Abstract
Interleukin-6 (IL-6) plays a contributory role in the progression and severity of many forms of cancer; it however remains unclear whether the relevance between circulating IL-6 and cancer is causal. We therefore meta-analyzed published articles in this regard using IL-6 gene -174G/C variant as an instrument. Seventy-eight and six articles were eligible for the association of -174G/C variant with cancer and circulating IL-6, respectively. Overall analyses failed to identify any significance between -174G/C and cancer risk. In Asians, carriers of the -174CC genotype had an 1.95-fold increased cancer risk compared with the -174GG genotype carriers (P = 0.009). By cancer type, significance was only attained for liver cancer with the -174C allele conferring a reduced risk under allelic (odds ratio or OR = 0.74; P = 0.001), homozygous genotypic (OR = 0.59; P = 0.029) and dominant (OR = 0.67; P = 0.004) models. Carriers of the -174CC genotype (weighted mean difference or WMD = −4.23 pg/mL; P < 0.001) and -174C allele (WMD = −3.43 pg/mL; P < 0.001) had circulating IL-6 reduced significantly compared with the non-carriers. In further Mendelian randomization analysis, a reduction of 1 pg/mL in circulating IL-6 was significantly associated with an 12% reduced risk of liver cancer. Long-term genetically-reduced circulating IL-6 might be causally associated with a lower risk of liver cancer.
As a multifactorial cytokine, interleukin-6 (IL-6) is widely believed to play a role in the progression and severity of many forms of cancer. Several observational studies have suggested that circulating IL-6 can explain inter-individual variability in predisposition to cancer. Heikkila and colleagues have written an excellent review, highlighting the involvement of elevated circulating IL-6 in human carcinogenesis1. However, it remains unclear whether the relevance between circulating IL-6 and cancer is causal as the issue of confounding or reverse causation is often unavoidable in observational epidemiology.
Ideally, randomized controlled trial of the intervention that alters circulating IL-6 is considered as the gold standard for evaluating this causal relevance, but in some circumstances it is neither practical nor ethical to randomize human beings for such trials. As an alternative, a more promising method termed as ‘Mendelian randomization’ has been developed to exploit the impact of long-term exposure differences on disease risk by using a genetic instrument to account for the potential biases due to confounding and reverse causation2. Like a randomized controlled trial, Mendelian randomization randomizes genotypes at conception according to Mendel’s second law3. This method has been successfully applied to a variety of genetic exposures such as obesity and alcohol consumption in causal susceptibility to cancer4.
The most immediate determinant of circulating IL-6 is its encoding gene IL-6. The genomic sequence of IL-6 gene is highly polymorphic and one of the most frequently evaluated variants is -174G/C (rs1800795) in the promoter region5,6. In vitro studies have found the allele-specific impact of -174G/C variant on IL-6 gene promoter activity, with the -174C allele corresponding to lower expression level7. Besides, carriers of the -174G allele had a higher level of circulating IL-6 than those with the -174CC genotype8,9. Based on these observations, we thereby develop a completing hypothesis that the association between circulating IL-6 and cancer is causally related. To test this hypothesis, we employed Mendelian randomization technique to meta-analyze all available published articles in this regard by using IL-6 gene -174G/C variant as an instrument.
Results
Eligible articles
The selection process of articles is schematized in Fig. 1. A total of 837 potentially relevant articles were identified after an initial literature search and 80 of them written in English language were finally analyzed5,6,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84. Article involving more than one independent study group was analyzed separately. Altogether, 78 articles with 87 study groups (45569 cancer patients and 57990 controls) were eligible for the association between IL-6 gene -174G/C variant and cancer6,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84, and 6 articles with 9 study groups (1727 study subjects) were eligible for circulating IL-6 changes across -174G/C genotypes5,8,9,13,16,52.
Figure 1. Flow diagram of search strategy and study selection.
Study characteristics
Table 1 summarizes the baseline characteristics of 87 eligible study groups for the association between IL-6 gene -174G/C variant and cancer. Supplementary Table S1 provides the quality assessment of 87 study groups and the genotype distributions of -174G/C variant. The quality score ranged from 4 to 11 and was averaged at 8.37. Cancer patients were older than controls (mean age: 59.26 versus 55.01 years, P = 0.0007). The percentage of smokers was slightly higher in cancer patients than healthy controls (42.24% versus 33.06%, P = 0.038). No significance was observed in mean body mass index and the percentages of male gender and drinkers between the two groups (P > 0.05).
Table 1. The baseline characteristics of all study groups in this meta-analysis.
| Author (year) | Ethnicity | Cancer type | Match | Source of controls | Study design |
Sample size |
Age (years) |
Males |
BMI (kg/m2) |
Smoking |
Drinking |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Cont’s | Cases | Cont’s | Cases | Cont’s | Cases | Cont’s | Cases | Cont’s | Cases | Cont’s | ||||||
| Slattery (2014) | Mixed | Breast | NA | Population | Prosp. | 3567 | 4157 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Mandal (2014) | Caucasian | Prostate | YES | Hospital | Retrosp. | 84 | 78 | 59.8 | 57.2 | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Mandal (2014) | African-American | Prostate | YES | Hospital | Retrosp. | 80 | 62 | 67.9 | 64.0 | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Cil (2014) | Mixed | Thyroid | YES | Hospital | Retrosp. | 190 | 216 | 47.2 | 46.0 | 0.23 | 0.26 | 25.55 | 25.70 | 0.34 | 0.32 | NA | NA |
| Tindall (2012) | Caucasian | Prostate | YES | Population | Prosp. | 818 | 734 | NA | NA | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Giannitrapani (2011) | Caucasian | Liver | YES | Population | Retrosp. | 95 | 98 | NA | NA | 0.53 | NA | NA | NA | NA | NA | NA | NA |
| Giannitrapani (2011) | Caucasian | Liver | YES | Population | Retrosp. | 105 | 98 | NA | NA | 0.63 | NA | NA | NA | NA | NA | NA | NA |
| Gaur (2011) | Mixed | Oral | YES | Hospital | Prosp. | 140 | 120 | 51.4 | 51.4 | 0.85 | 0.85 | NA | NA | NA | NA | NA | NA |
| Abuli (2011) | Caucasian | Colorectal | YES | Population | Prosp. | 1405 | 1388 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Cacev (2010) | Caucasian | Colorectal | NA | Population | Retrosp. | 160 | 160 | 64.5 | 63.1 | 0.53 | 0.54 | NA | NA | NA | NA | NA | NA |
| Ognjanovic (2010) | Mixed | Colorectal | YES | Population | Retrosp. | 271 | 539 | 62.5 | 62.0 | 0.68 | 0.69 | 26.90 | 26.70 | 0.14 | 0.10 | 0.45 | 0.41 |
