Abstract
Juvenile polyposis (JP) is an autosomal dominant syndrome in which affected patients develop upper- and/or lower-gastrointestinal (GI) polyps. A subset of families with JP have germline mutations in the SMAD4 (MADH4) gene and are at increased risk of GI cancers. To date, six families with JP have been described as having the same SMAD4 deletion (1244–1247delAGAC). The objective of the present study is to determine whether this deletion is a common ancestral mutation or a mutational hotspot. DNA from members of four families with JP, from Iowa, Mississippi, Texas, and Finland, that had this 4-bp deletion was used to genotype 15 simple tandem repeat polymorphism (STRP) markers flanking the SMAD4 gene, including 2 new STRPs within 6.3 and 70.9 kb of the deletion. Haplotypes cosegregating with JP in each family were constructed, and the distances of the closest markers were determined from the draft sequence of the human genome. No common haplotype was observed in these four families with JP. A 14-bp region containing the deletion had four direct repeats and one inverted repeat. Because no common ancestor was suggested by haplotype analysis and the sequence flanking the deletion contains repeats frequently associated with microdeletions, this common SMAD4 deletion in JP most likely represents a mutational hotspot.
Juvenile polyps were first recognized as comprising a distinct histopathologic entity in 1957 (Horrilleno et al. 1957), and a heritable form was described as “juvenile polyposis coli” in 1964 (McColl et al. 1964). In 1974, juvenile polyposis (JP [MIM 174900]) was divided into three different subtypes, including JP coli, JP of infancy, and generalized JP (Sachatello et al. 1974). In 1975, it was established that there was a significant risk of gastrointestinal (GI) malignancy in members of a large family with generalized JP, suggesting that the prevalent belief that these polyps were benign with no malignant potential was incorrect (Stemper et al. 1975). In 1998, genetic linkage to markers on chromosome 18q21.1 was found in the same family (Howe et al. 1998b), and germline mutations were identified in the SMAD4 gene (MIM 600993) (Howe et al. 1998c). In the latter report, three patients with JP were described who had the same 4-bp deletion in SMAD4 exon 9. In 1999, three additional unrelated patients with JP were reported as having this same deletion (Friedl et al. 1999; Roth et al. 1999).
Common ancestry has been implicated as the cause for the high prevalence of specific MLH1 (MIM 120436) mutations in Finland (Moisio et al. 1996), BRCA1 (MIM 113705) mutations in Quebec (Tonin et al. 1998), and BRCA2 (MIM 600185) mutations in northern Europe (Neuhausen et al. 1998). Another mechanism for common mutations in apparently unrelated families is that they result from mutational hotspots within genes (Cooper and Krawczak 1990; Krawczak and Cooper 1991). Among 60 unrelated patients with familial adenomatous polyposis of the colon (MIM 175100), 5 patients shared the same 5-bp deletion (3183–3187delACAAA), and 5 additional patients shared a different 5-bp deletion (3926–3930delAAAAG). Since 3 of these 10 patients' parents did not have these mutations, the high frequency of these specific changes could not be attributed entirely to founder mutations. The latter mutation occurred within a tandem repeat of 5 nt (AAAAGAAAAG), with one repeat being lost, which may have resulted from slippage of DNA polymerase (Groden et al. 1993). In multiple endocrine neoplasia type 2A (MIM 171400), 16 families with a C634R missense mutation were determined to have 11 separate haplotypes by use of six markers from a 318-kb region containing the RET gene (MIM 164761), suggesting that this common substitution was not due to a founder effect (Gardner et al. 1994). The objective of the present study is to apply haplotype analysis to the families with JP that shared a 4-bp deletion, to determine whether this is the result of either an ancestral mutation or a mutational hotspot.
Informed consent was obtained from family members for genetic studies, as well as for review of medical records and pathology specimens. Each of the four families in this report are white, have had GI cancers, and do not share surnames nor have known common relatives. The Iowa kindred with JP is an extended white family comprising 29 members with JP or GI cancer (Stemper et al. 1975). Eleven individuals had developed colorectal cancer and six had developed upper-GI (four stomach, one duodenum, and one pancreas) cancers (Howe et al. 1998a). The Mississippi kindred with JP has 11 members with documented JP, of whom one developed gastric cancer and another developed colorectal cancer (Subramony et al. 1994; Scott-Conner et al. 1995). Three other members of this family died prior to the present study and had had a history of unspecified GI malignancies. The Finnish kindred with JP consists of 11 affected individuals from five generations. Four individuals are known to have upper-GI polyps, one died of gastric cancer, and one died of metastatic colorectal cancer. One other member died of metastatic adenocarcinoma of the liver of unknown primary site and had gastric, esophageal, and colonic juvenile polyps. Three additional affected family members each had one of the following: leukemia, goiter, and a benign ovarian neoplasm of unknown histology. The Texas kindred has five known affected members from three generations. The matriarch of the family had rectal, colonic, gastric, and duodenal polyps and has four affected offspring who had colonic polyps but no history of upper-GI polyps. One of these offspring developed colon cancer at age 30 years.
