Abstract
A family with X-linked mental retardation characterized by severe mental retardation, speech and behavioral abnormalities, and seizures in affected male patients has been found to have a G1141C transversion in the creatine-transporter gene SLC6A8. This mutation results in a glycine being replaced by an arginine (G381R) and alternative splicing, since the G→C transversion occurs at the −1 position of the 5′ splice junction of intron 7. Two female relatives who are heterozygous for the SLC6A8 mutation also exhibit mild mental retardation with behavior and learning problems. Male patients with the mutation have highly elevated creatine in their urine and have decreased creatine uptake in fibroblasts, which reflects the deficiency in creatine transport. The ability to measure elevated creatine in urine makes it possible to diagnose SLC6A8 deficiency in male patients with mental retardation of unknown etiology.
X-linked mental retardation (XLMR) is composed of a heterogeneous group of entities that accounts for 5%–12% of all mental retardation, depending on the method of estimation (Stevenson et al. 2000). XLMR entities can be separated into two groups: syndromic (MRXS) and nonsyndromic (MRX) mental retardation (Neri et al. 1991). The separation is based on the presence (in MRXS) or absence (in MRX) of distinctive somatic, metabolic, or neurological features associated with the mental retardation. Both MRXS and MRX have been localized to all regions of the X chromosome (Lubs et al. 1999; Chiurazzi et al. 2001). One such region is Xq28, to which 11 MRXS and 8 MRX conditions have been mapped (Armfield et al. 1999; Chelly 2000). This region is also gene rich, which makes it difficult to choose candidate genes for screening in families with XLMR. Nonetheless, some success has been achieved by utilization of the positional cloning approach. L1CAM (L1 cell-adhesion molecule) was identified as the gene involved in X-linked hydrocephalus (Rosenthal et al. 1992), MASA syndrome (Vits et al. 1994), and X-linked agenesis of corpus callosum (Jouet et al. 1994). Mutations in the gene that encodes GDI1, a GDP-dissociation inhibitor, have been found to be responsible for MRX41 and MRX48 (D’Adamo et al. 1998). The gene that encodes MECP2, a methyl-CpG-binding protein, has also been shown to be involved in Rett syndrome (Amir et al. 1999) and MRX16 (Couvert et al. 2001) and has been implicated in some families with XLMR in which male patients have severe mental retardation (Meloni et al. 2000; Orrico et al. 2000; Couvert et al. 2001).
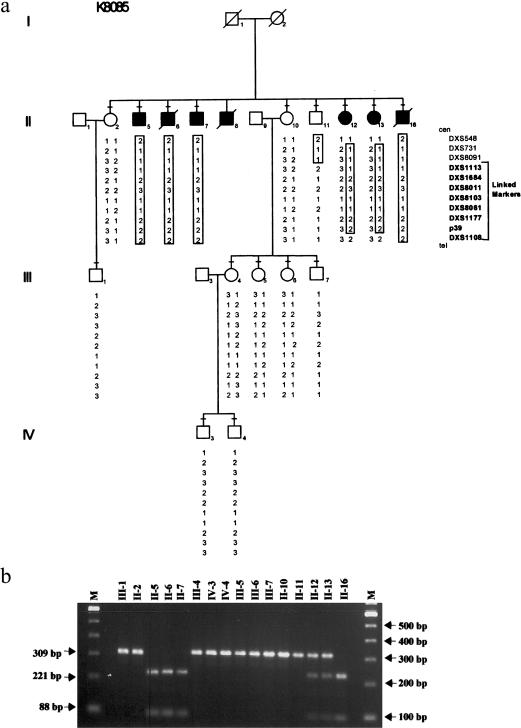
A partial pedigree of family K8085 is shown in figure 1a, with five affected male patients in one generation. All affected male patients, except II-8, underwent a clinical examination as described by Schwartz (1993). Clinical information on II-8 was taken from his medical records. Summaries of the clinical findings are given in table 1. All affected male patients had severe mental retardation with speech and behavioral abnormalities, as well as a history of seizures. Head circumference was normal in all affected male patients. Adult height (162.5–167.5 cm) was less than the adult height of the unaffected brother (175.5 cm). Midface hypoplasia, unfolded superior helices, soft skin, hyperextensible joints and hypotonia in the upper limbs, stub thumbs, and pes cavus occurred in some affected male patients. Gastrointestinal disturbances in the form of chronic constipation, megacolon, gastric and duodenal ulcer disease, ileus, and bowel perforation were common. The genitalia were normal, although inguinal hernias occurred in two affected male patients. Motor tone in the upper limbs was lower than that in the lower limbs, in two individuals. II-7 had combined spastic/dystonic gait and choreoathetoid/dystonic movements of the face and upper limbs, as well as a history of neuroleptic malignant syndrome. Two sisters (II-12 and II-13) of the affected male patients had mild cognitive impairment, and one (II-13) had chronic behavioral disturbances. No information was available on I-2, an obligate carrier.