| Hawken (2010) | Mixed | Colorectal | NA | Population | Retrosp. | 1133 | 1125 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Dossus (2010) | Mixed | Breast | YES | Population | Prosp. | 6292 | 8135 | 63.1 | 63.1 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Dossus (2010) | Mixed | Prostate | YES | Population | Prosp. | 8008 | 8604 | 68.4 | 68.4 | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Tsilidis (2009) | Mixed | Colorectal | YES | Population | Prosp. | 208 | 318 | 62.8 | 62.8 | 0.46 | 0.45 | 26.30 | 26.00 | 0.13 | 0.13 | NA | NA |
| Ozgen (2009) | Mixed | Thyroid | NA | Hospital | Retrosp. | 42 | 340 | 43.1 | 43.8 | 0.19 | 0.19 | NA | NA | NA | NA | NA | NA |
| Ognjanovic (2009) | Mixed | Liver | YES | Population | Retrosp. | 120 | 230 | 60.5 | 59.5 | 0.68 | 0.60 | NA | NA | 0.42 | 0.28 | 0.71 | 0.77 |
| Gangwar (2009) | Asian | Cervical | YES | Hospital | Retrosp. | 160 | 200 | 45.0 | 46.0 | 0.00 | 0.00 | NA | NA | 0.34 | 0.10 | 0.06 | 0.01 |
| Falleti (2009) | Caucasian | Liver | NO | Population | Retrosp. | 219 | 236 | 53.0 | 46.0 | 0.70 | 0.70 | NA | NA | NA | NA | NA | NA |
| Cherel (2009) | Caucasian | Breast | NA | Hospital | Prosp. | 293 | 112 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Vasku (2009) | Caucasian | Colorectal | YES | Hospital | Retrosp. | 102 | 101 | 68.0 | 68.1 | 0.77 | 0.58 | NA | NA | NA | NA | NA | NA |
| Talar-Wojnarowska (2009) | Caucasian | Pancreatic | YES | Hospital | Retrosp. | 97 | 50 | NA | NA | 0.57 | 0.57 | NA | NA | NA | NA | NA | NA |
| Slattery (2009) | Mixed | Colorectal | NA | Population | Retrosp. | 1839 | 2014 | 0.55 | 0.54 | NA | NA | NA | NA | NA | NA | ||
| Andrie (2009) | Caucasian | Lymphoma | YES | Hospital | Retrosp. | 81 | 81 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Aladzsity (2009) | Caucasian | Myeloma | YES | Hospital | Retrosp. | 97 | 99 | 65.0 | 68.0 | 0.35 | 0.45 | NA | NA | NA | NA | NA | NA |
| Birmann (2009) | Mixed | Myeloma | NA | Population | Prosp. | 82 | 159 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Wilkening (2008) | Caucasian | Colorectal | YES | Population | Prosp. | 308 | 585 | 56.8 | 56.8 | 0.44 | 0.44 | NA | NA | NA | NA | NA | NA |
| Vairaktaris (2008) | Caucasian | Oral | YES | Population | Retrosp. | 162 | 168 | 58.5 | 54.7 | 0.80 | 0.75 | NA | NA | NA | NA | NA | NA |
| Upadhyay (2008) | Asian | Esophageal | YES | Hospital | Retrosp. | 168 | 201 | 56.8 | 53.7 | 0.74 | 0.75 | NA | NA | 0.82 | NA | 0.37 | NA |
| Slattery (2008) b | Caucasian | Breast | YES | Population | Retrosp. | 1176 | 1330 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Slattery (2008) a | Mixed | Breast | YES | Population | Retrosp. | 576 | 727 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Kesarwani (2008) | Asian | Prostate | YES | Population | Retrosp. | 200 | 200 | 62.5 | 59.5 | 1.00 | 1.00 | NA | NA | 0.32 | 0.30 | 0.60 | 0.69 |
| Crusius (2008) | Caucasian | Gastric | YES | Population | Prosp. | 439 | 1138 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Colakogullari (2008) | Mixed | Lung | YES | Population | Prosp. | 44 | 58 | 60.0 | NA | 0.91 | 0.51 | NA | NA | NA | NA | NA | NA |
| Bao (2008) | Asian | Prostate | YES | Hospital | Retrosp. | 136 | 120 | 62.8 | 62.3 | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Vogel (2008) | Caucasian | Lung | YES | Population | Retrosp. | 403 | 744 | NA | NA | 0.54 | 0.56 | NA | NA | 0.73 | 0.35 | NA | NA |
| Kury (2008) | Caucasian | Colorectal | YES | Hospital | Retrosp. | 1023 | 1121 | 65.7 | 61.9 | 0.62 | 0.54 | NA | NA | NA | NA | NA | NA |
| Ennas (2008) | Caucasian | Leukaemia | NO | Population | Retrosp. | 40 | 113 | 61.8 | 56.5 | 0.73 | 0.40 | NA | NA | NA | NA | NA | NA |
| Ahirwar (2008) | Asian | Bladder | YES | Population | Prosp. | 136 | 200 | 61.6 | 58.3 | 0.88 | 0.88 | NA | NA | NA | NA | NA | NA |
| Vishnoi (2007) | Asian | Gallbladder | YES | Population | Retrosp. | 45 | 82 | 49.4 | 50.0 | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Vishnoi (2007) | Asian | Gallbladder | YES | Population | Retrosp. | 79 | 118 | 49.4 | 50.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Litovkin (2007) | Caucasian | Breast | YES | Population | Prosp. | 73 | 143 | 55.0 | 33.0 | 0.00 | 0.49 | NA | NA | NA | NA | NA | NA |
| Litovkin (2007) | Caucasian | Leiomyoma | YES | Population | Prosp. | 60 | 143 | 37.0 | 33.0 | 0.00 | 0.49 | NA | NA | NA | NA | NA | NA |
| Gonullu (2007) | Mixed | Breast | YES | Population | Retrosp. | 38 | 24 | 47.0 | 39.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Vogel (2007) | Caucasian | Breast | YES | Population | Prosp. | 361 | 361 | NA | NA | 0.00 | 0.00 | 25.00 | 25.00 | 0.34 | 0.36 | NA | NA |
| Vogel (2007) | Caucasian | Colorectal | YES | Population | Prosp. | 355 | 753 | 59.0 | 56.0 | 0.56 | 0.56 | 26.00 | 26.00 | 0.37 | 0.35 | NA | NA |
| Slattery (2007) | Mixed | Colorectal | YES | Population | Retrosp. | 1583 | 1979 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Slattery (2007) | Mixed | Colorectal | YES | Population | Retrosp. | 797 | 1011 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Nearman (2007) | Mixed | Leukaemia | NA | Hospital | Retrosp. | 28 | 362 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Gatti (2007) | Mixed | Gastric | NA | Hospital | Retrosp. | 56 | 56 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Duch (2007) | Mixed | Myeloma | YES | Hospital | Retrosp. | 52 | 60 | 58.5 | 59.3 | 0.42 | 0.60 | NA | NA | NA | NA | NA | NA |
| Deans (2007) | Caucasian | Gastric | NA | Population | Retrosp. | 203 | 224 | 71.0 | 39.2 | 0.66 | 0.53 | NA | NA | NA | NA | NA | NA |
| Berkovic (2007) | Caucasian | Gastric | YES | Hospital | Retrosp. | 80 | 162 | 80.0 | 46.5 | 0.48 | 0.48 | NA | NA | NA | NA | NA | NA |
| Vairaktaris (2006) | Caucasian | Oral | YES | Population | Retrosp. | 162 | 156 | 58.5 | 55.5 | 0.80 | 0.77 | NA | NA | NA | NA | NA | NA |
| Theodoropoulos (2006) | Caucasian | Colorectal | YES | Population | Prosp. | 222 | 200 | 64.7 | 62.7 | 0.58 | 0.60 | NA | NA | NA | NA | NA | NA |
| Nogueira (2006) | Mixed | Cervical | YES | Population | Retrosp. | 56 | 253 | 52.2 | 54.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Michaud (2006) | Mixed | Prostate | YES | Population | Prosp. | 503 | 652 | 67.1 | 66.6 | 1.00 | 1.00 | NA | NA | 0.08 | 0.11 | NA | NA |
| Kamangar (2006) | Caucasian | Gastric | YES | Population | Prosp. | 110 | 203 | 58.5 | 59.0 | 1.00 | 1.00 | NA | NA | 1.00 | 1.00 | NA | NA |
| Gonzalez-Zuloeta (2006) | Caucasian | Breast | NA | Population | Prosp. | 171 | 3651 | 67.8 | 70.8 | 0.00 | 0.00 | 26.70 | 27.10 | NA | NA | NA | NA |
| Balasubramanian (2006) | Caucasian | Breast | NA | Hospital | Prosp. | 197 | 490 | 63.0 | 57.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Rothman (2006) | Caucasian | Lymphoma | NA | Population | Retrosp. | 2658 | 3068 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Lan (2006) | Mixed | Lymphoma | YES | Population | Retrosp. | 518 | 597 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Gunter (2006) | Mixed | Colorectal | YES | Hospital | Retrosp. | 244 | 231 | 60.0 | 57.0 | 0.78 | 0.64 | 26.50 | 25.80 | 0.11 | 0.05 | NA | NA |
