Abstract
Gene families often consist of members with diverse expression domains reflecting their functions in a wide variety of tissues. However, how the expression of individual members, and thus their tissue-specific functions, diversified during the course of gene family expansion is not well understood. In this study, we approached this question through the analysis of the duplication history and transcriptional evolution of a rapidly expanding subfamily of insect Ly6 genes. We analyzed different insect genomes and identified seven Ly6 genes that have originated from a single ancestor through sequential duplication within the higher Diptera. We then determined how the original embryonic expression pattern of the founding gene diversified by characterizing its tissue-specific expression in the beetle Tribolium castaneum, the butterfly Bicyclus anynana, and the mosquito Anopheles stephensi and those of its duplicates in three higher dipteran species, representing various stages of the duplication history (Megaselia abdita, Ceratitis capitata, and Drosophila melanogaster). Our results revealed that frequent neofunctionalization episodes contributed to the increased expression breadth of this subfamily and that these events occurred after duplication and speciation events at comparable frequencies. In addition, at each duplication node, we consistently found asymmetric expression divergence. One paralog inherited most of the tissue-specificities of the founder gene, whereas the other paralog evolved drastically reduced expression domains. Our approach attests to the power of combining a well-established duplication history with a comprehensive coverage of representative species in acquiring unequivocal information about the dynamics of gene expression evolution in gene families.
Keywords: gene family evolution, gene duplication, Ly6 protein, regulatory evolution, Drosophila
Introduction
Expansion of gene families is an important driving force of genome evolution allowing for functional specialization and emergence of members with novel functions. Moreover, the increase in family size is associated with greater complexity at various levels of biological organization, from gene regulatory networks to morphology, metabolism, and environment sensing (Ohta 1991; McBride et al. 2007; Nei et al. 2008; Voordeckers et al. 2012; Holland 2013; Castillo-Morales et al. 2014). Recent findings that gene family size evolves even among closely related species point to the adaptive significance of this process both at the macro and microevolutionary time scales (Popesco et al. 2006; Hahn et al. 2007; Patel et al. 2012). At the core of gene family expansion is the process of gene duplication, which produces the raw material for natural selection to generate functional repertoires within every gene family.
In his seminal work, Ohno (1970) proposed three functional fates of new gene duplicates effectively retained in the genome (Hahn 2009). First, if an increased dosage or functional redundancy is beneficial, both duplicates can maintain the functions of the unduplicated gene. However, this fate is considered least likely, because one copy can be eliminated from the genome through accumulation of degenerative mutations. Second, the original functions carried out by the unduplicated gene may be divided between duplicates in a complementary fashion (subfunctionalization) (Force et al. 1999). Finally, one or both duplicates may acquire novel functions not present in the unduplicated ortholog (neofunctionalization). Subfunctionalization and neofunctionalization are not mutually exclusive processes and can operate both at the level of protein function, through alterations in the coding sequence, and at the level of tissue-specific function, through changes in cis-regulation (Ohno 1970; Castillo-Davis et al. 2004; He and Zhang 2005; Kassahn et al. 2009) as well as at other levels of regulation (Alonso and Wilkins 2005).
Various studies of individual duplicate pairs, as well as genome-wide analyses, have shown that functional divergence (subfunctionalization and neofunctionalization) at the level of tissue-specific expression is frequent after duplication, although conservation seems to be rarer (Force et al. 1999; Prince and Pickett 2002; Kassahn et al. 2009; Assis and Bachtrog 2013). Furthermore, some of these studies have examined the impact of different duplication mechanisms on the expression divergence (Cusack and Wolfe 2007; Katju 2013). Others have analyzed the temporal dynamics of expression divergence between duplicates relative to the duplication event (Huerta-Cepas et al. 2011; Pegueroles et al. 2013). However, despite these recent advances in our understanding of how duplicated pairs diverge in their tissue-specific functions, little is known about this process at the level of gene family evolution. For example, there is only a handful of studies addressing whether the expression diversity of extant gene families arose primarily by recurrent subfunctionalization of the ancestral tissue-specificities or by neofunctionalization of new family members (Huminiecki and Wolfe 2004; Freilich et al. 2006; Farré and Albà 2010). These genome-wide studies reported negative correlations between the size of gene families and the breadth of tissue-specific expression of an individual family member, an observation that was interpreted as a signature of subfunctionalization in expanding families (Huminiecki and Wolfe 2004; Freilich et al. 2006; Farré and Albà 2010). Interestingly, though, when Huminiecki and Wolfe (2004) analyzed the expression divergence of four of these families in their phylogenetic contexts, the proportion of neofunctionalization events increased, highlighting the importance of incorporating phylogeny for accurate inference of functional fates.
Therefore, inferring patterns of functional diversification of a multigene family requires a reliable knowledge of the duplication history as well as the functions of the founding ortholog and its duplicates in different stages of the expansion process. The increasing number of species with assembled genomes allows unprecedented levels of taxon sampling to address this issue, particularly at the level of tissue-specific expression. However, to date, few studies have taken full advantage of multiple species with sequenced genomes to carefully delineate the process of functional diversification in gene families (Voordeckers et al. 2012).
In this study, we focused on a subfamily of nine genes in Drosophila and their orthologs across insects, which have undergone extensive expression diversification in the last 250 Ma. These genes belong to the Ly6 gene superfamily whose members encode glycoproteins with small extracellular module(s) called three-finger-domains (TFD) (Galat et al. 2008). These domains, approximately 100 amino acid long, possess eight to ten highly conserved cysteine residues placed in stereotypical positions, and adopt a characteristic conformation with three protruding loops that interact with diverse targets (Galat et al. 2008). Present in most metazoans, the Ly6 proteins have been co-opted for a wide variety of physiological and developmental functions. For example, in the insect Drosophila, Ly6 members have been shown to participate in diverse processes such as the assembly of cell adhesion complexes (Hijazi et al. 2009; Nilton et al. 2010; Hijazi et al. 2011; Syed et al. 2011), the formation of cuticle (Moussian et al. 2005; Chaudhari et al. 2013), the modulation of motoneuron activity (Kim and Marqués 2012), or the regulation of circadian rhythms (Wu et al. 2010; Wu and Robinson 2014).
A key to the functional versatility of this protein superfamily may be the intrinsic flexibility of the TFD domain, indicated by highly divergent sequences of Drosophila and vertebrate Ly6 genes. Indeed, a salient feature of the Ly6 genes is their apparent tendency to undergo lineage-specific expansion and functional diversification in multiple groups of animals (Fry et al. 2003; Hijazi et al. 2009; Galat 2011; Vonk et al. 2013). An extreme example of this phenomenon is illustrated by a large set of Ly6 family toxins in the Elapid and Hydroid snakes. They evolved from a nontoxic ancestor after multiple rounds of gene duplication and are known to bind with different specificities to a wide array of targets in the prey, suggesting that their diversification confers strong selective advantages (Fry et al. 2003; Galat et al. 2008; Vonk et al. 2013).
The insect Ly6 gene superfamily also appears to have undergone lineage-specific expansion events, with the most prominent expansion occurring in the higher Diptera. The genome of Drosophila melanogaster (a higher dipteran) contains 36 family members (Hijazi et al. 2009), whereas those of the mosquito Anopheles gambiae (a lower dipteran), the beetle Tribolium castaneum (a coleopteran), and the honeybee Apis mellifera (a hymenopteran) contain 16, 26, and 15 members, respectively. In addition, the Ly6 genes in D. melanogaster have been shown to display both highly divergent coding sequences and tissue-specific expression in the embryo (Hijazi et al. 2009; Nilton et al. 2010). Thus, we reasoned that the dipteran Ly6 family could be an interesting model to study the process of gene family diversification.
In this work, we have focused on one episode of expansion, which produced a subfamily of Ly6 genes unique to the higher Diptera. By analyzing different insect genomes, we first reconstructed the duplication history of nine paralogs that have arisen from a single ancestral gene through sequential tandem duplications. We then characterized the embryonic tissue-specificities of the duplicates and their unduplicated orthologs to retrace the path of expression diversification during family expansion. We found a consistent pattern where the ancestral expression domains of the founding gene were sequentially inherited by one duplicate after each duplication event, whereas the other duplicate assumed divergent tissue specificities. Novel tissue-specificities were acquired frequently following duplication events as well as following speciation events and contributed to the present day diversity of tissue-specific expression of the insect Ly6 genes. Our work provides one of the first empirical studies addressing how tissue-specificities diversify in a rapidly evolving gene family.
Results
Phylogenetic Analysis of Cluster III and V Genes
A preliminary inventory of the TFD proteins present in insect genomes revealed that the size of the Ly6 superfamily varies significantly among different groups of insects. For instance, the genome of the coleopteran T. castaneum contains 26 genes in contrast with the 15 family members in the hymenopteran A. mellifera or the 16 genes in the lower dipteran An. gambiae. Among fully sequenced insect species, the largest number of paralogs is found in the higher dipterans with D. melanogaster harboring 36 genes (Hijazi et al. 2009). Interestingly, many members of the Drosophila Ly6 superfamily are arranged in clusters of contiguous loci with the conserved intron–exon structure, indicating that the recent expansion of this family involved multiple episodes of tandem gene duplication (Hijazi et al. 2009; Nilton et al. 2010).