Figure 1.
Linkage and segregation analysis of family K8085. a, Partial pedigree of family K8085, showing haplotype with 11 DNA loci located in Xq28. Microsatellite analysis was conducted using an automated laser fluorescent (ALF) sequencer (Amersham-Pharmacia), as described elsewhere (Stevenson et al. 1998). Boxed numbers indicates the at-risk haplotype associated with mental retardation. Blackened squares denote affected male patients, and blackened circles denote female carriers with phenotypic expression; a minus sign (−) above a symbol indicates that a blood sample was obtained for analysis. b, BsaAI analysis of the G1141C alteration in SLC6A8. Presence of the G1141C alteration is reflected in the presence of bands of 221 and 88 bp. Numbers above each lane refer back to the pedigree in panel a. L = size marker. For segregation analysis, intron-specific primers 8085IF (5′-AGTAAGCACCGCCGCCCTG-3′) and 8085IR (5′-TTTCAGCATTTCTATTGCATGTTC-3′) were designed to amplify exon 7 of SLC6A8, producing a product of 309 bp. PCRs were performed in a 30-μL volume that contained 52.5 ng genomic DNA, 1X Master Amp Buffer J (Epicentre), 50 μM dNTPs, 1 μM each primer, 1.5 U Sigma Taq polymerase (Boehringer Mannheim), and 0.33 μg TaqStart antibody (Clonetech). Annealing was done for 30 s at 58°C, extension for 30 s at 72°C, and the PCR was repeated for 30 cycles. PCR products were digested with BsaAI (New England Biolabs) for 16 h at 37°C with 10 U enzyme in buffer supplied by the manufacturer. Spermidine was added to a final concentration of 1 mM prior to digestion. Digested fragments were resolved by electrophoresis in a 2% agarose gel that contained ethidium bromide.
Table 1.
Summary of Clinical Findings[Note]
|
Male Patients |
||||||||
| Affected |
Female Patients |
|||||||
| II-5 | II-6 | II-7 | II-8 | II-16 | II-11(Unaffected) | II-12 | II-13 | |
| Age at examination (years) | 66 | 53 | 62 | 15 | 35 | 56 | 37 | 39 |
| Height (cm) [percentile] | 167.5 [10] | 162.5 [3] | 162.5 [3] | 162.5 [25] | NA | 175.5 [50] | NA | NA |
| Head circumference (cm) [percentile] | 55 [30] | 56.5 [60] | 54.5 [25] | 53.5 [25] | 55.3 [40] | 58.6 [90] | NA | NA |
| Ear length (cm) | 7.5 | 7.3 | 7.3 | NA | 6.3 | NA | NA | NA |
| Midface hypoplasia | + | + | + | NA | − | − | NA | NA |
| Stub thumb | + | + | + | NA | − | − | NA | NA |
| Hyperextensible joints | + | + | + | NA | − | − | NA | NA |
| Hypotonia | + | NA | + | NA | − | − | NA | NA |
| Gastrointestinal problem | + | + | + | − | + | − | 25 | NA |
| Seizures | + | + | + | + | + | − | − | − |
| Movement disorder | − | + | + | + | − | − | NA | NA |
| Speech problem | + | + | + | + | + | − | − | − |
| Behavior disturbance | + | + | + | + | + | − | + | − |
| Intelligence quotient | 16 | 4 | 4 | NA | 9 | NA | 69 | 67 |
| Urine creatine:creatinine ratioa | 2.5 | NA | 1.7 | NA | NA | .4 | NA | NA |
| Creatine uptakeb (pmol creatine/μg protein) | 6.2 | ND | 6.9 | ND | ND | ND | ND | ND |
Note.— NA = not available; ND = not determined.
