Abstract
Background
Aberrant DNA methylation at the 5-carbon on cytosine residues (5mC) in CpG dinucleotides is probably the most extensively characterized epigenetic modification in colon cancer. It has been suggested that the loss of adenomatous polyposis coli (APC) function initiates tumorigenesis and that additional genetic and epigenetic events are involved in colon cancer progression. We aimed to study the genome-wide DNA methylation profiles of intestinal tumorigenesis in Apcmin/+ mice.
Results
Methylated DNA immunoprecipitation (MeDIP) followed by next-generation sequencing was used to determine the global profile of DNA methylation changes in Apcmin/+ mice. DNA was extracted from adenomatous polyps from Apcmin/+ mice and from normal intestinal tissue from age-matched Apc+/+ littermates, and the MeDIP-seq assay was performed. Ingenuity Pathway Analysis (IPA) software was used to analyze the data for gene interactions. A total of 17,265 differentially methylated regions (DMRs) displayed a ≥ 2-fold change (log2) in methylation in Apcmin/+ mice; among these DMRs, 9,078 (52.6 %) and 8,187 (47.4 %) exhibited increased and decreased methylation, respectively. Genes with altered methylation patterns were mainly mapped to networks and biological functions associated with cancer and gastrointestinal diseases. Among these networks, several canonical pathways, such as the epithelial-mesenchymal transition (EMT) and Wnt/β-catenin pathways, were significantly associated with genome-wide methylation changes in polyps from Apcmin/+ mice. The identification of certain differentially methylated molecules in the EMT and Wnt/β-catenin pathways, such as APC2 (adenomatosis polyposis coli 2), SFRP2 (secreted frizzled-related protein 2), and DKK3 (dickkopf-related protein 3), was consistent with previous publications.
Conclusions
Our findings indicated that Apcmin/+ mice exhibited extensive aberrant DNA methylation that affected certain signaling pathways, such as the EMT and Wnt/β-catenin pathways. The genome-wide DNA methylation profile of Apcmin/+ mice is informative for future studies investigating epigenetic gene regulation in colon tumorigenesis and the prevention of colon cancer.
Keywords: DNA methylation, Epigenetic, MeDIP-seq, Wnt/β-catenin pathway, Epithelial-mesenchymal transition pathway
Introduction
It is widely accepted that the accumulation of genetic and epigenetic alterations contributes to cancer initiation and progression. Genetic alterations refer to mutations in tumor suppressor genes and oncogenes, whereas epigenetic modifications involve changes in chromatin structure that result in altered gene expression without primary changes to the DNA sequence [1]. The information conveyed by epigenetic modifications plays a vital role in regulating DNA-mediated processes, including transcription, DNA repair, and replication [2]. Specifically, aberrant DNA methylation at the 5-carbon on cytosine residues (5mC) in CpG dinucleotides is perhaps the most extensively characterized epigenetic modification in cancer. DNA methylation affects the rate of gene transcription and therefore regulates various biological processes, such as proliferation, apoptosis, DNA repair, cancer initiation, and cancer progression [3]. The genomic DNA methylation pattern is stably maintained in normal cells; however, aberrant alterations in the epigenome have been identified in tumor cells [4]. Evidence suggests that global hypomethylation and regional hypermethylation are characteristics of cancer cells [5]. Global genome-wide loss of methylation has been associated with increased genomic instability and proto-oncogene activation, whereas DNA hypermethylation of CpG islands in promoter regions silences tumor suppressor genes [6]. Unlike genetic mutations, the transcriptional repression of genes via epigenetic alterations can be reversed by further epigenetic modifications because these silenced genes remain genetically intact [7]. Thus, it is very important to profile the global DNA methylation changes that occur in early tumorigenesis.
Colorectal cancer (CRC) is the second leading cause of cancer-related death in western countries [8], and more than 80 % of CRC patients harbor a mutation in the adenomatous polyposis coli (APC) gene on chromosome 5q21 [9]. APC is a tumor suppressor gene that down-regulates the pro-proliferative Wnt-signaling pathway by promoting the destruction of β-catenin. Deleterious mutations in APC stabilize β-catenin, increase its translocation into the nucleus, promote its binding to the transcription factor TCF4, and activate target genes such as C-MYC and CCND1 [10, 11]. It has been suggested that the loss of APC function initiates tumorigenesis and that additional genetic and epigenetic events are involved in colon cancer progression [12]. Numerous genes that are silenced by epigenetic mechanisms have been identified in colon cancer, including CDKN2A [13], DKK1 [14], DLEC1 [15, 16], UNC5C [17], and SFRP [18]. However, the genome-wide profile of the aberrant methylation and the association of these methylation patterns with important signaling pathways and biological networks implicated in colon tumorigenesis remain unclear.
To address this issue, we examined the global DNA methylation profile in the well-established Apcmin/+ intestinal tumorigenesis mouse model using methylated DNA immunoprecipitation (MeDIP) and next-generation sequencing (MeDIP-seq). Apcmin/+ mice carry a heterozygous mutation in Apc and develop approximately 30 small intestinal adenomatous polyps following the somatic loss of functional Apc [19]. This mouse model of intestinal tumorigenesis is commonly used because the phenotype resembles that of patients with familial adenomatous polyposis (FAP) [20]. We analyzed adenomatous polyps from Apcmin/+ mice and not only identified genes with a modified methylation profile but also interpreted the data in the context of biological function, networks, and canonical signaling pathways associated with the methylation patterns.
Results
MeDIP-seq results
To identify changes in DNA methylation patterns during the progression of mouse intestinal polyps, whole-genome DNA methylation analysis was performed using the described MeDIP-seq method. The global differences in the DNA methylation profile between adenomatous polyps from Apcmin/+ mice and intestinal tissue from control mice are described in Fig. 1. We identified 12,761,009 mapped peaks and 2,868,549 non-mapped peaks from a total of 15,629,558 peaks in control mice and 11,470,541 mapped peaks and 2,262,073 non-mapped peaks from a total of 13,732,614 peaks in Apcmin/+ mice (Fig. 1a). A total of 17,265 differentially methylated regions (DMRs) had a ≥ 2-fold change (log2) in methylation in Apcmin/+ mice compared with control mice, of which 9,078 DMRs (52.6 %) exhibited increased methylation, and 8,187 (47.4 %) DMRs exhibited decreased methylation (Fig. 1b).
Fig. 1.
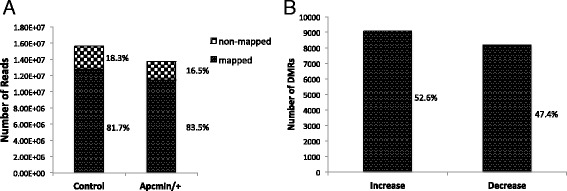
Global changes in the DNA methylation profile between Apc mutant adenomatous polyps and control tissue. a, Total number of peaks generated by MeDIP-seq. b, Number of DMRs with significantly increased or decreased changes in methylation (≥2-fold in log2) in polyps from Apcmin/+ mice
Functional and pathway analysis by IPA
To identify the biological function, networks, and canonical pathways that were affected by the differentially methylated genes, we performed Ingenuity Pathway Analysis (IPA) after the MeDIP-seq analysis. In the analysis of genes with altered methylation (≥2-fold in log2) in Apcmin/+ mice compared with control mice as determined by MeDIP-seq, IPA mapped 5,464 unique genes that were associated with its knowledge base. The top 50 genes with increased and decreased methylation levels based on log2 fold change are listed in Tables 1 and 2. The molecules with methylation changes were mainly categorized into 38 disease and biological functions. The five highest IPA-rated disease and biological functions were as follows: cancer, gastrointestinal disease, organismal injury and abnormalities, cellular growth and proliferation, and reproductive system disease (Fig. 2). Among the IPA-mapped genes with differential methylation patterns in polyps from Apcmin/+ mice, 3,299 were associated with cancer, and 1,668 were associated with gastrointestinal diseases. To examine the interaction networks that were affected by DNA methylation in Apc mutant polyps, IPA identified 25 networks with up to 35 focus molecules in each network. The five most affected gene networks as determined by IPA are shown in Table 3, and the detailed interactions in the most significant networks (cancer, cell cycle, and molecular transport) are presented in Fig. 3. In accordance with the most relevant biological functions as determined by IPA, genes with different methylation patterns predominantly mapped to the networks associated with cancer and gastrointestinal diseases. Taken together, these results suggested an important role for the altered methylation of genes associated with the development of cancer and gut disease in Apcmin/+ mice.
Table 1.
