Abstract
Our understanding of the pathways that regulate lymphocyte metabolism, as well as the effects of metabolism and its products on the immune response, is still limited. We report that a metabolic program controlled by the transcription factors hypoxia inducible factor-1α (HIF1-α) and aryl hydrocarbon receptor (AHR) supports the differentiation of type 1 regulatory (Tr1) cells. HIF1-α controls the early metabolic reprograming of Tr1 cells. At later time points, AHR promotes HIF1-α degradation and takes control of Tr1 cell metabolism. Extracellular adenosine triphosphate (eATP) and hypoxia, linked to inflammation, trigger AHR inactivation by HIF1-α and inhibit Tr1 cell differentiation. Conversely, CD39 promotes Tr1 cell differentiation by depleting eATP. CD39 also contributes to Tr1 suppressive activity by generating adenosine in cooperation with CD73 expressed by responder T cells and antigen presenting cells. These results suggest that HIF1-α and AHR integrate immunological, metabolic and environmental signals to regulate the immune response.
T-cell activation triggers metabolic changes required to support the adaptive immune response1-5. Indeed, the differentiation of cytotoxic and IL-17 producing (TH17) effector T cells requires a metabolic shift towards aerobic glycolysis that is controlled by the transcription factor HIF1-α6-8. Conversely, Foxp3+ regulatory T (Treg) cells and memory T cells are supported by oxidative phosphorylation6,9. In addition, besides supplying energy and biosynthetic precursors, the metabolism also provides molecules that modulate the immune response through feedback regulatory pathways3,10,11.
Type 1 regulatory T (Tr1) cells are Foxp3− regulatory CD4+ T cells that produce IL-10 and have non-redundant roles in the control of inflammation12-14. IL-27 is a growth and differentiation factor for Tr1 cells15-17. In addition, IL-21 produced by Tr1 cells acts in an autocrine manner to boost and stabilize their differentiation18,19. The transcription factor aryl hydrocarbon receptor (AHR) regulates IL-10 and IL-21 production in Tr1 cells20,21, but our understanding of the mechanisms that control the differentiation of Tr1 cells and metabolic processes within Tr1 cells is limited. Here we report that aerobic glycolysis supports Tr1 cell differentiation through a metabolic program controlled by HIF1-α and AHR. Moreover, we found that oxygen and extracellular adenosine triphosphate (eATP) regulate the differentiation of Tr1 cells through HIF1-α dependent mechanisms. Thus, our findings identify metabolic pathways that regulate the differentiation of Tr1 cells and provide potential targets for their therapeutic modulation in immune-mediated disorders.
Results
AHR and STAT3 control CD39 expression in Tr1 cells
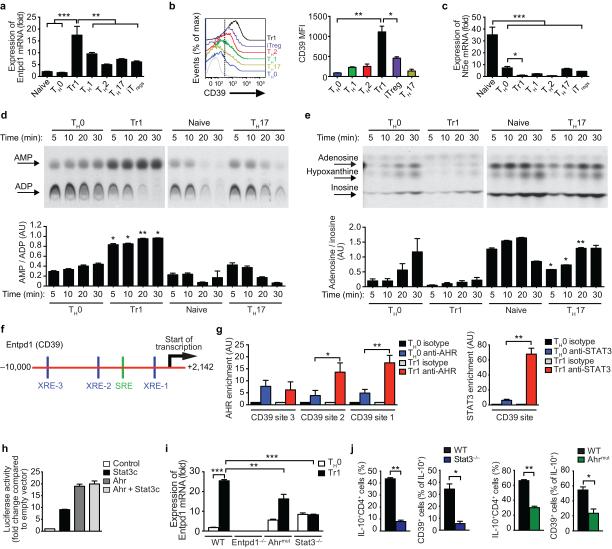
We detected the expression of Entpd1, which encodes the plasma membrane ectonucleotidase CD39, in Tr1 cells differentiated with IL-27 (Fig. 1a,b and Supplementary Fig. 1a-c). CD39 catalyzes the degradation of eATP into adenosine monophosphate (AMP)22. In Foxp3+ iTreg cells, CD39 is co-expressed with the ecto-5’-nucleotidase CD73 (encoded by Nt5e) that degrades AMP to adenosine23. However, we did not detect CD73 expression in Tr1 cells (Fig. 1c). In agreement with these findings, we detected CD39 but no CD73 enzymatic activity in Tr1 cells (Figs. 1d,e). We also detected CD39 expression in human CD4+ T cells activated in the presence of IL-2724 (Supplementary Fig. 1d).
Figure 1. CD39 is expressed in Tr1 cells.