DNA was extracted from peripheral blood by use of a salting-out technique (Miller et al. 1988). DNA from 2 affected members (IV-22 and V-2) of the Iowa kindred (Howe et al. 1998b) were amplified by PCR, along with DNA from 10 members (5 affected, 4 at risk, and 1 spouse) of the Mississippi kindred, 2 affected siblings from the Finnish kindred, and 7 members (4 affected and 3 unaffected) of the Texas kindred with JP. PCR was performed in a 10-μl volume that contained 25 ng DNA from each individual, 0.5 pmol of each primer, buffer (1.65 mM MgCl2, 50 mM KCl, 10 mM Tris, and 5% glycerol), 100 μmol each dNTP, and 0.2 U Taq DNA polymerase. Amplification was performed in a thermocycler for 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, for a total of 35 cycles. Samples were electrophoresed through 6% denaturing polyacrylamide gels for 2–3 h at 60 W, the gels were silver stained (Bassam et al. 1991), and genotypes were determined from the gels.
SMAD4 exons and their adjacent intron-exon boundaries were used to perform BLAST searches of the draft sequence of the human genome (International Human Genome Sequencing Consortium 2001) and the Celera genome sequence (Venter et al. 2001). The genomic structure of the SMAD4 gene was determined from the draft sequence by alignment of exons through use of the Sequencher program (Gene Codes). The genome-sequence segments that contained SMAD4 were then searched for the sequences of previously identified chromosome 18q21 STRPs. New STRPs were then sought by alignment of 20-bp sequences of different of dinucleotide-, trinucleotide-, and tetranucleotide-repeat elements. When repeats were identified in the genome sequence, flanking primer pairs were selected using the Primer3 program. New candidate STRPs were amplified from 51 control patients, to determine their allele frequencies and heterozygosity, and were then tested in families with JP. Genetic-mapping data for each known marker were obtained from the Center for Medical Genetics sex-averaged map and complemented physical-mapping data from the Whitehead Institute Center for Genome Research human physical-mapping project (Human BAC Mapping Data). Distances between each marker and the SMAD4 gene were determined by alignment of the human-genome sequences through use of the Sequencher program. Two novel polymorphic STRPs were identified from BAC clone RP11-729G3 (GenBank accession number AP001374) and Celera scaffold segments GA_x2KMHMRU853 and GA_x2KMHMRU8FE. SMAD4GATA (GenBank accession number AF364127) consists of 11 repeats in the BAC sequence and has three alleles (table 1). The other STRP is an AT/CA repeat (SMAD4AT/CA [GenBank accession number AF364126]), with 18 ATs, followed by 23 CAs in the BAC sequence, and has 14 alleles.
Table 1.
Haplotypes and Map Positions of Chromosome 18 Markers in Kindreds with JP
|
Alleles in Kindred from |
|||||||
| Marker | Iowa | Mississippi | Texas | Finland | Distancea(cM) | Distance fromSMAD4 Exon 1(kb) | Heterozygosity |
| D18S970 | 3 | 6 | 3 | 3 | 93.9 | … | .67b |
| D18S1099 | 4 | 3 | 3 | 3, 5c | 98.0 | … | .58d |
| D18S474 | 8 | 4 | 1 | 8, 10c | 98.0 | 121.2 | .82d |
| D18S1110 | 3 | 3 | 5 | 6 | 98.0 | 37.0 | .75d |
| SMAD4 exon 11 | 31.4 | … | |||||
| SMAD4AT/CA | 4 | 7 | 9 | 13 | 26.3 | .90 | |
| Deletion | 4 bp | 4 bp | 4 bp | 4 bp | 20.2 | … | |
| SMAD4GATA | 2 | 3 | 2, 3c | 3 | 50.7 | .55 | |
| D18S46 | 7 | 5 | 6 | 10 | <99.2e | 132.9 | .80d |
| GATA06 | 3 | 2 | 5 | 4 | 104.1 | … | .80b |
| D18S1156 | 3 | 2 | 2 | 2 | 104.1 | … | .54d |
| D18S851 | 3 | 2 | 5 | 4 | 105.5 | … | .55b |
| D18S484 | 1 | 1 | 3 | 3 | 105.5 | … | .72d |
| D18S539 | 3 | 2 | 5 | 4 | 105.5 | … | .63b |
| D18S487 | 5 | 5 | 6 | 6 | 106.3 | … | .81d |
| D18S846 | 2 | 2 | 2 | 2 | 107.5 | … | .38b |
| D18S977 | 1 | 4 | 4 | 4 | ∼110 | … | .92b |
Based on the Center for Medical Genetics sex-averaged map.