Top 50 annotated genes with increased methylation
| Rank | Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|---|
| 1 | ZNF330 | zinc finger protein 330 | 4.614 | Nucleus | other |
| 2 | ACTR3B | ARP3 actin-related protein 3 homolog B (yeast) | 4.540 | Other | other |
| 3 | CAV3 | caveolin 3 | 4.292 | Plasma Membrane | enzyme |
| 4 | NKX2-3 | NK2 homeobox 3 | 4.199 | Nucleus | transcription regulator |
| 5 | TLN2 | talin 2 | 4.199 | Nucleus | other |
| 6 | CPD | carboxypeptidase D | 4.100 | Extracellular Space | peptidase |
| 7 | CTNNBL1 | catenin, beta like 1 | 4.100 | Nucleus | other |
| 8 | Vmn2r1 | vomeronasal 2, receptor 1 | 4.100 | Plasma Membrane | other |
| 9 | Cmtm2a | CKLF-like MARVEL transmembrane domain containing 2A | 3.993 | Cytoplasm | transcription regulator |
| 10 | HPS6 | Hermansky-Pudlak syndrome 6 | 3.993 | Cytoplasm | other |
| 11 | KANK1 | KN motif and ankyrin repeat domains 1 | 3.993 | Nucleus | transcription regulator |
| 12 | RRP1 | ribosomal RNA processing 1 | 3.993 | Nucleus | other |
| 13 | SNX10 | sorting nexin 10 | 3.993 | Cytoplasm | transporter |
| 14 | UNC93A | unc-93 homolog A (C. elegans) | 3.993 | Plasma Membrane | other |
| 15 | Zfp932 | zinc finger protein 932 | 3.993 | Nucleus | other |
| 16 | ANKRD13D | ankyrin repeat domain 13 family, member D | 3.877 | Plasma Membrane | other |
| 17 | DACT1 | dishevelled-binding antagonist of beta-catenin 1 | 3.877 | Cytoplasm | other |
| 18 | DMRT2 | doublesex and mab-3 related transcription factor 2 | 3.877 | Nucleus | other |
| 19 | DSC3 | desmocollin 3 | 3.877 | Plasma Membrane | other |
| 20 | LDOC1 | leucine zipper, down-regulated in cancer 1 | 3.877 | Nucleus | other |
| 21 | LRRC8B | leucine rich repeat containing 8 family, member B | 3.877 | Other | other |
| 22 | SEPP1 | selenoprotein P, plasma, 1 | 3.877 | Extracellular Space | other |
| 23 | SMAD3 | SMAD family member 3 | 3.877 | Nucleus | transcription regulator |
| 24 | Smok2a | sperm motility kinase 2B | 3.877 | Other | other |
| 25 | TCEAL3 | transcription elongation factor A (SII)-like 3 | 3.877 | Other | other |
| 26 | TNS1 | tensin 1 | 3.877 | Plasma Membrane | other |
| 27 | TRHR | thyrotropin-releasing hormone receptor | 3.877 | Plasma Membrane | G-protein coupled receptor |
| 28 | WWC1 | WW and C2 domain containing 1 | 3.877 | Cytoplasm | transcription regulator |
| 29 | PER2 | period circadian clock 2 | 3.853 | Nucleus | other |
| 30 | BHLHE23 | basic helix-loop-helix family, member e23 | 3.752 | Nucleus | transcription regulator |
| 31 | GALNT13 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) | 3.752 | Cytoplasm | enzyme |
| 32 | KCNF1 | potassium voltage-gated channel, subfamily F, member 1 | 3.752 | Plasma Membrane | ion channel |
| 33 | MPP1 | membrane protein, palmitoylated 1, 55 kDa | 3.752 | Plasma Membrane | kinase |
| 34 | OPA1 | optic atrophy 1 (autosomal dominant) | 3.752 | Cytoplasm | enzyme |
| 35 | PTP4A1 | protein tyrosine phosphatase type IVA, member 1 | 3.752 | Cytoplasm | phosphatase |
| 36 | SGCZ | sarcoglycan, zeta | 3.752 | Plasma Membrane | other |
| 37 | ADCY7 | adenylate cyclase 7 | 3.614 | Plasma Membrane | enzyme |
| 38 | ALCAM | activated leukocyte cell adhesion molecule | 3.614 | Plasma Membrane | other |
| 39 | AR | androgen receptor | 3.614 | Nucleus | ligand-dependent nuclear receptor |
| 40 | C4orf33 | chromosome 4 open reading frame 33 | 3.614 | Other | other |
| 41 | CCNH | cyclin H | 3.614 | Nucleus | transcription regulator |
| 42 | CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 3.614 | Nucleus | kinase |
| 43 | CDV3 | CDV3 homolog (mouse) | 3.614 | Cytoplasm | other |
| 44 | COMT | catechol-O-methyltransferase | 3.614 | Cytoplasm | enzyme |
| 45 | CRYGC | crystallin, gamma C | 3.614 | Cytoplasm | other |
| 46 | FAM13A | family with sequence similarity 13, member A | 3.614 | Cytoplasm | other |
| 47 | IGF1R | insulin-like growth factor 1 receptor | 3.614 | Plasma Membrane | transmembrane receptor |
| 48 | IYD | iodotyrosine deiodinase | 3.614 | Plasma Membrane | enzyme |
| 49 | JAG1 | jagged 1 | 3.614 | Extracellular Space | growth factor |
| 50 | KCNMA1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 3.614 | Plasma Membrane | ion channel |
Table 2.
Top 50 annotated genes with decreased methylation
| Rank | Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|---|
| 1 | IRX1 | iroquois homeobox 1 | −5.897 | Nucleus | transcription regulator |
| 2 | OSBP2 | oxysterol binding protein 2 | −5.408 | Cytoplasm | other |
| 3 | CAPN5 | calpain 5 | −5.231 | Cytoplasm | peptidase |
| 4 | INTS9 | integrator complex subunit 9 | −4.837 | Nucleus | other |
| 5 | TRIML1 | tripartite motif family-like 1 | −4.837 | Other | other |
| 6 | CSMD1 | CUB and Sushi multiple domains 1 | −4.614 | Plasma Membrane | other |
| 7 | NCOR2 | nuclear receptor corepressor 2 | −4.272 | Nucleus | transcription regulator |
| 8 | C6orf89 | chromosome 6 open reading frame 89 | −4.167 | Other | other |
| 9 | TMEM242 | transmembrane protein 242 | −4.167 | Other | other |
| 10 | DCLRE1A | DNA cross-link repair 1A | −4.100 | Nucleus | other |
| 11 | EDNRA | endothelin receptor type A | −3.877 | Plasma Membrane | transmembrane receptor |
| 12 | GALNT11 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) | −3.877 | Cytoplasm | enzyme |
| 13 | PTPN11 | protein tyrosine phosphatase, non-receptor type 11 | −3.877 | Cytoplasm | phosphatase |
| 14 | AGPAT9 | 1-acylglycerol-3-phosphate O-acyltransferase 9 | −3.795 | Cytoplasm | enzyme |
| 15 | IER5 | immediate early response 5 | −3.795 | Other | other |
| 16 | PPM1D | protein phosphatase, Mg2+/Mn2+ dependent, 1D | −3.708 | Cytoplasm | phosphatase |
| 17 | RBBP6 | retinoblastoma binding protein 6 | −3.708 | Nucleus | enzyme |
| 18 | BLOC1S2 | biogenesis of lysosomal organelles complex-1, subunit 2 | −3.614 | Cytoplasm | other |
| 19 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 | −3.614 | Cytoplasm | other |
| 20 | ECI2 | enoyl-CoA delta isomerase 2 | −3.614 | Cytoplasm | enzyme |
| 21 | MMGT1 | membrane magnesium transporter 1 | −3.614 | Cytoplasm | transporter |
| 22 | NALCN | sodium leak channel, non-selective | −3.614 | Plasma Membrane | ion channel |
| 23 | RETNLB | resistin like beta | −3.614 | Extracellular Space | other |
| 24 | AMD1 | adenosylmethionine decarboxylase 1 | −3.515 | Cytoplasm | enzyme |
| 25 | C1orf198 | chromosome 1 open reading frame 198 | −3.515 | Other | other |
| 26 | DGKI | diacylglycerol kinase, iota | −3.515 | Cytoplasm | kinase |
| 27 | DYNLT3 | dynein, light chain, Tctex-type 3 | −3.515 | Cytoplasm | other |
| 28 | EPHA6 | EPH receptor A6 | −3.515 | Plasma Membrane | kinase |
| 29 | GABRA6 | gamma-aminobutyric acid (GABA) A receptor, alpha 6 | −3.515 | Plasma Membrane | ion channel |
| 30 | Gk2 | glycerol kinase 2 | −3.515 | Cytoplasm | other |
| 31 | GLT1D1 | glycosyltransferase 1 domain containing 1 | −3.515 | Extracellular Space | enzyme |
| 32 | HMGN2 | high mobility group nucleosomal binding domain 2 | −3.515 | Nucleus | other |
| 33 | KLHL17 | kelch-like family member 17 | −3.515 | Cytoplasm | other |
| 34 | Olfr266 | olfactory receptor 266 | −3.515 | Plasma Membrane | G-protein coupled receptor |
| 35 | Ott | ovary testis transcribed | −3.515 | Other | other |
| 36 | P2RX7 | purinergic receptor P2X, ligand-gated ion channel, 7 | −3.515 | Plasma Membrane | ion channel |
| 37 | PTER | phosphotriesterase related | −3.515 | Other | enzyme |
| 38 | Rnf213 | ring finger protein 213 | −3.515 | Cytoplasm | enzyme |
| 39 | SERPINC1 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | −3.515 | Extracellular Space | enzyme |
| 40 | TPD52L1 | tumor protein D52-like 1 | −3.515 | Cytoplasm | other |
| 41 | ZMAT4 | zinc finger, matrin-type 4 | −3.515 | Nucleus | other |
| 42 | RBM20 | RNA binding motif protein 20 | −3.462 | Nucleus | other |
| 43 | BEGAIN | brain-enriched guanylate kinase-associated | −3.408 | Nucleus | other |
| 44 | CHSY3 | chondroitin sulfate synthase 3 | −3.408 | Cytoplasm | enzyme |
| 45 | CKAP4 | cytoskeleton-associated protein 4 | −3.408 | Cytoplasm | other |
| 46 | DPF3 | D4, zinc and double PHD fingers, family 3 | −3.408 | Other | other |
| 47 | Ear2 | eosinophil-associated, ribonuclease A family, member 2 | −3.408 | Cytoplasm | enzyme |
| 48 | FAM135B | family with sequence similarity 135, member B | −3.408 | Other | enzyme |
| 49 | POT1 | protection of telomeres 1 | −3.408 | Nucleus | other |
| 50 | POU6F1 | POU class 6 homeobox 1 | −3.408 | Nucleus | transcription regulator |
Fig. 2.