(a) Entpd1 expression. (b) Flow cytometry analysis of CD39 expression in TH0, TH1, TH2, TH17, Tr1 cells and Foxp3+ Treg cells. Shown is CD39 expression and MFI data gated in cytokine (IFNγ, IL-4, IL-10 or IL-17) or Foxp3 positive T-cells. Total CD4+ T cells were used for the TH0 condition. (c) Nt5e expression. (d-e) CD39 (d) and CD73 (e) enzymatic activity in T cells. Quantification of AMP and adenosine expressed relative to ADP (bottom panel in d) and inosine (bottom panel in e), respectively. (f) AHR (blue; XRE-1, XRE-2 amd XRE-3) and STAT3 (green; SRE) binding sites in the Entpd1 promoter. (g) ChIP analysis of the interaction of AHR and STAT3 in the Entpd1 promoter. (h) Luciferase activity in HEK293 cells transfected with an Entpd1 luciferase reporter, alone (Control) or with a construct encoding constitutively activated STAT3 (Stat3c) or Ahr, separately or together (Ahr + Stat3c). (i) Expression of Entpd1 in T cells from WT, CD39-(Entpd1−/−) or STAT3-(Stat3−/−) deficient or Ahrmut mice after 72 h activation. (j) CD39 expression in Tr1 cells differentiated from Stat3−/− and Ahrmut mice analyzed by flow cytometry. Cells were gated in the CD4+Foxp3− population and the percentage of IL-10+CD39+ cells is shown. *P < 0.05; **P < 0.01; ***P < 0.001. Data are representative (b, d and e) or are the mean ± SEM (a-i) of three independent experiments. AU, arbitrary units.
To study the regulation of CD39 expression in Tr1 cells, we analyzed the Entpd1 promoter and identified three AHR responsive elements (XRE1, XRE2 and XRE3) and a STAT3 responsive element (SRE) (Fig. 1f). AHR binding to XRE-1 and XRE-2 and STAT3 binding to the SRE in the Entpd1 promoter was detected by chromatin immunoprecipitation assays (ChIP) in T cells activated under Tr1 polarizing conditions (Fig. 1g). Moreover, AHR and constitutively activated STAT3 (STAT3c) transactivated the Entpd1 promoter in reporter assays (Fig. 1h). Furthermore, using T cells harboring a hypomorphic Ahr allele (Ahrmut)25 or deficient in STAT3 (Stat3−/−)26, we found that CD39 is expressed in Tr1 cells in an AHR- and STAT3-dependent manner (Fig. 1i).
We also found that STAT3 and AHR are recruited to the Ahr promoter in T cells activated under Tr1 polarizing conditions (Supplementary Figs. 1e,f). Moreover, AHR and STAT3c transactivated the Ahr promoter in reporter assays, and the up-regulation of Ahr expression induced by IL-27 was abrogated in Stat3−/− T cells (Supplementary Figs. 1h-i). Taken together, these data show that IL-27 induces CD39 expression in Tr1 cells via AHR and STAT3 signaling, and identifies a positive feedback loop by which AHR in combination with STAT3 promotes Ahr expression.
CD39 contributes to the suppressive function of Tr1 cells
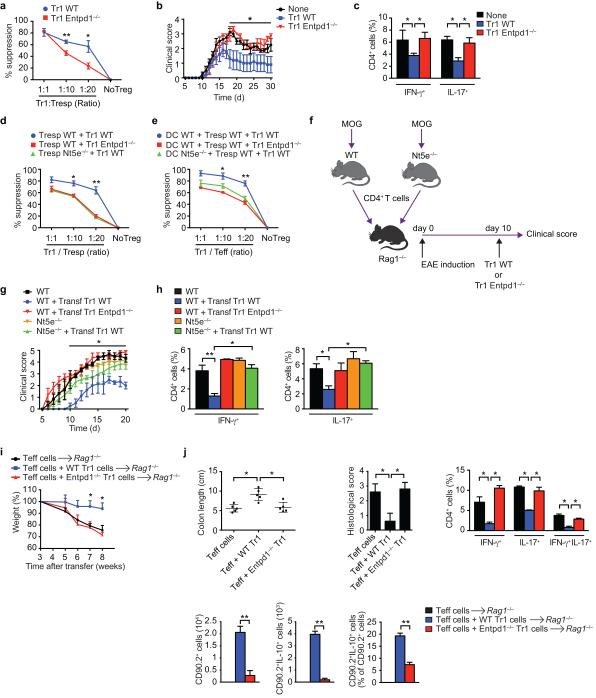
CD39 contributes to the suppressive activity of Foxp3+ Treg cells through its participation in the synthesis of adenosine21,27. We found that CD39-deficiency reduced the suppressive activity of Tr1 cells in vitro (Fig. 2a). To study the relevance of these findings for Tr1 cells in vivo, we transferred wild type (WT) or CD39-deficient IL-10:GFP+ Tr1 cells into C57BL/6 mice 10 days after the induction of EAE by immunization with MOG35-55; Tr1 cells did not express foxp3 (Supplementary Fig. 1j). WT, but not CD39-deficient Tr1 cells, reduced the EAE clinical score and the IFN-γ and IL-17 CD4+ T-cell recall response to MOG35-55 (Figs. 2b,c).
Figure 2. CD39 contributes to the suppressive function of Tr1 cells.
