Abstract
DMRT transcription factors are deeply conserved regulators of metazoan sexual development. They share the DM DNA binding domain, a unique intertwined double zinc-binding module followed by a C-terminal recognition helix, which binds to a pseudopalindromic target DNA. Here we show that DMRT proteins employ a unique binding interaction, inserting two adjacent antiparallel recognition helices into a widened DNA major groove to make base-specific contacts. Versatility in how specific base contacts are made allows human DMRT1 to employ multiple DNA binding modes (tetramer, trimer, dimer). ChIP-Exo indicates that multiple DNA binding modes also are used in vivo. We show that mutations affecting residues crucial for DNA recognition are associated with an intersex phenotype in flies and in male-to-female sex reversal in humans. Our results illuminate an ancient molecular interaction that underlies much of metazoan sexual development.
Keywords: DMRT protein, DM domain, sex determination, DMRT1
The sex of animals can be determined by varied cues in different species, including chromosomes, temperature, social status, and photoperiod1. A common feature of sexual regulation across much of the animal kingdom is the involvement of DMRT proteins2,3. These are transcription factors related to Doublesex (Dsx) and Male abnormal-3 (MAB-3), key sexual regulators of insects and nematodes, respectively, and they share the highly conserved DM DNA binding domain4,5.
Genetic studies have found that DMRT genes can control the primary sex determination decision or act subsequently in sexual differentiation or, in some species, do both2. DMRT genes are required for sexual development in planaria6, insects7, nematodes8, and vertebrates9, suggesting that their involvement in this process spans hundreds of millions of years. Vertebrates have six to seven DMRT genes and at least one of these appears to regulate testis development in most or possibly all species, with DMRT1 playing a leading role. In some vertebrate groups, including birds10,11 and some fish12 and amphibians13, a DMRT1 ortholog is located on a sex chromosome and plays a sex-determining role2. In mammals DMRT1 is crucial for many aspects of testicular development2. Deletions of human chromosome 9p that cause DMRT1 hemizygosity result in 46,XY gonadal dysgenesis, which can include sex reversal14,15. In the mouse, DMRT1 has been shown to regulate gonadal differentiation, and continuous DMRT1 expression is required to maintain the male cell fate of testicular Sertoli cells, preventing their transdifferentiation to female granulosa cells16. Moreover, DMRT1 overexpression in the mouse ovary can cause male sex determination or female-to-male cell fate transdifferentiation17,18.
DMRT1 appears to be a bifunctional transcription factor, activating or repressing transcription of target genes. We previously found in mice that DMRT1 binds and regulates genes known to play key roles in mammalian sexual development, activating the central male sex-determining gene Sox9, repressing the female sex-determining genes Wnt4 and Rspo1, and regulating many other genes involved in subsequent sexual differentiation16. Here we have combined genomic, molecular, biochemical, structural, and human genetics approaches to ask how DMRT1 recognizes target site DNA. We find that DMRT1 employs a unique type of protein-DNA interaction and can use multiple distinct stoichiometries to discriminate target sites with distinct DNA sequences. We also show that disrupting conserved residues in the DM domain that make base-specific contacts with DNA can severely reduce binding affinity and cause sex reversal in flies and in humans.
Results
In vivo DMRT1 binding site determination in mouse and human
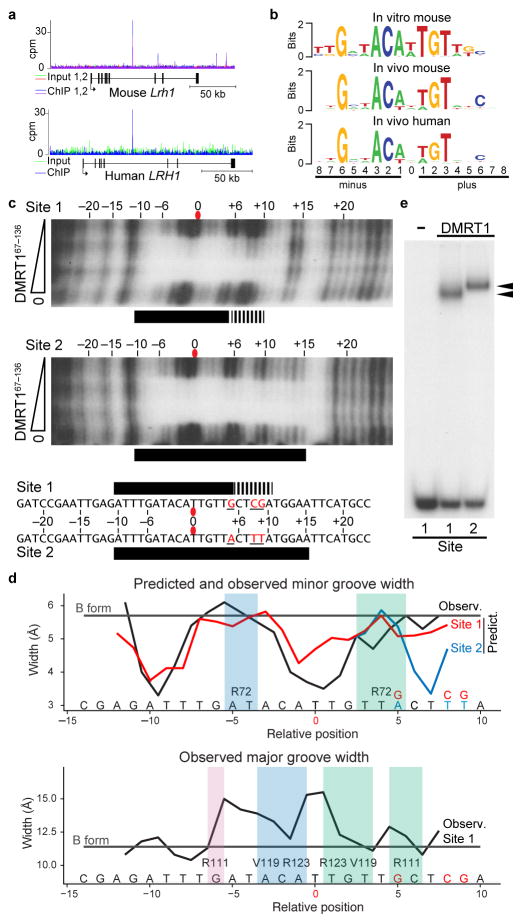
DMRT1 binds in vitro to a pseudopalindromic 13 base-pair DNA sequence19 but how the DM domain recognizes target DNAs is poorly understood as no composite protein-DNA structure has been described. For a genome-wide view of DMRT1 DNA binding sites, we first performed chromatin immunoprecipitation and sequencing (ChIP-Seq) in adult mouse and human testes. ChIP-Seq identified 8571 strongly-enriched sites in mouse and 7593 in human. Nine percent of human sites with synteny in mouse were bound in both species (an example is shown in Fig. 1a), typical for a tissue-specific transcription factor20,21. Motif searches revealed a DNA consensus element associated with in vivo binding in both species, which includes several nearly invariant nucleotides and resembles the in vitro consensus (Fig. 1b).
Figure 1. DMRT1 binds similar sites in vivo and in vitro.
(a) Examples of ChIP-Seq data showing binding of DMRT1 to the Lrh1 (Nr5a2) gene in mouse and human testes. (b) Consensus DMRT1 DNA binding motif derived in vitro, compared to motifs associated with in vivo binding in mouse and human testes. (c) DNase I footprinting showing protection by DMRT167–136 of the in vitro binding consensus top strand (Site 1, upper) and a modified DNA (Site 2, lower). Diagram at bottom summarizes protection by DMRT167–136. Solid bars indicate strong and dashed bars indicate weaker protection. (d) Top: predicted and observed minor groove width for DMRT1 binding sites. Horizontal line indicates width of canonical B form DNA minor groove, black trace is minor groove width of Site 1 observed in the structure of DMRT167–136 bound to Site 1 shown in Fig. 2a; red and blue lines are minor groove width of unbound Site 1 and Site 2 DNAs predicted using DNAshape36. Bottom: major groove width observed in structure of DMRT167–136 bound to Site 1. (e) EMSA analysis showing slower migrating complex (upper arrowhead) formed between full length DMRT1 and Site 2. Uncropped gels for all figures are shown in Supplementary Data Set 1.
To examine binding to the in vitro DNA consensus (“Site 1”) we next mapped DMRT1-DNA interactions by DNase I protection. A truncated DMRT1 protein containing the highly conserved human DM domain, DMRT167–136 (Supplementary Fig. 1), protected the top DNA strand beyond the central 13 bp (Fig. 1c). Protection was stronger on the left side, which is predicted22 to have a narrower minor groove (Fig. 1d). We made base changes to Site 1 that are expected to compress the right side minor groove (“Site 2”, Fig. 1d), and these resulted in extended protection and reduced electrophoretic mobility shift (EMSA) of full-length DMRT1 protein (Fig. 1c,e). The altered DNase I protection and electrophoretic mobility of Site 2 relative to Site 1 together suggest that DMRT1 can bind DNA with multiple stoichiometries or conformations.
DMRT1 inserts paired alpha helices into the DNA major groove
For a detailed view of the DM domain structure and insight into how DMRT1 interacts with DNA we employed X-ray crystallography, examining interaction between the DMRT1 DM domain and Site 1. Crystals of DMRT167–136 (Supplementary Fig. 1) and a 25 bp DNA corresponding to Site 1 yielded a 3.8 Å resolution structure (Table 1) containing three DM domain protomers bound to a single DNA molecule (protomers A-C; Fig. 2a,b). The overall resolution of the DMRT167–136 -DNA structure is not high; this likely reflects a combination of inherent flexibility of the complex, loose lattice contacts, and high solvent content and radiation sensitivity of the crystals. We were able to mitigate these issues by validating the registers of DNA and protein residues using crystals containing brominated DNA and selenomethionine-substituted protein (Supplementary Fig. 2, Table 1).