Urinary creatine was measured using capillary electrophoresis as described by Clark et al. (2001). A Beckman Pace MDQ capillary electrophoresis system was utilized for electrophoresis, and creatinine and creatine were monitored using a UV detector. Peak heights and migration times were calculated using Beckman software on an IBM 486 computer. A 50-mM phosphate buffer at pH 5.5 was used as the run buffer. The urine-dilution buffer consisted of the run buffer supplemented with 2% EDTA. The 0.020-mm capillary was 50 cm long (J&W Scientific). Each run included a 3-min wash with 1N NaOH followed by a 1-min wash with distilled water and a 3-min wash with run buffer. Each sample was injected for 5 s (20 psi) and was electrophoresed for 8 min at 25 kV. Absorbance was monitored at 214 nm. Urine was diluted 1:1 with the urine-dilution buffer and was electrophoresed. A set of standards for both creatine and creatinine were loaded with each sample run. Known concentrations were 0.5 mM, 1 mM, 2.5 mM, 5 mM, 7.5 mM, and 10 mM. For each sample, the concentration of both creatine and creatinine were calculated using standard curves derived from the set of standards. The creatine:creatinine ratio was calculated for both affected patients and the unaffected brother and control subjects.
Creatine uptake in fibroblasts was measured after a 24-h incubation in 500 mM creatine as described elsewhere (Salomons et al. 2001). The average uptake levels in two controls was 15.8 pmol creatine/μg protein.
Two-point linkage analysis (Lathrop and Lalouel 1984) by use of 34 DNA markers spanning the X chromosome indicated linkage of XLMR in family K8085 to Xq28, since seven markers had a LOD score of 2.40 at zero recombination (data not shown). Recombination occurred in an unaffected brother (II-11) between DXS8091 and DXS113, thereby establishing DXS8091 as the limiting proximal flanking marker in Xq28 (fig. 1a).
Once linkage to Xq28 was established, candidate-gene testing was initiated. Twelve genes (FRAXA, FRAXE, MECP2, LICAM, GLUR3, GABRA3, GABRE1, VATPS1, XAP3, XAP5, XAP6, and SYBL1) were tested. However, no causative mutations were found (C. E. Schwartz, unpublished data). After a report appeared that a nonsense mutation (R514X) in the creatine-transporter gene, SLC6A8 (MIM 300036; GenBank accession number NM_005629; Nash et al. 1994), was observed in DNA from a mildly retarded boy and in DNA from three female relatives—two of whom showed learning disabilities (Cecil et al. 2001; Salomons et al. 2001)—this gene was examined since it fell within the linkage interval.
cDNA analysis of lymphoblasts of male patient II-5 was undertaken. Primers were used to amplify a 1.6-kb fragment, which contained the translated region minus exon 1 (Salomons et al. 2001). This template was employed for a seminested PCR of 5′ and 3′ fragments of 1.1 kb and 530 bp, respectively. Direct sequence analysis of the sense strand of the 5′ fragment consisting of exons 2–8 indicated the apparent existence of a mixed population of amplicons, because of unreadability of the sequence after base 1130 (GenBank accession number NM_005629; data not shown). Primers were then designed to amplify a 250-bp cDNA product spanning nucleotides 1020–1269. Gel electrophoresis of this amplicon revealed the expected band of 250 bp, as well as a smaller band of 237 bp, in three members of family K8085. The 237-bp band was absent in a control sample (fig. 2a). The cDNA PCR product from patient II-5 was subcloned, allowing purification of the two different PCR fragments (fig. 2a). Sequence analysis of the 250-bp fragment revealed the presence of a G→C transversion at position 1141 (fig. 2b). The smaller fragment was found to contain a deletion of 13 bp (positions 1129–1141; fig. 2b). Sequencing of the genomic DNA from male patients II-3 and II-5 confirmed the G1141C alteration (data not shown). This G→C alteration gives rise to an arginine substitution for a glycine at position 381 (G381R).
Figure 2.