(a) Proliferation of responder CD4+ T cells (Tresp) stimulated in the presence of WT or Entpd1−/− Tr1 cells. The suppression of proliferation is shown relative to the control group. (b) IL-10:GFP+ Tr1 cells from WT or Entpd1−/− mice were transfered i.v. 10 days after EAE induction. The course of EAE is shown as the mean EAE score ± SEM (n = 5 mice per group). Effect of Tr1 genotype on disease course P<0.05 by two-way ANOVA. (c) Frequency of CD4+ splenic IFN-γ+, IL-17+ 21 days after EAE induction. (d) Proliferation of WT or CD73-deficient Tresp stimulated in the presence of WT or Entpd1−/− Tr1 cells. (e) Proliferation of WT or CD73-deficient Tresp stimulated in the presence of WT or Nt5e−/− DCs and WT or Entpd1−/− Tr1 cells. (f-h) WT or Nt5e−/− mice were immunized with MOG35–55 and draining lymph nodes and spleens were collected 7 d after immunization, cultured for 48 h with MOG. CD4+ cells were then transferred intravenously into Rag1-deficient mice. Recipient mice were immunized with MOG35–55 and on day 10, Tr1 cells from WT or Entpd1−/− mice were administered i.v. into recipient mice. (g) The course of EAE is shown as the mean EAE score ± SEM (n = 5 mice per group). (h) Frequency of CD4+ splenic IFN-γ+, IL-17+ 21 days after EAE induction. (i) Body weight of Rag1−/− mice transferred with WT CD90.1+CD4+CD25–CD45RBhi T cells alone or in combination with WT or CD39-deficient CD90.2+IL-10:GFP+ Tr1 cells. (j) Colon length, pathological scores and flow cytometry analysis of CD90.1+ IL-17 and IFN-γ producing cells and of CD90.2+ CD4+ T and IL-10+ CD90.2+ cells isolated from the lamina propria (LP) 8 weeks after colitis induction. Data are the mean ± SEM (a-h) of three independent experiments. *P<0.05; **P < 0.01.
CD73 is required for the suppressive activity of Foxp3+ Treg cells27. Since we did not detect CD73 expression in Tr1 cells, we studied whether CD73 in responder T cells and antigen presenting cells was needed for Tr1 cell-mediated suppression. We found that CD73 deficiency in responder T cells or DCs reduced the suppressive activity of Tr1 cells (Figs. 2d,e).
To study the role of CD73 in responder T cells for their regulation by Tr1 cells in vivo, we used a transfer EAE model28. Briefly, WT or CD73-deficient (Nt5e−/−) mice were immunized with MOG35-55 and 7 days later CD4+ T cells were isolated and transferred into Rag1−/− mice, which were then immunized with MOG35-55 to induce EAE. Ten days after EAE induction, WT or CD39-deficient Tr1 cells were transferred into the Rag1−/− recipients and disease development was monitored (Fig. 2f). WT Tr1 cells ameliorated EAE in mice reconstituted with WT T cells, but this amelioration was reduced in mice reconstituted with CD73-deficient T cells (Fig. 2g,h). These data suggest that adenosine produced by CD39 in Tr1 cells in cooperation with CD73 expressed by responder T cells and antigen presenting cells contributes to the suppressive activity of Tr1 cells.
We confirmed our findings on the role of CD39 on Tr1 suppressive activity using a T-cell transfer colitis model. Briefly, we co-transferred CD90.1+CD4+CD25−CD45RBhi effector T cells together with WT or CD39-deficient CD90.2+GFP:IL-10+ Tr1 cells into Rag1−/− recipients and monitored colitis development29. We found that CD39-deficient Tr1 cells failed to suppress the development of colitis (Figs. 2i-j). CD39 deficiency resulted in lower numbers of total and IL-10:GFP+ transferred CD90.2+ CD4+ T cells in the lamina propria, but did not lead to their expression of IL-17 or Foxp3 (Fig. 2j and not shown). Collectively, these data show that CD39 is needed for Tr1 cell stability and immunoregulatory activity.
CD39 promotes Tr1 cell differentiation by depleting eATP
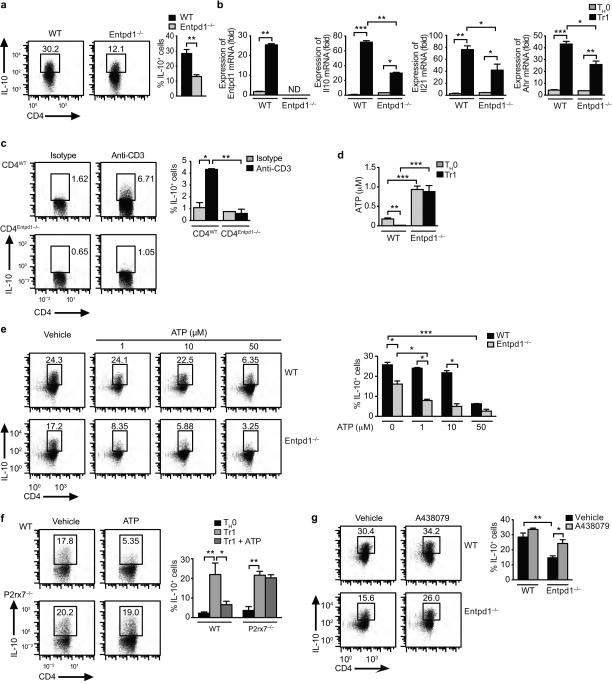
We also studied the role of CD39 on Tr1 cell differentiation. CD39 deficiency led to reduced Tr1 cell differentiation in vitro, as evidenced by the decreased expression of IL-10, IL-21, AHR, LAG-3 and CD49b30 (Fig. 3a,b and Supplementary Fig. 2a). In line with these in vitro findings, the differentiation of Tr1 cells was suppressed in Rag1−/− mice reconstituted with CD39-deficient T cells and treated with anti-CD3 antibodies as described20,31 (Fig. 3c). Hence, CD39 promotes Tr1 cell differentiation.
Figure 3. CD39 promotes Tr1 cell differentiation by limiting eATP levels.