Table 1.
Data collection and refinement statistics
| Native (Zn-SAD) | BrDNAb | SeMetb | |
|---|---|---|---|
| Data collection | |||
| Space group | I222 | I222 | I222 |
| Cell dimensions | |||
| a, b, c (Å) | 82.56, 138.49, 138.49 | 82.99, 138.31, 138.98 | 83.09, 139.48, 142.18 |
| α=β=γ (°) | 90 | 90 | 90 |
| Wavelength (Å) | 1.28 | 0.92 | 0.9795 |
| Resolution (Å) | 3.81 (3.95-3.81)a | 3.76 (3.96-3.76) | 4.93 (5.15-4.93) |
| Rmerge | 0.070 (1.02) | 0.052 (0.67) | 0.043 (0.395) |
| I / σI | 12.9 (1.9) | 16.7 (2.4) | 10.5 (2.4) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.8) | 97.0 (98.5) |
| Redundancy | 7.1 (7.3) | 6.6 (6.3) | 3.0 (3.1) |
| Refinement | |||
| Resolution (Å) | 3.81 | ||
| No. reflections | 8170 | ||
| Rwork / Rfree | 22.9 / 26.2 | ||
| No. atoms | 2534 | ||
| Protein/DNA | 2528 | ||
| Ligand/Ion | 6 | ||
| B-factor | 133.0 | ||
| Protein/DNA | 133.0 | ||
| Ligand/Ion | 120.0 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | ||
| Bond angles (°) | 0.90 | ||
Values in parentheses are for highest-resolution shell.
These datasets were not used for phasing but were used to guide the building of the model.
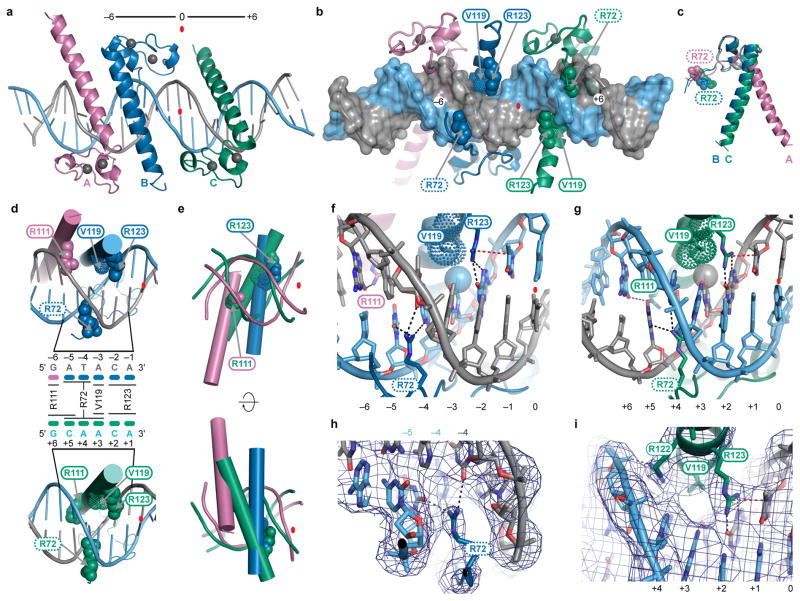
Figure 2. Major groove interactions and use of multiple DNA binding modes by DMRT proteins.
(a) Overview of structure, showing two DMRT167–136 protomers (A and B) inserted in DNA major groove on one side of Site 1 and one (C) in the major groove on the other side. Binding site symmetry is indicated (−6 and +6). Red oval: central basepair. Grey spheres: zinc ions. (b) Interaction of protomers A, B and C from back side, highlighting insertion of R72 sidechains of protomer B and C (dashed ovals) into minor groove. Amino acids labeled with solid ovals make major groove DNA contacts. (c) Overlay showing different orientation of R72 and different angle between zinc binding module and recognition helix of protomer A relative to those of B and C. (d) DNA base contacts. Middle diagram summarizes contacts made by each protomer. (e) Overlaid views of DMRT1 recognition helices bound to DNA, aligned at R111 and R123 and viewed from front and back. Right and left side DNAs are color-coded: pink is bound by protomers A and B; green by protomer C. (f) Major groove and minor groove interactions on left side of Site 1. (g) Major groove and minor groove hydrogen bond interactions on right side of Site 1. (h) Protomer B R72 interactions with minor groove. (i) Major groove interactions by protomer C. In h and I, blue mesh shows the sigma-A weighted 2Fo-Fc electron density map contoured at 1.5 σ Black dashed lines in f-i indicate hydrogen bonds. Red dashed line: arginine-thymine stacking interaction. Stippled spheres: van der Waals radii.
This first view of a DM domain bound to DNA revealed a unique type of DNA interaction. The zinc-binding module of each protomer spans the DNA minor groove primarily through phosphate backbone contacts, while a recognition helix inserts into the major groove, making base-specific contacts (Fig. 2a,b). Unexpectedly, recognition helices of protomers A (pink) and B (blue) lie antiparallel together in the major groove on one side of the consensus element while a third (C, green) lies in the major groove on the other side. We are unaware of any other protein that binds DNA by insertion of two adjacent α-helices into the same region of the major groove.
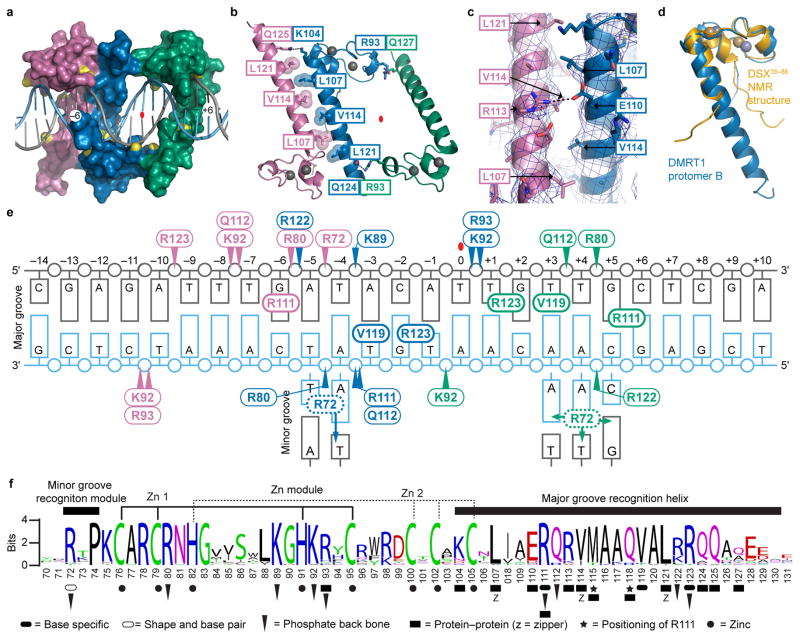
In the structure, protomers A and B bind DNA differently, reflecting different angles between their zinc-binding modules and recognition helices (Fig. 2c). Major-groove contacts on the left side of the binding site involve three amino acids (R111, V119, R123) that are provided by protomers A and B (Fig 2d, f). By contrast, while major groove contacts on the right side also involve these same three amino acids, all are provided by protomer C (Fig. 2d,g,i). The left side major groove is unusually wide (Fig. 1d) and accommodates protomers A and B, which sit more perpendicular to the helical axis than protomer C (Fig. 2d,e). In protomers B and C, R72, N-terminal to the zinc-binding module, inserts into the minor groove to hydrogen bond with base pairs (Fig. 2d,f-h); these interactions are consistent with use of arginine by other proteins to mediate minor groove contacts23. The hydrogen bond donor-acceptor pattern is almost indistinguishable in the minor groove for A-T vs T-A and G-C vs C-G base pairs24,25. Thus base readout by R72 likely involves base pair recognition rather than base recognition and may also involve shape readout25. All three DMRT1 protomers extensively contact the DNA backbone (Fig. 3a). They also interact with each other: the recognition helices of protomers A and B are held in close apposition by an interdigitating hydrophobic zipper, while protomers B and C are hydrogen-bonded (Fig. 3b,c). The overall folding pattern of the zinc-binding module is very similar to that of Dsx, previously determined by NMR (Fig. 3d)26. Critical DM domain amino acids and protein-DNA and protein-protein contacts are summarized in Fig. 3e and f. The contacts shown can explain virtually all of the conserved DM domain amino acids and DNA nucleotides in the binding site (Fig. 3f). Most conserved amino acids without functions indicated have structural roles to maintain the overall structure of the DM domain, for example by terminating helical domains, allowing bends, or mediating folding of the zinc binding module26.