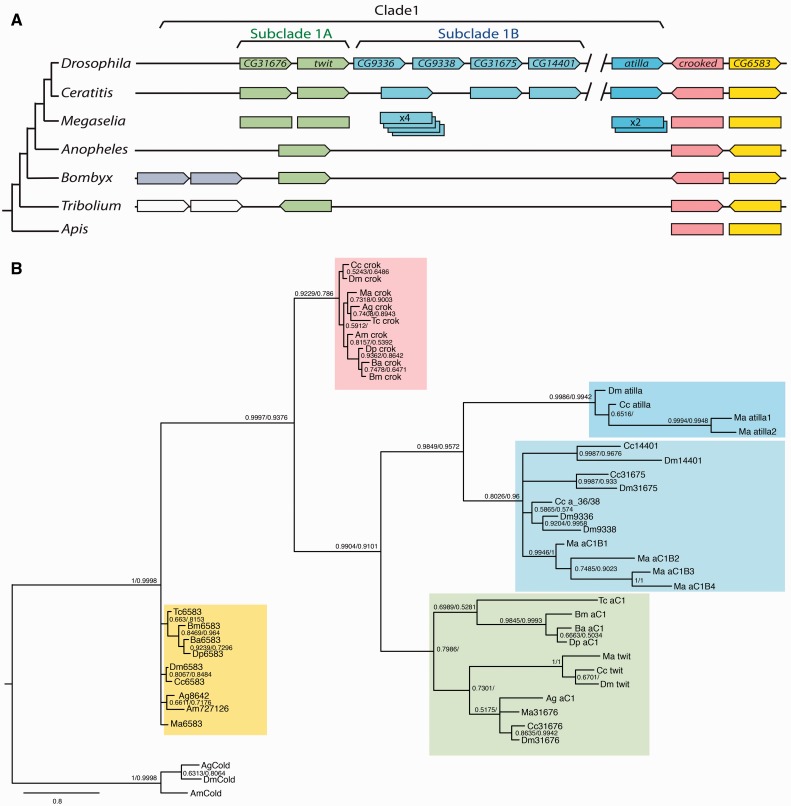
To describe one of these episodes in detail, we focused on the evolutionary history of a Drosophila Ly6 subfamily comprised of nine genes arranged in two clusters (Cluster III and V described in Hijazi et al. [2009]; fig. 1A). Cluster III is 6.5 kb in size and contains three contiguous Ly6 genes, CG6583, crok, and atilla, whereas Cluster V is a larger cluster (32 kb) with six genes: CG31675, twit, CG9336, CG9338, CG31675, and CG14401 (fig. 1A). We have searched for putative homologs of these nine genes among all the Ly6 genes identified above in different insect genomes. Both the amino acid sequence similarity and, where possible, the synteny of the candidate homologs were used to establish their orthology (see below and Materials and Methods for details). The results are summarized in supplementary figure S1A, Supplementary Material online. We found CG6583 and crok orthologs not only in all the holometabolous insects but also in distantly related arthropods such as crustaceans and chelicerates, indicating that they are ancient members of this family (fig. 1A, supplementary fig. S1A, Supplementary Material online). In comparison, the other seven members appear to have more recent origins, because in Coleoptera (T. castaneum), Lepidoptera (Bombyx mori, Danaus plexippus, Bicyclus anynana), and the lower Diptera (An. gambiae, Anopheles stephensi, Culex quinquefasciatus, and Aedes aegypti), we could only identify a single homolog, a gene related to CG31676 and twit (fig. 1A, supplementary fig. S1A, Supplementary Material online). This gene forms a single cluster with the crok and CG6583 orthologs in most assembled genomes (fig. 1A). We found two additional Ly6 genes in these clusters in the genomes of both T. castaneum and two lepidopteran species (B. mori and Da. plexippus) (fig. 1A). However, given their sequence divergence relative to other family members and their phylogenetic distribution, we considered these genes as lineage-specific duplicates unrelated to the history of the Drosophila genes.
Fig. 1.
Phylogenetic analysis of Cluster III and V Ly6 genes. (A) Numbers and cluster organizations of the Cluster III and V Ly6 genes in insect genomes. The phylogenetic relationship of the groups is shown on the left. Pointed ends on the genes indicate their orientations within the clusters. Genes in the same clusters are connected with lines. The members are further subdivided according to the phylogenetic analysis below (Clade1, Subclades A and B). In Megaselia, neither the cluster organization nor the orientations of the genes are known. Megaselia also has multiple copies of CG9336–CG9338 and atilla homologs. The Apis homologs are not in a cluster. (B) Bayesian consensus tree generated using BAli-Phy program. For each branch, posterior probabilities above 0.5 are shown on the left. Those obtained for the corresponding branches in the tree generated by MrBayes are shown on the right. A distantly related Ly6 gene coiled (cold) was used as an outgroup. Dm, Drosophila melanogaster; Cc, Ceratitis capitata; Ma, Megaselia abdita; Ag, Anopheles gambiae; Tc, Tribolium castaneum; Am, Apis mellifera; Dp, Danaus plexippus; Ba, Bicyclus anynana; and Bm, Bombyx mori.
In contrast, the higher dipterans harbor more homologs of this Ly6 subfamily (fig. 1A). We identified single CG31676 and twit orthologs in both M. abdita (Phoridae) and C. capitata (Tephritidae). In addition, C. capitata contains one ortholog of atilla and one gene highly related to CG9336 and CG9338. Megaselia abdita, in turn, contains two homologs of atilla and four homologs of the CG9336/CG9338 pair. Finally, we were only able to identify the orthologs of CG31675 and CG14401 in the C. capitata genome (fig. 1A, supplementary fig. S1A, Supplementary Material online). In this species, the homologs are found in the same orientation as in the Drosophila genome and, similarly, grouped in two separate clusters (fig. 1A). We could not determine the cluster organization in M. abdita due to the short length of the available genomic contigs.
Gene trees were estimated using the Bayesian-based phylogeny program BAli-Phy with the amino acid sequences of all homologs retrieved from nine holometabolan species (A. mellifera, T. castaneum, B. mori, Bi. anynana, Da. plexippus, An. gambiae, C. capitata, M. abdita, and D. melanogaster) to establish their relationships (see Materials and Methods) (fig. 1B). Using cold (an unrelated member of the Ly6 family) from three species as an outgroup (fig. 1B, supplementary fig. S1B and C, Supplementary Material online), we obtained multiple trees consistently displaying the same overall tree topology. Only minor differences arose within the clades, which do not qualitatively affect the conclusions (see below) and constitute good evidence for a strong phylogenetic signal. As is customary for phylogenetic trees computed with Bayesian methods, the consensus tree containing nodes with posterior probability values above 0.5 is shown (values on the left in fig. 1B). To validate the results obtained with BAli-Phy, we used the second Bayesian-based program MrBayes, which yielded trees with consistent overall topologies. Posterior probability values above 0.5 obtained from MrBayes are shown on the corresponding branches in the BAli-Phy-generated tree in figure 1B (values on the right).
We observed three major clades: One including all the CG6583 orthologs, another with the crok orthologs, and a third one grouping all the remaining genes (fig. 1B). The last large clade consisted of two major branches. One included CG31676 and CG9335, together with the single gene found in all nondipteran and lower dipteran species. The other branch contained the orthologs of atilla and all the Cluster V genes (CG9336, CG9338, CG31675, and CG14401). Within this branch, a well-supported clade exclusively contained the atilla orthologs. In its sister clade containing the four Cluster V genes, however, some of the internal nodes were either unresolved or weakly supported. Finally, the aC1B1–4 genes and atilla1 and 2 in M. abdita each formed their own clades indicating that they were produced through lineage-specific duplications (fig. 1B).
Reconstruction of the Cluster III and V Duplication History
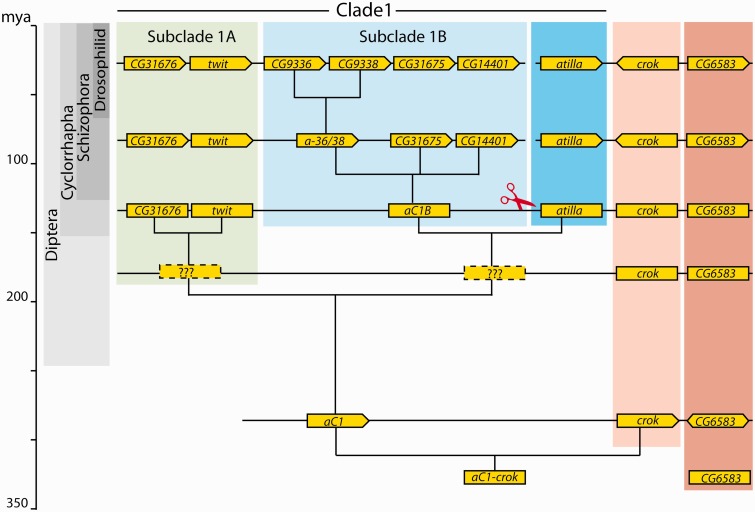
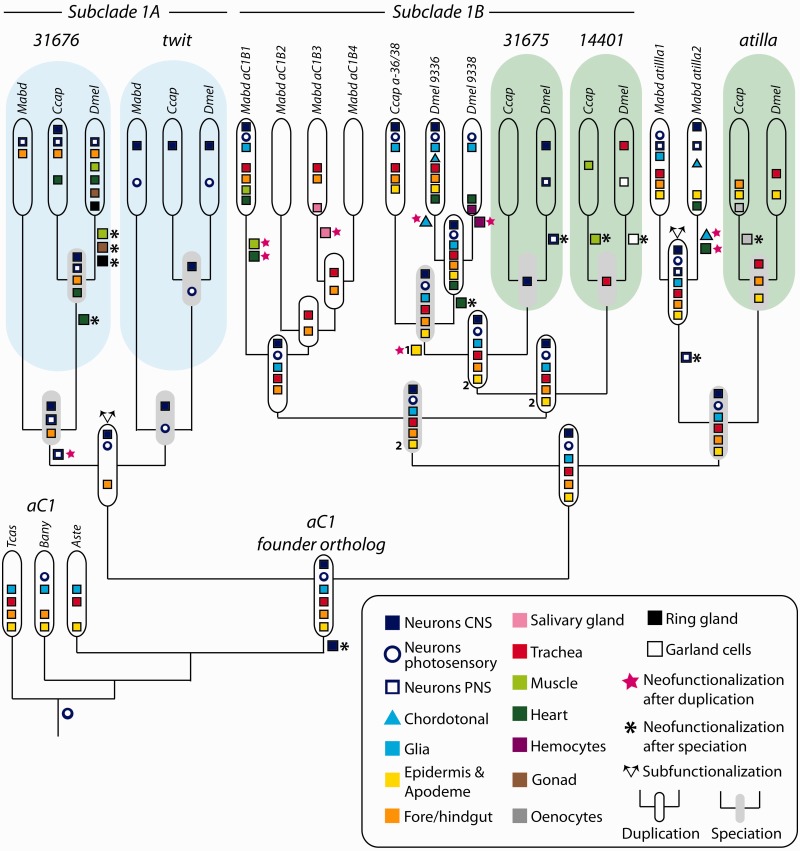
Combining the information derived from the phylogenetic analysis and the synteny blocks in the context of the species tree, we reconstructed the sequence of duplication events of this Ly6 subfamily (fig. 2). We could establish that the common ancestor of Holometabola had a cluster consisting of two genes, a state currently represented by some hymenopteran species. These genes were CG6583, which has never undergone duplication within the holometabolan lineage, and the crok-like gene, which duplicated to give rise to crok, and a third gene (hereafter called aC1, for ancestor of Clade1) after the split of Hymenoptera from the other Holometabola. Since then crok remained unduplicated in all the lineages analyzed in this study, whereas aC1 gave rise to the rest of the Cluster III and V genes, which we refer to as Ly6 Clade1 genes (figs. 1 and 2). At the base of the higher Diptera, aC1 underwent the first round of duplication to generate the ancestor of the CG31676–twit lineage (Subclade 1A) and a gene ancestral to the rest of the paralogs (fig. 2). The second round of duplications produced CG31676 and twit on one side and atilla and the ancestor of the CG9336, CG9338, CG31675, and CG14401 on the other side. We refer to the latter group of paralogs as Subclade 1B and to its ancestral gene as aC1B (fig. 2).
Fig. 2.
Evolutionary history of Clusters III and V Ly6 gene subfamily. The red scissor indicates the separation of the ancestral cluster into two distinct genomic locations. Note that in the ancestral cluster with three genes, CG6583 orthologs are found in different orientations in different groups. See text for details.