From the Cooperative Human Linkage Center.
The specific allele cosegregating with JP could not be determined.
From the Genome Database.
Map position of D18S363, which is 165.5 kb telomeric to D18S46 (see text).
The markers used to determine haplotypes are listed in table 1 according to their physical- and genetic-map positions. In addition, five STRPs mapped to BACs where human-genome draft sequence data were available, and their distances from SMAD4 could be calculated with precision. Uncertainties created by gaps and the lack of ordered segments in the BAC sequences were checked against the Celera sequence (Venter et al. 2001). SMAD4AT/CA was within the intron that is between exons 9 and 10, just 6.1 kb from the 4-bp deletion found in these four families. D18S363 was found to be 298 kb upstream from exon 1, which is consistent with the finding by Hahn et al. (1996) but discordant with that by Eppert et al. (1996), who placed D18S363 centromeric to SMAD4. The last 50 kb of Celera scaffold GA_x2KMHMRU853 contained exons 1–11 of the SMAD4 gene, and the following genes were more telomeric: SMAD4GATA (50.7 kb from exon 1), D18S46 (133 kb), and D18S363 (298 kb). The centromeric STRP D18S1110 was present on both RP11-729G3 and Celera scaffold GA_x2KMHMRU8FE; D18S474 was also present on GA_x2KMHMRU8FE but not on RP11-729G3. The 11 exons of the SMAD4 gene were distributed among 31,445 bp of genomic DNA, with the largest intron spanning 9,479 bp between exons 9 and 10, which contained the SMAD4AT/CA STRP. No matches were found for D18S1099, D18S470, D18S473, D18S970, D18S1156, and D18S851.
The results of haplotype analysis for the four families are summarized in table 1. For the five markers centromeric to the deletion, there was no shared haplotype between the families. The alleles in each family are all different for the SMAD4AT/CA marker, which is only 6.1 kb downstream from the common deletion. There was also no evidence of a shared haplotype with the markers that were telomeric to the deletion.
We evaluated the possibility that the 4-bp deletion could have been commonly inherited despite the absence of a shared haplotype. The decrease in disequilibrium over time decays as a function of (1-θ)n (Sham 1997), so that when a common haplotype is responsible for the mutations that are found in two families, the probability that both cases share the same haplotype is (1-θ)2n. The probability that none of the six possible haplotype pairs from four families are shared is then [1-(1-θ)2n]6. We set this probability to be .05 (the usual significance criterion) and consider the marker SMAD4AT/CA to be 0.006 cM from the 4-bp deletion. Solving for the number of generations, n, we find that, with ⩽5% probability, ⩾7,781 generations (233,430 years if one assumes 30 years per generation) must have passed for there not to be a shared haplotype among the cases if they had a common ancestor. Another mechanism for changes in alleles that originate from an ancestral allele is mutation at the microsatellite loci, which may vary in frequency from 0.2 to 3.3×10-3 (Sajantila et al. 1999). With the varied haplotypes flanking the common mutation seen in the present study, both recombination and mutation of microsatellites are less likely explanations than a mutational hotspot in exon 9 of the SMAD4 gene. Owing to the proximity of these markers, we conclude that any common ancestor would likely date from the dawn of the modern human species, which has been estimated to be between 171,000 and 479,000 years ago (Ingman et al. 2000).
Of the four families that we report, all include members with upper-GI juvenile polyps and colorectal cancer. In three of the four families, there are individuals with gastric cancer, and consistent extraintestinal phenotypic features have not been noted in these families. Friedl et al. (1999) also found two families with the same 4-bp deletion, one of which included three affected individuals with stomach cancer and two with colon cancer (Hofting et al. 1993). The other family was described as having two affected brothers and an asymptomatic carrier father. These findings suggest that this 4-bp deletion is associated with a more aggressive JP phenotype, with generalized JP and upper- and lower-GI cancers.