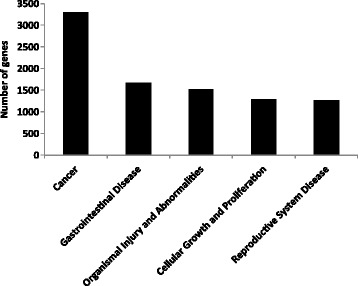
The 5 most significant biological functions and diseases related to changes in the methylation patterns. The number of molecules in the dataset associated with a known function was determined by IPA functional analysis
Table 3.
Ingenuity Pathway Analysis of gene networks
| Rank | Molecules in network | Score | Focus molecules | Top function |
|---|---|---|---|---|
| 1 | ↑AMOT,↑ANKRD26,↑CDKN1A,↓CEP152,↓CGGBP1,↓CPA3,↑CPVL, ↓DYNLL2, ↑EID2, ↓ELAC2, ↑EPB41L2, ↑EPB41L3, ↑FRMD6, ↑KIAA0195, ↑MAGEB1, ↑MBNL1, ↑MBNL2, ↑N4BP2L2, ↓NKD2, ↑NSA2, ↑RASSF4, ↓RASSF8, ↑RNF34, ↓SERPINI2, ↑SLC30A5, ↓SLC30A6, ↑SP110, ↑SP140, ↓SPAG5, ↑SYF2, ↓TROAP, ↑TXNDC11, ↑VGLL4, ↑WNT16, ↑WWC1 | 30 | 35 | Cancer, Cell Cycle, Molecular Transport |
| 2 | ↑ACACA, ↓ATRNL1, ↓BHMT, ↑CYP2A13, ↓Cyp2c70, ↑CYP3A43, ↓DCLRE1A, ↑E330013P04Rik, ↓FASN, ↑GPC6, ↑GSTP1, ↓HNMT, ↓IVNS1ABP, ↓Keg1, ↓Lcn4, ↑LRTM1, ↓MC4R, ↓Mill1, ↑MRGPRX3, ↑MT1E, ↑MTF1, ↑NR1H4, ↑RORA, ↑SLC13A1, ↑SLC16A7, ↓SLC29A4, ↓SLC30A1, ↓SLC38A4, ↑SULT1C3, ↓TMC6, ↓UCP1, ↑UPP2, ↓Xlr3c (includes others), ↑ZNF275, ↓ZNF292 | 30 | 35 | Renal Damage, Renal Tubule Injury, Molecular Transport |
| 3 | ↑ABTB2, ↑ALKBH8, ↑ALPK1, ↓BCKDHB, ↑BTBD7, ↑C11orf70, ↑C20orf194, ↑CAMKV, ↓CCDC39, ↑CUL2, ↓CUL3, ↑DCLK2, ↑EGFLAM, ↑FAM98A, ↓FARS2, ↑FBXO10, ↑FBXO34, ↑G2E3, ↓G3BP2, ↑HSP90AA1, ↓KCNG1, ↓KCNS3, ↓KCTD8, ↑KLHL10, ↓KLHL14, ↑KLHL29, ↑KLHL32, ↑KLHL36, ↑KRR1, ↑QDPR, ↑RCBTB1, ↓SEPHS1, ↓UST, ↓YWHAE, ↑ZBED4 | 30 | 35 | Hereditary Disorder, Respiratory Disease, Metabolic Disease |
| 4 | ↓ABCA6, ↓ABLIM3, ↓ABRA, ↑AIF1L, ↓AMBRA1, ↓ARAP2, ↓ARL6, ↓ATL2, ↓CAPN5, ↑CAPN6, ↓CASP12,CD80/CD86, ↑CLEC2D, ↑CLEC6A, ↑CRTAM, ↑GBP5, ↑Gbp8,Gbp6 (includes others), ↑GFM1, ↑GIMAP1-GIMAP5, ↑Gvin1 (includes others), ↑HERC6, ↑IFNG, ↓KIAA0226, ↓KIF16B,↑KLRB1, ↓KMO, ↓KY, ↑LAMP3, ↑LIX1, ↓Neurl3, ↑PCDH17, ↑Phb, ↑PILRB, ↓PMP2 | 28 | 34 | Endocrine System Disorders, Gastrointestinal Disease, Immunological Disease |
| 5 | ↑AFF2, ↑AP4S1, ↑ASAP2, ↓C21orf91, ↑C2orf88, ↑DLGAP1, ↑Eif2s3x, ↓FAM110A, ↑GNS, ↑GRB2, ↑HDGFRP3, ↓KCNH7, ↓KIRREL, ↑KRT83, ↑LRFN4, ↑MEPE, ↑NCK1, ↑NCKAP5, ↓PANX2, ↑PHACTR2, ↑RALGAPA2, ↓RALGPS1, ↑SEPN1, ↑SH2D4A, ↑SHANK2, ↓SHROOM2, ↓SLCO2A1, ↑SNX8, ↑SNX12, ↑SNX18, ↓SPRY, ↑TJAP1, ↑TTYH2, ↑WDR44, ↑ZNF32 | 28 | 34 | Cellular Assembly and Organization, Tissue Development, Cellular Function and Maintenance |
↑, increased methylation; ↓, decreased methylation
Fig. 3.
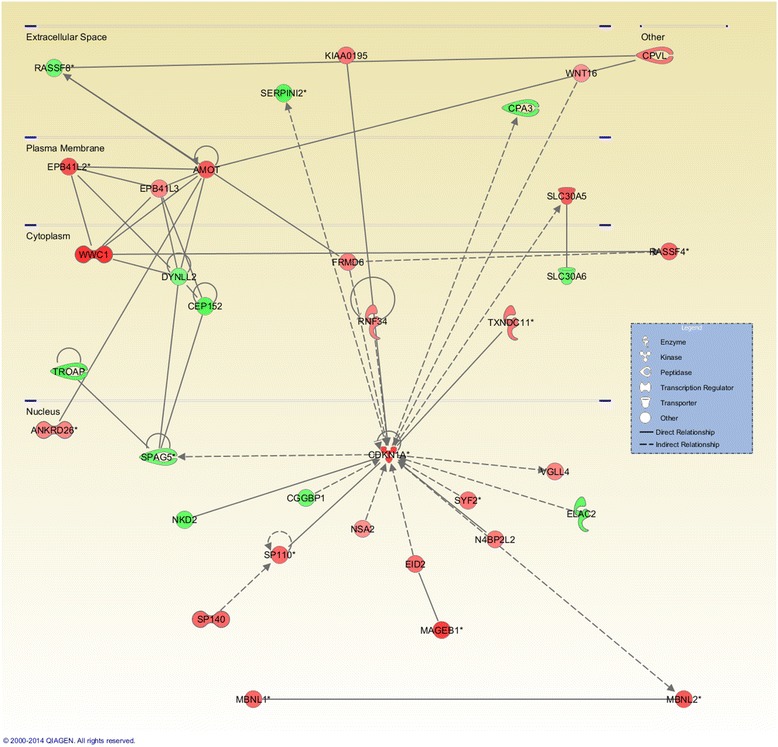
The most significant networks determined by IPA: cancer, cell cycle, and molecular transport. The IPA network analysis was conducted using the genes that were differentially methylated and their close relationships. IPA used triangle connectivity based on 30 focus genes and built the network according to the number of interactions between a single gene and others in the existing network. Red, increased methylation; green, decreased methylation
Canonical pathways associated with methylation changes in Apc mutant polyps were analyzed based on the ratio of the number of input genes to the total number of reference genes in the corresponding pathways in the IPA knowledge bases. Fisher’s exact test was employed to calculate the P values to determine whether the associations between the differentially methylated genes and the canonical pathways were significant or random. Using a cutoff value of P < 0.05, IPA identified 84 significant signaling pathways that contained genes with increased or decreased methylation. The 15 most significant pathways that correlated with methylation changes in polyps are presented in Fig. 4. Notably, regulation of the epithelial-mesenchymal transition (EMT) pathway was mapped by IPA and ranked as the 4th most significant canonical pathway associated with altered methylation. According to the IPA knowledge bases, the regulation of the EMT pathway includes 196 molecules. Among these molecules, 62 displayed greater than a 2 fold change (log2) in methylation in the polyps from Apcmin/+ mice by MeDIP-seq. The abnormal methylation changes in the EMT pathway included alterations in the methylation profiles of kinases, peptidases, phosphatases, transcription regulators, transmembrane receptors, and microRNAs. Tables 4 and 5 lists the genes involved in the EMT pathway that exhibited altered methylation (37 genes with increased methylation in Table 4; 25 genes with decreased methylation in Table 5). Signaling pathways, such as the Wnt/β-catenin, TGF-β, NOTCH, and receptor tyrosine kinase (RTK) pathways, can initiate an EMT program alone or in combination [21]. Although the genes that were determined to have differential methylation patterns in polyps by MeDIP-seq were not significantly associated with the TGF-β, NOTCH, and RTK signaling pathways, the Wnt/β-catenin pathway was identified as one of the most significant canonical pathways implicated based on methylation changes in the polyps (ranked 11th). Specifically, 53 out of 175 molecules in this pathway showed methylation changes of greater than 2-fold (log2) in polyps from Apcmin/+ mice; these molecules are listed in Tables 6 and 7 (30 genes with increased methylation in Table 6; 23 genes with decreased methylation in Table 7). Additionally, we found many shared genes in the EMT and Wnt/β-catenin pathways with altered methylation levels; these genes are shown in bold in Tables 4, 5, 6 and 7. To understand the role of DNA methylation in the crosstalk between the EMT and Wnt/β-catenin pathways in Apcmin/+ mice, IPA was utilized to predict the direct interaction of the differentially methylated genes in these two pathways based on the publication database (Fig. 5). The pathway analysis of the MeDIP-seq data suggested that cellular changes mediated via the EMT and Wnt/β-catenin pathways may be significantly associated with altered DNA methylation in polyps from Apcmin/+ mice.
