Abstract
Autosomal recessive polycystic kidney disease (ARPKD) is a severe form of polycystic kidney disease that presents primarily in infancy and childhood and that is characterized by enlarged kidneys and congenital hepatic fibrosis. We have identified PKHD1, the gene mutated in ARPKD. PKHD1 extends over ⩾469 kb, is primarily expressed in human fetal and adult kidney, and includes a minimum of 86 exons that are variably assembled into a number of alternatively spliced transcripts. The longest continuous open reading frame encodes a 4,074-amino-acid protein, polyductin, that is predicted to have a single transmembrane (TM)-spanning domain near its carboxyl terminus, immunoglobulin-like plexin-transcription–factor domains, and parallel beta-helix 1 repeats in its amino terminus. Several transcripts encode truncated products that lack the TM and that may be secreted if translated. The PKHD1-gene products are members of a novel class of proteins that share structural features with hepatocyte growth-factor receptor and plexins and that belong to a superfamily of proteins involved in regulation of cell proliferation and of cellular adhesion and repulsion.
Introduction
Autosomal recessive polycystic kidney disease (ARPKD [MIM 263200]) is a hereditary and severe form of polycystic kidney disease affecting the kidneys and biliary tract, with an estimated incidence of 1/20,000 live births (Zerres et al. 1998). The clinical spectrum is widely variable, with most cases presenting during infancy (Guay-Woodford 1996). The human fetal phenotypic features classically include enlarged and echogenic kidneys, as well as oligohydramnios secondary to a poor urine output (Reuss et al. 1990). Up to 50% of the affected neonates die shortly after birth, as a result of severe pulmonary hypoplasia and secondary respiratory insufficiency. Those who survive the perinatal period express widely variable disease phenotypes. In the subset that survived the perinatal period, morbidity and mortality are mainly related to severe systemic hypertension, renal insufficiency, and portal hypertension due to portal-tract fibrosis (Zerres et al. 1996).
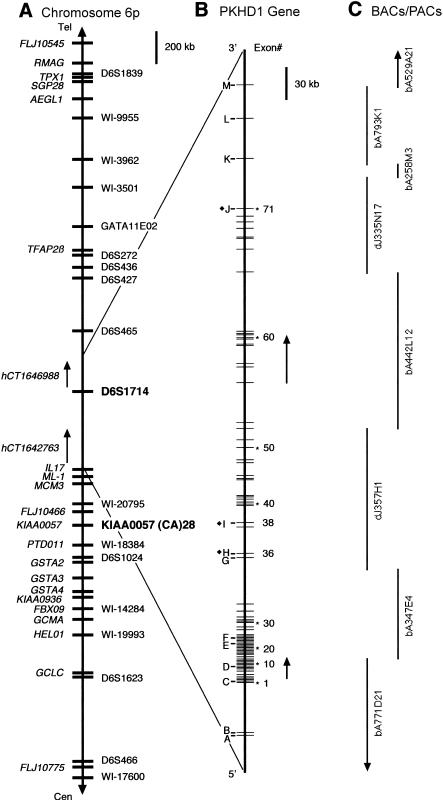
Mutations at a single locus, PKHD1 (polycystic kidney and hepatic disease 1), are responsible for all typical forms of ARPKD. In previous studies, we have mapped PKHD1 to 6p21.1-p12 (Zerres et al. 1994; Guay-Woodford et al. 1995). We subsequently constructed a series of physical and genetic maps that refine the localization of PKHD1 to a candidate region of ∼1 cM, delimited by D6S1714 and D6S1024 as the telomeric and centromeric flanking markers, respectively (Lens et al. 1997; Mucher et al. 1998; Park et al. 1999). More-recent recombination-mapping studies have further reduced the size of the interval, to 834 kb, with KIAA0057 (CA)28 as the new centromeric boundary (Onuchic et al., in press).
In the present study, we describe the identification of PKHD1, a novel gene encoded in a minimum of 86 exons that are assembled in a complex pattern of alternative splice variants. The predicted translation products are novel proteins that share homology to a superfamily of proteins involved in the regulation of cell proliferation and of cellular adhesion and repulsion.
Patients, Material, and Methods
Patients and Samples
The databases of patients used in this study are from University of Alabama at Birmingham and Rheinisch-Westfälische Technische Hochschule (Aachen, Germany). The diagnostic criteria were the same as those reported elsewhere (Zerres et al. 1998). The group of patients studied had clinical features representative of the entire ARPKD clinical spectrum. Pedigrees were recruited, and blood samples were obtained, with informed consent by the patients with ARPKD and by members of their families. Control DNA from 40 individuals also was obtained after informed consent had been given, and an additional 20 control DNA samples were purchased from the Coriell Cell Repository. DNA was extracted as described elsewhere (Eggermann et al. 1993).
Transcription Map
Database searches included a systematic surveillance of the UniGene, Sanger (see the Human Sequence Data Web site), TIGR (see the Tigr Databases Web site), Celera (public domain), and GenBank Overview Web sites. The gene-prediction algorithms FGenesh (see the Nucleotide Sequence Analysis Web site) (Salamov and Solovyev 2000) and GENSCAN (Burset and Guigo 1996; Burge and Karlin 1997) were used to annotate genomic sequence as it became available. Expressed sequences were confirmed by RT-PCR across putative splice junctions, with human adult kidney mRNA as template, by PCR-amplification using a panel of multiple-tissue cDNA samples as template (Origene) and by northern blot analysis using human multiple-tissue blots (Clontech).
PKHD1 cDNA Isolation
Most of the PKHD1 cDNA products were amplified with human adult kidney double-stranded cDNA (Marathon Ready cDNA, Clontech) used as template. A second set of products were generated by RT-PCR using either 20 ng of human adult kidney mRNA (Clontech) or 1.5–4.0 μg of human adult kidney total RNA as template. The total RNA was extracted by Trizol reagent (Invitrogen) and was reverse transcribed by random hexamer primers and Superscript reverse transcriptase (Gibco BRL). A third set of cDNA products was amplified by a 1:20 dilution of an oligo-dT–primed human adult kidney cDNA library (Gibco/BRL). The 5′ RACE and 3′ RACE experiments were performed according to the manufacturer’s instructions (Clontech). Primer sequences used to amplify the set of cDNA products are shown in table A1, in the Appendix.
Mutation Detection
PCR primers flanking individual exons and offset, by ∼20 bp, from intron-exon junctions were designed by the program Primer3 and were used to amplify 20 ng of genomic DNA from patients and controls. In cases of exons >400 bp, several overlapping primers were designed to ensure that the size of the amplicons remained <500 bp. All of the primer sequences used are provided in table A2, in the Appendix. Mutation detection was performed by the Transgenomic Wave denaturing high-performance liquid-chromatography system (DHPLC) (Transgenomic). PCR products were denatured at 98°C for 4 min and were allowed to reanneal; 8–12 ml of each amplicon were injected into the column and were eluted with a linear acetonitrile gradient, at a flow rate of 0.9 ml/min. The mobile phase consisted of a mixture of buffers A (0.1 M triethylammonium acetate and 1 mM EDTA) and B (25% acetonitrile in 0.1 M triethylammonium acetate). The buffer gradient for each amplicon was determined according to Wavemaker version 3.3 (and, subsequently, version 4.1) (Transgenomic) system-control software. The optimum denaturing temperature required for successful resolution of heteroduplexes was also determined by this software. If the resolution of the DHPLC profile was not adequate, a second temperature, typically either 2°C above or below the first temperature, was used to improve resolution. Samples showing altered elusion properties not present in controls were sequenced in both directions, and sequence variations were identified by visual inspection and comparison of the resulting electropherograms. When amplicons had altered elution properties in both control and patient DNA, they were sequenced in both samples, to confirm their identity at the sequence level.