After the split of the Phoridae (represented by M. abdita) and the Schizophora (represented by C. capitata and D. melanogaster), aC1B duplicated twice to give rise to CG31675, CG14401 and the parental genes of CG9336 and CG9338 (named a-36/38). However, the order of these two duplication events could not be resolved with high confidence in our analysis. By this time, a translocation event separated atilla, crok, and CG6583 from the rest of cluster. This notion is supported further by our synteny analysis (supplementary fig. S2, Supplementary Material online), which revealed that the 5′ neighbors of CG31676 in D. melanogaster and C. capitata are found on the 5′ side of aC1 in Ae. aegypti and Cu. quinquefasciatus. Finally after the split of the tephritid fly C. capitata, CG9336 and CG9338 arose through the last duplication event in Drosophilids. Meanwhile, within the Phoridae, aC1B underwent two rounds of duplication to give rise to four copies (aC1B1–4), and atilla duplicated once producing atilla1 and atilla2 (fig. 1).
Tissue-Specific Expression of the Founding Ortholog aC1
The Cluster III and V genes of D. melanogaster display diverse tissue-specific patterns suggesting extensive functional diversification during the course of family expansion (Hijazi et al. 2009; Nilton et al. 2010; Kim and Marqués 2012). To characterize this expression divergence process, the embryonic expression patterns of aC1 genes and its duplicates were characterized in six species representing different states of duplication. The full tissue-specific expression domains of all the paralogs are summarized in supplementary table S4, Supplementary Material online. Additionally, we have looked at the expression of crok, another ancient gene, which remained unduplicated for over 350 Ma in the species under study (supplementary fig. S3, Supplementary Material online). In all six species examined, the pattern of crok was similar showing generalized expression in the epidermis and the hindgut (supplementary fig. S3, Supplementary Material online).
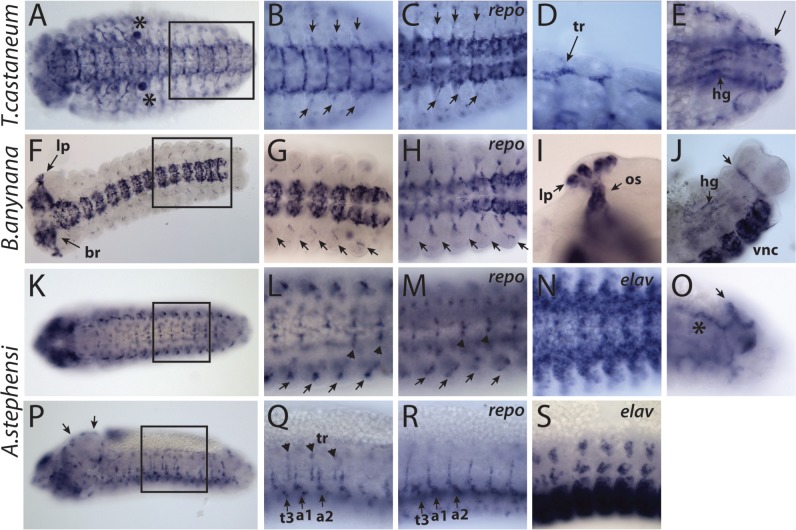
To establish the putative ancestral expression domains of the aC1 unduplicated ortholog, we first analyzed the expression patterns of this gene in the beetle T. castaneum, the butterfly Bi. anynana, and the mosquito An. stephensi. In T. castaneum, a prominent expression of aC1 transcripts was detected in a group of cells associated with the nervous system, specifically, in the ventral nerve cord (VNC), its exiting nerves and the brain (fig. 3A and B, supplementary fig. S4A, Supplementary Material online). In the VNC, these cells were found scattered over the surface and in the midline (fig. 3B). These groups of cells also stained positively for the T. castaneum repo gene (a known glial marker), indicating their glial identity (fig. 3C). aC1 expression was also observed in the dorsal and the leg trachea (fig. 3D) and in the hindgut (fig. 3E). In the late stage embryos, the expression was also observed in the epidermis of the ventral thorax and the proximal part of the legs (supplementary fig. S4B, Supplementary Material online).
Fig. 3.
Embryonic tissue-specificities of aC1 genes. (A–E) Tribolium castaneum. (A) Ventral view of the whole embryo. Asterisks indicate nonspecific labeling of the pleuripod. (B) Boxed area in (A), showing aC1 expression in the VNC and in the exiting nerves (arrows). (C) Glial cells in the VNC and the exiting nerves (arrows) visualized with the repo riboprobe. (D) Dorsal longitudinal trachea (tr). (E) Expression in the hindgut (hg) and anal structures (arrow). (F–J) Bicyclus anynana. (F) Ventral view showing the expression in the larval photoreceptor neurons (lp), the brain (br), and the VNC. (G) Boxed area in (F), showing aC1 expression in the VNC and in the PNS glia (arrows). (H) repo expression labels glial cells in both the CNS and PNS (arrows). (I) aC1 expression in the larval photoreceptor neurons and optic nerve glia (os). (J) Expression in the hindgut (hg), the anal structures (arrow), and the VNC. (K–S) Anopheles stephensi. (K–N) Ventral views. (L) Boxed area in (K), showing aC1 expression in the VNC (arrowheads) and the exiting nerves (arrows). (M) repo expression labeling glial cells associated with the exiting nerves (arrows) and the VNC (arrowheads). (N) Neurons in the VNC visualized with elav expression. (O) aC1 expression in the anal structures (arrow). Asterisk, Nonspecific staining. (P–S) Lateral views. (P) Lateral view of the whole embryo. Arrows, brain. (Q) Boxed area in (P), showing expression in the exiting nerves (arrows) and in trachea (arrowheads). t3, third thoracic segment; a1 and a2, first and second abdominal segments. (R) repo expression in the glia labeling exiting nerves (arrows) and PNS. (S) elav expression in the CNS and PNS neurons. Anterior is to the left in all figures.
In Bi. anynana, the aC1 expression was also most conspicuous in the VNC and the brain (fig. 3F and G). Additionally, several cells in what appeared to be the peripheral nervous system (PNS) displayed a prominent expression (fig. 3F and G). Labeling with probes against the Bi. anynana homologs of repo and the neuronal marker elav confirmed that the aC1 expression in the nervous system corresponded to glial cells (fig. 3H, supplementary fig. S4C, Supplementary Material online). As in T. castaneum, moderate expression was also observed in the hindgut and in the epidermis of the ventral thorax and the lateral body wall (fig. 3J, supplementary fig. S4D, Supplementary Material online). Finally, unique to this species, the neurons in the developing larval photosensory organ and the glia in the optic stalk strongly expressed aC1 (fig. 3I, supplementary fig. S4E–H, Supplementary Material online).
In An. stephensi, the aC1 transcript was also expressed in the nervous system (fig. 3K and L). However, unlike the other two species, the expression appeared stronger in the PNS than in the VNC (fig. 3P and Q). In the brain, only the cells on the surface appeared to express aC1 transcripts, suggesting that they might be perineural glia enveloping the brain (fig. 3P). We confirmed their glial identity and those of the VNC and PNS using riboprobes against the An. gambiae repo and the neuronal marker elav (fig. 3R and S). Other tissues expressing aC1 included the epidermis in the head and the terminal segments and the trachea (fig. 3O–Q).
In summary, the expression in the glia, the epidermis, and the hindgut appears to be the most conserved features of the aC1, whereas the other observed domains represent lineage-specific gains. Most likely, these conserved tissues-specificities were present in the founding ortholog of the Clade1 (fig. 2).
Tissue-Specific Expression of the Old Paralogs: CG31676, Twit, and Atilla
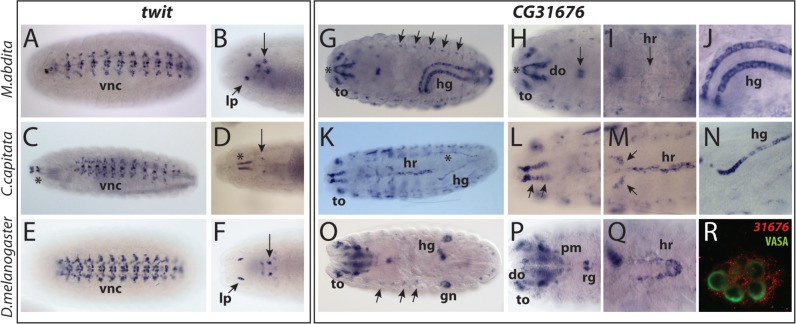
After the split between Brachycera and the lower dipterans, two duplication events produced four genes, CG31676, twit, atilla, and aCB1 (fig. 2). Of these, aC1B has undergone further duplications but the other three genes remained unduplicated without generating novel paralogs (except for atilla in M. abdita) (figs. 1A and 2). In Clade1, twit is the only gene that has been functionally characterized and its expression has also been analyzed in D. melanogaster (Kim and Marqués 2012). In the embryos of D. melanogaster, this gene is expressed in a subset of motor neurons in each segment of the VNC, small clusters of neurons in the brain, and the larval photosensory organs (Bolwig’s organs) (fig. 4E and F; Kim and Marqués 2012). The twit orthologs in C. capitata and M. abdita showed expression in what appears to be equivalent neuronal populations in the VNC and the brain (fig. 4A–D). The expression in the Bolwig’s organ, however, was not detected in C. capitata, suggesting that these expression domains may have been secondarily lost in this species.
Fig. 4.
Embryonic tissue-specificities of twit and CG31676 orthologs. (A–F) twit. (A, B) Megaselia abdita. (A) Ventral view showing expression in the VNC. (B) Dorsal view showing strong signal in the larval photoreceptors (lp) and a set of cells in the brain (arrow). (C, D) Ceratitis capitata. Asterisks label a nonspecific signal associated with the mouth hooks. (C) Ventral view showing expression in the VNC. (D) Dorsal view showing expression in distinct cells in the brain (arrow). (E, F) Drosophila melanogaster. (E) VNC expression. (F) Expression in the larval photoreceptors and the neurons in the brain (arrow). (G–R) CG31676. (G–J) Megaselia abdita dorsal views. (G). Transcript distribution in the hindgut (hg), terminal organ (to), and lateral sensory neurons (arrows). Asterisk labels nonspecific staining in the mouth hooks. (H) Expression in the dorsal organ (do) and in pharynx associated cells (arrow). (I) No detectable expression is observed in the heart (hr). (J) Expression in two lateral rows of cells in the hindgut. (K–N) Ceratitis capitata dorsal views. (K) Expression is detected in the terminal organ (to), anterior heart (hr), and the hindgut (hg). Asterisk labels nonspecific staining in the dorsal trachea. (L) Cephalic region showing expression in unidentified rows of cells dorsal to the pharynx (arrows). (M) Expression in the heart (hr) and an unidentified structure (arrows). (N) Expression in a single row of cells in the hindgut. (O–R) Drosophila melanogaster. (O) Expression in the terminal organ (to), lateral sensory neurons (arrows), gonads (gn), and a ring of cells in the hindgut (hg). (P) Details of the cephalic region, showing expression in the pharyngeal muscle (pm), dorsal organ (do), and ring gland (rg). (Q) Expression in the posterior heart (hr). (R) Germ cells labeled with VASA protein (green) are in contact with gonad mesodermal cells expressing CG31676 transcripts (red). In all images, anterior is to the left.