Figure 3. DNA backbone contacts, protein-protein interactions and binding summary.
(a) Molecular surface of DMRT167–136 bound to DNA with charged groups contacting DNA phosphate backbone indicated in yellow. (b) Amino acids mediating protomer-protomer contacts. Interdigitating hydrophobic zipper and a Q to K hydrogen bond link protomers A and B. Two Q to R hydrogen bonds link B and C. (c) Close-up view of interaction between protomers A and B. Leucines and valines of interdigitating hydrophobic zipper are shown. In addition, R113 of protomer A and E110 of protomer B appear to form a salt-bridge (dashed line). Blue mesh shows the sigma-A weighted 2Fo-Fc electron density map contoured at 1.0 σ (d) Overlay of DMRT1 protomer B structure with Dsx NMR structure26 showing similar fold of zinc-binding domain. (e) Summary of DMRT167–136-DNA interactions. Colors indicate which protomer makes each contact. Thin-lined ovals with arrowheads identify amino acids that make DNA backbone contacts and thick-lined ovals identify amino acids that contact DNA bases. (f) Conservation of metazoan DM domains. Structural motifs and functional amino acids revealed by DMRT1 structural analysis are indicated for the region resolved by crystallography. Additional interactions could exist, particularly those bridged by water molecules. Amino acids are colored according to their chemical properties: polar amino acids (G,S,T,Y,C) are green, basic (K,R,H) blue, acidic (D,E) red, hydrophobic (A,V,L,I,P,W,F,M) black and neutral amino acids (Q,N) are purple.
Sequence-specific binding is primarily via DNA major groove
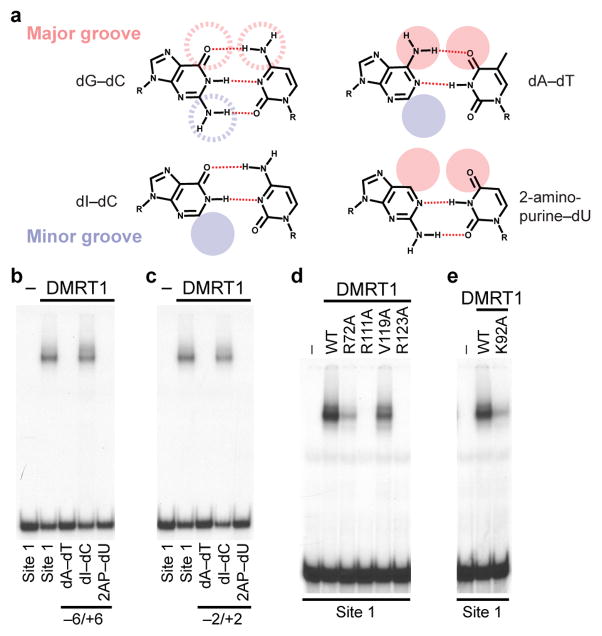
A prior study26 found that DM domain DNA binding tolerates extensive chemical modification of the DNA major groove but not the minor groove, and proposed on this basis that binding is mainly mediated by sequence-specific minor groove contacts. However, minor groove contacts can only distinguish A-T from G-C basepairs, not specific sequences. Indeed, while our structure revealed potential hydrogen bond interactions of R72 with the minor groove, the positions contacted by R72 do not show strong sequence conservation. By contrast, the structure revealed extensive sequence-specific major groove interactions. These interactions involved highly conserved DNA basepairs (−6 and +6, −2 and +2) that were not specifically tested in the previous study. To verify the importance of these base pairs we first changed the −6 and +6 positions from dG-dC to dA-dT (Fig. 4a), which strongly reduced DMRT1 binding (Fig. 4b). To query the minor groove at these positions, we substituted dI-dC base pairs, removing minor groove exo-cyclic amines without altering the major groove (Fig. 4a); these substitutions did not reduce binding (Fig. 4b). To query the major groove, we substituted 2-amino purine (2AP)-dU base pairs, inverting the carbonyl oxygen and removing the exocyclic amine from the major groove without altering minor groove structure (Fig 4a). 2AP-dU substitution virtually eliminated DMRT1 binding, demonstrating the importance of the major groove sequence identity at these positions (Fig. 4b). The same major groove modifications at the −2 and +2 positions also reduced binding but minor groove modifications at these positions did not (Fig. 4c). In summary, the −6 and +6 and the −2 and +2 positions are crucial for DNA binding and these positions are recognized primarily via the major groove.
Figure 4. Confirmation of critical protein-DNA contacts.
(a-c) Confirmation of major groove DNA contacts by chemical substitution in vitro. (a) Chemical structures of base pair analogs, with shaded circles indicating atoms altered in modified bases. (b) EMSA assay of DNAs modified at −6 and +6 positions, showing that major groove but not minor groove changes reduce DMRT1 binding. c, EMSA assay of DNAs modified at −2 and +2 positions, showing that major groove but not minor groove modifications reduce binding. (d,e) Confirmation of critical protein contacts by amino acid substitution. (d) EMSA assays showing effects of alanine substitution of DMRT1 amino acids making base contacts. (e) EMSA assay showing that substituting K92, which interacts with the DNA phosphate backbone, reduces DMRT1 DNA binding.
We also used protein sequence substitutions to assess the importance of amino acid sidechains that make major- or minor-groove contacts. Replacing R72, R111, V119 or R123 with alanine reduced or eliminated DMRT1 binding, indicating that these residues are crucial for DNA recognition (Fig. 4d, Supplementary Fig. 3a). This functional analysis was particularly important given the limited resolution of the X-ray structure. DNA backbone contacts also are important for DMRT1 DNA binding affinity, as K92A reduced binding (Fig. 4e, Supplementary Fig. 3a).
DMRT1 can bind DNA as a tetramer, trimer, or dimer
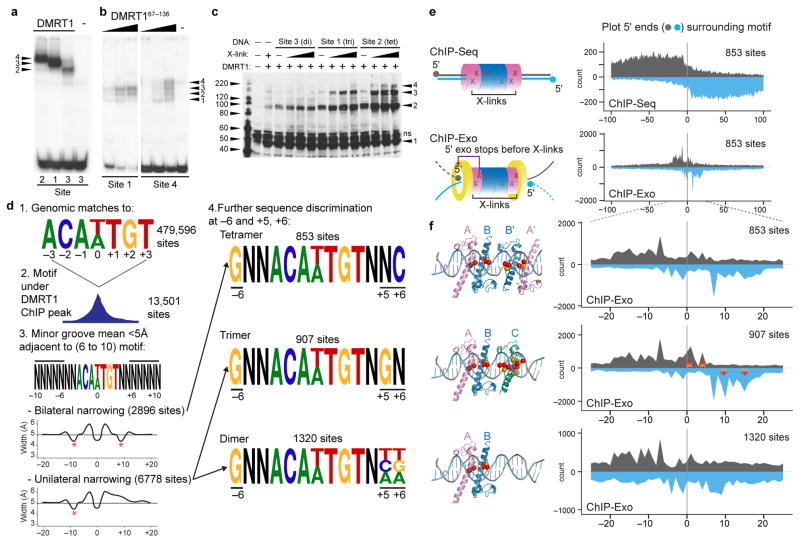
Next we further examined DNA binding stoichiometry. The structure shows that Site 1 can bind a DMRT1 trimer and DNase I protection showed that Site 2 is more extensively protected by DMRT1 than Site 1. These data suggest that the slower migrating EMSA complex on Site 2 (Figs. 1e, 5a) is a symmetric ABB′A′ tetramer. Site 2 differs from Site 1 at +5, a position that is uniquely recognized by protomer C (Fig. 2d; see also Fig. 7g), and it also has changes at +8 and +9 that are predicted to narrow the minor groove between +6 and +8 (Fig. 1d). These differences suggest that DNA sequence and shape may dictate protein-binding mode. We hypothesized that making other sequence changes to Site 1 guided by the structure might instead cause AB dimers to form. Indeed, modifying Site 1 at +2 and +6 to alter bases recognized specifically by protomer C (Site 3) generated a faster migrating EMSA complex that is consistent with an AB dimer (Fig. 5a, Supplemental Fig. 3b). To confirm that DMRT1 can bind DNA with multiple stoichiometries, we performed additional EMSAs. Instead of full-length DMRT1 as in Fig. 5a, we used DMRT167–136, which removes a multimerization domain (not shown) and reduces cooperative binding. DMRT167–136 formed three distinct complexes with Site 1 (Fig. 5b, left lanes), which we interpret as monomers, dimers, and trimers. Because binding to Site 2 was highly cooperative even with DMRT167–136, we also assayed a site with reduced affinity and cooperativity (Site 4). On Site 4, DMRT167–136 formed four complexes (Fig. 5b, right lanes), which we interpret as monomer through tetramer. To further confirm these stoichiometries we performed protein cross-linking using full-length DMRT1 bound to sites 1–3 (Fig 5c). As predicted, DMRT1 formed DNA-dependent complexes of different maximum stoichiometries. Dimers formed on Site 3, dimers and trimers formed on Site 1 with traces of tetramer, and dimers, trimers, and tetramers formed on Site 2. Together, the structure, DNase I protection, EMSA analyses, and protein cross-linking indicate that DMRT1 can bind DNA in vitro as a tetramer, trimer, or dimer, and, when the protein is truncated, as a monomer.