RT-PCR analysis of exons 7 and 8 of the SLC6A8 cDNA. a, Agarose gel electrophoresis of RT-PCR products generated with lymphoblasts from patients II-2, II-5, and II-7 and from a control subject. RT-PCR products from patient II-5 were subcloned, allowing purification of the two different amplicons (i.e., subclones A and B). b, Sequence analysis of RT-PCR product from a control subject, subclone A from patient II-5, and subclone B from patient II-5. The letters “G” and “C” atop a yellow screen, in the normal and subclone A diagrams, respectively, indicate the G→C transition at position 1141. The amino acids in boldface indicate the G381R missense mutation that results from the G1141C alteration. The arrow in the subclone B diagram indicates the location of the 13-bp deletion in the RT-PCR subclone from patient II-5. The two letters “G” atop a blue screen, in the normal and subclone B diagrams, indicate the position of the deletion relative to the normal exon 7 sequence. The novel amino acids that are substituted as a result of the 13-bp deletion are in italic. c, The alternative 5′ splice site in exon 7 predicted in family K8085. An arrow indicates the splice site. The predicted amino acid sequence for the altered SLC6A8 protein is given below the novel mRNA sequence. Total RNA was extracted from lymphoblastoid cell lines (∼3×106 cells) by use of TRIzol LS (Life Technologies) according to the manufacture’s protocol. Samples were treated with DNase I/Amp (Life Technologies) for 15 min at room temperature and were purified using the RNeasy Mini Kit (QIAGEN). Approximately 3 μg total RNA was reversed transcribed into cDNA by use of random hexamers (Super Script Preamplification System; Life Technologies). Primers 8085F (5′-GCCATCATCCTGGCTCTCATC-3′) and 8085R (5′-CCTCCACACCTACAAACTGGC-3′) were designed, flanking the region of the presumed cDNA alteration in family K8085, which produced an amplicon of 250 bp. PCR was performed in a 50-μL volume containing 105 ng cDNA, 1X Sigma buffer (Boehringer Mannheim), 50 μM dNTPs, 10 μM each primer, 1.5 U Sigma Taq polymerase, and 0.33 μg TaqStart antibody. Amplification was done in a 9600 Thermocycler (Perkin Elmer/Cetus) by use of the following parameters: 3 min at 96°C, 30 s at 94°C, annealing for 30 s at 67°C, extension for 30 s at 72°C for 30 cycles, and a final extension for 7 min at 72°C. PCR products were checked by electrophoresis on an 8% acrylamide/15% glycerol minigel (Hoefer). Products were purified using the Qiagen PCR Purification Kit (Qiagen). Amplicons were cloned into Escherichia coli One Shot INVαF′ competent cells using the Original TA Cloning Kit (Invitrogen). Clones containing the insertion were detected using LB ampicillin agar plates containing X-gal. Colonies containing the insertion were grown overnight in 5 ml LB broth containing ampicillin. Plasmid DNA was isolated using the Qiagen Spin Miniprep Kit (Qiagen). Direct sequence analysis of RT-PCR products was performed using Bigdye terminators and analysis on an ABI 310 machine (Perkin Elmer). Subcloned cDNA was sequenced using a Thermo Sequenase fluorescent-labeled primer cycle sequencing kit (Amersham) and FAM (5-fluorouracil, Adriamycin, and mitomycin C)–labeled M13 (−40) forward and reverse primers (Pharmacia Biotech) and the ALF DNA sequencer.
Additionally, the G at nucleotide position 1141 occupies the −1 base for the 5′ splice site of intron 7. Comparison of the sequence of the lower-sized transcript with the genomic sequence indicated that the 13-bp deletion likely resulted from utilization of an alternative upstream splice site within exon 7 (fig. 2c). The consensus value (CV) of this splice junction (AGgtggca) is 0.661, which is 93% of the CV of 0.712 for the splice junction (ACgtaggg) that contains the 1141 G→C transversion (Cooper and Krawczak 1995).
The G1141C change creates a BsaAI site in the mutant genomic DNA. BsaAI digestion of amplified genomic DNA was used to confirm the sequence data and to check for segregation of the mutation in family K8085. As seen in figure 1b, the mutation did segregate with XLMR in family K8085, and the presence of the BsaAI site was consistent with the haplotype data that was derived from the linkage analysis (fig. 1a). Furthermore, BsaAI digestion of 400 X chromosomes from unaffected male individuals failed to detect this alteration, indicating the G→C transversion was likely not a rare polymorphism.