Sequence Analysis and Protein Modeling
The genomic structure and the gene orientation were established by alignment of the confirmed expressed sequences and the interval genomic sequence, by BLAST2 (see the BLAST Web site). Sequence homologies were identified by the BLASTP/N/X programs (see the BLAST Web site). SMART (simple modular architecture research tool (Schultz et al. 1998; Letunic et al. 2002) and PROSITE (see the ExPASy Molecular Biology Server Web site) were used to identify domain architecture and protein motifs. All analyses were performed with the default parameters.
Northern Blot Analysis
Probes were amplified by use of cloned gene fragments as template. PCR products were gel purified, [32P]-labeled by the multiprime method, and hybridized to human adult and human fetal MTN blots (Clontech). Hybridizations were performed at either 68°C, with ExpressHyb (Clontech), or 42°C, with a formamide-based buffer, and were washed under stringent conditions (68°C for 1 h, in 0.1 SSC, 0.1% SDS). Images were obtained by a PhosphorImager (Molecular Dynamics).
Results
Transcription Map of the Minimal Interval
We assembled a transcription map of the minimal interval, using database searches, cDNA-library screening, genomic-sequence annotation with gene structure–prediction programs, RT-PCR, and northern blot analyses. A total of 35 nonoverlapping sets of genes, cDNA clones, ESTs, and RT-PCR products were mapped to the critical region (fig. 1A). The genes KIAA0057 (Onuchic et al. 1999), FLJ10466 (Onuchic et al., in press), MCM3 (Hofmann et al. 2000), Interleukin-17 (Rouvier et al. 1993), and ML-1 (Kawaguchi et al. 2001) have been described elsewhere. Each of these genes either has been excluded as a candidate for PKHD1 or, on the basis of its known function, has been deemed to be an unlikely candidate.
Figure 1.
Chromosomal localization and genomic organization of PKHD1. A, Schematic representation of chromosome 6p12. Currently known genes are identified on the far left (italics), and STSs/polymorphic markers are on the right. The closest flanking genetic markers that define the minimal PKHD1 interval are indicated (boldface). Not all of the 35 overlapping sets of expressed sequences described in the text are shown. B, Genomic organization of PKHD1. Exons identified by numbers have been shown to be part of PKHD1 transcripts. Letters indicate exons that belong to either hCT1642763 or hCT1646988 and that have not been confirmed by our analyses. The black diamond (♦) identifies, in hCT1642763 and hCT1646988, overlapping exons whose boundaries differ from those used in the present study. Arrows indicate the positions of the Gene Unit 6 and Gene Unit 442L12 transcripts described in the text. C, BACs and PACs sequenced by the Sanger Centre that cover the interval.
Transcripts with kidney expression were preferentially targeted for mutation analysis. One such transcript, a novel 1.82-kb expressed sequence, Gene Unit 442L12, identified by gene structure–prediction programs and confirmed by RT-PCR from kidney mRNA, mapped to BAC 442L12, within the distal portion of the critical interval (fig. 1B and C). A second novel transcript of 1.03 kb (Gene Unit 6), initially identified by similar methods and mapped to BACs 771D21 and 374E4 (fig. 1B and C), was subsequently found to share some, although not all, of its exons with both transcript hCT1642763 (Venter et al. 2001) and a single human EST (BF822430) derived from a kidney tumor library; Gene Unit 6 appeared to have a mouse ortholog in UniGene cluster Mm.25855, comprised of ESTs obtained from kidney and liver libraries.
The PKHD1 Transcript and Genomic Organization
As putative expressed sequences were identified in the PKHD1 region, they were systematically analyzed for mutations, by screening, by DHPLC, of 20 unrelated affected patients and 20 normal controls (see below). Virtually simultaneously, we discovered, in Gene Unit 6 and Gene Unit 442L12, two apparently unrelated expressed sequences, variants that appeared only in the samples from our patients and not in samples from the controls. Among hundreds of amplicons analyzed in the region up to that point, none had given a pattern of variation exclusive to affected individuals. We analyzed an additional 40 control individuals, to confirm that neither of these variants was present in 120 control chromosomes. Subsequent RT-PCR studies using kidney mRNA as template established that Gene Unit 6 and Gene Unit 442L12 are part of the same gene. We went on to determine both the structure of the complete transcript and its genomic organization.
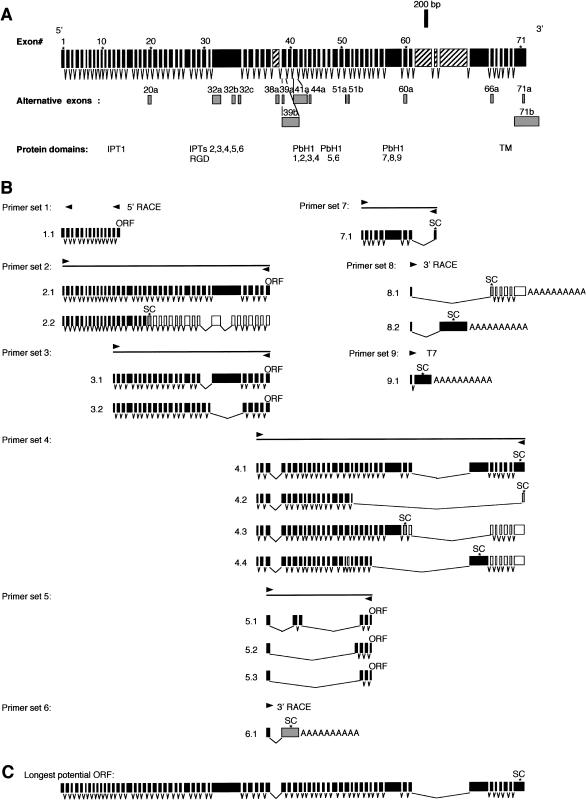
Northern blot analyses (fig. 2) suggested that the PKHD1 transcript was considerably longer and involved more complex splicing variations than was initially suggested by the Gene Unit 6 and Gene Unit 442L12 transcripts. We used a PCR-based approach with primers strategically positioned within Gene Unit 6 and Gene Unit 442L12 to determine the sequence of the longest open reading frame (ORF) of PKHD1, to elucidate its genomic structure and to define the complex pattern of exon assembly. Human kidney RNA, an human adult kidney cDNA library and human adult kidney double-stranded cDNA were used as templates (fig. 3). A number of primer combinations and end-clone amplifications, as well as 5′ RACE and 3′ RACE reactions, were required to establish the composite sequence of the full length gene (fig. A1 in the Appendix). A human adult kidney cDNA library, human kidney mRNA and total RNA and double-stranded cDNA served as templates for these studies. The sequences of all exons and splicing junctions were determined both by double strand sequencing of PCR products and by comparison with the publicly available genomic sequence of the interval (fig. 3).
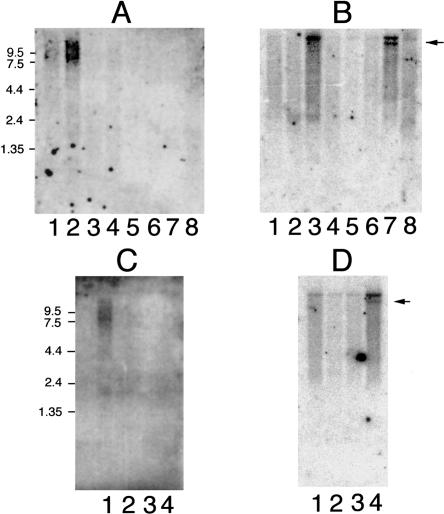
Figure 2.
PKHD1 expression profile. A, Human adult multiple-tissue northern blot probed with PKHD1 exon 59: lane 1, pancreas; lane 2, kidney; lane 3, skeletal muscle; lane 4, liver; lane 5, lung; lane 6, placenta; lane 7, brain; lane 8, heart. B, Same blot as in panel A, probed with PKD1. The arrow indicates the position of a known splicing variant of PKD1. C, Human fetal multiple-tissue northern blot probed with PKHD1: lane, 1, kidney; lane 2, liver; lane 3, lung; lane 4, brain. D, Same blot as in panel C, probed with PKD1.
Figure 3.