(a) Naive CD4+ cells from WT or CD39 (Entpd1−/−) deficient mice were activated under Tr1 polarizing conditions and the frequency of CD4+Foxp3−IL-10+ cells was analyzed by flow cytometry. (b) Expression of Entpd1, Il10, Il21 and Ahr in Tr1 cells from WT and Entpd1−/− mice. (c) Rag1−/− mice were reconstituted with WT or Entpd1−/− T cells and after 10 days treated with anti-CD3 or an isotype-matched control antibody, and the differentiation of Tr1 cells was analyzed in spleen as the percentage of CD4+Foxp3−IL-10+. (d) eATP in culture supernatants of Tr1 cells from WT or Entpd1−/− mice differentiated for 72 h. (e) Tr1 cells from WT and Entpd1−/− mice were differentiated in the presence of eATP and the percentage of CD4+Foxp3−IL-10+ analyzed by flow cytometry. (f) Tr1 cells from WT or P2x7r−/− mice were differentiated in the presence of eATP (50 µM) and the percentage of CD4+Foxp3−IL-10+ determined. (g) WT or CD39-deficient Tr1 cells were differentiated in the presence of the selective P2X7R antagonist A438079 (25 µM) and the frequency of CD4+Foxp3−IL-10+ T cells was determined. Shown is a representative experiment (of three) with n ≥ 5 mice/group. *P < 0.05, **P < 0.01 and ***P < 0.001.
T-cell activation increases eATP levels as a result of the controlled release of cellular ATP10, and eATP interacts with purinergic receptors to regulate T-cell function in an autocrine and paracrine manner10. CD39 catalyzes the degradation of eATP22. In support of a role for CD39 in limiting eATP levels during Tr1 cell differentiation, we detected higher amounts of eATP in culture supernatants from CD39-deficient T cells (Fig. 3d). Based on this finding, we analyzed the effects of eATP on Tr1 cell differentiation. eATP suppressed Tr1 cell differentiation in a dose-dependent manner (Fig. 3e) and independently of cell death (Supplementary Fig. 2b). The suppressive effects of eATP were increased in CD39-deficient T cells, suggesting that CD39 limits the inhibitory effects of eATP on Tr1 cell differentiation (Fig. 3e). eATP and CD39-blockade also inhibited IL-10 expression in human T cells activated in the presence of IL-27 24 (Supplementary Fig. 2c,d).
P2X7 and other purinergic receptors mediate the effects of eATP32-35. The inhibitory effects of eATP on Tr1 cell differentiation were abrogated in P2X7R-deficient, but not in P2Y2-deficient T cells or in cells treated with the P2X4R-specific inhibitor 5-BDBD (Fig. 3f and not shown). Indeed, the P2X7R selective inhibitor A438079 rescued the differentiation of CD39-deficient Tr1 cells to levels comparable to those observed in WT T cells (Fig. 3g). Taken together, these data demonstrate that P2X7R signaling triggered by eATP inhibits Tr1 cell differentiation in CD39-deficient T cells.
HIF1-α antagonizes AHR during Tr1 cell differentiation
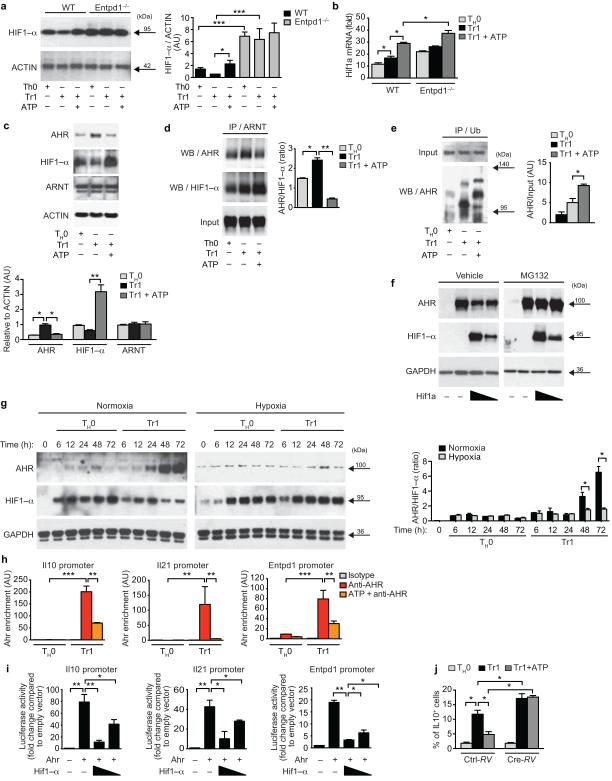
We then investigated the mechanism by which eATP interferes with Tr1 cell differentiation. P2X7R activation by eATP regulates the expression of HIF1-α responsive genes in tumor cells36,37. As HIF1-α regulates T cell differentiation6-8, we analyzed the effect of eATP on HIF1-α expression in Tr1 cells. HIF1-α is expressed after 72h of Tr1 cell differentiation (Fig. 4a); this expression was lower than that detected in TH17 cells and comparable to the observed in Foxp3+ iTreg cells (Supplementary Fig. 3a). However, supplementation with exogenous eATP and CD39 deficiency increased HIF1-α protein expression (Figs. 4a,b).
Figure 4. HIF1-α antagonizes AHR during Tr1 cell differentiation.