Fig. 4.
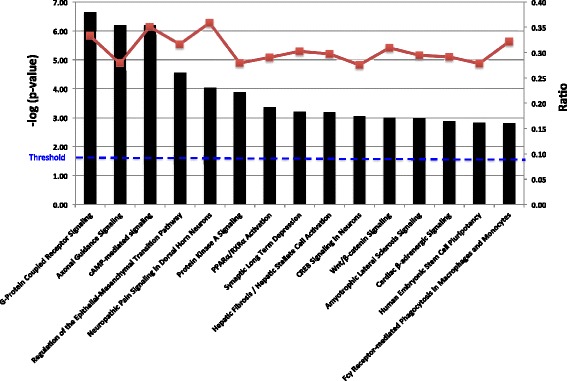
The 15 most significant canonical pathways related to changes in the methylation patterns. The left y-axis (bar graph) presents the data as the log (p-value) of each pathway using Fisher’s exact test. The right y-axis (line graph) corresponds to the ratio data for each pathway. The ratios were calculated as the number of input molecules mapped to a specific pathway divided by the total number of molecules in the given pathway
Table 4.
Genes with increased methylation that mapped to the regulation of the EMT pathway by IPA
| Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|
| SMAD3 | SMAD family member 3 | 3.877 | Nucleus | transcription regulator |
| JAG1 | jagged 1 | 3.614 | Extracellular Space | growth factor |
| WNT5A | wingless-type MMTV integration site family, member 5A | 3.292 | Extracellular Space | cytokine |
| FGF13 | fibroblast growth factor 13 | 3.100 | Extracellular Space | growth factor |
| WNT10A | wingless-type MMTV integration site family, member 10A | 3.100 | Extracellular Space | other |
| EGFR | epidermal growth factor receptor | 2.877 | Plasma Membrane | kinase |
| FGF7 | fibroblast growth factor 7 | 2.877 | Extracellular Space | growth factor |
| FGF14 | fibroblast growth factor 14 | 2.877 | Extracellular Space | growth factor |
| ID2 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 2.877 | Nucleus | transcription regulator |
| PIK3C2A | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha | 2.877 | Cytoplasm | kinase |
| FZD1 | frizzled class receptor 1 | 2.752 | Plasma Membrane | G-protein coupled receptor |
| CDH12 | cadherin 12, type 2 (N-cadherin 2) | 2.614 | Plasma Membrane | other |
| FGF8 | fibroblast growth factor 8 (androgen-induced) | 2.614 | Extracellular Space | growth factor |
| FZD8 | frizzled class receptor 8 | 2.614 | Plasma Membrane | G-protein coupled receptor |
| JAK2 | Janus kinase 2 | 2.614 | Cytoplasm | kinase |
| PIK3C2G | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma | 2.614 | Cytoplasm | kinase |
| ZEB1 | zinc finger E-box binding homeobox 1 | 2.614 | Nucleus | transcription regulator |
| GSC | goosecoid homeobox | 2.462 | Nucleus | transcription regulator |
| ADAM17 | ADAM metallopeptidase domain 17 | 2.292 | Plasma Membrane | peptidase |
| FGF9 | fibroblast growth factor 9 | 2.292 | Extracellular Space | growth factor |
| FGF11 | fibroblast growth factor 11 | 2.292 | Extracellular Space | growth factor |
| FGFR2 | fibroblast growth factor receptor 2 | 2.292 | Plasma Membrane | kinase |
| FRS2 | fibroblast growth factor receptor substrate 2 | 2.292 | Plasma Membrane | other |
| GRB2 | growth factor receptor-bound protein 2 | 2.292 | Cytoplasm | other |
| LOX | lysyl oxidase | 2.292 | Extracellular Space | enzyme |
| NCSTN | nicastrin | 2.292 | Plasma Membrane | peptidase |
| PARD6B | par-6 family cell polarity regulator beta | 2.292 | Plasma Membrane | other |
| PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma | 2.292 | Cytoplasm | kinase |
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | 2.292 | Cytoplasm | kinase |
| SOS2 | son of sevenless homolog 2 (Drosophila) | 2.292 | Cytoplasm | other |
| TGFB2 | transforming growth factor, beta 2 | 2.292 | Extracellular Space | growth factor |
| WNT2 | wingless-type MMTV integration site family member 2 | 2.292 | Extracellular Space | cytokine |
| MET | MET proto-oncogene, receptor tyrosine kinase | 2.180 | Plasma Membrane | kinase |
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | 2.100 | Cytoplasm | kinase |
| TWIST2 | twist family bHLH transcription factor 2 | 2.100 | Nucleus | transcription regulator |
| WNT2B | wingless-type MMTV integration site family, member 2B | 2.100 | Extracellular Space | other |
| WNT16 | wingless-type MMTV integration site family, member 16 | 2.029 | Extracellular Space | other |
Table 5.
Genes with decreased methylation that mapped to the regulation of the EMT pathway by IPA
| Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|
| PTPN11 | protein tyrosine phosphatase, non-receptor type 11 | −3.877 | Cytoplasm | phosphatase |
| PDGFD | platelet derived growth factor D | −3.167 | Extracellular Space | growth factor |
| RRAS2 | related RAS viral (r-ras) oncogene homolog 2 | −3.090 | Plasma Membrane | enzyme |
| FGF10 | fibroblast growth factor 10 | −2.877 | Extracellular Space | growth factor |
| FGF12 | fibroblast growth factor 12 | −2.877 | Extracellular Space | other |
| CDH2 | cadherin 2, type 1, N-cadherin (neuronal) | −2.708 | Plasma Membrane | other |
| ETS1 | v-ets avian erythroblastosis virus E26 oncogene homolog 1 | −2.708 | Nucleus | transcription regulator |
| mir-155 | microRNA 155 | −2.708 | Cytoplasm | microRNA |
| PIK3C3 | phosphatidylinositol 3-kinase, catalytic subunit type 3 | −2.708 | Cytoplasm | growth factor |
| PSEN2 | presenilin 2 | −2.708 | Cytoplasm | peptidase |
| TGFB3 | transforming growth factor, beta 3 | −2.708 | Extracellular Space | growth factor |
| SOS1 | son of sevenless homolog 1 (Drosophila) | −2.515 | Cytoplasm | other |
| WNT11 | wingless-type MMTV integration site family, member 11 | −2.515 | Extracellular Space | other |
| SMAD4 | SMAD family member 4 | −2.292 | Nucleus | transcription regulator |
| WNT7A | wingless-type MMTV integration site family, member 7A | −2.292 | Extracellular Space | cytokine |
| SMAD2 | SMAD family member 2 | −2.167 | Nucleus | transcription regulator |
| TCF7L1 | transcription factor 7-like 1 (T-cell specific, HMG-box) | −2.167 | Nucleus | transcription regulator |
| CLDN3 | claudin 3 | −2.029 | Plasma Membrane | transmembrane receptor |
| GAB1 | GRB2-associated binding protein 1 | −2.029 | Cytoplasm | other |
| HMGA2 | -- | −2.029 | Other | other |
| RAF1 | Raf-1 proto-oncogene, serine/threonine kinase | −2.029 | Cytoplasm | kinase |
| TCF7L2 | transcription factor 7-like 2 (T-cell specific, HMG-box) | −2.029 | Nucleus | transcription regulator |
| TWIST1 | twist family bHLH transcription factor 1 | −2.029 | Nucleus | transcription regulator |
| WNT7B | wingless-type MMTV integration site family, member 7B | −2.029 | Extracellular Space | other |
| WNT8B | wingless-type MMTV integration site family, member 8B | −2.029 | Extracellular Space | other |
Table 6.