CG31676 is the sister paralog of twit (fig. 1B), but the expression patterns of these genes are remarkably different (fig. 4G–R). In all the species examined, the CG31676 transcripts were detected in the hindgut and in the neurons of the terminal and dorsal organs in the head (fig. 4G, H, J, K, and N–P, supplementary fig. S5B, Supplementary Material online). In addition, we identified many lineage-specific expression domains. Both C. capitata and D. melanogaster showed expression in the heart (fig. 4K, M, and Q), whereas M. abdita and D. melanogaster shared expression in a row of dorsolateral neurons in the PNS (fig. 4G and O, supplementary fig. S5A, Supplementary Material online). Finally, CG31676 was expressed in D. melanogaster in the ring gland, the pharyngeal muscle, and the somatic cells of the gonad, indicating an acquisition of a novel expression in mesoderm-derived tissues in this lineage (fig. 4O, P, and R). In C. capitata, a species-specific expression was also present in what appears to be neurons located in the midline of the VNC, two unidentified rows of cells in the dorsal head and unidentified segmentally repeated structures in the ventrolateral part of the body (fig. 4L, supplementary fig. S5C, Supplementary Material online).
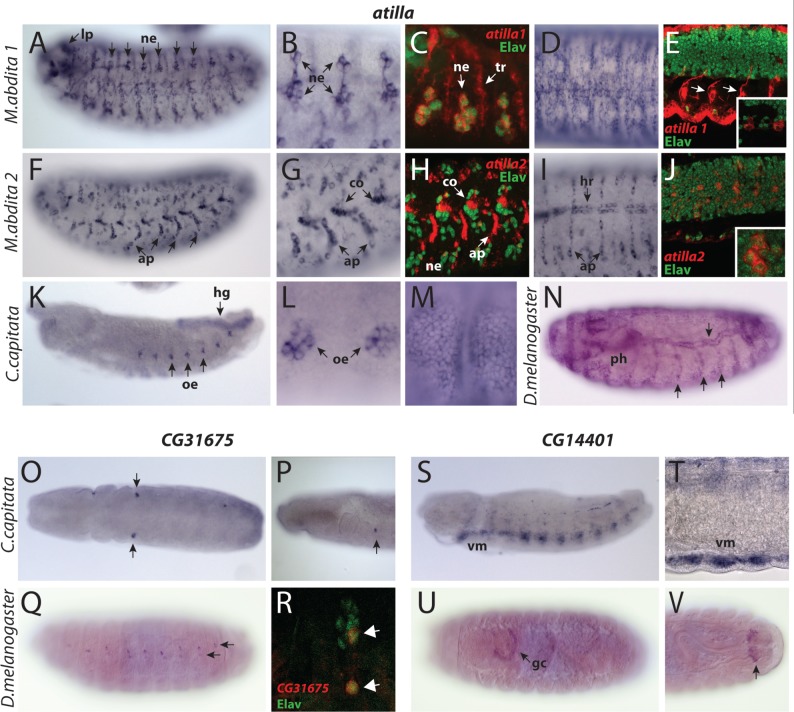
atilla, the sister paralog of Subclade 1B, is the third gene already present at the base of Cyclorrhapha (fig. 2). The expression of atilla in the D. melanogaster embryo became visible in epidermis, trachea, and pharynx of late embryos (fig. 5N). In C. capitata, this gene was also expressed at late stages in the epidermis and the pharynx (fig. 5M, supplementary fig. S5D, Supplementary Material online), but had additional expression in the oenocytes and in the hindgut starting in earlier stages (fig. 5K and L).
Fig. 5.
Embryonic tissue-specificities of atilla, CG14401, and CG31675 genes. (A–N) atilla genes. (A–E) Megaselia abdita atilla1. (A, B) Lateral views showing expression in the lateral sensory neurons (ne) and the larval photoreceptors (lp). (C) Fluorescent double staining showing atilla1 (red) and Elav protein (green) distribution on the lateral sensory neurons (ne). atilla1 expression is detected in both neurons and trachea (tr). (D) Expression in the dorsal epidermis. (E) Ventral view of atilla1—Elav double staining showing atilla1 expression in the Elav-negative glial cells associated with the exiting nerves (arrows) and within the VNC (inset). (F–J) Megaselia abdita atilla2. (F, G) Lateral views displaying expression in the muscle apodemes (ap) and the chordotonal organs (co). (H) Fluorescent staining of atilla2 (red) and Elav protein (green) on the lateral sensory organs. atilla2 transcripts are found both in nonneuronal components of the chordotonal organs (co) and in few Elav positive sensory neurons on the ventral side (ne). (I) Expression in the dorsal heart (hr) and apodemes (ap). (J) atilla2 is expressed in Elav positive cells in the VNC (magnified in inset). (K–M) Ceratitis capitata atilla. (K, L) Lateral views showing expression in the hindgut (hg) and the oenocytes (oe). (M) Late embryos show expression in the epidermis. (N) Drosophila melanogaster lateral view. atilla is expressed in the epidermis, trachea (arrows), and pharynx (ph). (O–R) CG31675 orthologs. (O, P) Ventral (O) and lateral (P) views of C. capitata embryo at the extended germband stage showing labeling of unidentified groups of cells posterior to the head (arrow). (Q, R) Drosophila melanogaster CG31675. (Q) Lateral view showing expression in the dorsal sensory organs. (R) CG31675 (red) is expressed in Elav-positive (green) lateral neurons (arrows). (S–V) CG14401 orthologs. (S, T) Ceratitis capitata. (S) Lateral view showing CG14401 expression in the ventral longitudinal muscles (vm). (T) Magnified view of the ventral muscle, seen from a ventrolateral perspective. (U, V). Drosophila melanogaster CG14401. (U) Ventral view showing expression in the garland cells (gc). (V) Dorsal view showing expression in a subset of cells associated with the posterior spiracles (arrow).
In M. abdita, atilla has undergone lineage-specific duplication to produce two copies, atilla1 and atilla2 (fig. 1A). atilla1 was prominently expressed in the developing trachea, in clusters of cells in the PNS including the Bolwig’s organs and other PNS sensory organs, and in a small segmentally repeated set of cells in the VNC (fig. 5A–C and E). Colabeling with the Elav antibody confirmed that almost all atilla1 positive cells in the PNS were neurons (including Bolwig’s organ and the dorsal and the terminal organs) (fig. 5C). The exception was a group of cells flanking the VNC, which were most likely glial cells of the exiting nerves (fig. 5E). We also detected expression in the hindgut (supplementary fig. S5E, Supplementary Material online) and, at later stages, in the epidermis (fig. 5D). Megaselia abdita atilla2 expression was quite distinct from that of atilla1 (fig. 5F–J). For example, prominent expression was observed in the heart, whereas it was completely absent in the PNS neurons (fig. 5G–J). Furthermore, the epidermal expression was strictly limited to the apodemes (muscle attachment sites) (fig. 5G–I). Finally, moderate level of expression was observed in many neurons of the VNC and in the nonneuronal components of chordotonal organs (fig. 5G, H, and J).
Tissue-Specific Expression of the Subclade aC1B Paralogs: a-36/38, CG31675, and CG14401
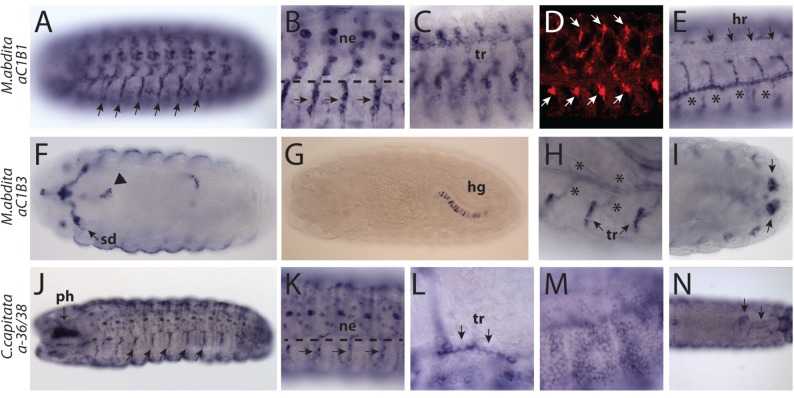
According to our analysis, the subclade aC1B founder gene (aC1B) was already present in the common ancestor of Cyclorrhapha (fig. 2). In M. abdita, our basal cyclorrhaphan species, aC1B underwent three rounds of lineage-specific duplications to produce four paralogs, aC1B1, aC1B2, aC1B3, and aC1B4 (figs. 1B and 8). During the embryonic development, however, only aC1B1 and aC1B3 had detectable expression, with the former gene being more widely expressed (fig. 6A–I, supplementary fig. S6A–E, Supplementary Material online). aC1B1 expression was visible in the glial cells of the exiting nerves, the trachea, the Bolwig’s organ, and the VNC, where a cluster of cells located in the ventral midline showed the highest level of expression (fig. 6A–D). Colabeling with the Elav antibody indicated that most of the cells in the VNC were neurons (supplementary fig. S6C, Supplementary Material online). A group of two to four cells in the leading end of the hindgut also showed expression (supplementary fig. S6E, Supplementary Material online). At late stages, the transcripts also appeared at a low level in the muscles and the heart (fig. 6D and E, supplementary fig. S6D, Supplementary Material online). aC1B3, on the other hand, was expressed in a single row of cells in the hindgut, in the salivary ducts and in the posterior spiracles as well as in a group of cells associated with the lateral branches of the trachea (fig. 6F–I). A small group of cells at the foregut–midgut border and an unidentified bilateral group of cells in the head also expressed aC1B3 (fig. 6F).
Fig. 8.
Parsimony analysis of evolution of the tissue-specificities in the aC1 lineage. The nodes with white genes represent duplication events, whereas the nodes with shaded genes indicate speciation. Tissue symbols appearing next to branches indicate acquisitions of new tissue-specificities (neofunctionalization) associated with either duplication events (stars) or speciation events (asterisks). Branching arrows above the genes indicate subfunctionalization. In this reconciliation tree, there are two equally parsimonious scenarios for evolution of epidermal expression (the yellow boxes 1 and 2). The first scenario yields nine instances of neofunctionalization after duplication and ten after speciation following the start of the Clade1 family expansion. The second scenario yields eight neofunctionalization after duplication and ten after speciation. For both scenarios, there are two cases of subfunctionalization.
Fig. 6.
Tissue specificities of aC1B and a-36/38 genes. (A–E) Megaselia abdita aC1B1. (A, B) Ventral view showing expression in the glia of exiting nerves (arrows) and VNC neurons (ne). The dashed line demarcates the boundary of the nerve cord. (C) Trachea (tr) on the lateral body wall. (D) aC1B1 (red) is expressed in muscles at a moderate level. Strong signal corresponds to glial cells (arrows). (E) Dorsal view showing low expression in the heart (hr). Asterisks indicate nonspecific staining in the cuticle of the tracheal lumen. (F–I) Megaselia abdita aC1B3. (F) Ventral view showing expression in the salivary ducts (sd) and at the fore–midgut junction (arrowhead). (G) Expression in a single row of cells belonging to the hindgut (hg). (H) Expression in the cells associated with the tracheal branches (tr). Asterisks indicate nonspecific labeling of the tracheal cuticle. (I) Expression in the posterior spiracles (arrow). (J–N) Ceratitis capitata a-36/38. (J, K) Ventral views showing expression in the pharynx-associated cells (ph) and the exiting nerves (arrows) and the VNC neurons (ne). (L) Tracheal expression in the anterior dorsal segment. (M) Expression in the epidermis. (N) Expression along the lateral sides and the tip of the hindgut is visible (arrows) in a dorsal view.