(a) Immunoblot and densitometric analysis of HIF1-α expression in WT and Entpd1−/− T cells differentiated in vitro in the presence eATP (50 µM). (b) Expression of Hif1a. (c) Immunoblot and densitometric analysis of AHR, HIF1-α and ARNT expression in T cells differentiated in vitro in the presence eATP. (d-e) CD4+ T cells were activated under Tr1-skewing conditions in the presence of eATP for 72h, and cell lysates were immunoprecipitated with antibodies to ARNT (d) or ubiquitin (e) antibodies, followed by SDS-PAGE and western blotting. AHR/HIF1-α ratio was calculated (d, right). (f) HEK293 cells were co-transfected with constructs encoding for AHR and HIF1-α and grown in the presence of the proteasome inhibitor MG132. Expression of AHR and HIF1-α was evaluated by western blot. (g) Expression of AHR, HIF1-α and AHR/HIF1-α ratio in Tr1 cells differentiated under normoxic or hypoxic conditions. (h) ChIP analysis of the interaction of AHR in the Il10, Il21 and Entpd1 promoter. (i) Luciferase activity in HEK293 cells transfected with an Il10, Il21 or Entpd1 (CD39) luciferase reporter, alone or with a construct encoding AHR or HIF1-α. (j) Frequency of CD4+Foxp3−IL-10+ cells in Hif1afl/fl CD4+ T cells differentiated under Tr1 polarizing conditions in the presence of eATP, and transduced with a cre-recombinase expressing retrovirus (Cre-RV) 24h after activation. *P < 0.05; **P < 0.01; ***P < 0.001. Data are representative (a, c, d,e,f,g and l) or are the mean ± SEM (a,b,d,e, h-n) of three independent experiments. AU, arbitrary units.
HIF1-α and AHR form a complex with the AHR nuclear translocator (ARNT) to regulate the expression of target genes11,38. AHR plays an important role in Tr1 cell differentiation11,20. Thus, we analyzed AHR, HIF1-α and ARNT expression following Tr1 cell differentiation in the presence of eATP. ARNT expression remained stable following eATP supplementation, but HIF1-α expression was increased concomitantly with a reduction on AHR levels (Fig. 4c, Supplementary Fig. 3b). We then studied whether the increase in HIF1-α protein expression induced by eATP interfered with AHR/ARNT complex formation. ARNT/AHR complexes decreased in the presence of exogenously added eATP (Fig. 4d, Supplementary Fig. 3c). Conversely, eATP increased the interaction of ARNT with HIF1-α, decreasing the AHR/HIF1-α binding ratio to ARNT (Fig. 4d, Supplementary Fig. 3c).
HIF1-α promotes Foxp3 ubiquitination and its degradation by the proteasome in TH17 cells6. Thus, we studied whether HIF1-α induction by eATP resulted in AHR ubiquitination and degradation in Tr1 cells. eATP treatment increased AHR ubiquitination (Fig. 4e, Supplementary Fig. 3d). Indeed, in co-transfection experiments, expression of a stabilized variant of HIF1-α decreased AHR protein expression (Fig. 4f, Supplementary Fig. 3e). This decrease in AHR protein levels was inhibited by treatment with the proteasome inhibitor MG132 (Fig. 4f, Supplementary Fig. 3e). Moreover, in agreement with a role for HIF1-α in the regulation of AHR protein levels in T cells, under Tr1 polarizing conditions HIF1-α expression peaked 24h after activation and decreased afterwards, whereas AHR expression peaked at 72h once HIF1-α protein expression was diminished (Fig. 4g). Taken together, these data show that the increase in HIF1-α induced by eATP interferes with the generation of AHR/ARNT complexes in Tr1 cells.
AHR interacts with genomic XRE sites in Il10, Il21 and Entpd1 to control their expression in Tr1 cells20 (Fig. 1). AHR, for example, is recruited to the Il10 promoter in Tr1 cells, but not in pathogenic Th17 cells which express AHR but not IL-10 (20,21 and Supplementary Fig. 3f). In agreement with our findings on the effects of HIF1-α on AHR stability, eATP supplementation decreased the recruitment of AHR to the Il10, Il21 and Entpd1 promoters during Tr1 cell differentiation (Fig. 4h). Moreover, the co-expression of a stabilized variant of HIF1-α led suppressed the transactivation of the Il10, Il21 and Entpd1 promoters by AHR in reporter assays (Fig. 4i).
Finally, to confirm that the effects of eATP on Tr1 cell differentiation were mediated by HIF1-α, we transduced Cre recombinase into Hif1afl/fl CD4+ T cells 24h after activation under Tr1 polarizing conditions in the presence of eATP. In agreement with our results on AHR destabilization by HIF1-α, Hif1a deletion resulted in increased Tr1 cell differentiation (Fig. 4j). Moreover, Hif1a deletion abrogated the suppression of Tr1 cell differentiation by eATP (Fig. 4j), demonstrating that HIF1-α mediates the inhibitory effects of eATP on Tr1 cell differentiation.