Figure 5. DMRT1 binds DNA with multiple stoichiometries in vitro and in vivo.
(a) EMSA showing binding of DMRT1 tetramer, trimer and dimer to Sites 2, 1, and 3, respectively. (b) EMSA of in vitro translated SUMO-DMRT167–136 binding to Sites 1 and 4, showing monomer through tetramer binding. (c) Protein crosslinking showing interaction of DMRT1 to form higher-order complexes. (d) Workflow testing DMRT1 binding stoichiometry by ChIP-Exo. Sites under ChIP peaks were grouped based on bilateral or unilateral minor groove narrowing and sequence at positions −6, +5, and +6 (see Methods). (e) Left, diagrams comparing ChIP-Seq and ChIP-Exo. Crosslinked protein blocks exonuclease digestion in ChiP-Exo within several bases of the crosslink. Colors indicate binding by different protomers; “X” illustrates potential crosslinks. Right, compilation of 5′ ends of ChIP-Seq (top) and ChIP-Exo (bottom) reads aligned to DMRT1 tetramer-binding consensus, showing higher resolution of ChIP-Exo. (f) ChIP-Exo analysis of DMRT1 binding in the mouse testis at sites sorted as indicated in panel d, showing three distinct patterns. Structural diagrams interpret DMRT1 binding modes based on ChIP-exo patterns and indicate potential crosslinks (red balls: crosslinkable DNA residues; yellow balls, crosslinkable protein residues). Note shared pattern in left side but differences on right side. Stars highlight prominent differences in ChIP-Exo pattern in putative trimer binding sites relative to tetramer sites. Based on the structure, trimers have more potential crosslinks with protomer C near center and right side of binding site due to its different position in the major groove.
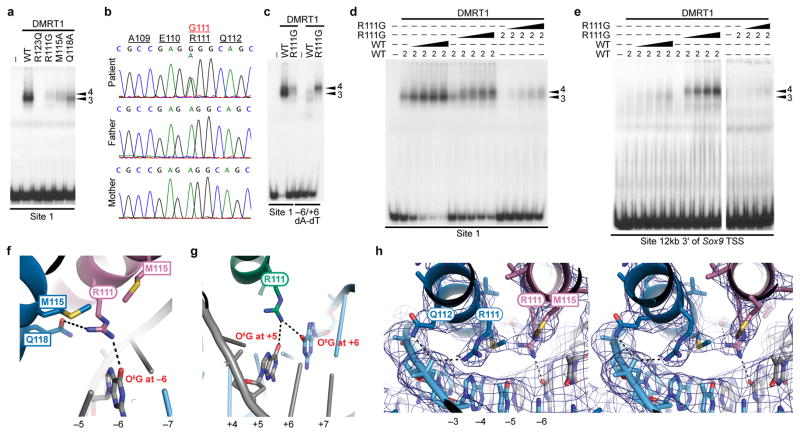
Figure 7. Disruption of crucial DMRT1 DNA contacts by a sex-reversing human mutation.
(a) EMSA assay showing importance of R123, R111 and three amino acids that position R111 of protomer A for sequence-specific DNA interaction. (b) Sanger sequencing chromatograms showing de novo A to G heterozygous sequence change causing R111G coding change in 46,XY female. (c) EMSA showing reduced binding affinity and altered binding specificity of R111G mutation. Binding to Site 1 is reduced, but DMRT1R111G binds Site 1 substituted at −6 and +6 better than wild-type DMRT1. (d) EMSA comparing DMRT1 and DMRT1R111G. Left lanes, wild type DMRT1 alone; middle, DMRT1 plus DMRT1R111G; right, DMRT1R111G alone. EMSAs contained 2 ul in vitro translated of protein; wedges indicate added increments up to a total of 6 ul. DMRT1R111G can convert wild type trimers on Site 1 into slower-migrating tetramers (arrowheads), likely by occupying the right side of the binding site. (e) Same experiment as in panel d, except DMRT1 binding site is from the Sox9 gene. (f) Protomer A R111 (pink) is positioned for hydrogen bonding with −6 guanine by M115 and Q118 of protomer B (blue) and M115 of protomer A (pink). Methionine methyl groups make van der Waals contacts with each other and R111. (g) In protomer C R111 can recognize +5 or +6. (h) Walleye stereo view of protomer A R111 interacting with −6G and protomer B R111 interacting with DNA backbone. Blue mesh: sigma-A weighted 2Fo-Fc electron density map contoured at 1.5 σ.
We next asked whether DMRT1 also binds DNA using multiple stoichiometries in vivo. For a higher resolution view of DMRT1-DNA interaction in vivo we used ChIP-Exo27, which employs strand-specific exonuclease digestion prior to sequencing to localize protein-DNA crosslinks with higher precision than ChIP-Seq. ChIP-Exo did not reveal exact binding details at individual sites so we used structural and in vitro DNA binding properties to group sites (Fig. 5d) and reveal their patterns of binding, a strategy that has also been used with other proteins28. We searched the genome for matches to the 7 bp core DMRT1 binding motif and selected those found under DMRT1 ChIP peaks. We then used minor groove width predictions29 to group peaks into those predicted to have bilateral narrowing of the minor groove (tetramers) and those with unilateral narrowing (dimers and trimers). Guided by the structure and EMSA analysis, we further selected sites based on the sequence at positions −6, +5, and +6, as indicated in Fig. 5d. Finally, we plotted the ChIP-Exo data in aggregate for each set of DMRT1 binding sites and compared the binding patterns, asking whether they differed and whether their differences were consistent with binding by each of the stoichiometries identified in vitro (Fig. 5 e,f). Comparison of the compiled ChIP-Exo data revealed a shared pattern on the left side in all three classes as expected, but distinct patterns on the right. Fig. 5f indicates the predicted crosslinking patterns for each binding mode, based on the structure, and these conform well to the observed crosslinking patterns for the different groups of sites (Fig. 5f; Supplementary Fig. 4a). Predicted tetramers had symmetrical ChIP-exo patterns, while those of trimers and dimers were asymmetric, as expected. In trimers, the protomer C recognition helix sits at an angle in the DNA major groove that allows contact with more bases than protomers A and B (Fig. 2d,e) and therefore has a higher density of potential crosslinks (Fig. 5f; Supplemental Fig. 4b); consistent with this prediction we observed stronger crosslinks on the right side of the binding site, where protomer C would bind. Compilation of the selected DMRT1 consensus sequences did not reveal additional sequence or shape preferences, suggesting that the primary determinant of stoichiometry is the sequence and shape at the DMRT1 binding site rather than presence of additional sequence motifs or DNA conformations (Supplemental Fig. 4c,d). Distinct patterns also were apparent in standard ChIP-Seq, at lower resolution (Supplementary Fig. 4a). In summary, ChIP-Exo suggests that DMRT1 binds as a tetramer, trimer or dimer in vivo, as in vitro, with the mode at each site determined by a combination of DNA sequence and shape.