| Gaustadnes (2006) | Caucasian | Colorectal | YES | Population | Retrosp. | 230 | 540 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Cozen (2006) | Mixed | Myeloma | YES | Population | Retrosp. | 150 | 112 | 61.0 | NA | 0.61 | 0.58 | NA | NA | NA | NA | NA | NA |
| Seifart (2005) | Caucasian | Lung | NA | Population | Retrosp. | 182 | 243 | 63.3 | 37.9 | 0.88 | 0.55 | NA | NA | 0.98 | 0.40 | NA | NA |
| Migita (2005) | Asian | Liver | NO | Hospital | Prosp. | 48 | 188 | 62.5 | 51.5 | 0.81 | 0.68 | NA | NA | NA | NA | NA | NA |
| Leibovici (2005) | Caucasian | Bladder | YES | Hospital | Prosp. | 465 | 450 | NA | NA | NA | NA | NA | NA | 0.74 | 0.53 | NA | NA |
| Hefler (2005) | Caucasian | Breast | YES | Hospital | Retrosp. | 269 | 227 | 54.9 | 53.3 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Basturk (2005) | Mixed | Renal cell | YES | Population | Retrosp. | 29 | 50 | NA | NA | 0.05 | 0.56 | NA | NA | NA | NA | NA | NA |
| Snoussi (2005) | Caucasian | Breast | NA | Population | Prosp. | 305 | 200 | 50.0 | 46.0 | 0.01 | 0.05 | NA | NA | NA | NA | NA | NA |
| Skerrett (2005) | Mixed | Breast | NA | Population | Retrosp. | 88 | 102 | 49.2 | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Mazur (2005) | Caucasian | Myeloma | NA | Hospital | Retrosp. | 54 | 50 | 62.0 | NA | 0.43 | 0.58 | NA | NA | NA | NA | NA | NA |
| Festa (2005) | Caucasian | Basal cell | NA | Population | Prosp. | 241 | 260 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Cordano (2005) | Caucasian | Lymphoma | YES | Population | Retrosp. | 408 | 349 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Campa (2005) | Caucasian | Lung | NA | Hospital | Retrosp. | 1995 | 1982 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Zhang (2004) | Caucasian | Basal cell | YES | Hospital | Retrosp. | 241 | 260 | 50.0 | 48.0 | 0.58 | 0.54 | NA | NA | NA | NA | NA | NA |
| Smith (2004) | Caucasian | Breast | NO | Population | Retrosp. | 144 | 263 | 59.6 | 40.3 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Campa (2004) | Caucasian | Lung | YES | Population | Prosp. | 250 | 214 | 63.1 | 64.8 | 0.71 | 0.75 | NA | NA | 0.73 | 0.91 | NA | NA |
| Bushley (2004) | Mixed | Ovarian | YES | Population | Retrosp. | 182 | 219 | NA | NA | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Landi (2003) | Mixed | Colorectal | YES | Hospital | Retrosp. | 377 | 326 | NA | NA | 0.60 | 0.53 | NA | NA | 0.15 | 0.18 | 0.67 | 0.56 |
| El-Omar (2003) | Mixed | Esophageal | YES | Population | Retrosp. | 161 | 210 | 65.5 | 66.0 | 0.87 | 0.85 | NA | NA | 0.40 | 0.24 | 0.87 | 0.85 |
| El-Omar (2003) | Mixed | Gastric | YES | Population | Retrosp. | 314 | 210 | 68.0 | 66.0 | 0.81 | 0.85 | NA | NA | 0.30 | 0.24 | 0.75 | 0.85 |
| Hwang (2003) | Asian | Gastric | NA | Hospital | Retrosp. | 30 | 30 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Hwang (2003) | Caucasian | Gastric | NA | Hospital | Retrosp. | 30 | 30 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Howell (2003) | Caucasian | Melanoma | NA | Hospital | Prosp. | 153 | 208 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Zheng (2000) | Caucasian | Myeloma | NA | Population | Retrosp. | 73 | 129 | 67.0 | NA | 0.45 | NA | NA | NA | NA | NA | NA | NA |
Abbreviations: BMI, body mass index; cont’s, controls; NA, not available; Prosp., prospective design; Retrosp., retrospective design.
Forty-six of 87 study groups were conducted in Caucasians, 31 in mixed populations, 9 in Asians and 1 in African-Americans. Sixteen study groups focused on colorectal cancer, 14 groups on breast cancer, 8 groups on gastric cancer, 7 groups on prostate cancer, 6 groups on myeloma, 5 groups respectively on lung and liver cancers, 4 groups on lymphoma, 3 groups on oral cancer, 2 groups respectively on esophageal, basal cell, bladder, cervical, leukatmia, gallbladder and thyroid cancers, 1 group respectively on melanoma, ovarian, renal cell, leiomyoma and pancreatic cancers. Thirty-three of 87 study groups had total sample size of at least 500. Age was reported to be matched in 60 study groups and unmatched in 4 study groups between cancer patients and controls. Fifty-seven study groups had controls enrolled from general populations and 30 study groups from hospitals. Fifty-nine and 28 study groups followed a retrospective and prospective design, respectively. Sixty-five of 87 study groups had genotype distributions of -174G/C variant in Hardy–Weinberg equilibrium at a significance level of 5%, 19 study groups in Hardy–Weinberg disequilibrium, and 3 study groups without mutation. The frequency of IL-6 gene -174C allele ranged from 0.0% to 52.94% in cancer patients and from 0.0% to 51.15% in healthy controls. By contrast, this frequency was significantly lower in Asians than in Caucasians (cancer patients: 0.0% to 28.0% versus 17.90% to 52.94%; healthy controls: 0.0% to 26.75% versus 13.33% to 51.15%).
Table 2 summarizes mean circulating IL-6 across -174G/C genotypes. All study groups involved Caucasians except for one with mixed descents. Seven of 9 study groups provided circulating IL-6 in cancer patients, and 1 study group respectively in healthy controls and in combined patients and controls.
Table 2. Mean circulating IL-6 across IL-6 gene -174G/C genotypes in this meta-analysis.
| Author (year) | Ethnicity | Status | Sample size |
Circulating IL-6 (pg/mL) across -174G/C genotypes |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (GG) | SD (GG) | Mean (GC) | SD (GC) | Mean (CC) | SD (CC) | ||||
| Malaponte (2013) | Caucasian | Cancer patients | 130 | 22.10 | 4.30 | 11.90 | 2.60 | 5.30 | 1.50 |
| Malaponte (2013) | Caucasian | Cancer patients | 190 | 7.20 | 2.20 | 12.00 | 1.70 | 4.50 | 1.10 |
| Malaponte (2013) | Caucasian | Healthy controls | 215 | 2.80 | 0.70 | 2.70 | 0.50 | 2.60 | 0.70 |
| Giannitrapani (2011) | Caucasian | Cancer patients | 67 | 2.20 | 2.86 | 2.30 | 1.70 | 2.90 | 0.60 |
| Giannitrapani (2011) | Caucasian | Cancer patients | 80 | 4.80 | 4.25 | 3.60 | 3.00 | 0.87 | 1.97 |
| Ognjanovic (2010) | Mixed | Cancer patients and controls | 806 | 1.90 | 0.05 | 2.23 | 2.69 | 2.35 | 1.91 |
| Talar-Wojnarowska (2009) | Caucasian | Cancer patients | 97 | 65.00 | 10.00 | 27.00 | 10.00 | 33.00 | 10.00 |
| Berkovic (2007) | Caucasian | Cancer patients | 80 | 3.07 | 1.03 | 4.31 | 2.98 | 7.51 | 4.33 |
| Belluco (2003) | Caucasian | Cancer patients | 62 | 0.32 | 0.62 | 0.14 | 0.13 | 0.14 | 0.13 |
Abbreviations: SD, standard deviation.
Prediction of -174G/C variant for cancer risk
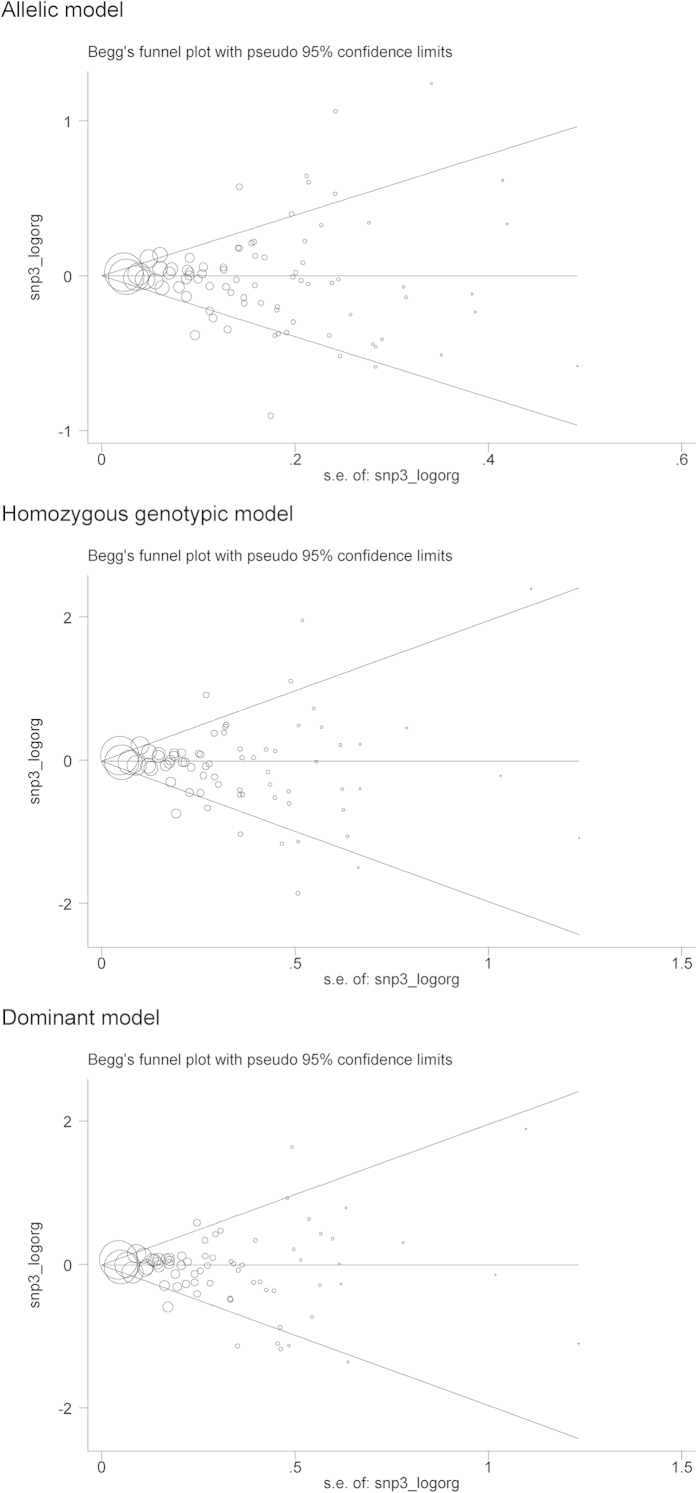
Overall and subgroup estimates of IL-6 gene -174G/C variant for cancer risk are provided in Table 3. Overall analyses failed to identify any significance for the -174C allele under allelic (OR = 1.02; 95% CI: 0.98 to 1.07; P = 0.290), homozygous genotypic (OR = 1.07; 95% CI: 0.99 to 1.16; P = 0.103) and dominant (OR = 1.02; 95% CI: 0.97 to 1.08; P = 0.465) models, with moderate heterogeneity (I2 = 66.6%, 54.0% and 65.1%, respectively). There was no indication of publication bias for three genetic models except for the homozygous genotypic model (Egger’s test: P = 0.075) (Fig. 2). After restricting study groups with Hardy–Weinberg equilibrium, there was no material change in effect estimates.