Hypoxia suppresses Tr1 cell differentiation
In normoxic conditions, prolyl hydroxylase domain (PHD) proteins (encoded by Egln1, Egln2, Egln3) promote the degradation of HIF1-α, while the HIF1-α inhibitor (encoded by Hif1an) inhibits its transcriptional activity38,39. However, under hypoxic conditions, these proteins are inactive resulting in the stabilization of HIF1-α and the induction of HIF1-α dependent cellular responses39. Hypoxia modulates T cell differentiation in a HIF1-α dependent manner6,7. Thus, we studied the effects of hypoxia on AHR and Tr1 cell differentiation. Hypoxia inhibited the differentiation of Tr1 cells and increased TH17 differentiation6 (Supplementary Figs. 3g-i). The inhibition of Tr1-cell differentiation was associated with increased HIF1-α protein expression at later time points and a concomitant decrease in AHR protein expression and recruitment to target promoters in Tr1 cells, such as Il10 and Entpd1 (Fig. 4g and Supplementary Fig. 3j). Hence, these results demonstrate that HIF1-α induced by eATP or hypoxia interferes with AHR-dependent signaling and limits Tr1 cell differentiation.
HIF1-α controls Tr1 cell early metabolic reprogramming
HIF1-α affects T-cell activation and polarization into specific lineages6-8,40. To further study the role of HIF1-α in Tr1 cells, we transduced cre-recombinase into Hif1afl/fl CD4 cells at different time points after during Tr1 cell differentiation (Supplementary Fig. 4a). Cre-recombinase transduction 24h after T-cell activation boosted Tr1 cell differentiation, but notably, transduction at earlier time points (6 or 12h after culture initiation) reduced the differentiation of Tr1 cells (Fig. 5a). Moreover, Tr1 cell differentiation was suppressed in CD4+ T cells from CD4cre Hif1a fl/fl mice (Fig. 5b). HIF1-α and AHR also affected the differentiation of Th17 and Foxp3+ i Treg cells (Supplementary Fig. 4b).
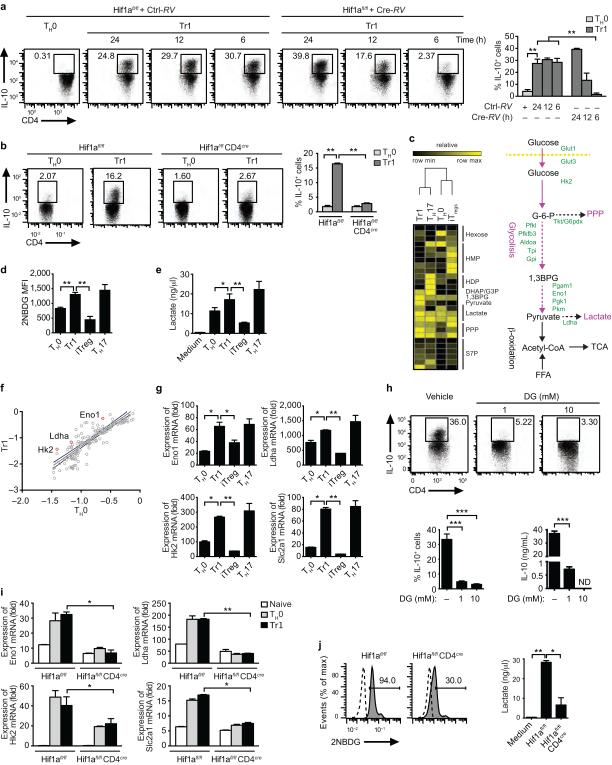
Figure 5. HIF1-α controls the early metabolic reprogramming of Tr1 cells.
(a) Frequency of CD4+Foxp3−IL-10+ cells in Hif1afl/fl CD4+ T cells differentiated under Tr1 polarizing conditions and transduced with a cre-recombinase expressing retrovirus (Cre-RV) at different time points (6, 12 and 24h). (b) Frequency of CD4+Foxp3−IL-10+ cells following activation of WT or Hif1afl/fl CD4cre T cells under Tr1 polarizing conditions. (c) Metabolomic analysis of T cells activated under Tr1, Th17 or Foxp3+ iTreg polarizing conditions and diagram of the glycolytic pathway. (d-e) Incorporation of 2NBDG (d) and lactate production (e) by Tr1, Th17 or Foxp3+ iTreg cells. (f) Expression relative to TH0 of genes associated with the control of glycolysis in Tr1 cells after 48 h of differentiation. (g) Expression of Eno1, Ldha, Hk2 and Slc2a1 in Tr1, Th17 or Foxp3+ iTreg cells after 48h of differentiation; expression is presented relative Gapdh. (h) Frequency of CD4+Foxp3−IL-10+ cells and IL-10 production by naive T cells differentiated under Tr1 conditions in the presence of vehicle or 2-DG (1 and 10 mM). (i-j) Hif1afl/fl CD4cre T cells were differentiated under Tr1 polarizing conditions for 12 h and the expression of Eno1, Ldha, Hk2 and Slc2a (i), the incorporation of 2NBDG and lactate production (j) were evaluated. Numbers adjacent to outlined areas indicate percentage of positive cells. *P < 0.05; **P < 0.01; ***P < 0.001. Data are representative of two independent experiments with similar results.
HIF1-α performs important functions in the regulation of cellular metabolism1-3. Thus, considering the metabolic functions of HIF1-α and its early up-regulation in Tr1 cells, we postulated that HIF1-α plays a dual role in Tr1 cells: at later time points HIF1-α destabilizes AHR, but at earlier time points HIF1-α might control their metabolic reprogramming . However, the metabolism of Tr1 cells is largely unknown.