Genes with increased methylation that mapped to the Wnt/β-catenin pathway by IPA
| Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|
| SOX11 | SRY (sex determining region Y)-box 11 | 3.614 | Nucleus | transcription regulator |
| TLE1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | 3.462 | Nucleus | transcription regulator |
| SOX2 | SRY (sex determining region Y)-box 2 | 3.292 | Nucleus | transcription regulator |
| WNT5A | wingless-type MMTV integration site family, member 5A | 3.292 | Extracellular Space | cytokine |
| WNT10A | wingless-type MMTV integration site family, member 10A | 3.100 | Extracellular Space | other |
| CDH5 | cadherin 5, type 2 (vascular endothelium) | 2.877 | Plasma Membrane | other |
| DKK3 | dickkopf WNT signaling pathway inhibitor 3 | 2.877 | Extracellular Space | cytokine |
| HDAC1 | histone deacetylase 1 | 2.877 | Nucleus | transcription regulator |
| PPP2R3A | protein phosphatase 2, regulatory subunit B”, alpha | 2.877 | Nucleus | phosphatase |
| RUVBL2 | RuvB-like AAA ATPase 2 | 2.877 | Nucleus | transcription regulator |
| UBD | ubiquitin D | 2.877 | Nucleus | other |
| FZD1 | frizzled class receptor 1 | 2.752 | Plasma Membrane | G-protein coupled receptor |
| CDH12 | cadherin 12, type 2 (N-cadherin 2) | 2.614 | Plasma Membrane | other |
| FZD8 | frizzled class receptor 8 | 2.614 | Plasma Membrane | G-protein coupled receptor |
| MYC | v-myc avian myelocytomatosis viral oncogene homolog | 2.614 | Nucleus | transcription regulator |
| SOX4 | SRY (sex determining region Y)-box 4 | 2.614 | Nucleus | transcription regulator |
| SOX6 | SRY (sex determining region Y)-box 6 | 2.614 | Nucleus | transcription regulator |
| APC2 | adenomatosis polyposis coli 2 | 2.292 | Cytoplasm | enzyme |
| APPL2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | 2.292 | Cytoplasm | other |
| CSNK2A1 | casein kinase 2, alpha 1 polypeptide | 2.292 | Cytoplasm | kinase |
| MMP7 | matrix metallopeptidase 7 (matrilysin, uterine) | 2.292 | Extracellular Space | peptidase |
| NR5A2 | nuclear receptor subfamily 5, group A, member 2 | 2.292 | Nucleus | ligand-dependent nuclear receptor |
| PIN1 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | 2.292 | Nucleus | enzyme |
| TGFB2 | transforming growth factor, beta 2 | 2.292 | Extracellular Space | growth factor |
| WNT2 | wingless-type MMTV integration site family member 2 | 2.292 | Extracellular Space | cytokine |
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | 2.100 | Cytoplasm | kinase |
| FRAT1 | frequently rearranged in advanced T-cell lymphomas | 2.100 | Cytoplasm | other |
| WNT2B | wingless-type MMTV integration site family, member 2B | 2.100 | Extracellular Space | other |
| WNT16 | wingless-type MMTV integration site family, member 16 | 2.029 | Extracellular Space | other |
Table 7.
Genes with decreased methylation that mapped to the Wnt/β-catenin pathway by IPA
| Symbol | Gene name | log2 Fold Change | Location | Type(s) |
|---|---|---|---|---|
| ACVR1C | activin A receptor, type IC | −3.029 | Plasma Membrane | kinase |
| GNAQ | guanine nucleotide binding protein (G protein), q polypeptide | −2.877 | Plasma Membrane | enzyme |
| SOX13 | SRY (sex determining region Y)-box 13 | −2.877 | Nucleus | transcription regulator |
| WIF1 | WNT inhibitory factor 1 | −2.877 | Extracellular Space | other |
| CDH2 | cadherin 2, type 1, N-cadherin (neuronal) | −2.708 | Plasma Membrane | other |
| PPP2R2A | protein phosphatase 2, regulatory subunit B, alpha | −2.708 | Cytoplasm | phosphatase |
| TGFB3 | transforming growth factor, beta 3 | −2.708 | Extracellular Space | growth factor |
| PPP2R1B | protein phosphatase 2, regulatory subunit A, beta | −2.614 | Plasma Membrane | phosphatase |
| CSNK1G3 | casein kinase 1, gamma 3 | −2.515 | Cytoplasm | kinase |
| WNT11 | wingless-type MMTV integration site family, member 11 | −2.515 | Extracellular Space | other |
| MARK2 | MAP/microtubule affinity-regulating kinase 2 | −2.292 | Cytoplasm | kinase |
| WNT7A | wingless-type MMTV integration site family, member 7A | −2.292 | Extracellular Space | cytokine |
| TCF7L1 | transcription factor 7-like 1 (T-cell specific, HMG-box) | −2.167 | Nucleus | transcription regulator |
| GJA1 | gap junction protein, alpha 1, 43 kDa | −2.029 | Plasma Membrane | transporter |
| PPP2R2B | protein phosphatase 2, regulatory subunit B, beta | −2.029 | Cytoplasm | phosphatase |
| PPP2R5A | protein phosphatase 2, regulatory subunit B’, alpha | −2.029 | Cytoplasm | phosphatase |
| SFRP2 | secreted frizzled-related protein 2 | −2.029 | Plasma Membrane | transmembrane receptor |
| SOX7 | SRY (sex determining region Y)-box 7 | −2.029 | Nucleus | transcription regulator |
| SOX14 | SRY (sex determining region Y)-box 14 | −2.029 | Nucleus | transcription regulator |
| TCF7L2 | transcription factor 7-like 2 (T-cell specific, HMG-box) | −2.029 | Nucleus | transcription regulator |
| TLE3 | transducin-like enhancer of split 3 | −2.029 | Nucleus | other |
| WNT7B | wingless-type MMTV integration site family, member 7B | −2.029 | Extracellular Space | other |
| WNT8B | wingless-type MMTV integration site family, member 8B | −2.029 | Extracellular Space | other |
Fig. 5.
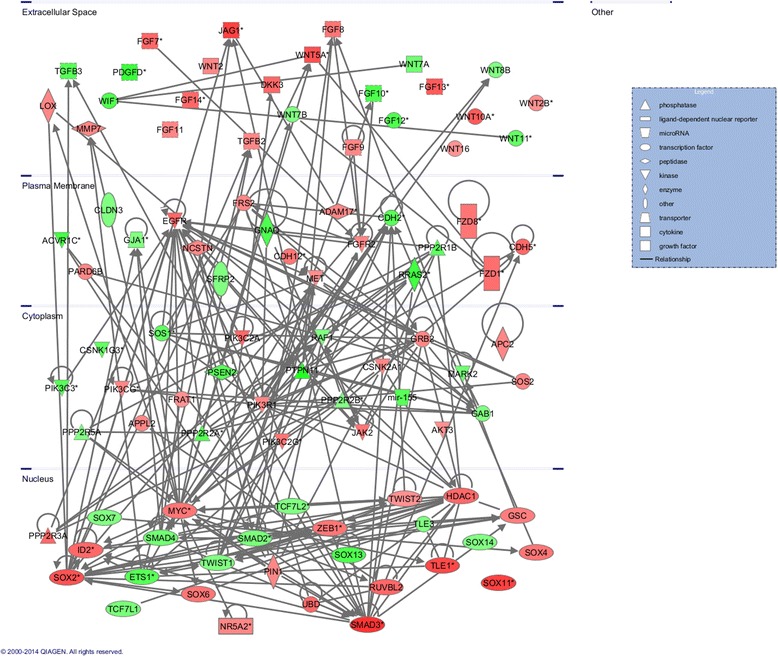
Predicted interactions between molecules with altered methylation that mapped to the EMT and Wnt/β-catenin pathways. IPA predicted direct interaction of the genes with altered methylation patterns in the EMT and Wnt/β-catenin pathways based on the publication database. Red, increased methylation; green, decreased methylation
Discussion
Global hypomethylation and hypermethylation of CpG islands in tumor suppressor genes occurs in human colon cancer cell lines and primary colon adenomatous tissues [12]. However, the global genomic distribution of aberrant methylation and the association of these methylation signatures with pivotal signaling pathways and biological networks in colon cancer remain unclear, mainly due to the limitations of the existing techniques for analyzing DNA methylation at specific sequences [22]. Recently, the development of the MeDIP-based approach has enabled the rapid and comprehensive identification of multiple CpG sites. MeDIP in conjunction with high-throughput sequence (MeDIP-seq) provides a genome-wide mapping technique that has been successfully used to profile the global DNA methylation patterns of many cancer models [23–26]. Notably, Grimm et al. used MeDIP-seq to identify a large number of DMRs with distinct methylation patterns in Apc mutant adenomas, which are partially conserved between intestinal adenomas in Apcmin/+ mice and human colon cancer [27]. In the present study, we used pathway analysis after MeDIP-seq to screen the global genomic methylation profile to identify genomic loci with aberrant methylation patterns in adenomatous polyps from Apcmin/+ mice and to determine the biological function, networks, and canonical pathways that were affected by the DNA methylation in Apc mutant adenomas.