Table 3. Overall and stratified risk estimates of IL-6 gene -174G/C variant for cancer risk under three genetic models.
| Groups | Studies |
Allelic model |
Genotypic model |
Dominant model |
|||
|---|---|---|---|---|---|---|---|
| OR; 95% CI; P | I2 | OR; 95% CI; P | I2 | OR; 95% CI; P | I2 | ||
| Overall | 87 | 1.02; 0.98–1.07; 0.290 | 66.6% | 1.07; 0.99–1.16; 0.103 | 54.0% | 1.02; 0.97–1.08; 0.465 | 65.1% |
| Ethnicity | |||||||
| Caucasian | 46 | 1.05; 0.98–1.12; 0.138 | 72.2% | 1.09; 0.97–1.23; 0.136 | 59.5% | 1.07; 0.98–1.17; 0.155 | 71.4% |
| Asian | 9 | 1.03; 0.75–1.42; 0.870 | 68.8% | 1.95; 1.19–3.20; 0.009 | 17.9% | 0.90; 0.66–1.22; 0.483 | 52.7% |
| Mixed | 31 | 0.98; 0.93–1.03; 0.374 | 50.6% | 0.97; 0.88–1.06; 0.450 | 27.5% | 0.97; 0.91–1.04; 0.430 | 53.0% |
| Sample size | |||||||
| <500 | 54 | 1.03; 0.92–1.15; 0.615 | 73.7% | 1.17; 0.94–1.45; 0.163 | 61.8% | 1.03; 0.89–1.20; 0.654 | 72.7% |
| >=500 | 33 | 1.01; 0.98–1.04; 0.730 | 43.5% | 1.02; 0.96–1.08; 0.615 | 29.2% | 1.01; 0.96–1.05; 0.817 | 41.4% |
| Cancer type | |||||||
| Myeloma | 6 | 1.06; 0.89–1.28; 0.496 | 0.0% | 1.13; 0.72–2.50; 0.592 | 0.0% | 1.09; 0.84–1.40; 0.521 | 0.0% |
| Gastric | 8 | 1.01; 0.79–1.28; 0.960 | 68.5% | 1.12; 0.81–1.54; 0.498 | 28.2% | 1.04; 0.72–1.51; 0.819 | 74.1% |
| Colorectal | 16 | 1.00; 0.93–1.07; 0.941 | 63.3% | 0.99; 0.88–1.13; 0.914 | 51.7% | 1.01; 0.92–1.11; 0.850 | 58.1% |
| Lung | 5 | 1.03; 0.95–1.11; 0.530 | 3.6% | 1.06; 0.91–1.22; 0.463 | 0.0% | 1.03; 0.86–1.25; 0.743 | 41.8% |
| Breast | 14 | 0.99; 0.93–1.05; 0.716 | 44.0% | 1.02; 0.89–1.16; 0.824 | 11.3% | 0.99; 0.89–1.09; 0.773 | 58.2% |
| Lymphoma | 4 | 1.00; 0.95–1.07; 0.888 | 0.0% | 1.01; 0.89–1.15; 0.855 | 0.0% | 1.00; 0.92–1.10; 0.940 | 0.0% |
| Liver | 5 | 0.74; 0.61–0.89; 0.001 | 0.0% | 0.59; 0.36–0.95; 0.029 | 0.0% | 0.67; 0.52–0.88; 0.004 | 21.5% |
| Prostate | 7 | 0.95; 0.80–1.14; 0.597 | 79.4% | 0.94; 0.66–1.34; 0.724 | 76.3% | 0.96; 0.81–1.13; 0.609 | 58.8% |
| Oral | 3 | 1.49; 0.58–3.81; 0.409 | 95.0% | 2.38; 0.34–16.93; 0.385 | 91.6% | 1.98; 0.54–7.26; 0.303 | 95.4% |
| Matched | |||||||
| NA | 23 | 1.00; 0.94–1.06; 0.961 | 43.1% | 0.99; 0.89–1.10; 0.821 | 18.1% | 1.00; 0.92–1.08; 0.926 | 41.1% |
| YES | 60 | 1.03; 0.98–1.09; 0.235 | 71.5% | 1.11; 1.00–1.23; 0.044 | 59.9% | 1.03; 0.96–1.11; 0.389 | 70.8% |
| NO | 4 | 0.99; 0.68–1.45; 0.976 | 70.4% | 1.00; 0.40–2.50; 0.999 | 75.0% | 0.97; 0.69–1.36; 0.846 | 35.1% |
| Control source | |||||||
| Population | 57 | 1.01; 0.97–1.06; 0.592 | 61.5% | 1.02; 0.94–1.11; 0.622 | 47.9% | 1.02; 0.96–1.08; 0.569 | 63.4% |
| Hospital | 30 | 1.02; 0.91–1.14; 0.701 | 73.3% | 1.17; 0.96–1.43;.0118 | 58.9% | 1.00; 0.86–1.16; 0.973 | 68.7% |
| Study design | |||||||
| Retrospective | 59 | 1.02; 0.96–1.08; 0.546 | 69.0% | 1.09; 0.97–1.23; 0.163 | 54.1% | 1.01; 0.93–1.10; 0.751 | 68.8% |
| Prospective | 28 | 1.03; 0.97–1.09; 0.316 | 61.6% | 1.06; 0.95–1.19; 0.328 | 55.7% | 1.04; 0.96–1.11; 0.350 | 54.8% |
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval; NA, not available.
Figure 2. The Begg’s funnel plots of IL-6 gene -174G/C variant for cancer under three genetic models.
Potential sources of heterogeneity were exploited by subgroup analyses according to ethnicity, cancer type, matching condition, source of controls, study design and sample size, respectively (Table 3). To avoid chance results, only subgroups with three or more study groups were considered in this meta-analysis. By ethnicity, no significance was attained in Caucasians under three genetic models, and contrastingly Asian carriers of -174CC genotype were observed to have 1.95-fold increased cancer risk compared with those with the -174GG genotype (95% CI: 1.95; 1.19 to 3.20; P = 0.009), even after the Bonferroni correction to control for the multiple testing (P < 0.05/3, here 3 refers to the total number of subgroups by ethnicity). There was no heterogeneity (I2 = 17.9%) for this significant association.