In Schizophora, aC1B followed a separate duplication history duplicating twice before the split of Tephritidae and Drosophilidae and once more within the latter group (figs. 1B and 2). As a consequence, C. capitata has three paralogs, whereas the basal condition for drosophilids is four. CG9336 and CG9338 in D. melanogaster are the most recently duplicated paralogs and derived from the unduplicated ortholog a-36/38 still found in C. capitata (fig. 2). Overall the C. capitata a-36/38 was expressed in the VNC neurons, in the glia of the exiting nerves, the Bolwig’s organ, and the hindgut (fig. 6J–L and N). This pattern is similar to that of M. abdita aC1B1, but, additionally, the C. capitata gene was also strongly expressed in the epidermis, in a group of cells under the pharynx, and in the anal pad (fig. 6J and M, supplementary fig. S6F, Supplementary Material online).
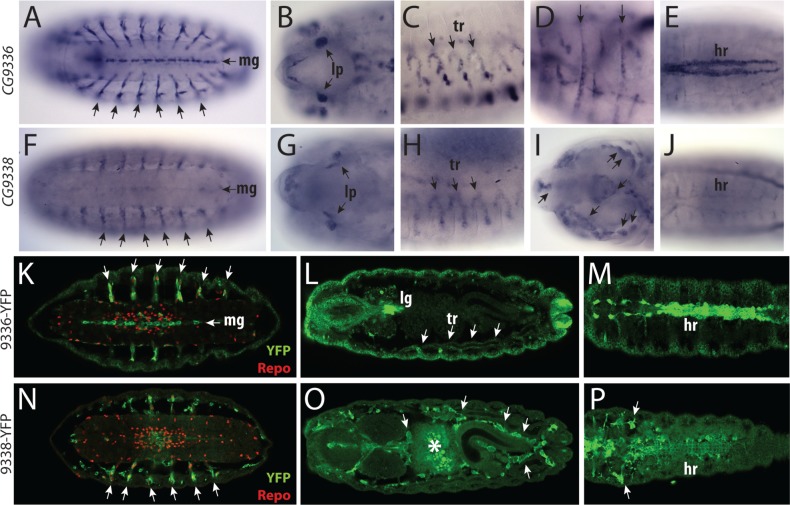
The recently duplicated paralogs CG9336 and CG9338 in D. melanogaster retain considerable nucleotide sequence similarities (72.6% identity between the coding sequences). To minimize the cross-reactivity during in situ hybridization, we designed riboprobes largely targeted against the 5′- or 3′-regions including the UTRs (61.7% and 67.2% identity between the probes, respectively). Although the observed staining patterns were different, we still saw some overlapping expression domains (fig. 7A–J). To exclude any ambiguity due to possible probe cross-reactivity, we have taken advantage of two Yellow Fluorescent Protein (YFP) trap lines available for CG9336 and CG9338 (Lowe et al. 2014). In these lines, the YFP is incorporated into the endogenous products, which remain under the control of their native cis-regulatory regions. We observed that the YFP distribution matched the staining pattern obtained with the riboprobes, with the exception of the epidermis and the hindgut (fig. 7K–P). Although the riboprobe only revealed expression in the apodemes in the anterior segments, CG9336-YFP showed a ubiquitous epidermal expression (fig. 7D, L, and M). In the case of the hindgut, the CG9336 riboprobe showed expression in the bilateral rows of cells in the late stages (the boundary cells) (supplementary fig. S6G, Supplementary Material online), whereas neither YFP lines had detectable signals in these cells. These discrepancies may reflect potential disruption of the endogenous regulatory elements by the protein-trap transposon insertion.
Fig. 7.
Drosophila melanogaster CG9336 and CG9338 expression. (A–E) D. melanogaster CG9336 in situ hybridization. (A) Ventral view showing prominent expression in the midline glia (mg) and the exit nerves glia (arrows). (B) Expression in the larval photoreceptor neurons (lp). (C) Expression in the trachea on the lateral body wall (tr). (D) Expression in the apodemes in anterior segments (arrows). (E) Dorsal view showing expression in the heart (hr). (F–J) D. melanogaster CG9338 transcripts detected by the riboprobe. (F) Ventral view showing expression in the glia in the exiting nerves (arrows) and weaker expression in the midline glia (compare with A). (G) Expression in the larval photoreceptors. (H) Trachea on the lateral body wall. (I) Expression in migrating hemocytes seen in the cephalic region (arrows). (J) Faint signal detected in the heart. Note that the two riboprobes share sequence similarity and may be cross-reacting. (K–M) The tissue distribution of CG9336-YFP (green) and the glial marker Repo (red) in the embryo homozygous for the CG9336-YFP protein trap insertion. (K) Ventral view. In both the exiting nerves and the VNC, the YFP signal is detected in Repo positive cells. Midline glia lacks Repo expression. (L) Dorsal view showing expression in the trachea (tr) and the lymph gland (lg). Note the ubiquitous epidermal expression, which is not visible with the riboprobes. (M) Dorsal view showing strong heart expression. (N–P) Embryos homozygous for the CG9338-YFP protein trap insertion, showing the distribution of CG9338-YFP (green) and Repo (red). (N) Ventral view showing expression in the glial cells associated with the exiting nerves, but no expression in the midline. (O) Dorsal view showing expression in the migrating hemocytes (arrows). Asterisk labels autofluorescent signal from the midgut yolk. (P) Dorsal view showing strong expression in the hemocytes (arrows) and weak expression in the heart (hr).
CG9336-YFP was most prominently expressed in various populations of glial cells in the embryo as indicated by coexpression with the Repo protein (fig. 7K). These glial populations included the midline glia in the VNC, the perineural glia and the cells in the exit nerves and the PNS (fig. 7K). Strong neuronal expression was observed in neurons of the Bolwig’s organ (supplementary fig. S6I, Supplementary Material online), whereas a lower level of expression was observed in neurons in the VNC on both sides of the midline (supplementary fig. S6H, Supplementary Material online). Prominent expression was also seen in the trachea, the heart, and the lymph gland (fig. 7L and M). Relative to CG9336-YFP, CG9338-YFP was expressed in a smaller number of tissues and at lower levels (fig. 7N–P). Although both genes were coexpressed in the Bolwig’s organ and in the glial cells in the exit nerve and the PNS, CG9338-YFP expression was not detected in the midline, epidermis, and trachea (fig. 7N). Instead a prominent expression was observed in the migrating hemocytes (fig. 7O). These results indicate that CG9336 and CG9338 have undergone extensive expression divergence despite their recent origin.
The orthologs of CG31675 and CG14401 are only found in D. melanogaster and in C. capitata. Their expression, quite distinct between the two species, was very limited in the embryos (fig. 5O–V). In C. capitata, CG31675 was transiently expressed in a pair of small cell clusters located posteriorly to the head at early stages (fig. 5O and P). In contrast, the D. melanogaster ortholog was found in two segmentally repeated neurons associated with the dorsolateral sensory organs and a few cells in the VNC (fig. 5Q and R). CG14401 was found only in the ventral muscles of C. capitata embryos (supplementary fig. 5S and T, Supplementary Material online), whereas in D. melanogaster, just the garland cells (a set of nephrocytes associated with the esophagus; fig. 5U) and the terminal portion of the trachea expressed this gene (fig. 5V).
Reconstruction of the Tissue-Specific Expression Diversification
To establish the tissue-specificities of the founding aC1 ortholog and to trace the probable path of expression diversification in Clade 1, we have conducted parsimony analyses of character evolution using reconciliation trees (see Materials and Methods). We have classified the tissue-specific expression domains into 16 characters (fig. 8, supplementary fig. S7, Supplementary Material online), including four tissues such as the chordotonal organs and the ring gland, which were either only present or identifiable in a subset of species. Although the inclusion of the latter tissues may have lead to an overestimation of the relative number of neofunctionalization events, it allowed us to better identify the divergence between the paralogs and between more closely related orthologs within the higher dipteran species. It should also be noted that by classifying the tissue expression into only 16 domains, we did not take into account spatial changes within each tissue, which represents another level of functional divergence (e.g., anterior heart vs. posterior heart, different subsets of neurons and glia). For our analysis, we defined neofunctionalization as acquisition of new tissue expression in a duplicate relative to the unduplicated ortholog; conservation as inheritance of a particular tissue-specific expression by both duplicates; subfunctionalization as complementary partitioning of the ancestral tissue-specificities with or without few overlaps (Hahn 2009). As one node in the tree (for a-36/9338, CG9336, CG9338, CG14401, and CG31675) contained a polytomy, we carried out the reconstruction for all three possible combinations of the node. For all three alternative reconciliation trees, the results of the character reconstruction were virtually identical and are summarized in figure 8 and supplementary figure S7, Supplementary Material online.
According to our reconstruction, the hypothetical unduplicated founder aC1 at the base of Cyclorrapha was most likely expressed in the neuronal and ectodermally derived tissues, namely, in the glia, the trachea, the epidermis, the hindgut, the photoreceptor neurons, and the CNS neurons. The expression in the first five tissues appeared as a pleisiomorphic characters of aC1 orthologs, whereas the expression in the CNS neurons was a lineage-specific acquisition after the split of lower and higher Diptera. aC1 then duplicated to give rise to the ancestors of Subclade 1A (CG31676-twit pair) and to the clade containing Subclade 1B and the atilla lineage (figs. 2 and 8).
The Subclade 1A ancestor retained the expression in the CNS neurons, the photosensory neurons, and the gut, but lost the expression in the glia, the trachea, and the epidermis. After the duplication into CG31676 and twit, the former inherited the gut expression and the CNS neuronal expression (only retained in C. capitata), while acquiring expression in the PNS neurons. Meanwhile, twit inherited only the neuronal tissue-specificity, representing a potential case of subfunctionalization. Although the twit expression pattern remained relatively stable throughout cyclorrhaphan evolution, that of CG31676 underwent many lineage-specific changes. These include novel expression in mesodermal derivatives such as the heart, the muscles, and the gonad as well as independent losses of the CNS expression in M. abdita and D. melanogaster.
In contrast to the Subclade 1A ancestral gene, the common ancestor of Subclade 1B and atilla inherited all the tissue-specificities of the founding ortholog aC1. Upon its duplication, it passed on all the expression domains to both atilla and the ancestor of Subclade 1B (aC1B in fig. 2). However, these two copies followed very different trajectories of functional divergence. On one hand, atilla underwent marked lineage-specific expression changes. In Schizophora (represented by C. Capitata and D. melanogaster), it lost many of its original tissue-specificities, whereas in M. abdita, it retained the ancestral expression domains. Moreover, the latter gene duplicated further to give rise to atilla1 and atilla2, after which some of the tissue-specificities have been partitioned between the two paralogs resulting in another case of subfunctionalization in our analysis.