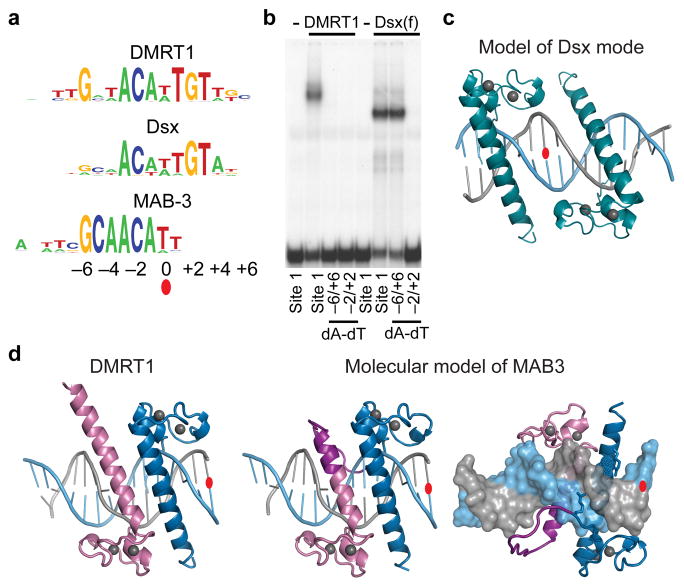
Modeling suggests different binding modes for Dsx and MAB-3
Different DNA binding modes likely are used by the invertebrate sexual regulators Dsx and MAB-3. In vitro, Dsx and DMRT1 bind similar motifs but Dsx has no sequence preference at −6 and +6 (Fig. 6a). EMSA confirmed that the −2 and +2 positions are important for binding of both Dsx and DMRT1, but −6 and +6 are only important for DMRT1 binding (Fig. 6b). This requirement for only the inner core of the binding motif suggests that Dsx binds as a symmetrical BB′-like dimer (modeled in Fig. 6c). C. elegans MAB-3 has tandem DM domains (Supplementary Fig. 5) and binds a site reminiscent of a DMRT1 half-site30 (Fig. 6a). Molecular modeling suggests that the MAB-3 tandem DM domains might be equivalent to a DMRT1 AB dimer, with the truncated first recognition helix allowing looping so that both helices can bind adjacent on one another in the major groove (Fig. 6d).
Figure 6. Modeling DNA interaction by Dsx and MAB-3 suggests different binding modes.
(a) In vitro DNA binding motifs for DMRT1, Dsx and MAB-3 showing that the Dsx site30 is symmetrical but lacks selection at −6 and +6 positions while the MAB-3 motif30 resembles the left side of DMRT1 motif. (b) EMSA assay showing that binding by the female Dsx isoform Dsx(f) requires specific DNA basepairs at the −2 and +2 but not the −6 and +6 DNA positions, consistent with the in vitro consensus. (c) Docking model of DMRT1 binding as a dimer to a previously determined Dsx binding site DNA structure37, illustrating likely Dsx binding mode. (d) A model of proposed interaction of MAB-3 DM domains with DNA illustrating binding of MAB-3 as a covalently-joined “internal dimer”. MAB-3 (center and right) is proposed to form a structure on its consensus element similar to DMRT1 protomers A and B bound to the left side of the DMRT1 consensus element (left). The first DM domain of MAB-3 (DMa) is predicted to have a truncated recognition helix, with the remainder forming a linker joining DMa to DMb (Supplementary Fig. 5).
DM domain point mutations affect DNA binding in fly and human sex reversal
dsx determines sex in insects7, and a number of dsx point mutations have been isolated that cause an intersex phenotype in Drosophila5. Most of these mutations alter residues required for zinc chelation but one, R91Q, affects a recognition helix residue equivalent to R123 in DMRT1 (Supplementary Fig. 5) and reduces DSX DNA binding5. We tested DMRT1R123Q by EMSA and found that, like DMRT1R123A (Fig. 4a), it eliminated DNA binding (Fig. 7a, Supplemental Fig. 3c). This result suggests that the DsxR91Q mutation disrupts a highly conserved sex-determining contact.
As discussed earlier, DMRT1 determines gonadal sex in some vertebrates2, but its role in human testis development has been less clear. In humans, primary XY male-to-female sex reversal results in female external genitalia and Mullerian structures (uterus and fallopian tubes), and undeveloped (“streak”) gonads. This condition is also called 46,XY complete gonadal dysgenesis, or 46,XY CGD31. Human genetics has implicated DMRT1 as a key regulator of testis development: chromosome 9p deletions that remove one copy of DMRT1 are associated with 46,XY feminization and gonadal dysgenesis, sometimes including 46,XY CGD15,32. While they suggest that DMRT1 is haploinsufficient for testicular development, these deletions usually remove other genes, including the neighboring DMRT2 and DMRT3. Also, most 9p deletions cause incomplete gonadal dysgenesis so it has been unclear whether hemizygosity of DMRT1 alone can cause full sex reversal. Although a DMRT1 deletion removing exons 3 and 4, downstream of the DM domain, was found in a strongly feminized 46,XY individual32, this deletion could have removed regulatory elements that affect other genes. Point mutations would help determine whether loss of DMRT1 alone can cause sex reversal but these have not been reported.
We therefore used exome resequencing to seek a DMRT1 point mutation. We were able to identify a 46,XY individual born fully feminized with complete gonadal dysgenesis (46,XY CGD) and carrying a heterozygous de novo point mutation (R111G) in the DMRT1 recognition helix (Fig. 7b, Supplementary Fig. 5a; Methods). Genetic analysis found normal ploidy, and fluorescent in situ hydridization confirmed two copies of the regions containing DMRT1 as well as the sex determining genes NR5A1, SOX9, WT1, and DAX1. No other potentially pathogenic mutations were apparent in the exome sequence and the DMRT1 mutation was not present in 240 ancestry-matched control individuals. Full details of the clinical and genetic characterization of this patient are provided in Methods. We conclude that the de novo DMRT1R111G mutation is the most likely cause of the complete gonadal dysgenesis and 46,XY sex reversal in this patient. To our knowledge this is the first human DMRT1 point mutation associated with 46,XY sex reversal. The phenotype is very similar to that caused by mutations in the testis-determining gene SRY33 and strongly suggests that DMRT1 is required for human sex determination.
We next examined the DNA binding properties of DMRT1R111G and found that the mutant protein had strongly reduced DNA affinity, similar to DMRT1R111A (Figs. 7a, 4d, Supplementary Fig. 3a,c). In the structure, R111 of protomer C interacts with the +5 and +6 positions of Site 1 (Fig. 7g). We found that DMRT1R111G had altered sequence specificity: it bound a site with −6 and +6 dG-dC to dA-dT substitutions weakly but better than wild-type DMRT1 (Fig. 7c). Moreover in an EMSA assay, when mixed with wild type DMRT1 the mutant protein could promote tetramer binding on Site 1, which normally is bound by trimers of wild type DMRT1 (Fig. 7d). We also tested binding of DMRT1R111G to in vivo DMRT1 binding sites from the Sox9 gene (activated by DMRT1) and Foxl2 gene (repressed by DMRT1)16,17. The Sox9 site is bound as a trimer by wild type protein (Fig. 7e). DMRT1R111G bound this site very weakly, but when mixed with wild type protein shifted the complex to a tetramer with much higher affinity. The Foxl2 site was bound as a tetramer by wild type DMRT1 and addition of DMRT1R111G had little or no effect on binding (Supplemental Fig. 6). Based on the ability of DMRT1R111G to alter binding stoichiometry of wild type DMRT1 on a biologically relevant site in vitro, we suggest that the DMRT1R111G mutation may combine severe loss-of-function and/or haploinsufficiency with a dominant disruption of normal binding stoichiometry at some DMRT1 binding sites. This combination of haploinsufficiency and dominant disruption may explain the severe phenotype caused by DMRT1R111G heterozygosity. In the structure, R111 of protomer A is positioned to contact −6 by its own M115 and by M115 and Q118 of protomer B (Fig. 7f,h). We found that mutating these residues also reduced DNA binding (Fig. 7a). In summary, the severe effects of the DMRT1R111G point mutation on DNA binding and its association with 46,XY male-to-female sex reversal strongly suggest that DMRT1 plays a role in human primary sex determination and identify another deeply conserved molecular interaction crucial for metazoan sexual development.