Subsequent biochemical analyses confirmed a defect in creatine metabolism in family K8085. The level of urinary creatine was substantially increased in affected male patients II-3 and II-5. Their urinary creatine:creatinine ratios were 2.5 and 1.73, respectively, whereas the ratio for their unaffected brother (II-9) was 0.39 (table 1). This ratio ranged from 0.3 to 0.7 in eight adult control subjects.
Creatine uptake in fibroblasts from patients II-5 and II-7 was measured after a 24-h incubation in the presence of either a physiological level of 25 mM or a higher level of 50 mM creatine (Salomons et al. 2001). No uptake could be measured in the presence of 25 mM creatine, and, at 500 mM, the level of uptake was approximately one-fourth that of control cells (table 1).
The G114C transversion in SLC6A8 has two effects:
-
1.
The base substitution gives rise to a missense mutation in which glycine is replaced by arginine at position 381 (G381R). G381 is part of a sequence of 12 invariant amino acids (VAASGPGLAFLA), which is conserved among related proteins in several species (fig. 3). This strong conservation likely reflects the important nature of this region of the gene.
-
2.
The G1141C transversion occupies the −1 position of the exon 7/intron 7 splice junction. Substitution of the cytosine results in a substantial reduction in the CV of the splice junction, from 0.847 to 0.712 (Cooper and Krawczak 1995). This, in turn, causes an alternative splice site upstream at position 1129 to be utilized part of the time, since its CV of 0.661 is close in value (ratio + 1.02). Utilization of the alternative splice site causes a deletion of 13 bp, which would cause a substitution of 14 novel amino acids and truncation of the protein at position 390 (fig. 2c). As a result, a large portion of the 3′ end of the protein, containing 145 amino acids that comprise transmembrane (TM) domains 8–12 as well as the carboxy end, would be absent from the SLC6A8 protein.
Figure 3.
Comparison of a partial amino acid sequence of SLC6A8 and with other members of the taurine-subfamily transporters. The presented amino acid sequence covers the extracellular domain between TM domains 7 and 8. The glycine at position 381 is shaded, and the 12 invariant amino acids are underlined.
The clinical presentation (i.e., severe mental retardation with speech and behavioral abnormalities, seizures, variable short stature, and midface hypoplasia) of the male patients with the G1141C mutation in SLC6A8 is more severe than the presentation by the mildly mentally retarded boy with a nonsense mutation (R514X) in SLC6A8, who has been described elsewhere (Cecil et al. 2001; Salomons et al. 2001). This may reflect a genotype-phenotype correlation. The R514X mutation results in a truncated protein missing TM domains 11 and 12, as well as the cytoplasmic carboxy end of the gene. The presence of creatine-transporter molecules that contain the G381R missense mutation may lead to a dominant negative effect and a more severe presentation of the disease in family K8085 when compared with the presence of only a nonsense mutation in the previously reported male patient with the R514X mutation. However, studies of in vitro creatine uptake in fibroblasts from two male patients in family K8085 and from the male patient with the R514X mutation showed equal impairment in creatine uptake, both when cultured in a physiological level of creatine of 25 mM and at a higher level of 500 mM. Alternatively, severe mental retardation in the men in family K8085 may reflect the possible progressive nature of long-term low cellular-creatine levels, although the limited history available on the male patients did not reveal any cognitive regression.
The presence of a mutation in SLC6A8 may result in muscle weakness or other symptoms of muscle impairment due to an inability to generate ATP from phosphocreatine. The hypotonia and myopathic facies, in one male patient, and ptosis, in another, are suggestive of muscle involvement. The hypotonia could be cerebral in origin, as are the extrapyramidal movements. Unfortunately, no muscle studies were conducted to determine if muscle pathology existed, and studies of muscle pathology and function were not conducted in the patient with a creatine transporter defect, who has been described elsewhere (Salomons et al. 2001). Since the 1930s, an excess of male patients has been noted in the population with mental retardation (Penrose 1938). This excess has been attributed to genes on the X chromosome (Lehrke 1974; Turner and Turner 1974; Stevenson et al. 1990). This concept has been supported by the clinical and molecular delineation of numerous syndromic and nonsyndromic forms of XLMR (Schwartz 1993; Chelly 2000). However, the recognized forms of XLMR do not account for the 20%–40% excess of male patients who have been reported in the major surveys conducted over the past 20 years. An excess of 30% should result in a frequency of 15% for all cases of mental retardation. However, the major surveys of mental retardation report <5% of cases resulting from XLMR (Moser and Wolf 1971; Kaveggia et al. 1975; Gustavson et al. 1977; Laxova et al. 1977; Hunter et al. 1980; Anderson et al. 1996).