On the other hand, in Subclade 1B, the ancestral expression domains have been retained by one paralog in all three species. In M. abdita, aC1B1 inherited most of the ancestral tissue-specificities, whereas the other three paralogs lost most or all of the embryonic expression. Among the Schizophora, the ancestral features were retained by a-36/38 in C. capitata and by CG9336 in D. melanogaster after two and three additional rounds of duplication events, respectively (in cases of the alternative tree in supplementary fig. S7A, Supplementary Material online, one and two duplication events, respectively).
After the final duplication step, which took place at the base of the drosophilid lineage, CG9336 inherited most of the tissue-specificities of a-36/38. CG9336 and CG9338 still share expression in the Bolwig’s organ and in a subset of glial cells, but only the former gene retained expression in the hindgut and the epidermis. Each duplicate also gained novel expression domains such as the heart in both duplicates and the hemocytes in CG9338. The other paralogs in Subclade 1B lost all or most of the ancestral expression domains. These include CG31675, virtually not expressed in C. capitata and only present in the dorsolateral PNS neurons in D. melanogaster and CG14401, found exclusively in novel tissues such as the muscles in C. capitata and the garland cells in D. melanogaster.
Overall, compared with the six tissue-specificities of the aC1 founding ortholog, this subfamily as a whole gained ten new expression domains including independent acquisitions in the same tissues (muscle, heart, and PNS neurons) by several paralogs. In two of the three alternative trees (fig. 8, supplementary fig. S7B, Supplementary Material online), out of 19 acquisitions of new expression domains after the start of expansion process (or 18 depending on the scenario), 9 (or 8) tissue-specificities were acquired following duplication events, and 10 were species-specific acquisitions. In these trees, there were 19 instances of conservation of tissue-specific expression and 31 instances of asymmetric inheritance after duplication (or depending the scenario, 18 and 30, respectively). For the third tree (supplementary fig. S7A, Supplementary Material online), out of 18 novel tissue-specificities, 8 were gained after duplication events and 10 after speciation. There were 19 conservation events and 27 asymmetric inheritance events. We found only two instances of subfunctionalization in two of the trees and three in the third tree (fig. 8, supplementary fig. S7, Supplementary Material online). Finally, there was only one potential case where the entire suit of expression domains was conserved in both duplicates (inferred ancestral genes of Subclade 1B and atilla).
Discussion
In this study, we investigated how expression patterns of a gene family evolved during the course of its expansion. Our approach consisted in determining the duplication trajectory of a defined set of paralogs and subsequently analyzing in toto their embryonic tissue-specificities across six different species.
We have chosen to restrict our analysis to embryonic development because at this stage the species considered are most amenable to whole mount in situ hybridization, which allows visualization of gene expression in the entire body. Also, the embryos are to a large extent anatomically comparable, although some features such as the larval visual organs and the legs are either highly reduced or lost in the dipteran species studied. However these differences do not affect our analysis, as these structures consist of tissues present in all species. Our exclusive examination of the embryonic stages may, however, result in a systematic underestimation of the extent of paralog divergence, as we ignore their postembryonic requirements and, thus, a large portion of the factors that could have sculpted their functional fates. Nevertheless, our embryonic data clearly show that the studied paralogs have undergone considerable divergence in their transcriptional regulation. We considered the tissue-specific expression as a proxy for function and, concomitantly postulate that expression differences, to a large extent, reflect functional diversification.
In comparison to genome-wide studies using microarray or RNA-seq data, our approach allows more precise detection of restricted and dynamic tissue-specific expression. Although the resolution of the high throughput studies is limited to the organ level, which in many cases consists of multiple tissue types, we are able to detect spatial changes within the tissues, even at the cellular level resolution. Consequently, our analysis is likely to reveal more cases of expression divergence (neofunctionalization and subfunctionalization) and provides better estimations of the evolutionary dynamics of transcriptional regulation upon gene duplication (Duarte et al. 2006; Liu et al. 2011; Johnson et al. 2013).
The members of the insect Ly6 gene family studied here also present very divergent amino acid sequences, indicating that changes in the protein structure have also played an important role during their functional diversification. Although many members of the Drosophila Ly6 gene superfamily are known to have genetically separable functions (Moussian et al. 2005; Hijazi et al. 2009; Nilton et al. 2010; Wu et al. 2010; Kim and Marqués 2012), we did not consider to what extent the nine paralogs in this analysis have diverged or remained redundant in their protein function. However, each member likely contributes to fitness either by carrying out separate genetic functions or by conferring phenotypic robustness through redundancy (Wagner 2005), as none of the genes was lost in the 12 fully sequenced species (divergence time estimated at 40 Ma). In addition, the results of z-tests for purifying selection clearly indicate that these duplicates have been maintained by this selective force (supplementary table S5, Supplementary Material online).
Evolutionary Diversification of Tissue-Specific Expression Patterns during Gene Family Expansion
A global comparison of the tissue-specificities of the founding ortholog aC1 and its duplicates revealed two striking patterns. First, the expansion of this Ly6 subfamily increased the number of tissues in which these genes are utilized from 6 to 16, including the species-specific variations. Not only did the expression domains expand within the ectoderm-derived tissues, but also to different mesodermal derivatives. Second, one paralog in each of the three cyclorrhaphan species (i.e., aC1B1, a-36/38, and CG9336) inherited most, if not all, of the six original expression domains of the founder gene aC1 (except in M. abdita in which atilla1 also appears to have inherited many of the original expression domains). In comparison, the other paralogs evolved much narrower expression breadths than the ancestral gene, indicating substantial loss of the original expression domains. This observation is consistent with previous findings from several genome-wide studies, which showed that individual members of larger gene families tend to have narrower expression breadths (Huminiecki and Wolfe 2004; Freilich et al. 2006; Farré and Albà 2010). Importantly, our parsimony analysis of the expression divergence process revealed that this trend is produced by a repeated asymmetric inheritance of the ancestral expression domains through at least four duplication events whereby one copy inherited the ancestral tissue-specificities, whereas the other copy lost all or most of the old expression domains.
Asymmetric Expression Divergence and Bias in Duplication Frequency
Although asymmetric divergence of expression domains as well as asymmetric sequence evolution after gene duplication was previously observed in genome-wide studies (Wagner 2002; Scannell and Wolfe 2008; Assis and Bachtrog 2013; Pegueroles et al. 2013), our study provides the first observation that the asymmetric inheritance persists through multiple rounds of duplication events.
This observation suggests that the copy maintaining the wide ancestral expression domain appears to act as a “seed” generating a copy with diverged expression. Is this predicted from previous theoretical and empirical studies? Theoretical models of expression divergence under neutral loss indicate 1) a pair of duplicates evolving asymmetric expression domains are more likely to be preserved (Wagner 2002) and 2) genes with a large number of cis-regulatory modules and expression in many tissues are more likely to be preserved after duplication (Lynch and Force 2000). Furthermore, Assis and Bachtrog (2013) found that genes expressed in greater number of tissues tended to produce a duplicate with novel tissue-specificity, which, in turn, would be more likely to be preserved under positive selection. If these theoretical and empirical evidences are extrapolated to multiple rounds of gene duplications in expanding gene families, one might predict to find one (or more) paralog with a broad tissue-specificity behaving like aCB1. It would be interesting to see whether other rapidly expanding gene families behave similarly, thus unveiling a general rule about the dynamics of gene family expansion and functional divergence.
Prevalence of asymmetric divergence during the expansion of this subfamily indicates an abundant loss of expression domains in copies that do not retain the ancestral expression. The observed frequency of expression loss after duplication exceeded that of expression gain in this Ly6 subfamily and was probably a major process contributing to its expression diversification. This is in agreement with the results of Oakley et al. (2006) showing that the rate of expression loss in Drosophila gene families was at least twice as high as that of expression gain. Further, their model suggested that gene families expanding more rapidly should have greater rates of expression loss, thereby, highlighting the important contribution of tissue-specific expression loss in diversification of gene families like the Ly6 subfamily. Taken together, the maintenance of the original expression domains in one copy and the rapid loss in the other copy after duplication events likely produced the observed pattern of asymmetric inheritance of the ancestral domains.
Neofunctionalization is Equally Frequent after Duplication Events and Speciation
In accordance with several recent studies concluding that neofunctionalization after gene duplication is a frequent outcome (and more common than subfunctionalization) (Kassahn et al. 2009; Assis and Batchtrog 2013), the duplication events in the Ly6 subfamily studied here were often accompanied by acquisitions of novel tissue-specificities. Remarkably, though, the instances of neofunctionalization occurred as often following duplication events as following speciation (8 or 9 after duplication vs. 10 after speciation) resulting in high interspecific variation between orthologs.
It has previously been proposed that expression divergence between interspecific orthologs of duplicated genes occurs more rapidly than between those of unduplicated genes. This was based on the premise that a duplicated pair retains certain degrees of functional redundancy, enabling one copy to adapt to species-specific functional requirements (Ohno 1970). In support of this hypothesis, two previous genome-wide studies reported higher divergence in temporal (Gu et al. 2004) and tissue-specific expression (Ha et al. 2009) between interspecific orthologs of duplicated genes. Considerable interspecific variations found in our study highlight the importance of examining the expression domains of orthologs and paralogs in multiple species for an accurate inference of their functional fates.
In agreement with the previous genome-wide studies reporting underrepresentation of subfunctionalization after duplication (Kassahn et al. 2009; Assis and Bachtrog 2013), we only found two to three potential cases of subfunctionalization in comparison to the eight clear instances of neofunctionalization associated with duplication events. In two of the three cases, the ancestral expression domains were not completely partitioned, but accompanied by conservation and neofunctionalization illustrating that these processes are not mutually exclusive (Huminiecki and Wolfe 2004; He and Zhang 2005).
The Mechanisms Behind cis-Regulatory Divergence in Expanding Gene Families
The nature of the genetic mechanisms causing duplications has been shown to influence the subsequent divergence of the duplicates due to their direct impact on cis-regulatory regions (Casneuf et al. 2006; Cusack and Wolfe 2007; Assis and Bachtrog 2013; Katju 2013). Whereas mechanisms such as transposition are thought to radically perturb original cis-regulatory regions (Cusack and Wolfe 2007; Duncan and Dearden 2010; Assis and Bachtrog 2013), whole genome or large-scale segmental duplications are more likely to allow enhancer conservation and, thus, original expression domains (Casneuf et al. 2006). Finally, small-scale processes such as tandem duplication are thought to have intermediate chances of altering the original regulatory landscape (Casneuf et al. 2006; Blount et al. 2012; Katju 2013).
Zhou et al. (2008) showed that 80% of the nascent paralogs within the drosophilid lineage originated as tandem duplicates. Likewise, the Clusters III and V Ly6 genes most likely appeared through this mechanism, as aC1 and its duplicates appear in contiguous positions in the genomes and share the same exon–intron structure. Our paralog alignments did not reveal the presence of chimeric proteins, suggesting that coding regions and most probably introns and proximal promoters could have duplicated as intact copies.
Despite this, only few overlapping patterns were identified when comparing intraspecific paralogs in the extant species. Thus, either remodeling of intergenic regulatory regions had an immediate impact on the divergence through processes such as enhancer loss and formation of chimeric enhancers (Rogers et al. 2010; Rogers and Hartl 2012), or expression divergence has gradually occurred after the duplication events. Interestingly, the latter should be the case for the ten instances of neofunctionalization following speciation reported here. Although these two different mechanisms explaining divergence are not mutually exclusive, our observations indicate that studying lineage-specific divergence of tandem duplicates could illuminate the process underlying their expression diversification.