Structure of full-length PKHD1 and its splicing variants. A, Set of 71 nonoverlapping exons that spans the entire length of PKHD1 (upper row) and 15 additional overlapping exons that use different splice sites (gray boxes, lower row). Exons, which are not present in the cDNA that encodes the longest ORF, are indicated by hatched boxes. The position of important protein domains is indicated. B, Approximate location of each primer set used to amplify various cDNAs, with representative set of amplified products (below each schema). White boxes indicate noncoding exons in the corresponding transcripts while gray boxes identify exons with alternative boundaries (A). The templates used for each amplification are as follows: human adult kidney double-stranded cDNA for primer sets 1–4, 6, and 8; human kidney mRNA and total RNA for primer sets 5 and 7; human adult kidney cDNA library for primer set 9. “SC” indicates approximate location of stop codons, and “ORF” indicates that an open reading frame extends throughout the length of the fragment. C, Longest ORF identified by RT-PCR/cDNA amplification. This ORF is the composite sequence of products 2.1 and 4.1 of panel B and includes a total of 67 exons.
These studies provided rigorous confirmation that the Gene Unit 6 and Gene Unit 442L12 were part of the same gene, but they also yielded a number of unanticipated results. We discovered that hCT1646988 (Venter et al. 2001), previously reported as an independent gene, was actually part of PKHD1. However, regardless of method used, we were not successful in linking the last three exons of hCT1646988 to the remainder of the PKHD1 transcript. We encountered similar problems with hCT1642763, as RT-PCR, cDNA amplification, and 5′ RACE could not confirm the existence of exons 1–3 within the PKHD1 transcript. These same methods did, however, identify two previously unknown exons at the putative 5′ end of PKHD1, one of which contained the predicted translation start site. We also identified a small number of errors in the sequence available in public databases. Although most were relatively minor, one would be predicted to alter the reading frame of the protein. We have verified the accuracy of our sequence for the regions in question by determining the sequence of both DNA strands in several templates.
These data indicate that PKHD1 spans ⩾469 kb of genomic sequence and, possibly—if the unverified 5′ and 3′ exons of hCT1642763 and hCT1646988, respectively, are included—as much as 643 kb. These results also show that the 3′ end of PKHD1 is positioned ⩾74-kb distal to the flanking genetic marker, D6S1714 (fig. 1), thus explaining why we found so few meiotic recombinations between this marker and the disease phenotype. The total number of exons that we identified in PKHD1 transcripts is 86 (fig. 3B). This may be a conservative estimate, since it is likely that at least some of the unverified exons in either hCT1642763 (exons 1–3, 9, 19, 23, 32, and 33), hCT1646988 (exons 9–11) or the EST database or predicted by computer algorithms will ultimately be experimentally confirmed if mRNA or cDNA from a suitable tissue is used as template.
In our attempt to assemble a complete cDNA, we identified a large number of distinct transcripts that had unique combinations of PKHD1 exons. Representative examples are presented in figure 3B. The absolute number of differentially spliced products is almost certainly higher, since we did not perform an exhaustive analysis of every primer combination. In numerous cases, a PCR reaction that appeared to yield a single amplification product was discovered, after cloning and sequencing, to include multiple, differentially spliced products of nearly identical size. We believe that these data provide a likely molecular explanation for the lack of a discrete message seen, in northern blot analysis, with labeled PKHD1-gene segments (fig. 2).
Mutation Analyses
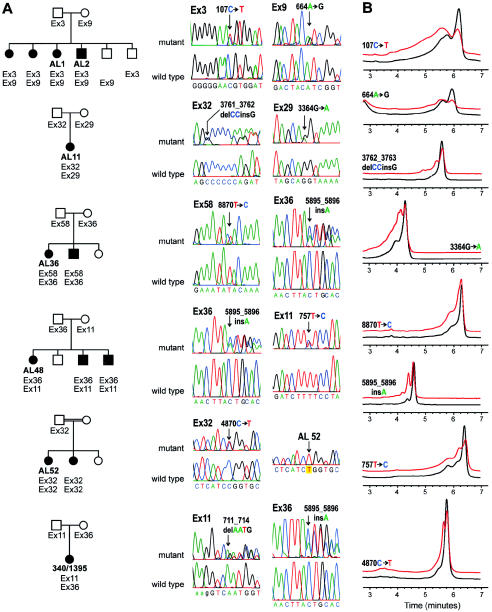
We performed mutation detection by using DHPLC across the 67 exons comprising the longest potential ORF (figs. 3 and 4 and table A2 in the Appendix ). We expanded our patient group to 25 individuals (50 disease chromosomes) and our control group to 60 individuals (120 chromosomes). We focused most of our mutation-detection efforts on individuals for whom we had family material enabling us to confirm segregation of alleles. The patients represented diverse nationalities and the complete spectrum of clinical disease (table 1). In all cases in which segregation could be established, the mutant alleles resided on separate chromosomes (fig. 4A). A minimum of 67 exons (longest ORF) was screened in each individual, and, in all cases, either 0, 1, or 2 putative pathogenic variants were found. We identified potentially pathogenic variants in 21 (42%) of 50 disease chromosomes; these were defined as variants detected in patients by DHPLC and confirmed by sequencing and not observed by DHPLC in any of the 120 control chromosomes. The finding of these mutations establishes this gene as PKHD1 (table 1).
Figure 4.
PKHD1 mutations in families studied. A, Representative family-segregation analyses of PKHD1 mutations for patients AL 1, AL 11, AL 36, AL 48, AL 52, and 340/1395 (see table 1). Sequence electropherograms showing wild-type and mutant sequences for amplicons containing the respective variants in each family are indicated on the right of each pedigree figure. Traces labeled “mutant” show heterozygous alterations in genomic PCR products. Segregation of the mutant allele (denoted by “Ex” followed by the exon number) is shown for each kindred studied. AL 1 has two missense changes, whereas AL 11, AL 36, and AL 48 each have a missense and a frameshifting mutation. Patient 340/1395 has two frameshifting mutations. AL 52 and her sibling, products of a consanguineous union, are homozygous for the Ex32 mutation (trace labeled AL 52). The Ex32 trace labeled “mutant” is from the heterozygous father. Black symbols denote affected individuals; white symbols denote unaffected individuals. B, Representative DHPLC profiles for several sequence variants in families with PKHD1 (see panel A and table 1). Black traces indicate control profiles; red traces indicate patient variant profiles (the red trace is displaced upward to facilitate comparison).
Table 1.
Mutations in PKHD1
| Patient (ARPKD Phenotype) and Nucleotide (ORF) Changea | Exonb | Parent in WhomVariant Is Identified | Country |
| AL 1 (later onset): | |||
| 107C→T (T36M) | 3 | Father | United States |
| 664A→G (I222V) | 9 | Mother | |
| AL 11 (perinatal onset): | |||
| 3761_3762delCCinsG (A1254Xfs1302) | 32 | Father | United States |
| 3364G→A (G1122S) | 29 | Mother | |
| AL 18 (later onset): | |||
| 8829_8830insC (I2944Xfs2949) | 58 | Father | South Africa (Afrikaner) |
| 664A→G (I222V) | 9 | Mother | |
| AL 36 (perinatal onset): | |||
| 8870T→C (I2957T) | 58 | Father | United States |
| 5895_5896insA (L1966Xfs1969) | 36 | Mother | |
| AL 45 (later onset): | |||
| 4870C→T (R1624W) | 32 | Father | Saudi Arabia |
| AL 47 (later onset): | |||
| 2279G→A (R760H) | 22 | Father | Saudi Arabia |
| 4870C→T (R1624W) | 32 | Mother | |
| AL 48 (later onset): | |||
| 5895_5896insA (L1966Xfs1969) | 36 | Father | United States |
| 757T→C (F253L) | 11 | Mother | |
| AL 52 (later onset):c | |||
| 4870C→T (R1624W) | 32 | Father | Saudi Arabia |
| 4870C→T (R1624W) | 32 | Mother | |
| 376/1559 (perinatal onset): | |||
| 1620_1621insAGTT (E541Xfs556) | 18 | Father | Germany |
| 9415G→T (D3139Y) | 59 | Mother | |
| 340/1395 (perinatal onset): | |||
| 711_714delAATG (S237Xfs244) | 11 | Father | United Kingdom |
| 5895_5896insA (L1966Xfs1969) | 36 | Mother | |
| 306/1272 (unknown): | |||
| 10075delG (G3359Xfs3399) | 61 | Mother | Turkey |
| 291/1207 (later onset): | |||
| 3306delT (Y1102X) | 29 | Mother | Turkey |
Nucleotide and codon numbers are based on the predicted 67-exon transcript of the longest ORF (fig. 3C; also see fig. A1 and fig. A2 in the Appendix). Nomenclature for the description of sequence variations is from the Nomenclature for the Description of Sequence Variations Web site. None of these variants were identified in 120 control chromosomes.