The top-ranked genes with increased and decreased methylation may provide information to facilitate the discovery of key genes, therapeutic targets, and biomarkers for the development, diagnosis, prognosis, and prevention of colon cancer. For example, CTNNBL1 [catenin (cadherin-associated protein) b-like 1] exhibited increased methylation in adenomatous polyp tissue (log2 fold change = 4.1, Table 1), as evidenced by MeDIP-seq. The CTNNBL1 gene is associated with obesity, a known risk factor for the development of CRC [28]. Recently, CTNNBL1 was reported to be a putative regulator of the canonical Wnt signaling pathway, and mutations in and dysregulation of this pathway are involved in CRC [29]. However, the potential epigenetic regulation of CTNNBL1 in colon cancer remains to be elucidated. To the best of our knowledge, this is the first report to suggest that CTNNBL1 might by aberrantly methylated in Apc mutant mice. Further experiments are necessary to investigate the epigenetic regulation of CTNNBL1 in colon cancer cells and patient specimens. CDKN1A (cyclin-dependent kinase inhibitor 1A, p21) showed increased methylation (log2 fold change = 3.6, Table 1) in adenomatous polyp tissue compared with control tissue. CDKN1A is a cyclin-dependent kinase inhibitor that plays a key role in regulating the cell cycle, especially the G1/S checkpoint, and its expression is lost in most cases of colon cancer. By analyzing 737 CRC samples, Ogino et al. concluded that the down-regulation of p21 inversely correlates with microsatellite instability and the CpG island methylator phenotype in colon cancer [30]. Here, we provided additional evidence by demonstrating potentially increased p21 methylation in Apcmin/+ polyps.
It is commonly believed that promoter hypermethylation is associated with silencing of tumor suppressor genes in carcinogenesis [31]. One study observed a significant increase in DNA methylation in primary colon adenocarcinoma samples relative to normal colon tissue by analyzing the DNA methylation data from Cancer Genome Atlas (TCGA) and found an inverse correlation between DNA methylation and gene expression: genes with cancer-specific DNA methylation showed decreased transcription activity in colon adenocarcinoma [32]. However, Grimm et al. reported that the correlation of gene expression and DNA methylation applies only to a small set of genes by analyzing the results from MeDIP-seq and RNA-seq in normal intestine tissues and Apc mutant adenomas. In addition, they analyzed the mRNA expression of 31 selected tumor suppressors, only 2 were found both promoter hypermethylated and transcriptionally silenced. Surprisingly, the majority of tumor suppressors examined in their study did not exhibit a decreased transcriptional activity in adenoma compared to normal intestine samples [27]. These results suggested that silencing of tumor suppressor genes by aberrant methylation may not be common events during early polyposis of Apc mutant mice. Nevertheless, it is possible that epigenetic changes mediated gene silencing arises during progression of adenoma to carcinoma [33]. Furthermore, it was reported that instead of directly intervene active promoters, DNA methylation affects genes that are already silent by other mechanisms such as histone modifications [34]. Thus, further studies are needed to elucidate the dynamic changes of DNA methylation, histone modifications, and gene transcription in different stages, such as initiation, progression, and metastasis during colon carcinogenesis.
This study aimed to discover functions and pathways associated with epigenomic alterations in colon cancer in addition to the individual affected molecules. We utilized IPA to interpret the MeDIP-seq data in the context of molecular interactions, networks, and canonical pathways. IPA revealed that the genes with altered methylation patterns in adenomatous tissues predominantly occupied the cancer and cell cycle networks (Table 3) and the cancer and gastrointestinal disease functional categories (Fig. 2). This information suggested that dynamic epigenetic modifications might occur in genes associated with cancer, cell cycle regulation, and gut disease development in Apcmin/+ mice.
Biological changes that lead to the switch from an epithelial to a mesenchymal cell phenotype, defined as EMT, play an important role in embryonic development and carcinogenesis [35]. In the context of tumorigenesis-associated EMT, neoplastic cells lose epithelial characteristics, such as cell-cell adhesion, cell polarity, and lack of motility, and acquire mesenchymal features, such as migratory ability, invasiveness, plasticity, and resistance to apoptosis [21]. The morphological alterations that occur during EMT enable neoplastic cells to escape from the basement membrane, migrate to neighboring lymph nodes, and eventually enter the circulation to establish secondary colonies at distant sites [36]. Thus, EMT program activation is considered a critical step in tumor growth, angiogenesis, and metastasis [37]. Chen et al. reported elevated expression of the mesenchymal marker vimentin in intestinal adenomas from Apcmin/+ mice and suggested that molecular alterations in the initial steps of EMT are involved in early tumorigenesis in Apcmin/+ mice; the early stages of intestinal tumorigenesis lack signs of invasion and metastasis [38]. These interesting observations highlighted the necessity to study the EMT process during early tumorigenesis. Although the molecular and biochemical mechanisms involved in the initiation and regulation of EMT in carcinogenesis are not yet fully understood, they appear to be associated with growth factor receptors (for example, RTKs), signaling pathways (for example, the Wnt/β-catenin, NOTCH, and TGF-β pathways), and stimuli (for example, oxidative stress) [39]. The involvement of epigenetic events in regulating the EMT proteome during carcinogenesis was recently demonstrated [40]. Using ChIP-seq (chromatin immunoprecipitation followed by sequencing) assays, Cieslik et al. showed that EMT is driven by the chromatin-mediated activation of transcription factors [41]. The current study identified many genes with increased or decreased methylation in the EMT pathway (Fig. 3, Tables 4 and 5), suggesting that aberrant DNA methylation may be associated with the activation of EMT during tissue remodeling in early tumorigenesis in Apcmin/+ mice. The present study also provided useful information regarding important molecules in the EMT pathway that undergo alterations in their methylation pattern during polyposis in Apcmin/+ mice. For example, SMAD3 (mothers against decapentaplegic homolog 3), a molecule that plays an essential role in TGF-β pathway-mediated EMT, was one of the genes that exhibited increased methylation (log2 fold change = 3.9, Table 4) in adenomas in Apcmin/+ mice. Interestingly, SMAD3 deficiency promotes tumor formation in the distal colon of Apcmin/+ mice [42]. EGFR (epidermal growth factor receptor), another important molecule that exhibited increased methylation, has been implicated in EMT in adenomas (log2 fold change = 2.9, Table 4). EGFR can induce EMT in cancer cells by up-regulating Twist [43], and promoter methylation of EGFR has been detected in metastatic tumors from patients with CRC [44]. The results of the current study indicated that aberrant methylation of EGFR may occur during early tumorigenesis in Apcmin/+ mice. Important transcription factors in the EMT pathway, including ZEB 1 and TWIST 2, also exhibited increased methylation in adenomas from Apcmin/+ mice (Table 4). Although the contribution of TWIST 2 to promoting EMT in breast cancer progression was recently reported [45], there is limited knowledge of the role of TWIST 2 in colon cancer; however, one study proposed that TWIST 2 is a potential prognostic biomarker for colon cancer [46]. Notably, aberrant methylation of TWIST 2 has been demonstrated in chronic lymphocytic leukemia [47] and acute lymphoblastic leukemia [48]. The present study is the first to suggest that methylation of the TWIST 2 gene may be involved in tumorigenesis in Apcmin/+ mice. Further studies are necessary to elucidate the role of DNA methylation in EMT pathway regulation in early tumorigenesis in Apcmin/+ mice.