By cancer type, effect estimates were significant only for liver cancer with the -174C allele conferring a reduced risk under allelic (OR = 0.74; 95% CI: 0.61 to 0.89; P = 0.001), homozygous genotypic (OR = 0.59; 95% CI: 0.36 to 0.95; P = 0.029) and dominant (OR = 0.67; 95% CI: 0.52 to 0.88; P = 0.004) models, and this significance was less likely to be interpreted by heterogeneity (I2 = 0.0%, 0.0% and 21.5%, respectively). Even after the Bonferroni correction, significance was still preserved for the allelic and dominant models (P < 0.05/9, here 9 refers to the total number of subgroups by cancer type). Effect estimates of -174G/C variant for cancer by source of controls, study design and sample size did not deviate significantly from the unity under three genetic models (all P > 0.05), and there was no material improvement in heterogeneity within these subgroups.
Changes of circulating IL-6 across -174G/C genotypes
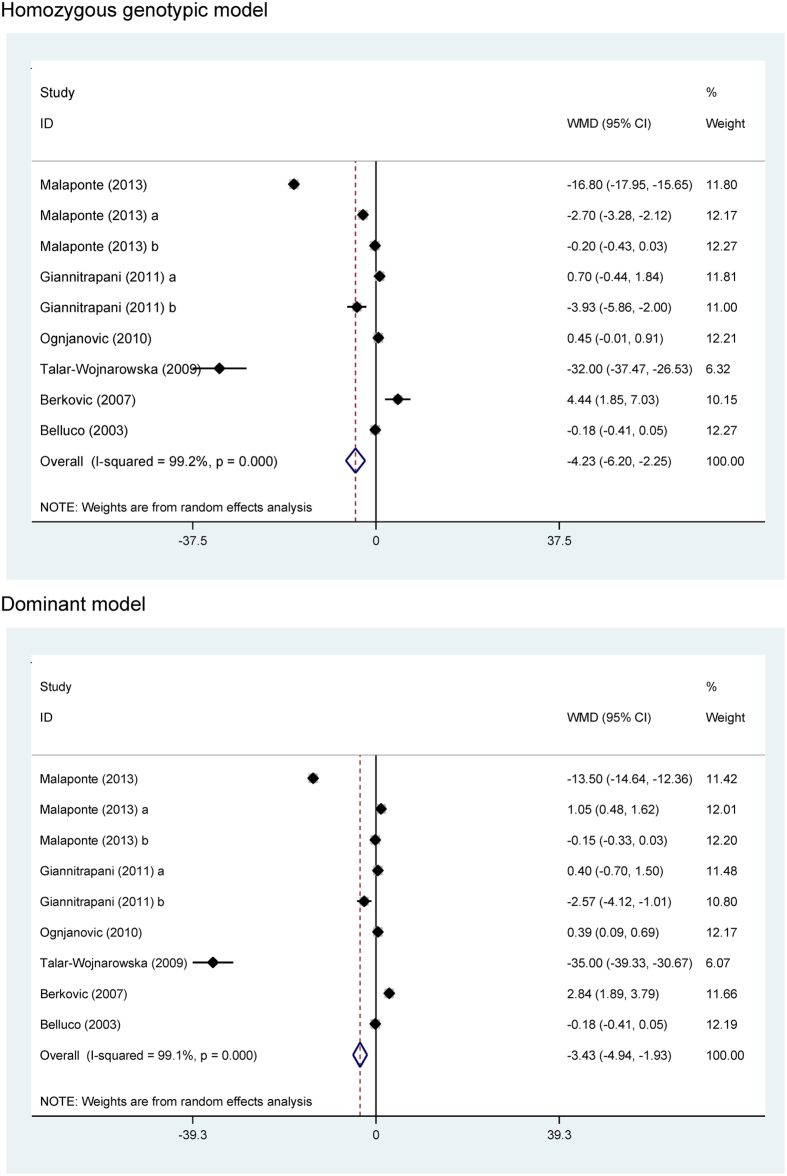
Carriers of the -174CC genotype (WMD = −4.23 pg/mL; 95% CI: −6.20 to −2.25; P < 0.001) and -174C allele (-174CC and -174GC genotypes) (WMD = −3.43 pg/mL; 95% CI: −4.94 to −1.93; P < 0.001) had significantly lowered circulating IL-6 when compared with the -174GG genotype carriers, yet with strong evidence of heterogeneity (I2 = 99.2% and 99.1%, respectively) (Fig. 3).
Figure 3. The funnel plots of circulating IL-6 changes across IL-6 gene -174G/C genotypes under homozygous genotypic and dominant models.
Predicted causality of circulating IL-6 for cancer
Under the principles of Mendelian randomization, a reduction of 1 pg/mL in circulating IL-6 was significantly associated with an 12% reduced risk of liver cancer (95% CI: 0.64 to 0.99). However in Asians, this association was totally reversed with 1 pg/mL reduced circulating IL-6 corresponding to an 17% increased cancer risk (95% CI: 1.03 to 1.68). Considering that the unity was not included by above 95% CIs, it is safe to the reject the null hypothesis of none causal relevance between circulating IL-6 and certain cancer subtypes.
Sensitivity analysis
Sensitivity analysis confirmed the overall differences in risk estimates for the prediction of IL-6 gene -174G/C variant for cancer risk and circulating IL-6 changes between -174G/C genotypes in both direction and magnitude by sequentially omitting each study once at a time and computing differential estimates for the remaining studies.
Discussion
In this meta-analysis of 80 qualified articles, we employed Mendelian randomization to test the completing hypothesis that the association between circulating IL-6 and cancer is causal. The most noteworthy finding of this study was that long-term genetically-reduced circulating IL-6 might be causally associated with a lower risk of liver cancer. To the authors’ knowledge, this is the first comprehensive meta-analysis assessing the impact of long-term differences in circulating IL-6 on cancer risk.
Currently, targeted anti-IL-6 antibody therapy has been successfully applied in several clinical trials and found to be well tolerated in cancer patients85. Evidence from epidemiological studies is accruing in favor of a contributory role of elevated circulating IL-6 in patients with advanced tumor stages of various cancers, such as non-small cell lung cancer, colorectal cancer and renal cell carcinoma86,87,88. Currently, whether the progression and severity of caner is due to elevated circulating IL-6 still remains an open question. Genetic association studies are deemed as more similar to randomized clinical trials than other types of observational epidemiological studies due to Mendelian randomization (Mendel’s second law)89. We therefore utilized Mendelian randomization to assess whether the relevance between circulating IL-6 and cancer is causal by selecting the most frequently evaluated variant -174G/C in IL-6 gene as a genetic instrument to minimize residual confounding and reverse causation.
In this meta-analysis, risk estimates of IL-6 gene -174G/C variant with cancer were heterozygous between Caucasians and Asians. Considering the multifactorial nature of cancer, divergent genetic backgrounds or linkage disequilibrium patterns might be the most likely explanation for such divergence90. This is well exemplified in the present study with regard to the frequency of -174C allele, which was exceedingly lower in Asians than in Caucasians (Table 1). Even in some Asian populations, the mutation rate of this allele was zero40,67,81. Generally, it is not uncommon for the same variant playing a different role in cancer susceptibility across different populations. This is the principal limitation of this Mendelian randomization meta-analysis, that is, if the other flanking variants within or near IL-6 gene related to cancer risk are in linkage disequilibrium with -174G/C variant we have examined, this will confound our findings. What’s more, such confounding is difficult to exclude completely; however it is unlikely that it would explain our finding that IL-6 gene -174CC genotype was associated with lowered circulating IL-6 without predicting a low cancer risk. There is also evidence that a variant may be in close linkage with another nearby causal locus in one ethnic population but not in another91. In view of this limitation, it is necessary to establish an ethnicity-specific database of candidate genes and variants in susceptibility to cancer92. Another limitation of this study is that excluding the pleiotropy of IL-6 gene -174G/C variant seems impractical for us since data on other inflammatory factors across -174G/C genotypes are rarely provided from most eligible articles, necessitating further confirmation using additional genetic variants and/or exposure outcomes.
It is also worth noting that IL-6 gene -174G/C variant exhibited heterozygous association with different forms of cancer in this meta-analysis. For example, the -174C allele was observed to confer a significantly protective effect against liver cancer, yet a risk effect for oral cancer with no attainable significance. The identification of -174G/C variant affecting the significant risk of liver cancer allowed us to employ Mendelian randomization to account for potential biases due to residual confounding and reverse causation. Consistent with the findings of other studies8,9, carriers of the -174C allele or -174CC genotype had lower circulating IL-6 than the non-carriers, supporting the plausibility of causal relevance between circulating IL-6 and liver cancer. Given the insufficient statistical power of this meta-analysis in some subgroups, far larger sample sizes than studied here will be required to produce enough power to evaluate the causality between circulating IL-6 and various forms of cancer.