Numerous factors may be responsible for the difference between the estimated prevalence and the detectable prevalence of XLMR: First, many XLMR entities are nonsyndromic (i.e., MRXS), and, thus, physical examination provides no clues for clinical diagnosis. Second, if a significant proportion of XLMR is due to new mutations, the pedigree information will not suggest X-linked inheritance. Third, very few entities have associated metabolic abnormalities, which might bring them to attention.
There are seven genes associated with MRXS—IRL1RAP, TM4SF2, OPHN1, PAK3, ARHGEF6, FMR2, and GDI1 (Chelly and Mandel 2001)—and three genes associated with MRX—RSK2 (Merienne et al. 1999), MECP2 (Meloni et al. 2000; Orrico et al. 2000; Couvert et al. 2001), and FGD1 (Lebel et al., in press). Perhaps with the exception of RSK2, defects in these genes are not detectable by use of a biochemical or metabolic assay. Identification of a XLMR entity that is associated with a mutation in the creatine transporter will allow testing for SLC6A8 mutations in male patients with mental retardation of unknown etiology.
This report, in conjunction with the that of publication of a report of two female siblings with arginine:glycine amidinotrasferase (AGAT) deficiency (Item et al. 2001) and those of the already known guanidinoacetate methyltransferase (GAMT) deficiency (Stöckler et al. 1996; Von Figura et al. 2000), clearly indicates the importance of creatine metabolism in brain function. In all three errors of creatine metabolism, mental impairment of varying degree is observed. This would be in agreement with already-existing literature that creatine metabolism plays a significant role in brain function beyond its use as a high-energy phosphate store. There is evidence that there exists direct coupling of creatine kinase with neurotransmitter release (Wallimann and Hemmer 1994), with growth cone migration (Wang et al. 1998) and restoration of ion gradients before and after depolarization (Wallimann and Hemmer 1994). Thus, brain function appears to be linked in a number of ways with creatine metabolism, although the relationships are not yet fully understood.
All three errors of creatine-metabolism conditions lend themselves to biochemical tests, since guanidinoacetate and creatine levels can be measured in either urine or plasma. As a result, diagnosis of these inborn-error disorders can be made, which is important, since two of them, AGAT and GAMT deficiencies, may be amenable to oral creatine supplementation (Stöckler et al. 1996; Item et al. 2001).
Acknowledgments
We are very appreciative of the cooperation of members of family K8085 in our XLMR research. Dr. Ruth Amir (Baylor University, Houston), did the mutation analyses of GLUR3, GABRA3, GABRE1, VATPS1, and SYBL1. Dr. Joe Gleeson (University of Califormia–San Diego) did the mutation analyses of XAP3 and XAP6. Susan Daniels and Tonya Moss of the Core Facility of the Center for Molecular Studies, Greenwood Genetic Center assisted with sequencing and isolation of RNA from lymphoblasts respectively. Mrs. Silvy, J. M. van Dooren, and Dana Bunea provided technical assistance in the Metabolic Unit, VU University Medical Center, Amsterdam. This work was supported, in part, by National Institute of Child Health and Human Development grant HD26202 (to C.E.S. and H.A.L.), National Institute of Mental Health grant MH57840 (to R.E.S. and C.E.S.), and a grant from the South Carolina Department of Disabilities and Special Needs.
