 601518], ELAC2/HPC2 [MIM 605367], PCAP [MIM 602759], HPCX [MIM 300147], CAPB [MIM 603688], HPC20 [MIM 176807], and RNASEL [MIM 180435])
References
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, et al (2002) Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet 30:181–184 [DOI] [PubMed] [Google Scholar]
- Castelli J, Wood KA, Youle RJ (1998) The 2-5A system in viral infection and apoptosis. Biomed Pharmacother 52:386–390 [DOI] [PubMed] [Google Scholar]
- Cytel Software (1999) StatXact-4, version 4.0.1. Cytel Software, Cambridge, MA [Google Scholar]
- Dong B, Silverman RH (1999) Alternative function of a protein kinase homology domain in 2′,5′-oligoadenylate dependent RNase L. Nucleic Acids Res 27:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH (1993) A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J 12:3297–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Stanford JL (2000) Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet 67:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentyala SN, Lee J, Hsieh K, Waltzer WC, Trocchia A, Musacchia L, Rebecchi MJ, Khan SA (2000) Prostate cancer: a comprehensive review. Med Oncol 17:85–105 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Walker AH, Zeigler-Johnson C, Weisburg S, Martin AM, Nathanson KL, Wein AJ, Malkowicz SB (2000) Association of HPC2/ELAC2 genotypes and prostate cancer. Am J Hum Genet 67:1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rökman A, Ikonen T, Mononen N, Autio V, Matikainen MP, Koivisto PA, Tammela TL, Kallioniemi OP, Schleutker J (2001) ELAC2/HPC2 involvement in hereditary and sporadic prostate cancer. Cancer Res 61:6038–6041 [PubMed] [Google Scholar]
- Schleutker J, Matikainen M, Smith J, Koivisto P, Baffoe-Bonnie A, Kainu T, Gillanders E, Sankila R, Pukkala E, Carpten J, Stephan D, Tammela T, Brownstein M, Bailey-Wilson J, Trent J, Kallioniemi OP (2000) A genetic epidemiological study of hereditary prostate cancer (HPC) in Finland: frequent HPCX linkage in families with late-onset disease. Clin Cancer Res 6:4810–4815 [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed] [Google Scholar]
- SPSS (1999) SPSS 9.0 for Windows. SPSS, Chicago [Google Scholar]
- Suarez BK, Gerhard DS, Lin J, Haberer B, Nguyen L, Kesterson NK, Catalona WJ (2001) Polymorphisms in the prostate cancer susceptibility gene HPC2/ELAC2 in multiplex families and healthy controls. Cancer Res 61:4982–4984 [PubMed] [Google Scholar]
- Syvänen AC (1998) Solid-phase minisequencing as a tool to detect DNA polymorphism. Methods Mol Biol 98:291–298 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, et al (2001) A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27:172–180 [DOI] [PubMed] [Google Scholar]
- Vesprini D, Nam RK, Trachtenberg J, Jewett MA, Tavtigian SV, Emami M, Ho M, Toi A, Narod SA (2001) HPC2 variants and screen-detected prostate cancer. Am J Hum Genet 68:912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, McDonnell SK, Elkins DA, Slager SL, Christensen E, Marks AF, Cunningham JM, Peterson BJ, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN (2001) Role of HPC2/ELAC2 in hereditary prostate cancer. Cancer Res 61:6494–6499 [PubMed] [Google Scholar]
- Xu J (2000) Combined analysis of hereditary prostate cancer linkage to 1q24-25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet 66:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Carpten JD, Nupponen NN, Robbins CM, Mestre J, Moses TY, Faith DA, Kelly BD, Isaacs SD, Wiley KE, Ewing CM, Bujnovszky P, Chang B, Bailey-Wilson J, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB (2001) Evaluation of linkage and association of HPC2/ELAC2 in patients with familial or sporadic prostate cancer. Am J Hum Genet 68:901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH (1993) Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753–765 [DOI] [PubMed] [Google Scholar]