Why Does the Ly6 Family Expand and Diversify So Rapidly?
A comprehensive explanation of why the insect Ly6 genes underwent such episodes of rapid expansion and expression diversification must also take into account their genetic requirements and the nature of their cellular functions. Unfortunately, we do not know at present the precise biochemical function of any of these proteins. However, we propose that they are likely to have roles in terminal tissue differentiation or physiology and are not expected to play prominent roles in early developmental processes. These assumptions are supported by several observations. First, their expression initiates relatively late during embryonic development and assumes patterns of expression often restricted to specific differentiated cell types. We also detected substantial variation in tissue-specificity among orthologs, indicating that their function is not deeply wired within essential developmental processes. Finally, the analysis of twit null mutant, the only Clade1 mutant reported in Drosophila, has shown that this gene is not essential for viability (Kim and Marqués 2012).
Interestingly, it has been proposed that “nonessential” genes tend to leave more duplicates in the genome (He and Zhang 2006; Woods et al. 2013). Genes can be nonessential due to their subtle fitness effects or genetic redundancy. For genes with subtle phenotypic effect such as twit, having extra copies may only cause slight dosage effects, which would allow retention of duplicated copies and expression in new tissues without incurring negative fitness consequences (He and Zhang 2006; Makino et al. 2009). In the case of functionally redundant genes, one copy would be allowed to lose expression in the original tissue and diverge. It will thus be interesting to analyze in the future the functional roles of the rest of the paralogs to unravel their contribution to fitness and the degree of genetic redundancy between them.
Some of the rapidly expanding gene families known in higher Diptera contain many members which are seldom identified in genetic screens, perhaps reflecting their subtle phenotypic effects or redundancy (Fradkin et al. 2002; Patel et al. 2012). In a striking parallel to the Ly6 superfamily, members of these families, some of which have arisen as tandem duplicates, have highly diverse tissue-specific expression (Fradkin et al. 2002; Patel et al. 2012). In the methuselah gene family, a subfamily consisting of 12 genes in D. melanogaster (Patel et al. 2012) displays a multitude of embryonic tissue-specific expression patterns. These genes appear to have arisen from a singleton ortholog still found in T. castaneum, whose expression is restricted to few tissues (Patel et al 2012). Thus, it appears that the nonessentiality of the gene products and the semiconservative mechanism of duplication, which has the potential to immediately alter tissue-specific regulation, may be common factors underlying lineage-specific expansion and marked expression divergence found in rapidly expanding gene families across insect genomes.
Materials and Methods
Sequence Retrieval
Ly6 sequences were retrieved from publicly available genomic databases using the iterative PSI-BLAST algorithm (Altschul et al. 1997). For this, the amino acid sequences of all 36 D. melanogaster Ly6 genes (Hijazi et al. 2009) were first used as queries to recover the full complement of Ly6 superfamily members in each species. Subsequently, the sequences retrieved from each species were used as queries for additional rounds of search to ensure that no species-specific members were missed. Alternatively, the TBLASTN program was used for the transcriptomic sequences (Jiménez-Guri et al. 2013) and the unreleased genome assembly of M. abdita, using as queries the amino acid sequences of the D. melanogaster and An. gambiae proteins. Where possible, the syntenic organizations of the sequences from each genome were characterized. Finally, among these sequences, putative homologs of the nine D. melanogaster Ly6 genes in the study were identified based on sequence similarities and conserved synteny. The accession numbers of the identified Cluster III and V homologs are available in supplementary figure S1A, Supplementary Material online. For subsequent sequence analyses, we have selected the region between the first and the tenth conserved cysteine residues present in the TFD of each gene product.
Multiple Sequence Alignment and Phylogenetic Tree Inference
The amino acid sequences of the proteins in this study were particularly challenging for conventional multiple sequence alignment (MSA) and phylogenetic tree inference methods due to the relatively short length of the TFD domains and their typically high sequence divergence. To accommodate this challenge, we opted to use BAli-Phy program (Redelings and Suchard 2005; Suchard and Redelings 2006), a Bayesian Markov-chain Monte Carlo (MCMC) based program, which simultaneously estimates the MSA and phylogenetic tree relating the sequences. The advantages of BAli-Phy are 1) the MSA is independent of a potentially inaccurate single guide-tree, 2) accurate gap placement using a phylogenetic Indel model, 3) direct incorporation of MSA uncertainty in the inference of the phylogenetic tree and vice versa, and 4) the ease by which the Bayesian paradigm can incorporate prior knowledge of sequences in the form of alignment constraints. The last point was exploited by forcing the homology of the cysteine residue containing sites, which are known to be essential for the structure and the function of the Ly6 proteins. However, with or without this constraint, the results of MSA of the 47 Ly6 proteins were nearly identical (supplementary fig. S1B, Supplementary Material online)
All BAli-Phy runs used the site rate heterogeneity model, Γ4 + Inv, and the Indel model RS072, and consisted of five independent chains of 90,000 iterations after a burnin of 9,999 iterations. No subsampling was used. Analyses were run with different substitution matrices as well as with or without a distantly related Ly6 protein Coiled as an outgroup (Nilton et al. 2010; Hijazi et al. 2011; Syed et al. 2011). The outputs are shown in supplementary table S1, Supplementary Material online. The runs obtaining the highest likelihood values either with or without Cold as an outgroup are highlighted in yellow. A third substitution model (JTT [Jones, Taylor, and Thorton]) has also been tested, but consistently resulted in lower likelihood values and was therefore not considered further (data not shown).
To further confirm the tree topology obtained from BAli-Phy (a combined Bayesian inference of phylogenetic tree and MSA), we applied the Bayesian tree inference program MrBayes on the MSAs (Huelsenbeck and Ronquist 2001). This corresponds to fixing the MSAs obtained by BAli-Phy, neglecting the alignment uncertainty computed and used by BAli-Phy for the inference of the trees (supplementary fig. S1B, Supplementary Material online). Each run comprised five independent Metropolis-coupled MCMC chains, each consisting of three heated and one cold chain, running for 500,000 generations with a relative burnin of 10%. Trees were sampled every 500 generations. The outputs of MrBayes are summarized in supplementary table S2, Supplementary Material online.
Analysis of Character Evolution
To trace the diversification of the tissue-specificities, we combined information from the species phylogeny, the synteny, and the gene tree, and manually built a reconciliation tree for the genes whose expression patterns were examined. Reconstruction of the ancestral tissue-specificities of the unduplicated genes was carried out using the parsimony method with MacClade 4.08a software (Maddison DR and Maddison WP 2006). We opted to use this more conservative approach because the reconciliation tree lacked the basis for justified branch length estimates, which are required for using the maximum-likelihood method. Individual expression domains were coded as a character with binary states (expression present or absent). Step matrix was specified where no changes cost zero steps, loss of expression one step, and gain of expression two steps. The relative costs of gain and loss were based on the ratio between frequencies of expression loss and gain after gene duplication reported by Oakley et al. (2006). To circumvent one polytomy in the tree, the reconstruction was carried out for all three possible branch configurations (fig. 8, supplementary fig. S7A and B, Supplementary Material online).
Animal Husbandry
Tribolium castaneum (San Bernadino strain, a kind gift from Dr Gregor Bucher, Georg August University) was raised at 29 °C on the whole wheat flour supplemented with 5% Brewer’s yeast. The C. capitata culture was generously provided by Dr Andrew Jessup (IAEA Seibersdorf, Austria). The adults were reared on a diet of sugar and hydrolyzed yeast protein and the larvae on a mixture of bran, sugar, and yeast. Megaselia abdita, kind gift of Dr Johannes Jaeger (CRG, Barcelona), was raised according to Rafiqi et al. (2011a). Bi. anynana and An. stephensi eggs were kindly provided by Dr Patrícia Beldade (IGC, Portugal) and Dr Maria Mota (IMM, Portugal), respectively. Drosophila melanogaster strains were reared on standard cornmeal food at 25 °C and included the Oregon R wild type strain and the transgenic Cambridge Protein Trap Insertion lines CG9336CPTI001654 and CG9338CPTI100000 provided by the Kyoto DGRC Stock Center (Lowe et al. 2014).
In situ Hybridization and Immunocytochemistry
To make riboprobes for in situ hybridization, partial fragments of each gene were cloned from embryonic or larval cDNA and used as templates. The sequences of the cloning primers used to make the riboprobes and the accession numbers of repo and elav orthologs of T. castaneum, Bi. anynana, and An. stephensi are available in supplementary figure S8, Supplementary Material online.
Drosophila melanogaster, C. capitata, and M. abdita embryos were dechorionated and fixed according to Tautz and Pfeifle (1989). Tribolium castaneum embryos were processed as in Schinko et al. (2009) and removed from the eggs prior to the in situ hybridization step. Bicyclus anynana embryos were dechorionated in 50% bleach for 3 min and rinsed in DEPC-treated phosphate-buffered saline (PBS) three times. The eggs were poked with forceps to make a small opening, then fixed overnight in 3.7% formaldehyde in PBS, stored in methanol at −20 °C, and dissected prior to in situ hybridization. Anopheles stephensi embryos were fixed using the protocol developed for M. abdita by Rafiqi et al. (2011b) and their serosa was removed as in Clemons et al. (2010). In situ hybridization was carried out as in Panganiban et al. (1995) following the protocol of Tautz and Pfeifle (1989) with the following modifications. Bicyclus anynana, An. stephensi, and C. capitata embryos were incubated for 3 min and T. castaneum embryos for 5 min in 4 µg/ml proteinase K at 37 °C, and the hybridization buffer included heparin instead of glycogen. Hybridization was carried out at 55 °C or 65 °C. Embryos were mounted in 70% glycerol in PBS and observed under the Leica DM LB2 upright microscope.
For immunocytochemistry, the embryos were blocked in 5% normal goat serum in PBT (PBS with 0.1% Tween) for 30 min followed by overnight incubation in primary antibodies and secondary antibodies, respectively. The antibody concentrations used were 1:10 rat anti-Elav antibody (7E8A10, DSHB), 1:50 mouse anti-Repo (8D12, DSHB), 1:250 rabbit anti-VASA (kind gif of P. Lasko, McGill University), 1:500 rabbit anti-GFP (Torrey Pines Biolabs), 1:000 Alexa488 anti-rat, 1:200 TRIC anti-mouse, and 1:200 FITC anti-rabbit (all from Invitrogen).
For simultaneous detection of the Elav or VASA protein and the Ly6 mRNA transcripts, we performed the immunocytochemistry protocol after the in situ hybridization protocol. Instead of NBT/BCIP color substrates, FastRed (Sigma) was used for developing the color reaction.
Fluorescent images were collected under the Leica SP5 inverted confocal microscope. All images were processed using the Fiji software (Schindelin et al. 2012) and Adobe Photoshop (Adobe Systems).