Based on 71 exons shown in figure 3A.
Consanguineous union.
Eight different nonconservative missense changes accounted for mutations in 12 of 21 disease chromosomes for which we found mutations. Among these, 9415G→T (D3139Y) corresponded to the initial variant discovered in Gene Unit 442L12, and 107C→T (T36M) corresponded to the initial variant found in Gene Unit 6; six different frameshifting mutations accounted for the remaining 9 disease chromosomes. One individual, 340/1395 (table 1 and fig. 4), had frameshifting mutations in both alleles. This individual provides perhaps the clearest proof of that we have identified PKHD1. This finding is also the most consistent with the notion that PKHD1 causes disease by a loss-of-function mechanism. One frameshifting variant, 5895_5896insA, occurred in three unrelated individuals (AL36, AL 48, and 340/1395). Two missense variants also recurred: variant 664A→G (I222V) was seen in two individuals who were not known to be related and who are of distinct national origins; variant 4870C→T (R1624W), on the other hand, was identified in a family with known consanguinity and in two other individuals from the same geographical region. This variant may represent a founder allele in this population. With the exception of the consanguineous family, all other individuals are compound heterozygous for mutations in which both variants were identified. In individuals in whom only one or no mutations were found, it is likely that the DHPLC mutation screen as applied has failed to detect the second (or either) variant.
Expression Features of PKHD1
Commercially acquired human adult and human fetal multiple-tissue northern blots were hybridized with two different probes (exon 59 and exons 66–70; fig. 3A), to determine the expression pattern of PKHD1. Rather than a single, discrete message, both probes detected a smear that ranged from ∼8.5 kb to ∼13 kb (fig. 2). The highest level of expression was observed in the human fetal and human adult kidney samples, consistent with a role of this gene in kidney development and function. In the human adult specimen, the peak signal was observed as two diffuse bands, of ∼9 kb and ∼12 kb. In human fetal kidney, the size distribution of the transcripts appeared to be somewhat lower and more uniform. PKHD1 is also present in the pancreas, but at much lower levels. PKHD1 is barely detectable in human fetal and human adult liver. The remaining tissue samples had no visible signal. Given the diffuse signal from the transcripts, however, we cannot exclude a low level of expression in other organs. When the identical blots were hybridized with a probe recognizing exons 43–46 of human PKD1, discrete, high-molecular-weight bands of the correct size were produced, excluding nonspecific degradation of high-molecular-weight mRNA in the samples as being an explanation for the PKHD1 results in northern blot analysis.
The PKHD1 Product: a Membrane-Anchored Protein with Multiple Immunoglobulin-Like Plexin-Transcription Factor (IPT) Domains (SMART Accession Number SM0429) and Parallel Beta-Helix 1 (PbH1) Repeats (SMART Accession Number SM0710)
The composite cDNA that yielded the longest continuous ORF, experimentally amplified, from kidney cDNA, as two overlapping fragments (fig. 3B), is 12.6 kb in length, includes 67 exons, and is predicted to encode a protein of 4,074 amino acids (fig. A2 in the Appendix). This novel protein, which we have named “polyductin,” is predicted, by SMART, to be an integral membrane protein with a 3,858-amino-acid extracellular amino terminus, a single transmembrane (TM)-spanning domain , and a short carboxyl terminus (fig. 5).
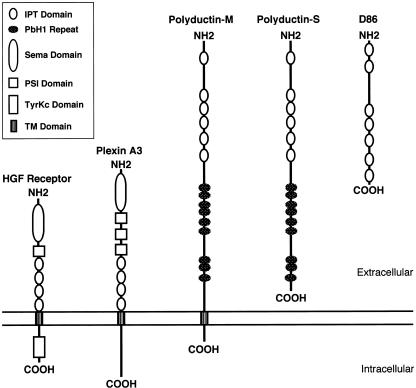
Figure 5.
Structure of polyductin and related proteins. Multiple tandemly repeated IPT domains are common features of the group. Polyductin-M shares the general structure of the HGFR and plexin A3, in having a long extracellular domain, a single TM domain, and a short cytoplasmic carboxyl terminus, whereas polyductin-S is more like D86.
BLASTP (see the BLAST Web site) analysis revealed that polyductin has highest homology to murine protein D86. The region of homology begins near the amino terminus of both molecules and stretches over most of the entire length (1,944 amino acids) of D86. The function of D86 is not known, but it is described, in GenBank, as a novel protein secreted from lymphocytes (see the GenBank Overview Web site). Significant homologies were also observed for two expressed sequences, KIAA1412 and TM protein 2 (accession number AAF21348 [see the Nomenclature for the Description of Sequence Variations Web site]), that encode identical novel proteins of unknown function (Nagase et al. 2000; Scott et al. 2000).
Several short segments of polyductin have weak homology to other proteins whose functions are known, including the hepatocyte growth-factor receptor (HGFR [accession number P08581]) and several plexins. Using SMART, we determined that these sequences encode IPT domains. The structure of several IPT-containing proteins has been determined (Cramer et al. 1997), but their function remains unknown. IPT domains consist of an immunoglobulin-like fold, and proteins that contain IPT domains generally belong to one of two classes: intracellular DNA transcription factors (the Rel family) or single-pass cell-surface receptors that are members of the Sema superfamily of proteins (i.e., HGFR, Ron, and the large family of plexins). Although all members of the Rel family have a single IPT unit that is involved in DNA binding, virtually all of the receptor proteins contain multiple IPT domains, often tandemly arranged. Topology predictions indicate that polyductin contains six IPT domains within its extracellular segment (fig. 5). The overall similarity between polyductin and receptor molecules such as HGFR and plexin A3 (accession number P51805 [see the Nomenclature for the Description of Sequence Variations Web site]) suggests a similar function for polyductin. However, there are differences in structure that suggest that polyductin is unique. Polyductin lacks the Sema domain (SMART accession number SM0630) and the plexin/semaphorin/integrin (PSI) domain (SMART accession number SM0423), which are common to all other members of the Sema superfamily. It also lacks (a) an intracellular kinase domain present in HGFR and (b) other conserved cytoplasmic sequences present in plexin subclasses.
SMART analysis identified a second motif within polyductin, a motif that might provide additional functional clues. The program revealed a minimum of 9 (and, possibly, 10) PbH1 repeats clustered within three groups between the last IPT domain and the TM domain (fig. 5). PbH1 repeats are most commonly associated with polysaccharidases, and, within this enzyme class, bacterial polysaccharidases are the most extensively studied. These bacterial enzymes serve as important virulence factors for plant pathogens, since they allow bacteria to degrade plant cell-wall polysaccharides. The PbH1 repeats are essential for enzyme function, forming both the ligand-binding and catalytic sites. The presence of multiple PbH1 domains within polyductin suggests that polyductin may have similar catalytic properties.
Motif analysis by the PROSITE program identified multiple potential N-glycosylation sites and a single arginine-glycine-aspartate (RGD) domain. This motif is found in fibronectin and numerous other proteins, where it has been shown to play a role in cell adhesion. In addition, three putative cAMP/cGMP phosphorylation sites were identified within the cytoplasmic carboxyl terminus. No tyrosine phosphorylation consensus sites were recognized within this cytoplasmic tail, further distinguishing between polyductin and members of the plexin family.