Discussion
We have undertaken a structural analysis of DMRT protein-DNA interaction. We used ChIP-seq to define the DNA binding preference of DMRT1 in mouse and human, and then employed X-ray crystallography to determine a DMRT1-DNA structure. The structure revealed that binding of the human DMRT1 DM domain to DNA involves the recognition of specific bases primarily in the DNA major groove. We confirmed this finding using chemical substitutions that selectively altered the major or minor groove of the DNA at key base pairs. A previous report26 concluded, based on DNA substitutions, that Dsx binds DNA primarily via the minor groove. Based on our structural analysis it is apparent that the minor groove modifications that reduced binding likely limited the ability of the major groove to expand and accommodate the DM domain recognition helix rather than affecting sequence-specific base contacts; thus the previous data are in accord with our structure.
Binding of DMRT1 to DNA has two particularly noteworthy features. First, binding involves the insertion of paired recognition helices together into a widened DNA major groove. To our knowledge this is the only example of two closely neighboring alpha helices inserting into the same section of a major groove. Second, DMRT1 can bind DNA using different stoichiometries. The basis of this versatility is that binding involves a small number of amino acid sidechains that can make distinct sets of DNA interactions. As a result, different DNA sites can bind distinct configurations of protomers, ranging from dimers to tetramers. ChIP-exo analysis suggests that DMRT1 also binds in vivo with differing stoichiometries. Our ability to predict stoichiometry based on DNA sequence preference and conformation (Fig. 5), suggests that the stoichiometry at a specific bindings site is determined largely by the sequence and shape of that site. A key remaining question is what biological significance the DMRT1 binding stoichiometries may have. Possibilities include association with transcriptional activation or repression or binding to different classes of regulatory elements (eg promoters or enhancers), or interaction with other regulatory proteins. Consistent with the third possibility, we previously found that a subset of DMRT1 binding sites contain overlapping GATA1 and SOX9 consensus elements34. Distinguishing among these possibilities is an important goal but this will require cell type-specific approaches, as DMRT1 has cell type-specific functions in germ cells and Sertoli cells.
While a number of deletions removing part or all of DMRT1 have been found in patients with 46,XY sex reversal, the DMRT1R111G mutation we report here is, to our knowledge, the only DMRT1 mutation shown to affect an essential functional domain. The severely reduced DNA binding affinity of DMRT1R111G combined with the complete sex reversal and gonadal dysgenesis of the patient strongly suggest that DMRT1 plays a role in human sex determination. Our finding that the mutant protein can interfere with binding stoichiometry of wild type DMRT1 further suggests that the mutant protein may behave at least partially as a dominant negative. A dominant effect of DMRT1R111G may help explain why the phenotype of this point mutation is more severe than that of most 9p deletions that completely remove DMRT1. The highly specific nature of point mutations such as DMRT1R111G that can alter function of the remaining wild type allele also may explain why DMRT1 point mutations able to cause sex reversal are so rare. Because we also observed reduced DNA binding specificity, we cannot exclude the possibility that DMRT1R111G also binds and misregulates genes that are not normally controlled by DMRT1. However, we consider this unlikely given the very low DNA binding affinity of DMRT1R111G on its own. An animal model of the DMRT1R111G may help elucidate the in vivo effects of this mutation.
In summary, we have obtained a detailed view of how DMRT proteins recognize and associate with target DNA. We have defined crucial conserved atomic interactions that mediate DNA binding and found that these are required for sex determination in flies and humans. DMRT proteins have directed metazoan sexual differentiation for hundreds of millions of years2,3. Reproduction is the crucible of natural selection35 and the long-term involvement of DMRT genes in sexual development suggests they have substantially shaped metazoan evolution.
Online Methods
Vertebrate Animals
Experimental protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. Mice were adult males of mixed C57BL/6J and 129S1 genetic background. No statistical method was used to predetermine sample size. The experiments were not randomized and were not performed with blinding to the conditions of the experiments.
Figure Preparation
Figures were prepared using Adobe Photoshop, Adobe Illustrator, and Pymol software.
ChIP and ChIP-exo
Chromatin immunoprecipitation (ChIP) was performed as described34 except that tissue was disaggregated with a Virtis Virtishear homogenizer (#225318) in phosphate buffered saline (PBS) containing 1% paraformaldehyde. Sonication times were extended to allow for smaller average size products suitable for Illumina sequencing. Crosslinks were reversed overnight at 55°C. Illumina sequencing libraries were prepared according to manufacturer’s protocol except that end polishing of the ChIP fragments was by DNA terminator (Lucigen Corp.) and adapters were diluted 1:50 prior to ligation. For ChIP-exo, chromatin precipitation was performed as above and prior to elution of complexes from protein-A-sepharose beads. ChIP-exo libraries were prepared as described27 except that primer sequences were modified to be compatible with the Illumina sequencing platform adapted for Illumina sequencing.
Primers for ChIP-Exo library preparation:
-
P2 adapter:
5′ P-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-OH 3′
annealed to 5′ OH-AGATCGGAAGAGCGTCGTGTAG-OH 3′
-
Primer extension oligonucleotide:
OH-ACACTCTTTCCCTACACGAC-OH
-
P1 adapter:
OH-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-OH
annealed to OH-AGATCGGAAGAGCACACGTCTG-OH
-
PCR amplification was performed with primers P1 and P1:
P1: CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGC
P2: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGC
Bold text in P1 indicates the sequence that was varied for multiplexing.
Human tissue for ChIP
Fresh testicular tissue from an orchiectomy was provided by the University of Minnesota Tissue Procurement Facility under IRB supervision and with informed consent. Normal histology of Bouin’s fixed subsamples was confirmed by hematoxylin/eosin staining of paraffin-embedded sections.
DNA binding substrates
EMSA assays to assay stoichiometry of DMRT1 binding used shorter (27 base pair) DNA duplexes for better resolution of complexes:
| Site 1 top strand | 5′-gagatttgatacattgttgctcgatgg-3′ |
| Site 2 top strand | 5′-gagatttgatacattgttactttatgg-3′ |
| Site 3 top strand | 5′-gagatttgatacattattaatttatgg-3′ |
| Site 4 top strand | 5′-ttgctatgatacattgtatcttgctgg-3′ |
| Sox9 site top strand | 5′-gtggctgggcaccctgcagagacaatgtttccagctgcaggtcaggtct-3′ |
| Foxl2 site top strand | 5′-gtggctgggcacaactctgtaacattgtttccaaggggaggtcaggtct-3′ |
EMSA to evaluate DNA and protein mutant effects on binding used longer (49 base pair) DNA duplexes based on the Site 1 DNA duplex:
5′-GTGGCTGGGCAgagatttgatacattgttgctcgatggAGGTCAGGTCT-3′
Mutations were incorporated into hDMRT1 by overlap extension PCR38 using a T7-hDMRT1 (pDZ142) plasmid clone as template. The mutated products were sub-cloned back into pDZ142 and translated in vitro with the TNT Quick Coupled transcription/translation system (Promega).
In vitro DNA binding
Electrophoretic mobility shift analysis (EMSA) was performed as described19 except that substrates were end-labeled using T4 polynuceotide kinase (NEB). DNAse I footprint analysis was performed with highly purified bacterially expressed hDMRT1S67–P136 protein as described39 except that following the DNase I digestion step, the sample was phenol/chloroform extracted to remove protein prior to precipitation.
Protein cross-linking
Proteins were in vitro translated as for EMSA. Complexes were formed under the same conditions as EMSA except with five times as much DNA at room temperature for ten minutes prior to addition of glutaraldehyde to 0.0075% final concentration. Cross-linking was stopped at indicated times by the addition of glycine to 0.125M final concentration. Complexes were resolved on 4–12% NuPage Novex Bis-Tris mini gels (Invitrogen) and DMRT1 was detected by immunoblotting.