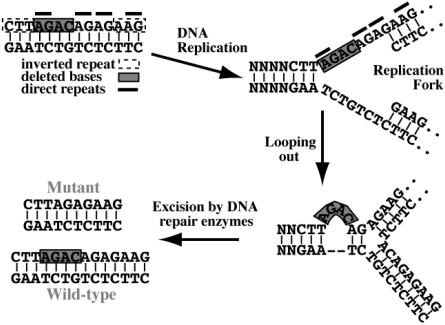
Ancestral mutations are relatively common in some diseases, such as hereditary nonpolyposis colorectal cancer (Moisio et al. 1996), cystic fibrosis (Morral et al. 1994), and breast/ovarian cancer (Neuhausen et al. 1998; Tonin et al. 1998). However, in many other diseases, new mutations appear to arise from an increased susceptibility due to specific DNA sequences. In this 4-bp deletion, the same mutant sequence could result if the deletion began at any of four consecutive bases. Krawczak and Cooper (1991) examined 60 deletions of <20 bp that were known to cause human diseases, to identify sequence characteristics that might predispose genes to microdeletion. They found that direct repeats flanked or spanned the deletion in two-thirds of cases, with the most common repeat length being 2–4 bp. In the deletion that we report, a direct repeat of AG is present in the first 2 bp of the deletion, and another follows the deleted AGAC. There is another AG repeat immediately after the second AG, and there is a fourth AG repeat 2 bp downstream from this. One proposed mechanism of this deletion would be slippage of DNA polymerase through these repeat sequences. Another proposed mechanism is slipped mispairing, in which the second repeat pairs with the complement of the first repeat at a DNA replication fork, which results in the repeat and intervening sequence looping outward and being excised by repair enzymes (fig. 1). Palindromic (i.e., inverted repeat) sequences are another mechanism that causes loops on a single DNA strand and were present flanking the deletion in this case (CTT 2 bp 5′ and AAG 2 bp 3′ to the deletion).
Figure 1.
Possible mechanism of the 4-bp deletion through slipped mispairing. At the replication fork, the second AG direct repeat can mispair with the complement of the first AG repeat, causing a loop on the upper strand that is excised by DNA repair enzymes. The resultant upper-strand copies will have the 4-bp deletion, whereas the lower-strand copies will be the wild-type sequence.
In the present study, the finding of different haplotypes in four families with markers in close proximity to the SMAD4 deletion effectively excludes the possibility of a common ancestral mutation. The presence of direct and inverted repeats flanking the 4-bp deletion, which are features commonly observed in microdeletions, suggest that this area is instead a mutational hotspot. This mutation leads to a more virulent form of JP, with a high incidence of gastric and colonic polyposis, as well as to gastric and colorectal cancer.
Acknowledgments
The present study was supported by a generous grant from the Roy J. Carver Charitable Trust, the American College of Surgeons Owen H. Wangensteen Faculty Research Fellowship, National Institutes of Health grant CA62924, and American Cancer Society grant RPG9903001. We thank Jennifer Bair and Mary Anderson for technical assistance and Dr. Sanjay Shete for helpful statistical discussions.
Electronic-Database Information
Accession numbers and URLs for data in this report are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Celera Publication Site, http://public.celera.com/cds/login.cfm (for human genome sequence)
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html (for comprehensive human genetic maps)
- Cooperative Human Linkage Center, The, http://www.chlc.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SMAD4 cDNA [accession number NM_005359], BAC clone RP11-729G3 [accession number AP001374], SMAD4GATA [accession number AF364127], and SMAD4AT/CA [accession number AF364126])
- Genome Database, The, http://gdbwww.gdb.org/
- Human BAC Mapping Data, http://www-genome.wi.mit.edu/seq/mapping.html (for the Whitehead Institute Center for Genome Research human physical-mapping project)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for JP [MIM 174900], SMAD4 [MIM 600993], MLH1 [MIM 120436], BRCA1 [MIM 113705], BRCA2 [MIM 600185], adenomatous polyposis of the colon [MIM 175100], multiple endocrine neoplasia type II [MIM 171400], and RET [MIM 164761])
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi/
References
- Bassam BJ, Caetano-Anolles G, Gresshoff P (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83 [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M (1990) The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet 85:55–74 [DOI] [PubMed] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L (1996) MADR2 maps to 18q21 and encodes a TGFB-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 86:543–552 [DOI] [PubMed] [Google Scholar]