Finally, we examined how the various splicing arrangements might affect the protein(s) structure. If, in fact, some of the alternatively spliced products are also translated, then the gene products are predicted to fall into two broad groups: one group, which includes the longest continuous ORF but which may also include molecules lacking some middle domains, has a single TM element and is likely to be associated with the plasma membrane (polyductin-M); the other group lacks a TM domain, and thus its members may be secreted (polyductin-S) (fig. 5).
Discussion
We have reported here the initial description and characterization of a novel gene, PKHD1, implicated in all typical forms of ARPKD. Multiple lines of evidence strongly support the pathogenic role that mutations in this gene play in ARPKD. First, the genomic structure of this candidate extends over nearly 50% of the critical PKHD1 interval defined by recombination mapping. This observation alone provides a high prior probability that this gene is the disease-susceptibility locus. Second, this gene is expressed predominantly in the kidney, an organ invariably involved in this disorder. Third, we have identified a large number of protein-truncating mutations and missense variants that are found only in affected individuals and not in a large number of controls. For individuals in whom we have identified two mutations, we have shown that the mutations occur on separate haplotypes and that they segregate with the disease chromosomes. In no case did we identify an individual who had more than two putative pathogenic variants.
The panel of patient material used included both the severe perinatal and milder, later-onset disease phenotypes (table 1). The limited sample size leaves open any conclusions regarding genotype-phenotype correlations, but it does permit a few preliminary hypotheses. Among individuals in whom both mutations were identified, the only individual with two chain-terminating mutations (i.e., individual 340/1395) had the severe phenotype. The three individuals with missense mutations on both alleles (i.e., individuals AL 1, AL 47, and AL 52) had the later-onset phenotype. Of the five individuals with both a chain-terminating frameshifting mutation and a missense mutation, two had the later-onset phenotype, and three had the severe phenotype; none shared the same missense allele. It is possible that not all missense variants are functionally equivalent—some may result in hypomorphic alleles that allow for a clinically milder course. An expanded study of genotype-phenotype correlations should clarify this point. In light of the complex splicing pattern and multiple transcripts, identification of pathogenic mutations is one means of identifying those exons whose presence in a transcript is essential for the function of PKHD1 in the kidney and liver. The current analysis suggests that exons 3, 9, 11,18, 22, 29, 32, 36, 58, 59, and 61 (of the 67 exons in the longest ORF) are essential for normal polyductin function. These data also highlight the lack of evidence for clustering of mutations either in any one region of the gene or in any functional domain of the putative protein.
The PKHD1 gene and its translation products have several distinctive features that warrant special note. First, with a genomic size of ⩾469 kb, PKHD1 is among the largest human genes characterized to date. Second, the gene encodes a complex and extensive array of splice variants discovered by RT-PCR and cDNA cloning and confirmed by northern blot analysis. We excluded the possibility that the diffuse signal observed on northern blots results from degradation of RNA, by the presence of an intact 14-kb PKD1 transcript on the same blots. Moreover, the multiplicity of different transcripts discovered in public databases, revealed by RT-PCR of kidney mRNA, and amplified from aliquots of double-stranded cDNA as well as provided by a cDNA library, correlates well with the results of northern blot analysis. It is important to note that almost all of the exons exhibit consensus donor and acceptor splice sites, further supporting the conclusion that these are legitimate transcripts.
The multiplicity of splicing variants observed for PKHD1 is an uncommon feature of mammalian genes. Preliminary studies of mouse tissue suggest that the complicated splicing pattern is likely conserved (Y.N., unpublished observations), indicating a functional role for this property. The abundance of these splice variants in poly-A–enriched samples indicates that many, if not all, are fully processed to include a poly-A tail. It is not presently known how many of the transcripts are actually translated into protein. In the event that most of the mRNAs are translated, it could mean that this single gene might encode numerous distinct polypeptides. Similar findings have been reported for the neurexin family of genes (Missler and Sudhof 1998). Just three genes may encode >1,000 isoforms that, through alternative splicing, differ in size and amino acid sequence. Interestingly, the general structure of the largest gene products is similar to that of polyductin-M. Likewise, a subset of the transcripts is predicted to contain stop codons and to produce secreted proteins without a TM region. The neurexin family of proteins is expressed in neurons, where they function as receptors important for neuronal-cell recognition.
Such a complicated pattern of splicing poses a significant challenge to prediction of the functional consequences of putative pathogenic mutations. For most genes, the implications of protein-truncating mutations are relatively easily defined. Loss of critical domains usually results in either constitutive activation or functional loss. In the case of PKHD1, many of the normal splicing products are predicted to yield truncated proteins that lack critical domains, including the TM region and cytoplasmic tail of polyductin. Similar outcomes are predicted for many of the mutations described herein, yet the diseases caused by these mutations are a de facto bioassay for normal polyductin function. We suggest two potential explanations to reconcile these findings. First, all of the observed mutations are predicted to alter the sequence of the largest ORF. This may suggest that a critical amount of the full-length protein is necessary for normal function. A second, alternative possibility is that mutations disrupt a critical functional stoichiometric or temporal balance between the different protein products that is normally maintained by elaborate, tightly regulated splicing patterns.
The data of northern blot analysis suggest that PKHD1 is predominantly expressed in the kidney, consistent with the observed phenotype in ARPKD. Much lower transcript expression was detected in liver—not an unexpected finding, given that biliary ductules, which are abnormal in ARPKD, constitute only a small fraction of the total tissue. The human fetal expression pattern of PKHD1 is consistent with both the observation that renal and hepatic abnormalities develop in utero and the hypothesis that disease pathogenesis involves a defect in terminal epithelial differentiation (Calvet 1993). Continued expression of PKHD1 in human tissues suggests an additional, undefined role for its gene product in mature, terminally differentiated organs. PKHD1 expression slightly greater than that observed in the liver was also observed in the pancreas. A disease-associated phenotype has not been described in this organ. This situation is not dissimilar to that found in dominant polycystic kidney disease, in which pancreatic cysts were an underappreciated manifestation of the disease until the role of the polycystin genes in pancreatic development became apparent on the basis of mouse studies (Lu et al. 1997; Wu et al. 1998). Pancreatic cysts do not result in clinical symptoms in dominant polycystic disease (Nicolau al. 2000).
We propose that the transcript with the longest ORF is the likeliest gene product of PKHD1, since it is the only transcript that would be altered by all of the mutations that we have described. The product that it is predicted to encode, polyductin, shares some structural features with both the Ron class of tyrosine kinase receptors and the plexin superfamily and thus may also function to regulate either cell-cell recognition or cell motility. However, the PKHD1-gene product(s) lacks key structural elements of these protein classes, suggesting that its mechanism of action will differ from that seen in the other classes. The presence of multiple PbH1 repeats in polyductin suggests a possible role for this molecule in carbohydrate recognition and modification. Targets for binding could include carbohydrate moieties present either in glycoproteins on the cell surface or in the matrix of the basement membrane; interactions with polyductin may modulate cell-cell or cell-matrix attachments. One intriguing possibility is that the variable number of IPT and PbH1 domains encoded by some of the shorter transcripts could potentially result in products with different specificities or binding affinities for target factors, as has been postulated for the neurexins (Missler and Sudhof 1998). We presently are unable to determine whether polyductin serves primarily as a receptor, ligand, or membrane-associated enzyme.
Polyductin has a unique combination of structural features not previously observed in a single molecule. The discovery of a second protein, D86, with a very similar pattern suggests that the gene products of the D86 and PKHD1 loci may be prototypes of a novel class of proteins. From a structural perspective, D86 is most similar to the polyductin-S family of polypeptides. By analogy with polyductin, we propose the possible existence of a larger, membrane-associated form of D86. The fact that D86 is described as a secreted protein further supports our hypothesis that polyductin-S may have the same properties.