X-ray crystallography
hDMRT167–136 was expressed as SUMO-fusion in E. coli Rosetta2(DE3) and purified by metal-affinity chromatography. The His6-tagged SUMO was removed by cleavage with the SUMO protease Ulp1. To form the protein-DNA complex, purified hDMRT167–136 (~1 mM) in 20 mM Tris-HCl (pH7.4), 0.2 M NaCl, 10 μM ZnCl2, and 2 mM β-mercaptoethanol, was mixed with a blunt-ended 25 bp target DNA (Site 1: 5′-CGAGATTTGATACATTGTTGCTCGA-3′ and its complement) at a protein:DNA molar ratio of 2:1. The complex (~0.6 mM protein) was crystallized at 20°C by the hanging drop vapor diffusion method with a reservoir solution [100 mM Bis-Tris (pH 6.5), 4~12% polyethylene glycol 3,350, 4~10 % 2-methyl-2,4-pentanediol (MPD), 2–10 mM dithiothreitol]. Crystals containing SeMet-labeled protein or the 5-Br-dU-labeled oligonucleotide were grown under conditions similar to that for the native complex. The crystals were transferred in a stepwise fashion to the reservoir solution with increasing concentrations of glycerol (final concentration of 15%) and flash-cooled in liquid nitrogen for X-ray data collection.
X-ray diffraction data were collected at the Advanced Photon Source Northeastern Collaborative Access Team beamlines (24-ID-C/E) and the Advanced Light Source Molecular Biology Consortium (4.2.2) beamlines and processed using RAPD (https://rapd.nec.aps.anl.gov/rapd), HKL200040 or XDS41. X-ray wavelength corresponding to the K-absorption edge of Zn, Se, and Br was used respectively for the native, SeMet, and 5-Br-dU labeled crystals. The structure was determined by SAD phasing with a 3.81 Å resolution data set from a native crystal (Table 1) using PHENIX42. Six zinc sites were found, from which the structure factor phases were calculated with a mean figure of merit of 0.66. The atomic model was built in COOT43 and refined using REFMAC44 and PHENIX, with Ramachandran and DNA restraints to maintain geometries for protein, DNA base-pairs, and base-stacking. The model building was facilitated by the Se and Br anomalous difference Fourier peaks, which showed the correct register of amino acids and nucleotides, respectively. Protein residues with poor side-chain electron density were modeled as alanines. The Ramachandran plot for the final model was generated by MolProbity45, with 89.0%, 8.2%, and 2.8% of residues in favored, allowed, and outlier regions, respectively. The DNA structure was analyzed using 3DNA46, the minor groove width of the unbound DNA was estimated using DNAshape36, and the molecular graphics images were produced using PYMOL (www.pymol.org).
Identification of patient with DMRT1R111G mutation
Overview of study
The study was approved by the ethical board of Institut Pasteur (RBM 2003/8) and informed consent was obtained. Patient ancestry was determined by self reporting, based on responses to a personal questionnaire, which asked questions pertaining to the birthplace, languages and ethnicity of the participants, their parents and grandparents. The control panel consisted of 240 unrelated 46,XY males of French ancestry who are either normospermic or have fathered at least two children and have no history of testicular anomalies (determined by self reporting). All samples used for this study were collected with proper informed consent. Sequencing of the coding region of DMRT1 gene was performed as described previously47.
Whole Exome sequencing
Exon enrichment was performed using Agilent SureSelect Human All Exon V4. Paired-end sequencing was performed on the Illumina HiSeq2000 platform using TruSeq v3 chemistry. Read files (Fastq) were generated from the sequencing platform via the manufacturer’s proprietary software. Reads were mapped using the Burrows-Wheeler Aligner48 and local realignment of the mapped reads around potential insertion/deletion (indel) sites was carried out with the GATK version 1.649. Duplicate reads were marked using Picard version 1.62 (http://picard.sourceforge.net). Additional BAM file manipulations were performed with Samtools (0.1.18)50. SNP and indel variants were called using the GATK Unified Genotyper for each sample. SNP novelty was determined against dbSNP138. Novel variants were analyzed by a range of web-based bioinformatics tools using the EnsEMBL SNP Effect Predictor (http://www.ensembl.org/homosapiens/userdata/uploadvariations). All variants were screened manually against the Human Gene Mutation Database Professional Biobase (http://www.biobase-international.com/product/hgmd). In silico analysis was performed to determine the potential pathogenicity of the variants. Potentially pathogenic mutations were verified using classic Sanger sequencing
Characterization of patient
The patient has two healthy brothers and a sister. A routine fetal karyotype was performed as part of protocol for pregnancy with advanced maternal age. The karyotype was 46,XY whereas the ultrasound showed a completely female foetus. The baby, born by cesarean section, was completely feminine. At day 1, serum testosterone levels were 57 ng/dl, dihydrotestosterone 12 ng/dl, Adrenocorticotropic hormone (ACTH) 70.8 ng/ml and Anti-Müllerian hormone (AMH) 0.02 ng/ml. The hormonal profile was consistent with gonadal dysgenesis. At 3 months of age serum LH levels were 0.08 UI/l, FSH 15.1 UI/l, inhibin B <15 ng/ml, AMH <0.15 ng/ml, testosterone <0.05 ng/dl, androstenedione 11 ng/dl and ACTH 11 ng/dl. At this time ultrasound revealed the presence of a uterus and an apparent absence of gonads. At 18 months gonadectomy revealed bilateral streak gonads with a gonadoblastoma on the right side. Histology of the gonads revealed ovarian-like stroma with no evidence of any testicular material. The diagnosis was 46,XY complete gonadal dysgenesis.
Genetic analysis
At three months of age the karyotype of the patient on peripheral blood lymphocytes was 46,XY (50 cells). FISH analysis on lymphocyte spreads indicated two copies of the regions 9p24 (DMRT1), 9q22 (NR5A1), 11p13 (SOX9), 17q24 (WT1) and Xp21 (DAX1). Direct sequencing of the SRY and NR5A1 genes revealed wild-type sequences. Array comparative genomic hybridization using the Agilent 44k platform confirmed a normal ploidy in the patient.
Whole exome sequencing was performed on the parents and patient. The number of paired-end reads were 24,328,671 (father), 26,580,579 (mother) 20,566,862 (child) with a mean coverage of 61.09, 68.01 and 53.41 respectively. The percentage of target bases with >x10 coverage was 96.26%, 96.97% and 95.59%. The number of variants with predicted serious (involving an essential splice site, a stop codon gained or lost, a complex indel, a frameshift mutation in the coding sequence or a non-synonymous change with predicted deleterious effect on protein function) consequences for the father, mother and child was 11,930, 11,944 and 11,758 respectively.
Analyses of the datasets revealed several de novo mutations that were predicted by PolyPhen251 and/or SIFT52 to be deleterious substitutions for protein function. These were the novel heterozygous mutations c.644A>G (p.Glu215Gly) in C2CD4C (ENST00000332235), c.1309G>A (p.Glu437Lys) in CEP104 (ENST00000378230), c.761G>C (p.Ala761Pro) in DLGAP3 (ENST00000235180), c.331A>G (p.Arg111Gly) in DMRT1 (ENST00000382276), c.3779G>C (p.Gly1260Ala) in HSPG2 (ENST00000374695), c.58C<G (p.Leu20Val) in MECR (ENST00000263702) and c.560G>A (p.Gly187Asp) in MPST (ENST00000397225).
Assuming a recessive or X-linked model of inheritance and after filtering to remove variants with a minor allelic frequency of 0.05, there was only a single remaining gene with a serious mutation. This was a hemizygous c.262G>A (p.Arg88Trp) mutation in the X chromosome gene MID2 (ENST00000262843). This variant has previously been reported (rs375584547) with an allelic frequency of 1:6727 in individuals of European-American ancestry. With the exception of DMRT1, there is an absence of a clear functional relationship between the variants in these genes and the absence of testis formation seen in the patient.