Electronic-Database Information
Accession numbers and URLs for data in this report are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SLC6A8 [accession number NM_005629] and the sequence after base 1130 [accession number NM_005629])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLC6A8 [MIM 300036])
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 [DOI] [PubMed] [Google Scholar]
- Anderson G, Schroer RJ, Stevenson RE (1996) Mental retardation in South Carolina. II. Causation. Proc Greenwood Genet Cent 15:32–44 [Google Scholar]
- Armfield K, Nelson R, Lubs HA, Hane B, Schroer RJ, Arena F, Schwartz CE, Stevenson RE (1999) X-linked mental retardation syndrome with short stature, small hands and feet, seizures, cleft palate, and glaucoma is linked to Xq28. Am J Med Genet 85:236–242 [DOI] [PubMed] [Google Scholar]
- Cecil KM, Salomons GS, Ball WS Jr, Wong B, Chuck G, Verhoeven NM, Jakobs C, DeGrauw TJ (2001) Irreversible brain creatine deficiency with elevated serum and urine creatine: a creatine transporter defect? Ann Neurol 49:401–404 [DOI] [PubMed] [Google Scholar]
- Chelly J (2000) MRX review. Am J Med Genet 94:364–366 [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL (2001) Monogenic causes of X-linked mental retardation. Nat Rev Genet 2:669–680 [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Hamel BC, Neri G (2001) XLMR genes: update 2000. Eur J Hum Genet 9:71–81 [DOI] [PubMed] [Google Scholar]
- Clark EA, Fanguy JC, Henry CS (2001) High-throughput multi-analyte screening for renal disease using capillary electrophoresis. J Pharm Biomed Anal 25:795–801 [DOI] [PubMed] [Google Scholar]
- Cooper D, Krawczak M (1995) Human gene mutation. Bios Scientific, Oxford, p 224 [Google Scholar]
- Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre AC, Souville I, Steffann J, des Portes V, Ropers HH, Yntema HG, Fryns JP, Briault S, Chelly J, Cherif B (2001) MECP2 is highly mutated in X-linked mental retardation. Hum Mol Genet 10:941–946 [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Menegon A, Lo Nigro C, Gasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, Tandon A, Valtorta F, Balch WE, Chelly J, Toniolo D (1998) Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet 19:134–139 [DOI] [PubMed] [Google Scholar]
- Gustavson K-H, Hagberg B, Hagberg G, Sars K (1977) Severe mental retardation in a Swedish county. II. Etiologic and pathogenetic aspects of children born 1959–1970. Neuropädiatrie 8:293–304 [DOI] [PubMed] [Google Scholar]
- Hunter AG, Evans JA, Thompson DR, Ramsay S (1980) A study of institutionalized mentally retarded patients in Manitoba. I: Classification and preventability. Dev Med Child Neurol 22:145–162 [DOI] [PubMed] [Google Scholar]
- Item CB, Stockler-Ipsiroglu S, Stromberger C, Muhl A, Alessandri MG, Bianchi MC, Tosetti M, Fornai F, Cioni G (2001) Arginine:glycine amidinotransferase deficiency: the third inborn error of creatine metabolism in humans. Am J Hum Genet 69:1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 [DOI] [PubMed] [Google Scholar]
- Kaveggia EG, Durkin MV, Pendleton E, Opitz JM (1975) Diagnostic/genetic studies on 1224 patients with severe mental retardation. In: Primrose DAA (ed) Proceedings of the 3rd International Congress of the International Association for the Scientific Study of Mental Deficiency. Polish Medical, Warsaw, pp 82–93 [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Laxova R, Ridler MA, Bowen-Bravery M (1977) An etiological survey of the severely retarded Hertfordshire children who were born between January 1, 1965 and December 31, 1967. Am J Med Genet 1:75–86 [DOI] [PubMed] [Google Scholar]
- Lebel RR, May M, Pouls S, Lubs HA, Stevenson RE, Schwartz CE. Nonsyndromic X-linked mental retardation associated with a missense mutation (P312L) in the FGD1 gene. Clin Genet (in press) [DOI] [PubMed] [Google Scholar]
- Lehrke RG (1974) X-linked mental retardation and verbal disability. Birth Defects Orig Art Ser 10:1–100 [PubMed] [Google Scholar]