We have identified the gene responsible for ARPKD and have determined that it is a novel, large, and complex gene. Although this complexity may pose some challenges with respect to the implementation of DNA-based diagnostic testing, the discovery of PKHD1 should provide important biologic insights into epithelial differentiation and organogenesis. In addition, these new insights should help to establish a platform for development of targeted therapeutic interventions for patients with this often devastating disease.
Acknowledgments
The authors would like to thank the many families—and their health-care providers—who have cooperated with these studies. This work was supported by National Institutes of Health grant R01 DK51259 and Fundacao de Amparo a Pesquisa do Estado de Sao Paulo grant 2000/00280-3 and by the Deutsche Forschungsgemeinschaft. We thank Gabi Muecher, Jutta Becker, and Kirstin Mangasser-Stephan for their work
Appendix
Table A1.
Primer Pairs Used to Amplify PKHD1 cDNA Fragments from Human Kidney mRNA and Total RNA, Human Kidney Double-Stranded cDNA, and a Library of Human Adult Kidney cDNA (Fig. 3)
|
Primer(5′→3′) |
||
| Primer Set | Forward | Reverse |
| 1: | ||
| First primer | 5′-CCTGTGACCAAAACCCAAATG-3′ | AP1 (Clontech) |
| Second primer | 5′-ATCCACGTTCCCCCTGCAAG-3′ | AP2 (Clontech) |
| 2 | 5′-GAAAGGATCATTTCTCCCTTGAGTC-3′ | 5′-CAGGAACTTGACTCCATAGGGAAAG-3′ |
| 3 | 5′-TGTCTCCCAGGAAGATTGAGTGC-3′ | 5′-CAGGAACTTGACTCCATAGGGAAAG-3′ |
| 4 | 5′-GCCCAATCAGCCAATTACCG-3′ | 5′-TCTTCTTAGTTGTCCCAGCAGGAC-3′ |
| 5 | 5′-GCACACGCCATCCTTGTTTC-3′ | 5′-CAATATCAGCTCCAAGATGTGTCC-3′ |
| 6: | ||
| First primer | 5′- TCCTTGTTTCTGATGGTGG -3′ | AP1 |
| Second primer | 5′- CAACTCCCTTCTTTCCCTATG -3′ | AP2 |
| 7 | 5′-GATCTTCCATTCTTCAAAGGGC-3′ | 5′-GGATATAAGGGTCTGTTAAACTC-3′ |
| 8: | ||
| First primer | 5′- ACCAAATGCAGAGAACAGTGG -3′ | AP1 |
| Second primer | 5′-CTCTTCAAGGATCTGGATGGG-3′ | AP2 |
| 9 | 5′-CTCTTCAAGGATCTGGATGGG-3′ | T7 |
Table A2.
Primer Pairs Used to Amplify PKHD1 Exons for Mutation Analysis[Note]
|
Primer(5′→3′) |
||
| Exon | Forward | Reverse |
| 1 | 5′-TACACATTTGGCTGGGACAC-3′ | 5′-GGACAAAGTTGCAAGGCACT-3′ |
| 2 | 5′-CAGAACAGCAAAATAATCGTATGT-3′ | 5′-TCAAGGTAACCTATTGTGTTCTTACC-3′ |
| 3 | 5′-CTGGTGATTCTGAGGCAGGT-3′ | 5′-GCACTTGGTAAAACCCCAAC-3′ |
| 4 | 5′-TGACAATTCTATGCAGCCTGA-3′ | 5′-GGCAATTGAATCACAGCAAA-3′ |
| 5 | 5′-GCTAGCTTTGGGAATTCATG-3′ | 5′-GACACGCTGGCTCATTTACA-3′ |
| 6 | 5′-GTCTGAGGAAAGGCTTGTGC-3′ | 5′-TTTCAAAACAGCATCAAAGAAGA-3′ |
| 7 | 5′-CATTGAGTTTGAGCTAAGTCCATTA-3′ | 5′-AGCAATTCTGTGCCAACTGC-3′ |
| 8 | 5′-TTCATAGCCATGTTTCCTCTGA-3′ | 5′-TGGTGAGTGGGGAGAGAAAG-3′ |
| 9 | 5′-TTTTGCTTTCTACTTTCCTGGTT-3′ | 5′-ACAGAGAAAGAAATGGATAAGACTTT-3′ |
| 10 | 5′-CCCAGAAGACTCGTGCAGAT-3′ | 5′-TCAAGGAGTTATATGTGGTCTCA-3′ |
| 11 | 5′-TTCAGGCAGCCGACTAAAAA-3′ | 5′-TCATCAAGAAATGGCCAAAA-3′ |
| 12 | 5′-TCATATTCTGGTCTATATTTGGAAGC-3′ | 5′-TGGTCTCAACCAAACACATGA-3′ |
| 13 | 5′-TGTATACCTACACACACACACATACA-3′ | 5′-ACCCAGGGCTCACTGAGTA-3′ |
| 14 | 5′-TCTGGGTCATCTTTCACCTTG-3′ | 5′-CCTGGAAATCTTGATACCTTCC-3′ |
| 15 | 5′-CAGTGATTCAGGCCTTGGTT-3′ | 5′-CTTCATGGGTATGGGACTGG-3′ |
| 16 | 5′-ATGCCTGGAAGCTGCATAGT-3′ | 5′-TCAGTGCTCCTGCTACATGG-3′ |
| 17 | 5′-TTGAGGAGGAATGTCCTGTG-3′ | 5′-CACTCCCCTCCCTCATTTTT-3′ |
| 18 | 5′-GAGAACTGGGGGTGGTCTTT-3′ | 5′-GGAAATGGGGATTGTTCTCA-3′ |
| 19 | 5′-CGGGGGTTTTCTTAAGTGAG-3′ | 5′-CTACCCACCTGACCCAGAAG-3′ |
| 20 | 5′-GGATGTGGACTCCTCACTACG-3′ | 5′-AATCCTCCCAGCTGACTGAA-3′ |
| 21 | 5′-TAACCGGAGAGGACTGCAAG-3′ | 5′-CCCCTAGCAGGAAAGTTTGA-3′ |
| 22 | 5′-ACTGGGATTTTCCACACAGC-3′ | 5′-CTCAAGGCCAACAAGCATTC-3′ |
| 23 | 5′-CCCCAACCCAGACGTTAATA-3′ | 5′-GAATTGTTGCAACCCCAATC-3′ |
| 24 | 5′-GGATGAAACTCTGTAAGGTGGA-3′ | 5′-TCCATGCCACTAGAAGGGATA-3′ |
| 25 | 5′-TTCGGTTCCATGACAGAATTT-3′ | 5′-ACTGGAGCTTGCACTTAGGG-3′ |
| 26 | 5′-TGGAAAAGAAATTTTGCCTCA-3′ | 5′-GGCCTCTAACAAAATCACTGC-3′ |
| 27 | 5′-TCTCCTTTAAGATGGTGACATCAA-3′ | 5′-TATGGAATTCATTACACAGAAGGA-3′ |
| 28 | 5′-TTGTCTGCCTGTATGGTTGG-3′ | 5′-CAGTGGTCACTCACCCAGAG-3′ |
| 29 | 5′-TGGAGTTCTCTTCCCTTAAGTCAG-3′ | 5′-TGCCCTTTTTATAGGACCAATG-3′ |
| 30 | 5′-CCCCACATGTCAGAGGCTAT-3′ | 5′-AGCTGAATGCTAGACCATCAAA-3′ |
| 31 | 5′-TGGCTCTTCTGGCTTACTCTTT-3′ | 5′-AACAAAATCTGAATATCCAGCAAA-3′ |
| 32a | 5′-GGACTTCCACAGGTGCTATGA-3′ | 5′-TCCTTGCATGGCAGTGACTA-3′ |
| 32b | 5′-GGCAGCAACACCAGTAGTCA-3′ | 5′-ACCCGAACTGACCTCCTTCT-3′ |
| 32c | 5′-GCAGTGATGCCTCGGATAAT-3′ | 5′-GTGGTGGCTAACCTCTGACC-3′ |
| 32d | 5′-GGGGAATTGCACTCTTTTCA-3′ | 5′-CGTGGTGTTCTGTCCTCTCA-3′ |