The de novo missense mutation in DMRT1 was confirmed by direct sequencing of the DMRT1 gene. No other potentially pathogenic mutations were identified in the exome sequencing dataset in other genes known to cause 46,XY gonadal dysgenesis (e.g. WT1, NR5A1, SRY). The DMRT1 mutation was not observed in direct sequence analysis of 240 unrelated ancestry-matched control individuals.
Bioinformatics work flow
ChIP libraries were sequenced on the Illumina HiSeq2000 platform with 50 cycles single end (ChIP-Seq) or 50 cycles paired-end (ChIP-Exo) reads. Illumina fastq files were adapter trimmed (Trimmomatic 0.32)53 and mapped (Burrows-Wheeler Aligner - Mem, 0.7.10-r789)48 to the mm9 or hg19 genome builds. Samtools (0.1.18)50 was used to sort and convert aligned reads and to extract the R2 mate pairs in the paired-end ChIP-Exo dataset. Peaks with a p-value less than 0.05 were identified in the ChIP-Seq datasets (MACS 2.1.0.20140616)54. Peaks present in both replicates available for the mouse ChIP-Seq were used for further analysis. The peaks that scored in the top sextile of the mouse and human peaklists contained 8571 and 7593 peaks respectively. 100bp of DNA sequence surrounding the summits of these top peaks was analyzed by MEME (4.10.0)55 to identify enriched sequence motifs. Genomic regions with DNA sequences compatible with dimer, trimer and tetramer binding modes found underneath ChIP-Seq peaks were used to aggregate read counts from the 5′ positions of the ChIP-Exo reads using RSamtools (1.18.2; http://bioconductor.org/packages/release/bioc/html/Rsamtools.html) and GenomicRanges (3.0)56 using custom R (3.1.1; http://www.R-project.org/) scripts available at https://github.com/micahgearhart/exotools and at Nature Protocol Exchange XXX.
Analysis of ChIP-exo stoichiometry
Non-overlapping 51 base-pair mouse genomic locations centered around the sequence ACA(A/T)TGT were identified using the BioStrings package in Bioconductor57. Sites coinciding with DMRT1 occupancy in the mouse 8-week ChIP-Seq dataset were selected for further analysis. The predicted minor groove width at these locations was retrieved from the GBShape database29. The mean of the minor groove width at positions 6–10 on either side of the motif was used to classify sites containing either a unilateral or bilateral narrowing of the minor groove. A second classification based on the nucleotides at positions −6, +5 and +6 was used to predict which sites were likely bound by DMRT1 dimers, trimers, or tetramers, as indicated in Fig. 5d.
Supplementary Material
Acknowledgments
We thank K. Shi and J. Nix for help with crystallization trials and data collection, the University of Minnesota Supercomputing Institute for computational resources, Linda Amble, the University of Minnesota Tissue Procurement Facility, and anonymous donors for human testis tissue, and D. Greenstein, C. Kim, M. Slattery, B.F. Pugh, J. Simon, H. Towle and members of our laboratories for helpful comments on the manuscript, and T. Gamble for help with phylogenetic analysis. X-ray data were collected at the Advanced Photon Source (APS) NE-CAT beamlines, which are supported by the US National Institute of General Medical Science (P41 GM103403. APS is a US Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02-06CH11357. This work was funded by the US National Institutes of Health (GM59152 and GM50399 to D.Z.; AI087098 and GM095558 to H.A.), COST Action DSDnet BM1303 (to K.M.) and Program Blanc Assistance-Publique-Institut Pasteur (to K.M.).
Footnotes
Accession Codes
The atomic coordinates and the structure factors have been deposited in the Protein Data Bank under the accession code 4YJ0. Sequencing data have been deposited in the Gene Expression Omnibus under the accession code GSE64892.
Author contributions:
M.W.M. performed and, with V.J.B, D.Z. and M.D.G., analyzed in vitro and in vivo DNA binding studies. M.D.G. performed bioinformatic analysis of ChIP data. J.K.L., K.K., and H.A. performed protein purification and crystallization. J.K.L, S.B., and H.A. collected X-ray diffraction data. J.K.L. processed the X-ray data and built and refined the atomic model. M.W.M., J.K.L., M.D.G., D.Z., H.A. and V.J.B. analysed the structure and prepared the figures. G.L. coordinated the patient clinical studies. A.B. and K.M. designed the human genetic studies and with S.R. analyzed the exome datasets. A.B. and S.R. performed Sanger sequencing. D.Z and V.J.B wrote the manuscript. M.W.M., M.D.G., H.A., A.B., K.M. and S.A. edited the manuscript. The first four authors made equivalent contributions.
The authors declare no competing financial interest.
References
- 1.Gamble T, Zarkower D. Sex determination. Current biology : CB. 2012;22:R257–262. doi: 10.1016/j.cub.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp A. Dmrt genes in the development and evolution of sexual dimorphism. Trends in genetics : TIG. 2012;28:175–184. doi: 10.1016/j.tig.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raymond CS, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 5.Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. The EMBO journal. 1993;12:527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong T, Collins JJ, 3rd, Brubacher JL, Zarkower D, Newmark PA. A sex-specific transcription factor controls male identity in a simultaneous hermaphrodite. Nature communications. 2013;4:1814. doi: 10.1038/ncomms2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- 9.Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 11.Lambeth LS, et al. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev Biol. 2014;389:160–172. doi: 10.1016/j.ydbio.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda M, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veitia R, et al. Deletions of distal 9p associated with 46,XY male to female sex reversal: definition of the breakpoints at 9p23.3-p24.1. Genomics. 1997;41:271–274. doi: 10.1006/geno.1997.4648. [DOI] [PubMed] [Google Scholar]
- 15.Tannour-Louet M, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PloS one. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindeman RE, et al. Sexual Cell-Fate Reprogramming in the Ovary by DMRT1. Current biology : CB. 2015;25:764–771. doi: 10.1016/j.cub.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L, Svingen T, Ng ET, Koopman P. Female-to-male sex reversal in mice caused by transgenic overexpression of Dmrt1. Development. 2015 doi: 10.1242/dev.122184. [DOI] [PubMed] [Google Scholar]
- 19.Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annual review of biochemistry. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slattery M, et al. Absence of a simple code: how transcription factors read the genome. Trends in biochemical sciences. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, et al. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starick SR, et al. ChIP-exo signal associated with DNA-binding motifs provide insights into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome research. 2015 doi: 10.1101/gr.185157.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu TP, et al. GBshape: a genome browser database for DNA shape annotations. Nucleic acids research. 2015;43:D103–109. doi: 10.1093/nar/gku977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- 31.Ostrer H. In: GeneReviews(R) Pagon RA, et al., editors. 2009. [Google Scholar]
- 32.Ledig S, Hiort O, Wunsch L, Wieacker P. Partial deletion of DMRT1 causes 46,XY ovotesticular disorder of sexual development. European journal of endocrinology / European Federation of Endocrine Societies. 2012;167:119–124. doi: 10.1530/EJE-12-0136. [DOI] [PubMed] [Google Scholar]
- 33.Jager RJ, Anvret M, Hall K, Scherer G. A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature. 1990;348:452–454. doi: 10.1038/348452a0. [DOI] [PubMed] [Google Scholar]
- 34.Murphy MW, et al. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13360–13365. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darwin C. The Descent of Man, and Selection in Relation to Sex. John Murray; 1871. [Google Scholar]
- 36.Zhou T, et al. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic acids research. 2013;41:W56–62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayana N, Weiss MA. Crystallographic analysis of a sex-specific enhancer element: sequence-dependent DNA structure, hydration, and dynamics. Journal of molecular biology. 2009;385:469–490. doi: 10.1016/j.jmb.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 39.Connaghan-Jones KD, Moody AD, Bain DL. Quantitative DNase footprint titration: a tool for analyzing the energetics of protein-DNA interactions. Nature protocols. 2008;3:900–914. doi: 10.1038/nprot.2008.53. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. Section D, Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. Acta crystallographica. Section D, Biological crystallography. 2010. MolProbity: all-atom structure validation for macromolecular crystallography; pp. 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nature protocols. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vinci G, et al. Association of deletion 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Molecular human reproduction. 2007;13:685–689. doi: 10.1093/molehr/gam045. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu P, et al. OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PloS one. 2014;9:e97507. doi: 10.1371/journal.pone.0097507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS computational biology. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.