A metabolomics analysis of T cells activated under Tr1 polarizing conditions detected a metabolite profile associated to aerobic glycolysis, similar to that of TH17 cells and different from the oxidative phosphorylation associated with Foxp3+ Treg cells (Fig. 5c and Supplementary Fig. 4c). In support of a role for glycolysis in Tr1 cell differentiation, we detected increased glucose uptake and lactate production in T cells activated in the presence of IL-27 (Figs. 5d,e). T cell activation under Tr1 polarizing conditions did not affect mitochondria-dependent fatty acid β -oxidation or pyruvate oxidation through the tricarboxylic acid cycle (not shown).
The analysis of 84 metabolism genes detected increased expression of genes linked to glycolysis under Tr1 polarizing conditions. These transcripts included enolase 1 (Eno1), lactate dehydrogenase (Ldha), hexokinase 2 (Hk2) and the glucose transporter Glut1 (encoded by Slc2a1) (Figs. 5f,g), suggesting that aerobic glycolysis contributes to Tr1 cell differentiation similarly to what has been described for Th17 cells40. Indeed, inhibition of glycolysis with 2-deoxyglucose (2-DG) suppressed the differentiation of Tr1 and Th17 cells, while it boosted Foxp3+ Treg cell differentiation (Fig. 5h, Supplementary Fig. 4d). Thus, aerobic glycolysis is required for Tr1 cell differentiation.
To investigate the role of HIF1-α on the early metabolic reprograming of Tr1 cells we measured Eno1, Ldha, Hk2 and Slc2a1 expression in WT and HIF1-α deficient T cells (Hif1afl/fl CD4cre), 24h after activation under Tr1 polarizing conditions. HIF1-α deficiency suppressed the up-regulation of these transcripts, as well glucose uptake and lactate production (Fig. 5i,j). Taken together, these data suggest that HIF1-α participates in the control of glycolysis during the early stages of Tr1 cell differentiation.
AHR maintains the metabolic program of Tr1 cells
HIF1-α expression peaked 24h after activation under Tr1 polarizing conditions, but was decreased at later time points, concomitant with increased AHR expression (Fig. 4g). To study the regulation of HIF1-α and AHR expression in Tr1 cells we constructed a mathematical model that considered the regulation of AHR stability by HIF1-α, the ability of AHR to induce Ahr expression and HIF1-α degradation by PHD proteins38,39. This model did not fit our experimental observations, because it predicted a continued increase of HIF1-α expression (Model 1, Fig. 6a and Supplementary Fig. 5b). Thus, we hypothesized that AHR may limit HIF1-α expression, and generated a revised model in which AHR promotes HIF1-α degradation by inducing the expression of PHD proteins. The predictions of this model were in agreement with our experimental data (Model 2, Fig. 6a and Supplementary Fig. 5b), suggesting that AHR limits HIF1-α in Tr1 cells.
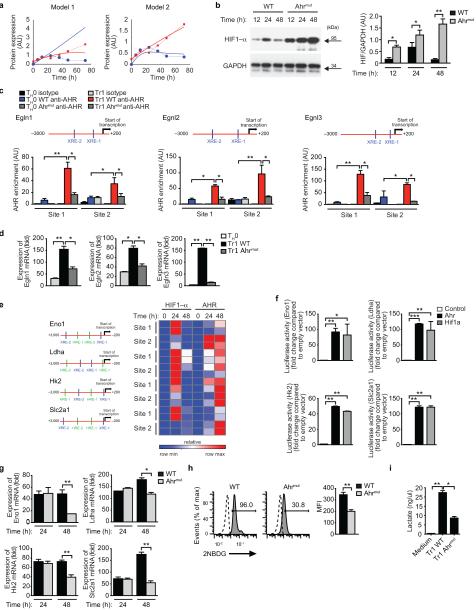
Figure 6. AHR maintains the metabolic program of Tr1 cells.
(a) HIF1-α (blue) and AHR (red) protein expression during Tr1 cell differentiation normalized to actin (full lines) and protein levels predicted by the model including Egln1 expression (dashed lines). (b) Immunoblot (left) and densitometric analysis (right) of HIF1-α expression in WT and Ahrmut T cells activated under Tr1-polarizing conditions. (c) ChIP analysis of the interaction of AHR with the Egln1, Egln2 and Egln3 promoters after 48h under TH0 or Tr1 polarizing conditions. (d) qPCR analysis of Egln, Egln2 and Egln3 expression in Tr1 cells from WT and Ahrmut mice after 48h of differentiation; expression is presented relative to Gapdh. (e) Heatmap depicting the recruitment of HIF1-α and AHR to Eno1, Ldha, Hk2 and Slc2a1 promoters at 24 and 48 h differentiation. Two independent experiments are shown. AHR-binding sites (blue; XRE-1, XRE-2) and HIF1-α binding sites (green; HRE-1, HRE-2) are shown on the left. (f) Luciferase activity in HEK293 cells transfected with an Eno1, Ldha, Hk2 or Slc2a1 luciferase reporter, alone (Control) or with a construct encoding Ahr or Hif1a. (g-i) Ahrmut T cells were differentiated under Tr1 polarizing conditions for 48 h (or 24 h when indicated), and the expression of of Eno1, Ldha, Hk2 and Slc2a (g), the incorporation of 2NBDG (h) and lactate production (i) were evaluated. Numbers adjacent to outlined areas indicate percentage of positive cells. *P < 0.05; **P < 0.01; ***P < 0.001. Data are representative of three independent experiments with similar results.