- Friedl W, Kruse R, Uhlhaas S, Stolte M, Schartmann B, Keller KM, Jungck M, Stern M, Loff S, Back W, Propping P, Jenne DE (1999) Frequent 4-bp deletion in exon 9 of the SMAD4/MADH4 gene in familial juvenile polyposis patients. Genes Chromosomes Cancer 25:403–406 [PubMed] [Google Scholar]
- Gardner E, Mulligan LM, Eng C, Healey CS, Kwok JB, Ponder MA, Ponder BA (1994) Haplotype analysis of MEN 2 mutations. Hum Mol Genet 3:1771–1774 [DOI] [PubMed] [Google Scholar]
- Groden J, Gelbert L, Thliveris A, Nelson L, Robertson M, Joslyn G, Samowitz W, Spirio L, Carlson M, Burt R (1993) Mutational analysis of patients with adenomatous polyposis: identical inactivating mutations in unrelated individuals. Am J Hum Genet 52:263–272 [PMC free article] [PubMed] [Google Scholar]
- Hahn SA, Shamsul Hoque ATM, Moskaluk CA, da Costa LT, Scutte M, Rozenblum E, Seymour AB, Weinstein CL, Yeo CJ, Hruban RH, Kern SE (1996) Homozygous deletion map at 18q21.1 in pancreatic cancer. Cancer Res 56:490–494 [PubMed] [Google Scholar]
- Hofting I, Pott G, Schrameyer B, Stolte M (1993) Familiare juvenile polyposis mit vorwiegender magenbeteiligung. Z Gastroenterol 31:480–483 [PubMed] [Google Scholar]
- Horrilleno EG, Eckert C, Ackerman LV (1957) Polyps of the rectum and colon in children. Cancer 10:1210–1220 [DOI] [PubMed] [Google Scholar]
- Howe JR, Mitros FA, Summers RW (1998a) The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol 5:751–756 [DOI] [PubMed] [Google Scholar]
- Howe JR, Ringold JC, Summers RW, Mitros FA, Nishimura DY, Stone EM (1998b) A gene for familial juvenile polyposis maps to chromosome 18q21.1. Am J Hum Genet 62:1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IPM, Houlston R, Bevan S, Mitros FA, Stone EM, Aaltonen LA (1998c) Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280:1086–1088 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Paabo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Cooper DN (1991) Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet 86:425–441 [DOI] [PubMed] [Google Scholar]
- McColl I, Bussey HJR, Veale AMO, Morson BC (1964) Juvenile polyposis coli. Proc R Soc Med 57:896–897 [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisio AL, Sistonen P, Weissenbach J, de la Chapelle A, Peltomaki P (1996) Age and origin of two common MLH1 mutations predisposing to hereditary colon cancer. Am J Hum Genet 59:1243–1251 [PMC free article] [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Gimenez J, Reis A, Varon-Mateeva R, Macek M Jr, Kalaydjieva L (1994) The origin of the major cystic fibrosis mutation (ΔF508) in European populations. Nat Genet 7:169–175 [DOI] [PubMed] [Google Scholar]
- Neuhausen SL, Godwin AK, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, et al (1998) Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet 62:1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Sistonen P, Salovaara R, Hemminki A, Loukola A, Johansson M, Avizienyte E, Cleary KA, Lynch P, Amos CI, Kristo P, Mecklin JP, Kellokumpu I, Jarvinen H, Aaltonen LA (1999) SMAD genes in juvenile polyposis. Genes Chromosomes Cancer 26:54–61 [DOI] [PubMed] [Google Scholar]
- Sachatello CR, Hahn IL, Carrington CB (1974) Juvenile gastrointestinal polyposis in a female infant: Report of a case and review of the literature of a recently recognized syndrome. Surgery 75:107–114 [PubMed] [Google Scholar]
- Sajantila A, Lukka M, Syvanen AC (1999) Experimentally observed germline mutations at human micro- and minisatellite loci. Eur J Hum Genet 7:263–266 [DOI] [PubMed] [Google Scholar]
- Scott-Conner CEH, Hausmann M, Hall TJ, Skelton DS, Anglin BL, Subramony C (1995) Familial juvenile polyposis: Patterns of recurrence and implications for surgical management. J Am Coll Surg 181:407–413 [PubMed] [Google Scholar]
- Sham P (ed) (1997) Statistics in human genetics. Arnold, London, pp 146–147 [Google Scholar]
- Stemper TJ, Kent TH, Summers RW (1975) Juvenile polyposis and gastrointestinal carcinoma. Ann Intern Med 83:639–646 [DOI] [PubMed] [Google Scholar]
- Subramony C, Scott-Conner CEH, Skelton D, Hall TJ (1994) Familial juvenile polyposis. Study of a kindred: evolution of polyps and relationship to gastrointestinal carcinoma. Am J Clin Pathol 102:91–97 [DOI] [PubMed] [Google Scholar]
- Tonin PN, Mes-Masson AM, Futreal PA, Morgan K, Mahon M, Foulkes WD, Cole DE, Provencher D, Ghadirian P, Narod SA (1998) Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am J Hum Genet 63:1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]