Several limitations should be acknowledged in this meta-analysis. Firstly, only published articles were retrieved and the ‘grey’ literature (articles written in languages other than English) was not covered, leading to the possible existence of publication bias. However, the influence of publication bias on the gene-disease association is expected to result in an overestimation, rather than an underestimation. Secondly, as the majority of involved studies in this meta-analysis recruited cancer patients aged over 50 years for whom environmental factors are likely to contribute more prominently than a genetic component to the development of cancer, more large studies in a younger cancer population will of great interest. Thirdly, given the possible impact of drug regimens on circulating IL-6, the relationship between circulating IL-6 changes and IL-6 gene -174G/C genotypes might be biased, calling for further validation of this relationship in healthy controls. Fourthly, nearly all involved studies had circulating IL-6 measured only once, which cannot reflect its long-term level in the development of cancer. Fifthly, this meta-analysis was based on summarized data, rather than individual participant data, precluding further gene-to-environment interaction. Sixthly, only one variant in IL-6 gene was selected, and investigation on other candidate genes or polymorphisms involved in IL-6 regulation was highly encouraged, leaving a challengeable task to test whether this variant integrated with other risk determinants will enhance cancer risk prediction. The jury therefore must refrain from drawing a firm conclusion until large, well-designed studies to confirm our findings.
To sum up, there is possible evidence for causal association between long-term genetically-reduced circulating IL-6 and reduced risk of liver cancer. For practical reasons, we hope that this study will advance our understanding of the role of circulating IL-6 leading to the progression and severity of liver cancer. Further studies to elucidate the specific role of IL-6 in cancer pathogenesis are required.
Methods
The implementation of this meta-analysis complied with the guidelines outlined in the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement (Please see the supplementary PRISMA Checklist)93.
Search strategy
PubMed and Embase (excerpta medica database) were searched for potentially relevant articles from the earliest possible year to August 4, 2014. The key terms included ‘interleukin-6’, ‘interleukin 6’, ‘IL6’, ‘IL-6’, ‘IL 6’, ‘cancer’, ‘carcinoma’, ‘neoplasia’, ‘tumor’, ‘adenoma’, ‘neoplasm’, ‘myeloma’, ‘melanoma’, ‘lymphoma’, ‘leukaemia’, ‘leiomyoma’, in combination with ‘level’, ‘concentration’, ‘polymorphism’, ‘variant’, ‘variation’, ‘mutation’, ‘SNP’. Citations from retrieved potential articles and reviews were also checked for eligibility.
The titles and abstracts of all retrieved relevant articles were independently reviewed by two investigators (Chunhua Yang and Xuri Li). In case of uncertainty for rejection, the full text and supplementary materials if available were downloaded to check whether information on the topic of interest was provided. If more than one article from a study group was published, data from the most recent or complete article were abstracted. The eligibility of each retrieved article was assessed in duplicate and independently by two authors (Chunhua Yang and Xuri Li). Any uncertainty over the eligibility was adjudicated by further joint inspection of the articles.
Inclusion/exclusion criteria
Inclusion criteria were to test the hypothesis that IL-6 gene -174G/C variant was associated with cancer or circulating IL-6 and to provide detailed genotype or allele counts of this variant between cancer patients and healthy controls or the mean levels of circulating IL-6 across -174G/C genotypes. Articles were excluded if they assessed the progression, severity, phenotypic modification and response to treatment or survival of cancer, or if they lacked patients or controls, or if they were conference abstracts or proceedings, case reports or series, editorials, narrative reviews, and non-English articles.
Data extraction
Two authors (Chunhua Yang and Xuri Li) independently abstracted the following data from each qualified article according to a fixed protocol, including the first author’s last name, publication year, ethnicity, cancer type, matching condition, study design, sample size, the genotype or allele counts of the -174G/C variant between patients and controls, mean level of circulating IL-6 for each genotype carriers expressed as mean ± standard deviation, as well as some baseline characteristics of study populations where available, including age, gender, body mass index, the percentages of smokers and drinkers. The unit of circulating IL-6 was uniformly transferred into pg/mL in this meta-analysis.
Quality assessment
Criteria for quality assessment of the association between IL-6 gene -174G/C variant and cancer risk were in agreement with the standards formulated by Thakkinstian et al.94 There were 7 criteria in total and summarized as a quality score, ranging from 0 (the worst) to 12 (the best). Quality assessment was independently conducted by two authors (Chunhua Yang and Xuri Li), and any disagreement was resolved by consensus.
Statistics
Risk estimates for the association of IL-6 gene -174G/C variant with cancer were expressed as odds ratio (OR) and its corresponding 95% confidence interval (95% CI), and for the changes of circulating IL-6 between genotypes of this variant as weighted mean difference (WMD) and its 95% CI. Hardy–Weinberg equilibrium was tested by Chi-squared test. Differences at P < 0.05 were accepted as statistically significant. A random-effects model was employed to bring individual effect-size estimates together by using the DerSimonian and Laird method.
Heterogeneity between studies was quantified by the inconsistency index (I2) statistic, which ranges from 0% to 100%. This statistic is defined as the percentage of the observed between-study variability that is due to heterogeneity rather than chance. A threshold of over 50% for I2 statistic was treated as statistically significant heterogeneity.
Predefined subgroup analyses were explored to identify the potential sources of between-study heterogeneity according to ethnicity, cancer type, matching condition, source of controls (population-based controls and hospital-based controls), study design (prospective study and retrospective study) and sample size (<500 subjects and ≥500 subjects). To assess the contribution of each individual studies to pooled effect estimates, sensitivity analyses were undertaken by sequentially omitting each study one at a time and computing differential estimates for remaining studies.
Publication bias was assessed by the Begg’s funnel plot and the Egger regression asymmetry test. The Egger test can identify the asymmetry of funnel plots by determining whether the intercept deviates significantly from zero in regressing the standardized effect estimates against their precision. A value of P < 0.10 was used to indicate statistical significance for Egger’s test.
Risk estimates in Mendelian randomization analysis were calculated as the ratio of the coefficient of the association between IL-6 gene -174G/C variant and cancer to that of the association between -174G/C variant genotypes and circulating IL-6 as a reflection of the potential causal impact of circulating IL-6 on cancer.
All statistical analyses described above were completed with the StataCorp STATA version 12.0.
Additional Information
How to cite this article: Tian, G. et al. Circulating interleukin-6 and cancer: A meta-analysis using Mendelian randomization. Sci. Rep. 5, 11394; doi: 10.1038/srep11394 (2015).
Supplementary Material
Acknowledgments
Grant support: Taishan Scholars Construction Engineering; National Natural Science Foundation of China (81400771 and 81171303), Shandong provincial natural science foundation (ZR2014HL028 and ZR2010HM091) and Binzhou Medical University Scientific Research Funds (BY2013KYQD17, BY2013KYQD18).
Footnotes
Author Contributions G.T. and B.W. conceived and designed the experiments; G.T., J.M. and X.D.W. performed the experiments; X.D.W. and C.H.Y. analyzed the data; X.R.L., X.L.L., D.M.Z., L.Y.Q. and S.P.Z. contributed materials/analysis tools; G.T., C.H.Y. and B.W. wrote and revised the manuscript. All authors reviewed and approved the manuscript prior to submission.