| 32e | 5′-CGTCAGTCTATATTGACCAGCAG-3′ | 5′-CAATTCATTTACATAAAGAAAGTGTGC-3′ |
| 33 | 5′-TGGGTAGTGAGGAAGCAAATG-3′ | 5′-TGTGGCATCTTTACTCACCATC-3′ |
| 34 | 5′-GGTCAAAGGAGGACACAGGA-3′ | 5′-CCCGAGAATGATGGAAACAT-3′ |
| 35 | 5′-AGCTAATGGCTTGCAATGAT-3′ | 5′-TCGCTGCCATTTGGACTAA-3′ |
| 36 | 5′-CCAACCCAGCTCTCTGTTTT-3′ | 5′-CAGAAAGTTTCCCTCCTCCA-3′ |
| 37 | 5′-CAAAACGGTAAGCCTTATCC-3′ | 5′-TTTCCACTGCTAGACACAGCTC-3′ |
| 39 | 5′-AAATTATCTGGACAACTTTTCCTC-3′ | 5′-AGACCCCAATACAAATGTCCA-3′ |
| 40 | 5′-TTTGGAGTGATGTCCTCAGTTCT-3′ | 5′-GCAATGCCATCTATCATCAGAC-3′ |
| 41 | 5′-CATGCTTTAGGTTCTCTGGACTT-3′ | 5′-TGCCTTAAACATGGGAGAAA-3′ |
| 42 | 5′-CAACAGAATCTCAGGAGCCATA-3′ | 5′-TTGGGGAGAATTCATTGTGA-3′ |
| 43 | 5′-TGCAGCATCTCTTGTTTTCC-3′ | 5′-ATTTTGCCATCAGGCTTGTC-3′ |
| 44 | 5′-TGAAATGCATTATGTGAACTCTGA-3′ | 5′-TGCATTTTATGTGTGACCCAAT-3′ |
| 45 | 5′-CCAGCAACCTTATCATACATGG-3′ | 5′-AATGCCCAAAGTGCTCTCAT-3′ |
| 46 | 5′-TTGGCTTAATTTCATTCTGTTTCA-3′ | 5′-GGGCAAGTCAATCCCATTTA-3′ |
| 47 | 5′-AACCAAGCCTCAGACCTTTG-3′ | 5′-GGCCCAGCACATGTAATTTT-3′ |
| 48 | 5′-TCCAGTTTTCTTATTTTGCTTTCA-3′ | 5′-TGGGCCTTATTTATCATCTGTTC-3′ |
| 49 | 5′-TTGCTATTTGTGCCATTGTG-3′ | 5′-CCATCGGCAAGCTAAAAAGT-3′ |
| 50 | 5′-TGCAATTGATGAAAGAGGAATG-3′ | 5′-TCAACCCATTATCTCAAACAAAGA-3′ |
| 51 | 5′-AGGGTGGTGTGGTGGAGTAG-3′ | 5′-TGGAATTGAAGGGTGATTGG-3′ |
| 52 | 5′-CACATGCAAGTTGTGGCTTT-3′ | 5′-CTGGAGGACATTCTGCCATA-3′ |
| 53 | 5′-GAATTTGGAAGTTATCACAATGGA-3′ | 5′-GGGTTCAGCCTGTCTGTGAT-3′ |
| 54 | 5′-TGGGAGAACCCCAGAATAAA-3′ | 5′-GCAACCTGCTTGATGATACC-3′ |
| 55 | 5′-TGCAATTTCTCCCTCTCTTTC-3′ | 5′-TTGCCTAAAAGGGTGTTTGG-3′ |
| 56 | 5′-CACAACAGCCTCTGGAATGA-3′ | 5′-TCAAGCAGAAGCAACCTAAACA-3′ |
| 57 | 5′-TTCTCTGGTGGGATATGTTGG-3′ | 5′-TCCCCAGCTAGGTTACCAAA-3′ |
| 58 | 5′-GGGTCCCCAGCTAGTGATTT-3′ | 5′-CAGTTGGGATTTCTGGGTGT-3′ |
| 59a | 5′-GCCATTCTTCAGCCTTTTGT-3′ | 5′-CCATGGAGGTTGATGTCCTT-3′ |
| 59b | 5′-TTGTGGTTCTGATGACACAGC-3′ | 5′-GACGGAATTTTGTGGAGCAG-3′ |
| 59c | 5′-GCTCCACAAAATTCCGTCAA-3′ | 5′-TGGACAGCAAGCACTAGACC-3′ |
| 60 | 5′-TTCGATATTTGTGGCTGGTG-3′ | 5′-TGTTTAGGGTTTGAAGAATTGC-3′ |
| 61 | 5′-CAATGTATATTTTCTTCTTGCTGCTT-3′ | 5′-TCAGTGAGCACAGCAAAACC-3′ |
| 65a | 5′-GATGATGGCAAGAGTGATGG-3′ | 5′-GGCTCTGGAGCTCATGGTAG-3′ |
| 65b | 5′-AGAGCCCCCACGTCTTCTTA-3′ | 5′-TCCATCATGAGAGGCCTACG-3′ |
| 65c | 5′-CCTGGCCATGAAGAGACCT-3′ | 5′-AATCAGCCCTCATTTGGATG-3′ |
| 66 | 5′-GAAACACAGGCAAATGCAAA-3′ | 5′-GGCTGAATGCTACATGCTACTT-3′ |
| 67 | 5′-TTTTCTTCCTCCCTCCTCCT-3′ | 5′-GGAGTTTCTTCCCAAATTACTGA-3′ |
| 68 | 5′-TTGTCCAAATGTCTGCCTTC-3′ | 5′-CATGTGCTCTTTCTTCTCAAGC-3′ |
| 69 | 5′-GCATCTTATTAACATATCATTGAAACA-3′ | 5′-GTCTTTGGGGAAAGAAACAGAA-3′ |
| 70 | 5′-TGCTGATGGTCCCACTTACA-3′ | 5′-GGGGCTATAAACCAAATTGC-3′ |
| 71 | 5′-AAATCCATTTCTTCCCCTATTTC-3′ | 5′-CCCTGATCACAGTTGCTCCT-3′ |
Note.— Primers for exons <500 bp were positioned in flanking intronic sequences, allowing analysis of splicing junctions. Larger exons were evaluated as a set of overlapping fragments that included flanking intronic sequences.
Figure A1.
PKHD1 cDNA sequence encoding the longest potential reading frame. Colored bases establish the exon-exon boundaries: blue denotes the last base of the upstream exon; red, the first base of the downstream exon; green, the translation start site; pink, the stop codon.
Figure A2.
Amino acid sequence of polyductin encoded by the longest ORF of PKHD1. Blue denotes IPT domains; green, PbH1 repeats; red, TM domain; yellow, RGD domain.
Note added in proof.—
Since submission of this article, another group has reported similar findings (Ward et al. 2002). In that publication, the PKHD1-gene product described is essentially identical, in both size and sequence, to polyductin and was named “fibrocystin.”
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/blast/
- Celera, http://public.celera.com/cds/login.cfm
- ExPASy Molecular Biology Server, http://ca.expasy.org/
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for sequences of all 86 exons and the composite cDNA with the longest ORF [accession number AF480064])
- GENSCAN, http://bioweb.pasteur.fr/seqanal/interfaces/genscan.html
- Human Sequence Data, http://www.sanger.ac.uk/HGP/sequence/
- Nomenclature for the Description of Sequence Variations, http://www.dmd.nl/mutnomen.html (for PbH1 repeat [accession number SM0710], Sema domain, PSI domain, IPT domain, KIAA1412 and TM protein 2 [accession number AAF21348], HGFR [accession number PO8581], and plexin A3 [accession number P51805])
- Nucleotide Sequence Analysis, http://genomic.sanger.ac.uk/gf/gf.shtml (for the FGenesh algorithm).