Apcmin/+ mice are thought to have a hyperactive Wnt/β-catenin pathway [10], but the epigenetic modifications of the Wnt/β-catenin pathway are still not fully understood. IPA identified the Wnt/β-catenin pathway as one of the most significant canonical pathways that contained genes with increased or decreased methylation, suggesting an important role for epigenetic alterations in the Wnt/β-catenin pathway in tumorigenesis. Some of the molecules with increased or decreased methylation patterns that were mapped to this pathway in the present study are consistent with the findings of previous publications. For example, Dhir et al. analyzed tissue samples from inflammatory bowel disease (IBD) and colon cancer patients and demonstrated that aberrant methylation of Wnt/β-catenin signaling genes is an early event in IBD-associated colon cancer. Aberrant methylation of APC2 (adenomatousis polyposis coli 2), SFRP1 (secreted frizzled-related protein 1), and SFRP2 (secreted frizzled-related protein 2) is associated with the progression from colitis to neoplasia [49]. In the current study, we observed increased methylation of APC2 and decreased methylation of SFRP2 in adenomas in Apcmin/+ mice (Tables 6 and 7). Wang et al. demonstrated that black raspberries can prevent colonic ulceration in a DSS-induced model and in interleukin-10 knockout mice by epigenetically modifying genes with hypermethylated promoters in the Wnt/β-catenin pathway, such as DKK3 (dickkopf-related protein 3), APC, SFRP1, and SOX17 [SRY (sex determining region Y)-box 17] [50, 51]. In the present study, DKK3 consistently displayed increased methylation (log2 fold change = 2.9, Table 6) in adenomas from Apcmin/+ mice compared with normal tissue. Furthermore, we provided additional information regarding the genes with altered methylation in the Wnt/β-catenin pathway in polyps from Apcmin/+ mice, potentially facilitating future research on the involvement of aberrantly methylated Wnt/β-catenin pathway components in colon cancer development and on potential targets for epigenetic modification for the prevention of colon cancer. Intestinal adenoma in mouse originated from intestinal stem cells (ISC), a small fraction of cells in proliferative crypts [52]. Interestingly, Grimm and co-workers demonstrated that the adenoma-specific methylation signatures are not acquired from ISC by showing that the methylation patterns were similar in ISC, proliferative crypt cells, and differentiated villus cells, but are distinct in adenoma tissue [27]. Since ISC are responsive to Wnt signaling and we identified Wnt/β-catenin pathway as one of the most significant pathways associated with DNA methylation in polyps from Apcmin/+ mice, it would be important to understand the mechanisms underlying the acquisition of aberrant DNA methylation patterns in Wnt/β-catenin pathway in adenoma and how the hypermethylated genes involved in Wnt/β-catenin pathway influence the neoplastic transformation from ISC to adenoma. Furthermore, the Wnt/β-catenin pathway is intimately associated with EMT pathway [53]. The present study provided valuable information regarding the potential crosstalk between the EMT and Wnt/β-catenin pathways, which are both affected by DNA methylation in Apcmin/+ mice (Fig. 5). Further studies are needed to understand the role of the complex crosstalk between multiple signaling pathways in the progression of colon cancer.
In addition to DNA methylation, histone modification and non-coding RNA are major epigenetic mechanisms that regulate gene transcription in carcinogenesis [54]. It is currently accepted that these epigenetic modifications are linked to one another in the modulation of the epigenome landscape [55, 56]. For example, these epigenetic modifications may work in combination in carcinogenesis [57]. It was found that DNA hypermethylation in Apc mutant adenomas preferentially target the polycomb repressive complex 1/2 (PRC 1/2) target genes, suggesting an interplay of DNA methylation and histone modification in Apcmin/+ mice [27]. On the other hand, different epigenetic mechanisms may cross-regulate each other in the regulation of cellular activity. For instance, the expression of certain microRNAs is potentially controlled by DNA methylation or histone modification. However, some microRNAs can target epigenetic-modifying enzymes, such as DNMTs (DNA methyltransferases) and EZH2 (enhancer of zeste homolog 2) [58]. Furthermore, Tahara, et al. found that 74 chromatin regulatory genes are mutated more frequently in CpG island methylator phenotype - high CRC in the TCGA dataset [59]. Changes in the methylation patterns of several genes encoding microRNAs, histone modification enzymes, and proteins that function in chromatin remodeling were identified using MeDIP-seq. For example, we discovered decreased methylation of microRNA-155 (log2 fold change = −2.7, Table 5), which mapped to the EMT pathway; microRNA-155 expression promotes the migration and invasion of several CRC cell lines [60]. Moreover, HDAC1 (histone deacetylase 1) was mapped to the Wnt/β-catenin pathway with a 2.9-fold (log2) increase in methylation in Apc mutant polyps (Table 6). In addition, we observed an increased methylation in the gene coding for chromodomain-helicase-DNA-binding protein 1 (CHD1) in Apc mutant polyps (data not shown). CHD1 protein is known to be involved in transcription-related chromatin remodeling [61]. Taken together, our data indicated that epigenetic alterations may be complex and may occur at multiple levels during tumorigenesis in Apcmin/+ mice.
Conclusions
In conclusion, polyps from Apcmin/+ mice exhibited extensive, aberrant DNA methylation. The methylation changes in the genes detected using the MeDIP-seq assay were mainly attributed to functions and networks in cancer, the cell cycle, and gastrointestinal diseases. These differentially methylated genes were situated in several canonical pathways that are important in colon cancer, such as the EMT and Wnt/β-catenin signaling pathways.
Materials and methods
Mouse strains
C57BL/6 J male mice that are heterozygous for the Apc allele (Apcmin/+) and their wild type littermates (Apc+/+) were originally obtained from Jackson Laboratories (Bar Harbor, ME, USA). The animals were housed in the Animal Care Facility at Rutgers University with a 12 h-light/12 h-dark cycle and were provided ad libitum access to food and water. The Apcmin/+ and control mice were sacrificed by CO2 inhalation at 20 weeks of age. Polyp and intestine samples were collected as previously described [62]. Briefly, after sacrificing the mice, the gastrointestinal tract was removed, opened longitudinally, and rinsed thoroughly with saline. Intestinal adenomatous polyps were excised from the intestines carefully. The normal intestine tissue and polyps were snap frozen and stored at −80 °C for future use.
DNA extraction
Genomic DNA was isolated from adenomatous polyps from three Apcmin/+ mice and from normal intestinal tissue from three Apc+/+ littermates using a DNeasy Kit (Qiagen, Valencia, CA, USA). Prior to fragmentation by Covaris (Covaris, Inc., Woburn, MA, USA), the quality of the extracted genomic DNA was confirmed by agarose gel electrophoresis and OD ratio. After fragmentation, the genomic DNA was further assessed for size distribution using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The fragmented genomic DNA concentrations were measured with a Nanodrop spectrophotometer.
MeDIP-seq
MeDIP was performed using a MagMedIP kit (Diagenode, Denville, NJ, USA) as previously described [63]. Briefly, immunoprecipitations were performed using a monoclonal antibody against 5-methylcytidine (Diagenode, Denville, NJ, USA) to separate the methylated DNA fragments from the unmethylated fragments. The captured DNA was used to create the Illumina libraries using NEBNext reagents (catalog# E6040; New England Biolabs, Ipswich, MA, USA). After the quality of the libraries was evaluated, the samples were sequenced using an Illumina HiSeq 2000 machine. The results were analyzed for data quality and exon coverage using the platform provided by DNAnexus (DNAnexus, Inc., Mountain View, CA, USA). Subsequently, the samples were subjected to Illumina next-generation sequencing (Otogenetics Corporation, Norcross, GA, USA). After downloading the BAM files for analysis, MeDIP alignments were compared with control samples using Cuffdiff 2.0.2 as previously described [64, 63]. To judge the quantitative enrichment in MeDIP samples versus control samples in Cuffdiff, the overlapping regions of sequence alignment common to the MeDIP and control samples were used. Significant peaks at a 5 % false discovery rate (FDR) with a minimum of a 4-fold difference in R (Cummerbund package) were selected. The peaks were matched with adjacent annotated genes using ChIPpeakAnno as previously described [65].
Ingenuity Pathway Analysis (IPA)
To investigate the significance of the altered methylation observed by MeDIP-seq, we analyzed genes that exhibited greater than a 2-fold change (log2) in methylation (Apcmin/+ polyps vs. control) using IPA (IPA 4.0, Ingenuity Systems, www.ingenuity.com). IPA utilized gene symbols that were identified as neighboring enriched methylation peaks by ChIPpeakAnno for all of the analyses. IPA mapped the input genes to its knowledge bases and identified the most relevant biological functions, networks, and canonical pathways related to the altered methylation profiles in the Apc mutant polyps.
Acknowledgments
The authors thank the members of Dr. Tony Kong’s laboratory for their helpful discussions. This work was supported by R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS).
Footnotes
Yue Guo and Jong Hun Lee contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YG, JHL, LS, and YH conceived the research design, carried out the experiments, and analyzed the data. YG, JHL, LS, WL, CZ, AYY, SB, and ANTK wrote, reviewed, and revised this manuscript. AP participated in the design of the study and performed the experiments. RH, MV, and ANTK provided administrative, technical, and material support. All the authors read and approved the final version of the manuscript.
Contributor Information
Yue Guo, Email: yue.guo166@rutgers.edu.
Jong Hun Lee, Email: foodguy@cha.ac.kr.