References
- Heikkila K., Ebrahim S. & Lawlor D. A. Systematic review of the association between circulating interleukin-6 (IL-6) and cancer. Eur J Cancer 44, 937–945 (2008). [DOI] [PubMed] [Google Scholar]
- Bochud M. & Rousson V. Usefulness of Mendelian randomization in observational epidemiology. Int J Environ Res Public Health 7, 711–728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson N. J. et al. Does greater adiposity increase blood pressure and hypertension risk?: Mendelian randomization using the FTO/MC4R genotype. Hypertension 54, 84–90 (2009). [DOI] [PubMed] [Google Scholar]
- Brennan P. et al. Obesity and cancer: Mendelian randomization approach utilizing the FTO genotype. Int J Epidemiol 38, 971–975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluco C. et al. -174 G>C polymorphism of interleukin 6 gene promoter affects interleukin 6 serum level in patients with colorectal cancer. Clin Cancer Res 9, 2173–2176 (2003). [PubMed] [Google Scholar]
- Mandal S., Abebe F. & Chaudhary J. -174G/C polymorphism in the interleukin-6 promoter is differently associated with prostate cancer incidence depending on race. Genet Mol Res 13, 139–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira P. R. et al. Interleukin-6 expression and gene polymorphism are associated with severity of periodontal disease in a sample of Brazilian individuals. Clin Exp Immunol 148, 119–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaponte G. et al. IL-6-174 G>C and MMP-9-1562 C > T polymorphisms are associated with increased risk of deep vein thrombosis in cancer patients. Cytokine 62, 64–69 (2013). [DOI] [PubMed] [Google Scholar]
- Talar-Wojnarowska R. et al. Clinical significance of interleukin-6 (IL-6) gene polymorphism and IL-6 serum level in pancreatic adenocarcinoma and chronic pancreatitis. Dig Dis Sci 54, 683–689 (2009). [DOI] [PubMed] [Google Scholar]
- Slattery M. L. et al. Genetic variants in interleukin genes are associated with breast cancer risk and survival in a genetically admixed population: the Breast Cancer Health Disparities Study. Carcinogenesis 35, 1750–1759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cil E. et al. Interleukin-10-1082 gene polymorphism is associated with papillary thyroid cancer. Mol Biol Rep 41, 3091–3097 (2014). [DOI] [PubMed] [Google Scholar]
- Tindall E. A. et al. Interleukin-6 promoter variants, prostate cancer risk, and survival. Prostate 72, 1701–1707 (2012). [DOI] [PubMed] [Google Scholar]
- Giannitrapani L. et al. IL-6 -174G/C polymorphism and IL-6 serum levels in patients with liver cirrhosis and hepatocellular carcinoma. OMICS 15, 183–186 (2011). [DOI] [PubMed] [Google Scholar]
- Gaur P., Mittal M., Mohanti B. & Das S. Functional variants of IL4 and IL6 genes and risk of tobacco-related oral carcinoma in high-risk Asian Indians. Oral Dis 17, 720–726 (2011). [DOI] [PubMed] [Google Scholar]
- Abuli A. et al. Case-control study for colorectal cancer genetic susceptibility in EPICOLON: previously identified variants and mucins. BMC Cancer 11, 339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ognjanovic S. et al. Serum CRP and IL-6, genetic variants and risk of colorectal adenoma in a multiethnic population. Cancer Causes Control 21, 1131–1138 (2010). [DOI] [PubMed] [Google Scholar]
- Hawken S. J. et al. The utility and predictive value of combinations of low penetrance genes for screening and risk prediction of colorectal cancer. Hum Genet 128, 89–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossus L. et al. PTGS2 and IL6 genetic variation and risk of breast and prostate cancer: results from the Breast and Prostate Cancer Cohort Consortium (BPC3). Carcinogenesis 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacev T., Jokic M., Loncar B., Krizanac S. & Kapitanovic S. Interleukin-6-174 G/C polymorphism is not associated with IL-6 expression and susceptibility to sporadic colon cancer. DNA Cell Biol 29, 177–182 (2010). [DOI] [PubMed] [Google Scholar]
- Vasku A., Vokurka J. & Bienertova-Vasku J. Obesity-related genes variability in Czech patients with sporadic colorectal cancer: preliminary results. Int J Colorectal Dis 24, 289–294 (2009). [DOI] [PubMed] [Google Scholar]
- Tsilidis K. K. et al. Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control 20, 1739–1751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M. L. et al. Colon tumor mutations and epigenetic changes associated with genetic polymorphism: insight into disease pathways. Mutat Res 660, 12–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgen A. G. et al. The (-174) G/C polymorphism in the interleukin-6 gene is associated with risk of papillary thyroid carcinoma in Turkish patients. J Endocrinol Invest 32, 491–494 (2009). [DOI] [PubMed] [Google Scholar]
- Ognjanovic S., Yuan J. M., Chaptman A. K., Fan Y. & Yu M. C. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis 30, 758–762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar R., Mittal B. & Mittal R. D. Association of interleukin-6 -174G>C promoter polymorphism with risk of cervical cancer. Int J Biol Markers 24, 11–16 (2009). [DOI] [PubMed] [Google Scholar]
- Falleti E. et al. Interleukin-6 polymorphisms and gender: relationship with the occurrence of hepatocellular carcinoma in patients with end-stage liver disease. Oncology 77, 304–313 (2009). [DOI] [PubMed] [Google Scholar]
- Cherel M. et al. Molecular screening of interleukin-6 gene promoter and influence of -174G/C polymorphism on breast cancer. Cytokine 47, 214–223 (2009). [DOI] [PubMed] [Google Scholar]
- Birmann B. M. et al. Insulin-like growth factor-1- and interleukin-6-related gene variation and risk of multiple myeloma. Cancer Epidemiol Biomarkers Prev 18, 282–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrie E. et al. Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol 83, 334–342 (2009). [DOI] [PubMed] [Google Scholar]
- Aladzsity I. et al. Comparative analysis of IL6 promoter and receptor polymorphisms in myelodysplasia and multiple myeloma. Leuk Res 33, 1570–1573 (2009). [DOI] [PubMed] [Google Scholar]
- Wilkening S. et al. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis 29, 1202–1206 (2008). [DOI] [PubMed] [Google Scholar]
- Vogel U. et al. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res 639, 89–100 (2008). [DOI] [PubMed] [Google Scholar]
- Vairaktaris E. et al. Gene expression polymorphisms of interleukins-1 beta, –4, –6, –8, –10, and tumor necrosis factors-alpha, -beta: regression analysis of their effect upon oral squamous cell carcinoma. J Cancer Res Clin Oncol 134, 821–832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R., Jain M., Kumar S., Ghoshal U. C. & Mittal B. Association of interleukin-6 (-174G>C) promoter polymorphism with risk of squamous cell esophageal cancer and tumor location: an exploratory study. Clin Immunol 128, 199–204 (2008). [DOI] [PubMed] [Google Scholar]
- Slattery M. L. et al. Modifying effects of IL-6 polymorphisms on body size-associated breast cancer risk. Obesity (Silver Spring) 16, 339–347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani P., Ahirwar D. K., Mandhani A. & Mittal R. D. Association between -174 G/C promoter polymorphism of the interleukin-6 gene and progression of prostate cancer in North Indian population. DNA Cell Biol 27, 505–510 (2008). [DOI] [PubMed] [Google Scholar]
- Ennas M. G. et al. Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol 26, 98–103 (2008). [DOI] [PubMed] [Google Scholar]
- Crusius J. B. et al. Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST). Ann Oncol 19, 1894–1902 (2008). [DOI] [PubMed] [Google Scholar]
- Colakogullari M. et al. The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct 26, 283–290 (2008). [DOI] [PubMed] [Google Scholar]
- Bao S., Yang W., Zhou S. & Ye Z. Relationship between single nucleotide polymorphisms in -174G/C and -634C/G promoter region of interleukin-6 and prostate cancer. J Huazhong Univ Sci Technolog Med Sci 28, 693–696 (2008). [DOI] [PubMed] [Google Scholar]
- Ahirwar D., Kesarwani P., Manchanda P. K., Mandhani A. & Mittal R. D. Anti- and proinflammatory cytokine gene polymorphism and genetic predisposition: association with smoking, tumor stage and grade, and bacillus Calmette-Guerin immunotherapy in bladder cancer. Cancer Genet Cytogenet 184, 1–8 (2008). [DOI] [PubMed] [Google Scholar]
- Vogel U. et al. Peroxisome proliferator-activated [corrected] receptor-gamma2 [corrected] Pro12Ala, interaction with alcohol intake and NSAID use, in relation to risk of breast cancer in a prospective study of Danes. Carcinogenesis 28, 427–434 (2007). [DOI] [PubMed] [Google Scholar]
- Vogel U. et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat Res 624, 88–100 (2007). [DOI] [PubMed] [Google Scholar]
- Vishnoi M. et al. Do TNFA -308 G/A and IL6 -174 G/C gene polymorphisms modulate risk of gallbladder cancer in the north Indian population? Asian Pac J Cancer Prev 8, 567–572 (2007). [PubMed] [Google Scholar]
- Litovkin K. V., Domenyuk V. P., Bubnov V. V. & Zaporozhan V. N. Interleukin-6 -174G/C polymorphism in breast cancer and uterine leiomyoma patients: a population-based case control study. Exp Oncol 29, 295–298 (2007). [PubMed] [Google Scholar]