Supplementary Material
Supplementary tables S1–S5 and figures S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Johannes Jaeger and Dr Eva Jiménez-Guri for providing Megaselia abdita resources including the prereleased genome sequence, Gregor Bucher for the Tribolium castaneum culture, Patrícia Beldade for Bicyclus anynana eggs and comments on the manuscript, Maria Mota and António Mendes for Anopheles stephensi eggs, Andrew Jessup (IAEA Seibersdorf, Austria) for the Ceratitis capitata culture, Al Handler (USDA, Florida) for sharing the prereleased C. capitata genome sequence, Mohamed Noor (Duke University) for blasting Megaselia scalaris sequences, and Paul Lasko (McGill University) for the VASA antibody. They also acknowledge Dr Marc Haenlin and Dr Lucas Waltzer for sharing their lab space and Luis González for his technical assistance. They also thank the DRGC (Kyoto), the DSHB (Iowa), and the Toulouse RIO Imaging Platform for making available reagents and facilities. K.T. is supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal (SFRH/BPD/75139/2010). This work was supported by Fundação Calouste Gulbenkian/ Instituto Gulbenkian de Ciência and by Agence Nationale de la Recherche (ANR) (France) and FCT (Portugal) through ANR-13-ISV7-0001-01 to F.R. and ANR-13-ISV7-0001-02 and FCT-ANR/BIA-ANM/0003/2013 to E.S.
References
- Alonso CR, Wilkins AS. The molecular elements that underlie developmental evolution. Nat Rev Genet. 2005;6:709–715. doi: 10.1038/nrg1676. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 2013;110:17409–17414. doi: 10.1073/pnas.1313759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL, Achaz G. cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Morales A, Monzón-Sandoval J, Urrutia AO, Gutiérrez H. Increased brain size in mammals is associated with size variations in gene families with cell signalling, chemotaxis and immune-related functions. Proc Biol Sci. 2014;281:20132428. doi: 10.1098/rspb.2013.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari SS, Arakane Y, Specht CA, Moussian B, Kramer KJ, Muthukrishnan S, Beeman RW. Retroactive maintains cuticle integrity by promoting the trafficking of Knickkopf into the procuticle of Tribolium castaneum. PLoS Genet. 2013;9:e1003268. doi: 10.1371/journal.pgen.1003268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons A, Haugen M, Flannery E, Kast K, Jacowski C, Severson D, Duman-Scheel M. Fixation and preparation of developing tissues from Aedes aegypti. Cold Spring Harb Protoc. 2010;2010:pdb.prot5508. doi: 10.1101/pdb.prot5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol. 2007;24:679–686. doi: 10.1093/molbev/msl199. [DOI] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- Duncan EJ, Dearden PK. Evolution of a genomic regulatory domain: the role of gene co-option and gene duplication in the enhancer of split complex. Genome Res. 2010;20:917–928. doi: 10.1101/gr.104794.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré D, Albà MM. Heterogeneous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol. 2010;27:325–335. doi: 10.1093/molbev/msp242. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin LG, Kamphorst JT, DiAntonio A, Goodman CS, Noordermeer JN. Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc Natl Acad Sci U S A. 2002;99:13663–13668. doi: 10.1073/pnas.212511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Massingham T, Blanc E, Goldovsky L, Thornton JM. Relating tissue specialization to the differentiation of expression of singleton and duplicate mouse proteins. Genome Biol. 2006;7:R89. doi: 10.1186/gb-2006-7-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, Wüster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57:110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- Galat A. Common structural traits for cystine knot domain of the TGFβ superfamily of proteins and three-fingered ectodomain of their cellular receptors. Cell Mol Life Sci. 2011;68:3437–3451. doi: 10.1007/s00018-011-0643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A, Gross G, Drevet P, Sato A, Ménez A. Conserved structural determinants in three-fingered protein domains. FEBS J. 2008;275:3207–3225. doi: 10.1111/j.1742-4658.2008.06473.x. [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li W-H. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim E-D, Chen ZJ. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc Natl Acad Sci. 2009;106:2295–2300. doi: 10.1073/pnas.0807350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Higher duplicability of less important genes in yeast genomes. Mol Biol Evol. 2006;23:144–151. doi: 10.1093/molbev/msj015. [DOI] [PubMed] [Google Scholar]
- Hijazi A, Haenlin M, Waltzer L, Roch F. The Ly6 protein coiled is required for septate junction and blood brain barrier organisation in Drosophila. PLoS One. 2011;6:e17763. doi: 10.1371/journal.pone.0017763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi A, Masson W, Augé B, Waltzer L, Haenlin M, Roch F. Boudin is required for septate junction organisation in Drosophila and codes for a diffusible protein of the Ly6 superfamily. Development. 2009;136:2199–2209. doi: 10.1242/dev.033845. [DOI] [PubMed] [Google Scholar]
- Holland LZ. Evolution of new characters after whole genome duplications: insights from amphioxus. Semin Cell Dev Biol. 2013;24:101–109. doi: 10.1016/j.semcdb.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Huynen MA, Gabaldón T. Evidence for short-time divergence and long-time conservation of tissue-specific expression after gene duplication. Brief Bioinform. 2011;12:442–448. doi: 10.1093/bib/bbr022. [DOI] [PubMed] [Google Scholar]
- Huminiecki L, Wolfe KH. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 2004;14:1870–1879. doi: 10.1101/gr.2705204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Guri E, Huerta-Cepas J, Cozzuto L, Wotton KR, Kang H, Himmelbauer H, Roma G, Gabaldón T, Jaeger J. Comparative transcriptomics of early dipteran development. BMC Genomics. 2013;14:123. doi: 10.1186/1471-2164-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Atallah J, Plachetzki DC. The importance of tissue specificity for RNA-seq: highlighting the errors of composite structure extractions. BMC Genomics. 2013;14:586. doi: 10.1186/1471-2164-14-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: Comparative analyses in vertebrates. Genome Res. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V. To the beat of a different drum: determinants implicated in the asymmetric sequence divergence of Caenorhabditis elegans paralogs. BMC Evol Biol. 2013;13:73. doi: 10.1186/1471-2148-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NC, Marqués G. The Ly6 neurotoxin-like molecule target of wit regulates spontaneous neurotransmitter release at the developing neuromuscular junction in Drosophila. Dev Neurobiol. 2012;72:1541–1558. doi: 10.1002/dneu.22021. [DOI] [PubMed] [Google Scholar]
- Liu S-L, Baute GJ, Adams KL. Organ and cell type-specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in. Arabidopsis thaliana. Genome Biol Evol. 2011;3:1419–1436. doi: 10.1093/gbe/evr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe N, Rees J, Roote JS, Ryder E, Armean IM, Johnson G, Drummond E, Spriggs H, Drummond J, Magbanua JP, et al. Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development. 2014;141:3994–4005. doi: 10.1242/dev.111054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. 2005. MacClade 4: Analysis of phylogeny and character evolution. Version 4.08a. http://macclade.org, Sunderland (MA): Sinauer Associates, Inc. [Google Scholar]
- Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, O’Meara BC. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Söding J, Schwarz H, Nüsslein-Volhard C. Retroactive, a membrane-anchored extracellular protein related to vertebrate snake neurotoxin-like proteins, is required for cuticle organization in the larva of Drosophila melanogaster. Dev Dyn. 2005;233:1056–1063. doi: 10.1002/dvdy.20389. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Nilton A, Oshima K, Zare F, Byri S, Nannmark U, Nyberg KG, Fehon RG, Uv AE. Crooked, coiled and crimpled are three Ly6-like proteins required for proper localization of septate junction components. Development. 2010;137:2427–2437. doi: 10.1242/dev.052605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley TH, Ostman B, Wilson AC. Repression and loss of gene expression outpaces activation and gain in recently duplicated fly genes. Proc Natl Acad Sci U S A. 2006;103:11637–11641. doi: 10.1073/pnas.0600750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin (Germany): Springer-Verlag; 1970. [Google Scholar]
- Ohta T. Multigene families and the evolution of complexity. J Mol Evol. 1991;33:34–41. doi: 10.1007/BF02100193. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Sebring A, Nagy L, Carroll SB. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- Patel MV, Hallal DA, Jones JW, Bronner DN, Zein R, Caravas J, Husain Z, Friedrich M, Vanberkum MFA. Dramatic expansion and developmental expression diversification of the methuselah gene family during recent Drosophila evolution. J Exp Zool Part B Mol Dev Evol. 2012;318:368–387. doi: 10.1002/jez.b.22453. [DOI] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Albà MM. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 2013;30:1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Rafiqi AM, Lemke S, Schmidt-Ott U. The scuttle fly Megaselia abdita (Phoridae): a link between Drosophila and Mosquito development. Cold Spring Harb Protoc. 2011a;2011:pdb.emo143. doi: 10.1101/pdb.emo143. [DOI] [PubMed] [Google Scholar]
- Rafiqi AM, Lemke S, Schmidt-Ott U. Megaselia abdita: fixing and devitellinizing embryos. Cold Spring Harb Protoc. 2011b;2011:pdb.prot5602. doi: 10.1101/pdb.prot5602. [DOI] [PubMed] [Google Scholar]
- Redelings BD, Suchard MA. Joint Bayesian estimation of alignment and phylogeny. Syst Biol. 2005;54:401–418. doi: 10.1080/10635150590947041. [DOI] [PubMed] [Google Scholar]
- Rogers RL, Bedford T, Lyons AM, Hartl DL. Adaptive impact of the chimeric gene Quetzalcoatl in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2010;107:10943–10948. doi: 10.1073/pnas.1006503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, Hartl DL. Chimeric genes as a source of rapid evolution in Drosophila melanogaster. Mol Biol Evol. 2012;29:517–529. doi: 10.1093/molbev/msr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Wolfe KH. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinko J, Posnien N, Kittelmann S, Koniszewski N, Bucher G. Single and double whole-mount in situ hybridization in red flour beetle (Tribolium) embryos. Cold Spring Harb Protoc. 2009;2009:pdb.prot5258. doi: 10.1101/pdb.prot5258. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Redelings BD. BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics. 2006;22:2047–2048. doi: 10.1093/bioinformatics/btl175. [DOI] [PubMed] [Google Scholar]
- Syed MH, Krudewig A, Engelen D, Stork T, Klämbt C. The CD59 family member leaky/coiled is required for the establishment of the blood-brain barrier in Drosophila. J Neurosci. 2011;31:7876–7885. doi: 10.1523/JNEUROSCI.0766-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJR, Kerkkamp HME, Vos RA, Guerreiro I, Calvete JJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, Verstrepen KJ. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012;10:e1001446. doi: 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Wagner A. Distributed robustness versus redundancy as causes of mutational robustness. BioEssays. 2005;27:176–188. doi: 10.1002/bies.20170. [DOI] [PubMed] [Google Scholar]
- Woods S, Coghlan A, Rivers D, Warnecke T, Jeffries SJ, Kwon T, Rogers A, Hurst LD, Ahringer J. Duplication and retention biases of essential and non-essential genes revealed by systematic knockdown analyses. PLoS Genet. 2013;9:e1003330. doi: 10.1371/journal.pgen.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Robinson JE. SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr Biol. 2014;24:621–629. doi: 10.1016/j.cub.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, Li X, Ding Y, Yang S, Wang W. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.