- Lubs H, Chiurazzi P, Arena J, Schwartz C, Tranebjaerg L, Neri G (1999) XLMR genes: update 1998. Am J Med Genet 83:237–247 [PubMed] [Google Scholar]
- Meloni I, Bruttini M, Longo I, Mari F, Rizzolio F, D’Adamo P, Denvriendt K, Fryns JP, Toniolo D, Renieri A (2000) A mutation in the Rett syndrome gene, MECP2, causes X-linked mental retardation and progressive spasticity in males. Am J Hum Genet 67:982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Jacquot S, Pannetier S, Zeniou M, Bankier A, Gecz J, Mandel JL, Mulley J, Sassone-Corsi P, Hanauer A (1999) A missense mutation in RPS6KA3 (RSK2) responsible for non-specific mental retardation. Nat Genet 22:13–14 [DOI] [PubMed] [Google Scholar]
- Moser HW, Wolf PA (1971) The nosology of mental retardation: including the report of a survey of 1378 mentally retarded individuals at the Walter E. Fernald State School. Birth Defects Orig Art Ser 7:117–134 [PubMed] [Google Scholar]
- Nash SR, Giros B, Kingsmore SF, Rochelle JM, Suter ST, Gregor P, Seldin MF, Caron MG (1994) Cloning, pharmacological characterization, and genomic localization of the human creatine transporter. Receptors Channels 2:165–174 [PubMed] [Google Scholar]
- Neri G, Gurrieri F, Gal A, Lubs HA (1991) XLMR genes: update 1990. Am J Med Genet 38:186–189 [DOI] [PubMed] [Google Scholar]
- Orrico A, Lam C, Galli L, Dotti MT, Hayek G, Tong SF, Poon PM, Zappella M, Federico A, Sorrentino V (2000) MECP2 mutation in male patients with non-specific X-linked mental retardation. FEBS Lett 481:285–288 [DOI] [PubMed] [Google Scholar]
- Penrose LS (1938) A clinical and genetic study of 1280 cases of mental defect. In: Special report series, Medical Research Council, no 229. Her Majesty’s Stationery Office, London [Google Scholar]
- Rosenthal A, Jouet M, Kenwrick S (1992) Aberrant splicing of neural cell adhesion molecule L1 mRNA in a family with X-linked hydrocephalus. Nat Genet 2:107–112 [DOI] [PubMed] [Google Scholar]
- Salomons GS, van Dooren SJ, Verhoeven NM, Cecil KM, Ball WS, Degrauw TJ, Jakobs C (2001) X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am J Hum Genet 68:1497–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CE (1993) X-linked mental retardation: in pursuit of a gene map. Am J Hum Genet 52:1025–1031 [PMC free article] [PubMed] [Google Scholar]
- Stevenson RE, Arena JF, Ouzts E, Gibson A, Shokeir MH, Vnencak-Jones C, Lubs HA, May M, Schwartz CE (1998) Renpenning syndrome maps to Xp11. Am J Hum Genet 62:1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RE, Goodman HO, Schwartz CE, Simensen RJ, McLean WT Jr, Herndon CN (1990) Allan-Herndon syndrome. I. Clinical studies. Am J Hum Genet 47:446–453 [PMC free article] [PubMed] [Google Scholar]
- Stevenson RE, Schwartz CE, Schroer RJ (2000) X-linked mental retardation. New York: Oxford University Press [Google Scholar]
- Stöckler S, Hanefeld F, Frahm J (1996) Creatine replacement therapy in guanidinoacetate methyltransferase deficiency, a novel inborn error of metabolism. Lancet 348:789–790 [DOI] [PubMed] [Google Scholar]
- Turner G, Turner B (1974) X-linked mental retardation. J Med Genet 11:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vits L, Van Camp G, Coucke P, Fransen E, De Boulle K, Reyniers E, Korn B, Poustka A, Wilson G, Schrander-Stumpel C, et al (1994) MASA syndrome is due to mutations in the neural cell adhesion gene L1CAM. Nat Genet 7:408–413 [DOI] [PubMed] [Google Scholar]
- Von Figura K, Hanefeld F, Isbrandt D, Stöckler-Ipsiroglu S (2000) Guanidinoacetate methyltransferase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. McGraw-Hill, New York, pp 1897–1908 [Google Scholar]
- Wallimann T, Hemmer W (1994) Creatine kinase in non-muscle tissues and cells. Mol Cell Biochem 133–134:193–220 [DOI] [PubMed] [Google Scholar]
- Wang Y-ME, Esbensen P, Bentley D (1998) Arginine kinase expression and localization in growth cone migration. J Neurosci 18:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]