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/ (for ARPKD [MIM 263200])
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
- SMART, http://smart.embl-heidelberg.de/ (for PbH1 repeat [accession number SM0710], Sema domain [accession number SM0630], PSI domain [accession number SM0423], and IPT domain [accession number SM0429])
- Tigr Databases, http://www.tigr.org/tdb/
- Unigene, http://www.ncbi.nlm.nih.gov/UniGene/
References
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94 [DOI] [PubMed] [Google Scholar]
- Burset M, Guigo R (1996) Evaluation of gene structure prediction programs. Genomics 34:353–367 [DOI] [PubMed] [Google Scholar]
- Calvet JP (1993) Polycystic kidney disease: primary extracellular matrix abnormality or defective cellular differentiation? Kidney Int 43:101–108 [DOI] [PubMed] [Google Scholar]
- Cramer P, Larson CJ, Verdine GL, Muller CW (1997) Structure of the human NF kappaB p52 homodimer-DNA complex at 2.1 A resolution. EMBO J 16:7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Nothen MM, Propping P, Schwanitz G (1993) Molecular diagnosis of trisomy 18 using DNA recovered from paraffin embedded tissues and possible implications for genetic counselling Ann Genet 36:214–216 [PubMed] [Google Scholar]
- Guay-Woodford L M (1996) Autosomal recessive disease: clinical and genetic profiles. In: Torres V, Watson M (eds) Polycystic kidney disease. Oxford University Press, Oxford, pp 237–267 [Google Scholar]
- Guay-Woodford LM, Muecher G, Hopkins SD, Avner ED, Germino GG, Guillot AP, Herrin J, Holleman R, Irons DA, Primack W, Thomson PD, Waldo FB, Lunt PW, Zerres K (1995) The severe perinatal form of autosomal recessive polycystic kidney disease maps to chromosome 6p21.1-p12: implications for genetic counseling. Am J Hum Genet 56:1101–1107 [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y, Becker J, Wright F, Avner E, Mrug M, Guay-Woodford L, Somlo S, Zerres K, Germino GG, Onuchic LF (2000) Genomic structure of the gene for the human P1-protein (MCM3) and its exclusion as a candidate gene for autosomal recessive polycystic kidney disease. Eur J Hum Genet 8:163–166 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Onuchic LF, Li X-D, Essayan DM, Schroeder J, Xiao H-Q, Liu MC, Germino G, Huang SK (2001) Identification of a novel cytokine and its expression in subjects with asthma. J Immunol 167:4430–4435 [DOI] [PubMed] [Google Scholar]
- Lens XM, Onuchic LF, Wu G, Hayashi T, Daoust M, Mochizuki T, Santarina LB, Stockwin JM, Mucher G, Becker J, Sweeny WE Jr, Avner ED, Guay-Woodford L, Zerres K, Somlo S, Germino GG (1997) An integrated genetic and physical map of the autosomal recessive polycystic kidney disease region. Genomics 41:463–466 [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt l, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30:242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J (1997) Perinatal lethality with kidney and pancreas defects in mice with a targeted Pkd1 mutation. Nat Genet 17:179–181 [DOI] [PubMed] [Google Scholar]
- Missler M, Sudhof TC (1998) Neurexins: three genes and 1001 products. Trends Genet 14:20–26 [DOI] [PubMed] [Google Scholar]
- Mucher G, Becker J, Knapp M, Buttner R, Moser M, Rudnik-Schoneborn S, Somlo S, Germino G, Onuchic L, Avner E, Guay-Woodford L, Zerres K (1998) Fine mapping of the autosomal recessive polycystic kidney disease locus (PKHD1) and the genes MUT, RDS, CSNK2 beta, and GSTA1 at 6p21.1-p12. Genomics 48:40–45 [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Ishikawa KI, Hirosawa M, Ohara O (2000) Prediction of the coding sequences of unidentified human genes. XVI. The complete sequences of 150 new cDNA clones from brain which code for large proteins in vitro. DNA Res 7:65–73 [DOI] [PubMed] [Google Scholar]
- Nicolau C, Torra R, Bianchi L, Vilana R, Gilabert R, Darnell A, Bru C (2000) Abdominal sonographic study of autosomal dominant polycystic kidney disease. J Clin Ultrasound 28:277–282 [DOI] [PubMed] [Google Scholar]
- Onuchic LF, Mrug M, Hou X, Nagasawa Y, Furu L, Eggermann T, Bergmann C, Muecher G, Avner ED, Zerres K, Somlo S, Germino GG, Guay-Woodford LM. Refinement of the autosomal recessive polycystic kidney disease (PKHD1) interval and exclusion of an EF hand-containing gene as PKHD1 candidate gene. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Onuchic LF, Mrug M, Lakings AL, Muecher G, Becker J, Zerres K, Avner ED, Dixit M, Somlo S, Germino GG, Guay-Woodford LM (1999) Genomic organization of the KIAA0057 gene that encodes a TRAM-like protein and its exclusion as a polycystic kidney and hepatic disease 1 (PKHD1) candidate gene. Mamm Genome 10:1175–1178 [DOI] [PubMed] [Google Scholar]
- Park JH, Dixit MP, Onuchic LF, Wu G, Goncharuk AN, Kneitz S, Santarina LB, Hayashi T, Avner ED, Guay-Woodford L, Zerres K, Germino GG, Somlo S (1999) A 1-Mb BAC/PAC-based physical map of the autosomal recessive polycystic kidney disease gene (PKHD1) region on chromosome 6. Genomics 57:249–255 [DOI] [PubMed] [Google Scholar]
- Reuss A, Wladimiroff JW, Stewart PA, Niermeijer MF (1990) Prenatal diagnosis by ultrasound in pregnancies at risk for autosomal recessive polycystic kidney disease. Ultrasound Med Biol 16:355–359 [DOI] [PubMed] [Google Scholar]
- Rouvier E, Luciani MF, Mattei MG, Denizot F, Golstein P (1993) CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol 150:5445–5456 [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Drury S, Sundstrom RA, Bishop J, Swiderski RE, Carmi R, Ramesh A, Elbedour K, Srikumari Srisailapathy CR, Keats BJ, Sheffield VC, Smith RJ (2000) Refining the DFNB7-DFNB11 deafness locus using intragenic polymorphisms in a novel gene, TMEM2. Gene 246:265–274 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC (2002) The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30:259–269 [DOI] [PubMed] [Google Scholar]
- Wu G, D'Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, Maeda Y, Le TC, Hou H Jr, Kucherlapati R, Edelmann W, Somlo S (1998) Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93:177–188 [DOI] [PubMed] [Google Scholar]
- Zerres K, Mucher G, Bachner L, Deschennes G, Eggermann T, Kaariainen H, Knapp M, Lennert T, Misselwitz J, von Muhlendahl KE (1994) Mapping of the gene for autosomal recessive polycystic kidney disease (ARPKD) to chromosome 6p21-cen. Nat Genet 7:429–432 [DOI] [PubMed] [Google Scholar]
- Zerres K, Mucher G, Becker J, Steinkamm C, Rudnik-Schoneborn S, Heikkila P, Rapola J, Salonen R, Germino GG, Onuchic L, Somlo S, Avner ED, Harman LA, Stockwin JM, Guay-Woodford LM (1998) Prenatal diagnosis of autosomal recessive polycystic kidney disease (ARPKD): molecular genetics, clinical experience, and fetal morphology. Am J Med Genet 76:137–144 [PubMed] [Google Scholar]
- Zerres K, Rudnik-Schoneborn S, Deget F, Holtkamp U, Brodehl J, Geisert J, Scharer K (1996) Autosomal recessive polycystic kidney disease in 115 children: clinical presentation, course and influence of gender. Acta Paediatr 85:437–445 [DOI] [PubMed] [Google Scholar]