Limin Shu, Email: lmnshu@gmail.com.
Ying Huang, Email: huangyingms@gmail.com.
Wenji Li, Email: wl365@scarletmail.rutgers.edu.
Chengyue Zhang, Email: cz141@scarletmail.rutgers.edu.
Anne Yuqing Yang, Email: yy217@rutgers.edu.
Sarandeep SS Boyanapalli, Email: sboyana@rutgers.edu.
Ansu Perekatt, Email: aperekatt@gmail.com.
Ronald P Hart, Email: rhart@rutgers.edu.
Michael Verzi, Email: verzi@dls.rutgers.edu.
Ah-Ng Tony Kong, Email: kongt@pharmacy.rutgers.edu.
References
- 1.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Su Z-Y, Kong A-NT. Current perspectives on epigenetic modifications by dietary chemopreventive and herbal phytochemicals. Curr Pharmacol Rep. 2015;1–13. doi:10.1007/s40495-015-0023-0. [DOI] [PMC free article] [PubMed]
- 8.Harrison S, Benziger H. The molecular biology of colorectal carcinoma and its implications: a review. Surgeon. 2011;9(4):200–10. doi: 10.1016/j.surge.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–70. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 10.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 11.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 12.Pancione M, Remo A, Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Pathol Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: cohort study and literature review. Int J Cancer J Int du Cancer. 2011;128(5):1080–94. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belshaw NJ, Pal N, Tapp HS, Dainty JR, Lewis MP, Williams MR, et al. Patterns of DNA methylation in individual colonic crypts reveal aging and cancer-related field defects in the morphologically normal mucosa. Carcinogenesis. 2010;31(6):1158–63. doi: 10.1093/carcin/bgq077. [DOI] [PubMed] [Google Scholar]
- 15.Ying J, Poon FF, Yu J, Geng H, Wong AH, Qiu GH, et al. DLEC1 is a functional 3p22.3 tumour suppressor silenced by promoter CpG methylation in colon and gastric cancers. Br J Cancer. 2009;100(4):663–9. doi: 10.1038/sj.bjc.6604888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Shu L, Zhang C, Su ZY, Kong AN. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem Pharmacol. 2015;94(2):69–78. doi: 10.1016/j.bcp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hibi K, Mizukami H, Shirahata A, Goto T, Sakata M, Saito M, et al. Aberrant methylation of the UNC5C gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009;29(1):271–3. [PubMed] [Google Scholar]
- 18.Li X, Lu P, Li B, Zhang W, Luo K. Effects of iodine-125 seeds on the methylation of SFRP and P16 in colorectal cancer. Exp Ther Med. 2013;6(5):1225–8. doi: 10.3892/etm.2013.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halberg RB, Katzung DS, Hoff PD, Moser AR, Cole CE, Lubet RA, et al. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. P Natl Acad Sci USA. 2000;97(7):3461–6. doi: 10.1073/pnas.97.7.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 21.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 23.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alisch RS, Wang T, Chopra P, Visootsak J, Conneely KN, Warren ST. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med Genet. 2013;14:18. doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang WJ, Zheng Y, Wu LM, Ke QH, Shen H, Yuan Y, et al. Genome-wide analysis of aberrant DNA methylation for identification of potential biomarkers in colorectal cancer patients. Asian Pac J Cancer Prev: APJCP. 2012;13(5):1917–21. doi: 10.7314/APJCP.2012.13.5.1917. [DOI] [PubMed] [Google Scholar]
- 26.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm C, Chavez L, Vilardell M, Farrall AL, Tierling S, Bohm JW, et al. DNA-methylome analysis of mouse intestinal adenoma identifies a tumour-specific signature that is partly conserved in human colon cancer. PLoS Genet. 2013;9(2):e1003250. doi: 10.1371/journal.pgen.1003250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan LJ, Zhu H, He H, Wu KH, Li J, Chen XD, et al. Replication of 6 obesity genes in a meta-analysis of genome-wide association studies from diverse ancestries. PLoS One. 2014;9(5):e96149. doi: 10.1371/journal.pone.0096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huhn S, Ingelfinger D, Bermejo JL, Bevier M, Pardini B, Naccarati A, et al. Polymorphisms in CTNNBL1 in relation to colorectal cancer with evolutionary implications. Int J Mol Epidemiol Genet. 2011;2(1):36–50. [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Kawasaki T, Kirkner GJ, Ogawa A, Dorfman I, Loda M, et al. Down-regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol. 2006;210(2):147–54. doi: 10.1002/path.2030. [DOI] [PubMed] [Google Scholar]
- 31.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 32.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21(5):655–67. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5(2):676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 35.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–9. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Halberg RB, Burch RP, Dove WF. Intestinal adenomagenesis involves core molecular signatures of the epithelial-mesenchymal transition. J Mol Histol. 2008;39(3):283–94. doi: 10.1007/s10735-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K, Nelson CM. New insights into the regulation of epithelial-mesenchymal transition and tissue fibrosis. Int Rev Cell Mol Biol. 2012;294:171–221. doi: 10.1016/B978-0-12-394305-7.00004-5. [DOI] [PubMed] [Google Scholar]
- 40.Kiesslich T, Pichler M, Neureiter D. Epigenetic control of epithelial-mesenchymal-transition in human cancer. Mol Clin Oncol. 2013;1(1):3–11. doi: 10.3892/mco.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cieslik M, Hoang SA, Baranova N, Chodaparambil S, Kumar M, Allison DF, et al. Epigenetic coordination of signaling pathways during the epithelial-mesenchymal transition. Epigenetics Chromatin. 2013;6(1):28. doi: 10.1186/1756-8935-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodir NM, Chen X, Park R, Nickel AE, Conti PS, Moats R, et al. Smad3 deficiency promotes tumorigenesis in the distal colon of ApcMin/+ mice. Cancer Res. 2006;66(17):8430–8. doi: 10.1158/0008-5472.CAN-06-1437. [DOI] [PubMed] [Google Scholar]
- 43.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67(19):9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scartozzi M, Bearzi I, Mandolesi A, Giampieri R, Faloppi L, Galizia E, et al. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br J Cancer. 2011;104(11):1786–90. doi: 10.1038/bjc.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z, et al. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene. 2011;30(47):4707–20. doi: 10.1038/onc.2011.181. [DOI] [PubMed] [Google Scholar]
- 46.Yu H, Jin GZ, Liu K, Dong H, Yu H, Duan JC, et al. Twist2 is a valuable prognostic biomarker for colorectal cancer. World J Gastroenterol: WJG. 2013;19(15):2404–11. doi: 10.3748/wjg.v19.i15.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raval A, Lucas DM, Matkovic JJ, Bennett KL, Liyanarachchi S, Young DC, et al. TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23(17):3877–85. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 48.Thathia SH, Ferguson S, Gautrey HE, van Otterdijk SD, Hili M, Rand V, et al. Epigenetic inactivation of TWIST2 in acute lymphoblastic leukemia modulates proliferation, cell survival and chemosensitivity. Haematologica. 2012;97(3):371–8. doi: 10.3324/haematol.2011.049593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhir M, Montgomery EA, Glockner SC, Schuebel KE, Hooker CM, Herman JG, et al. Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointestinal Surg Off J Soc Surg Alimentary Tract. 2008;12(10):1745–53. doi: 10.1007/s11605-008-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang LS, Kuo CT, Huang TH, Yearsley M, Oshima K, Stoner GD, et al. Black raspberries protectively regulate methylation of Wnt pathway genes in precancerous colon tissue. Cancer Prev Res. 2013;6(12):1317–27. doi: 10.1158/1940-6207.CAPR-13-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang LS, Kuo CT, Stoner K, Yearsley M, Oshima K, Yu J, et al. Dietary black raspberries modulate DNA methylation in dextran sodium sulfate (DSS)-induced ulcerative colitis. Carcinogenesis. 2013;34(12):2842–50. doi: 10.1093/carcin/bgt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 53.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 54.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miao CG, Yang YY, He X, Xu T, Huang C, Huang Y, et al. New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cell Signal. 2013;25(5):1118–25. doi: 10.1016/j.cellsig.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 57.Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J. 2009;50(4):455–63. doi: 10.3349/ymj.2009.50.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tahara T, Yamamoto E, Madireddi P, Suzuki H, Maruyama R, Chung W, et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146(2):530–38 e5. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31(6):1375–80. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 61.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28(4):665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khor TO, Cheung WK, Prawan A, Reddy BS, Kong AN. Chemoprevention of familial adenomatous polyposis in Apc(Min/+) mice by phenethyl isothiocyanate (PEITC) Mol Carcinog. 2008;47(5):321–5. doi: 10.1002/mc.20390. [DOI] [PubMed] [Google Scholar]
- 63.Yang AY, Lee JH, Shu L, Zhang C, Su ZY, Lu Y, et al. Genome-wide analysis of DNA methylation in UVB- and DMBA/TPA-induced mouse skin cancer models. Life Sci. 2014;113(1-2):45–54. doi: 10.1016/j.lfs.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]