- Gonullu G. et al. Association of breast cancer and cytokine gene polymorphism in Turkish women. Saudi Med J 28, 1728–1733 (2007). [PubMed] [Google Scholar]
- Slattery M. L., Wolff R. K., Herrick J. S., Caan B. J. & Potter J. D. IL6 genotypes and colon and rectal cancer. Cancer Causes Control 18, 1095–1105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nearman Z. P. et al. Immunogenetic factors determining the evolution of T-cell large granular lymphocyte leukaemia and associated cytopenias. Br J Haematol 136, 237–248 (2007). [DOI] [PubMed] [Google Scholar]
- Gatti L. L. et al. Interleukin-6 polymorphisms, Helicobacter pylori infection in adult Brazilian patients with chronic gastritis and gastric adenocarcinoma. Arch Med Res 38, 551–555 (2007). [DOI] [PubMed] [Google Scholar]
- Duch C. R. et al. Analysis of polymorphism at site -174 G/C of interleukin-6 promoter region in multiple myeloma. Braz J Med Biol Res 40, 265–267 (2007). [DOI] [PubMed] [Google Scholar]
- Deans C. et al. Host cytokine genotype is related to adverse prognosis and systemic inflammation in gastro-oesophageal cancer. Ann Surg Oncol 14, 329–339 (2007). [DOI] [PubMed] [Google Scholar]
- Berkovic M. C. et al. IL-6-174 C/G polymorphism in the gastroenteropancreatic neuroendocrine tumors (GEP-NETs). Exp Mol Pathol 83, 474–479 (2007). [DOI] [PubMed] [Google Scholar]
- Vairaktaris E. et al. Strong association of interleukin-6 -174 G>C promoter polymorphism with increased risk of oral cancer. Int J Biol Markers 21, 246–250 (2006). [DOI] [PubMed] [Google Scholar]
- Theodoropoulos G. et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol 12, 5037–5043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira de Souza N. C. et al. Interleukin-6 polymorphisms and the risk of cervical cancer. Int J Gynecol Cancer 16, 1278–1282 (2006). [DOI] [PubMed] [Google Scholar]
- Michaud D. S. et al. Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL-8, and IL-10 and risk of prostate cancer. Cancer Res 66, 4525–4530 (2006). [DOI] [PubMed] [Google Scholar]
- Kamangar F. et al. Polymorphisms in inflammation-related genes and risk of gastric cancer (Finland). Cancer Causes Control 17, 117–125 (2006). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Zuloeta Ladd A. M. et al. Interleukin 6 G-174 C polymorphism and breast cancer risk. Eur J Epidemiol 21, 373–376 (2006). [DOI] [PubMed] [Google Scholar]
- Balasubramanian S. P. et al. Interleukin gene polymorphisms and breast cancer: a case control study and systematic literature review. BMC Cancer 6, 188 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman N. et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol 7, 27–38 (2006). [DOI] [PubMed] [Google Scholar]
- Lan Q. et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood 107, 4101–4108 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter M. J. et al. Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol Biomarkers Prev 15, 1126–1131 (2006). [DOI] [PubMed] [Google Scholar]
- Gaustadnes M., Orntoft T. F., Jensen J. L. & Torring N. Validation of the use of DNA pools and primer extension in association studies of sporadic colorectal cancer for selection of candidate SNPs. Hum Mutat 27, 187–194 (2006). [DOI] [PubMed] [Google Scholar]
- Cozen W. et al. Interleukin-6-related genotypes, body mass index, and risk of multiple myeloma and plasmacytoma. Cancer Epidemiol Biomarkers Prev 15, 2285–2291 (2006). [DOI] [PubMed] [Google Scholar]
- Snoussi K., Strosberg A. D., Bouaouina N., Ben Ahmed S. & Chouchane L. Genetic variation in pro-inflammatory cytokines (interleukin-1beta, interleukin-1alpha and interleukin-6) associated with the aggressive forms, survival, and relapse prediction of breast carcinoma. Eur Cytokine Netw 16, 253–260 (2005). [PubMed] [Google Scholar]
- Seifart C. et al. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers 21, 157–165 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita K. et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection–association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol 42, 505–510 (2005). [DOI] [PubMed] [Google Scholar]
- Leibovici D. et al. Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. J Clin Oncol 23, 5746–5756 (2005). [DOI] [PubMed] [Google Scholar]
- Hefler L. A. et al. Interleukin-1 and interleukin-6 gene polymorphisms and the risk of breast cancer in caucasian women. Clin Cancer Res 11, 5718–5721 (2005). [DOI] [PubMed] [Google Scholar]
- Basturk B. et al. Cytokine gene polymorphisms as potential risk and protective factors in renal cell carcinoma. Cytokine 30, 41–45 (2005). [DOI] [PubMed] [Google Scholar]
- Skerrett D. L., Moore E. M., Bernstein D. S. & Vahdat L. Cytokine genotype polymorphisms in breast carcinoma: associations of TGF-beta1 with relapse. Cancer Invest 23, 208–214 (2005). [DOI] [PubMed] [Google Scholar]
- Mazur G. et al. IL-6 and IL-10 promoter gene polymorphisms do not associate with the susceptibility for multiple myeloma. Immunol Lett 96, 241–246 (2005). [DOI] [PubMed] [Google Scholar]
- Festa F. et al. Basal cell carcinoma and variants in genes coding for immune response, DNA repair, folate and iron metabolism. Mutat Res 574, 105–111 (2005). [DOI] [PubMed] [Google Scholar]
- Cordano P. et al. Effect of IL-6 promoter polymorphism on incidence and outcome in Hodgkin's lymphoma. Br J Haematol 128, 493–495 (2005). [DOI] [PubMed] [Google Scholar]
- Campa D. et al. Lack of association between polymorphisms in inflammatory genes and lung cancer risk. Cancer Epidemiol Biomarkers Prev 14, 538–539 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. Use of pyrosequencing to detect clinically relevant polymorphisms of genes in basal cell carcinoma. Clin Chim Acta 342, 137–143 (2004). [DOI] [PubMed] [Google Scholar]
- Smith K. C., Bateman A. C., Fussell H. M. & Howell W. M. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur J Immunogenet 31, 167–173 (2004). [DOI] [PubMed] [Google Scholar]
- Campa D. et al. Association of a common polymorphism in the cyclooxygenase 2 gene with risk of non-small cell lung cancer. Carcinogenesis 25, 229–235 (2004). [DOI] [PubMed] [Google Scholar]
- Bushley A. W. et al. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol 95, 672–679 (2004). [DOI] [PubMed] [Google Scholar]
- Landi S. et al. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res 63, 3560–3566 (2003). [PubMed] [Google Scholar]
- Hwang I. R. et al. Interleukin-6 genetic polymorphisms are not related to Helicobacter pylori-associated gastroduodenal diseases. Helicobacter 8, 142–148 (2003). [DOI] [PubMed] [Google Scholar]
- Howell W. M., Turner S. J., Theaker J. M. & Bateman A. C. Cytokine gene single nucleotide polymorphisms and susceptibility to and prognosis in cutaneous malignant melanoma. Eur J Immunogenet 30, 409–414 (2003). [DOI] [PubMed] [Google Scholar]
- El-Omar E. M. et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124, 1193–1201 (2003). [DOI] [PubMed] [Google Scholar]
- Zheng C. et al. Interleukin 6, tumour necrosis factor alpha, interleukin 1beta and interleukin 1 receptor antagonist promoter or coding gene polymorphisms in multiple myeloma. Br J Haematol 109, 39–45 (2000). [DOI] [PubMed] [Google Scholar]
- Puchalski T., Prabhakar U., Jiao Q., Berns B. & Davis H. M. Pharmacokinetic and pharmacodynamic modeling of an anti-interleukin-6 chimeric monoclonal antibody (siltuximab) in patients with metastatic renal cell carcinoma. Clin Cancer Res 16, 1652–1661 (2010). [DOI] [PubMed] [Google Scholar]
- Guo Y., Xu F., Lu T., Duan Z. & Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev 38, 904–910 (2012). [DOI] [PubMed] [Google Scholar]
- Jacobson C. M., Rosenfeld B., Pessin H. & Breitbart W. Depression and IL-6 blood plasma concentrations in advanced cancer patients. Psychosomatics 49, 64–66 (2008). [DOI] [PubMed] [Google Scholar]
- Mitsunaga S. et al. Serum levels of IL-6 and IL-1beta can predict the efficacy of gemcitabine in patients with advanced pancreatic cancer. Br J Cancer 108, 2063–2069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti G., Sanderson S. & Higgins J. P. Obstacles and opportunities in meta-analysis of genetic association studies. Genet Med 7, 13–20 (2005). [DOI] [PubMed] [Google Scholar]
- Zhou X. C., Dowdy S. C., Podratz K. C. & Jiang S. W. Epigenetic considerations for endometrial cancer prevention, diagnosis and treatment. Gynecol Oncol 107, 143–153 (2007). [DOI] [PubMed] [Google Scholar]
- Yu K. et al. Methionine synthase A2756G polymorphism and cancer risk: a meta-analysis. Eur J Hum Genet 18, 370–378 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H. et al. Contributory role of five common polymorphisms of RAGE and APE1 genes in lung cancer among Han Chinese. PLoS One 8, e69018, 10.1371/journal.pone.0069018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J. & Altman D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339, b2535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkinstian A. et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol 162, 201–211 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.