In support of a role for AHR in limiting HIF1-α cellular levels, we detected increased HIF1-α expression in Ahrmut T cells (Fig. 6b). Indeed, we detected AHR binding to the Egln1, Egln2 and Egln3 promoters under Tr1 polarizing conditions, and the expression of these genes was decreased in Ahrmut T cells (Fig. 6c,d). Egln1, Egln2 and Egln3 were expressed by Foxp3+ iTreg cells but not by Th17 cells, concomitant with a lack of AHR recruitment to the promoter of these genes in Th17 cells (Supplementary Fig 5c,d). These data are in agreement with the promoting and inhibitory roles, respectively, of HIF1-α in Th17 cell and Foxp3+ iTreg cell differentiation6,40 and suggest that lineage specific factors control promoter accessibility to AHR.
HIF1-α participates in the control of glycolysis during the early stages of Tr1 cell differentiation, but its decreased expression at later time points suggests that other factors participate in the control of Tr1 cell metabolism. The expression of c-Myc and estrogen-related receptor-α (ERRα), which have been linked to the regulation of glycolysis in T cells41,42, did not differ between Tr1 or TH0 polarizing conditions (Supplementary Fig. 5e). However, we identified AHR binding sites in the promoters of Eno1, Ldha, Hk2 and Slc2a1 (Fig. 6e). HIF1-α was recruited to these promoters 24h after the initiation of Tr1 cell cultures, but by 48h of activation the recruitment of HIF1-α was decreased, concomitant with an increased recruitment of AHR (Fig. 6e). Indeed, AHR transactivated the Eno1, Ldha, Hk2 and Slc2a1 promoters to similar levels than HIF1-α (Fig. 6f). Moreover, Ahrmut T cells activated under Tr1 polarizing conditions for 48h showed decreased expression of genes linked to glycolysis, decreased glucose uptake and lactate production (Figs. 6g-i). We did not observe differences in glycolytic genes in Ahrmut T cells activated under Tr1 polarizing conditions for 24h (Figs. 6g). We also failed to detect an increase in the recruitment of c-Myc to Eno1 and Ldha in Tr1 cells (Supplementary Fig. 5f). Collectively, these data show that AHR participates in the control of metabolism during the late stages of Tr1 cell differentiation.
Discussion
Metabolic reprogramming associated to T-cell activation matches the bioenergetic needs of the immune system during the immune response. In addition, immunometabolites participate in immunoregulatory feedback loops. We found that through their effects on HIF1-α, eATP and hypoxia inhibit Tr1 cell differentiation. Both increased eATP and hypoxia characterize sites of inflammation10,22,43. Thus, our data identifies a potential mechanism through which metabolites in the microenvironment of the inflamed tissue modulate the immune response. This mechanism might be co-opted by other immune cells. Indeed, the production of IL-10 by both IL-27 induced Tr1 cells and TH1 cells has been linked to the transcription factor c-Maf18,20,44, and CD39 also contributes to the control of inflammation by Foxp3+ Treg cells and dendritic cells27,34. Thus, the crosstalk between HIF1-α and AHR provides a pathway to modulate the immune response in response to immunological, metabolic and environmental signals.
The metabolism of effector CD4+ and CD8+ T cells displays a strong bias towards glycolysis, while Foxp3+ Treg cells rely on oxidative phosphorylation to match their bioenergetic needs7,9,40. Our data demonstrate that regulatory Tr1 cells are supported by glycolytic metabolism. These metabolic similarities between effector and regulatory Tr1 cells likely reflect common metabolic needs associated with proliferative T cells45 and the participation of STAT3-dependent transcriptional programs in the differentiation of Tr1 and TH17 cells15,16,46,47. Conversely, the different metabolic programs associated with Tr1 cells and Foxp3+ Treg cells might reflect the functional compartmentalization of these immunoregulatory cell populations in vivo. Moreover, these findings suggest therapeutic interventions to preferentially promote the differentiation of Tr1 or Foxp3+ Treg cells.
Online Methods
Animals
Ahrmut, Entpd1−/−, Stat3−/− and Hif1afl/fl mice have been previously described6,25,48,49. Eight-week old C57BL/6, Rag1-, (Nt5e−/−) CD73- and (P2x7r−/−) P2X7R-deficient female mice were purchased from the Jackson Laboratories. All mice strains were C57BL/6 background. Mice were kept in a conventional, pathogen-free facility at the Harvard Institutes of Medicine. All mouse experiments were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
Human samples
Samples were collected as described 50 from healthy controls (25–41 years of age) after informed consent was provided. All procedures were approved by the institutional review board of Brigham and Women’s Hospital.
In vitro T-cell differentiation
Naive CD4+ T cells (CD4+ CD44lo CD62Lhi CD25−) were purified by flow cytometry from the spleen and lymph nodes; the purity of isolated T-cell populations routinely exceeded 98%. Naive T cells were stimulated with plate-bound anti-CD3 (2 µg/ml; 14-0031-86; eBioscience) and anti-CD28 (2 µg/ml; PV-1; Abcam), in the presence of IL-27 (20 ng/ml) for the generation of Tr1 cells; IL-12 (30 ng/ml) and anti-IL-4 (2.5 µg/ml; C17.8; Biolegend) for the generation of TH1 cells; IL-4 (30 ng/ml) and anti-IFN-γ (5 µg/ml; XMG1.2; Biolegend) for the generation of TH2 cells; or IL-6 (30 ng/ml), TGF-β1 (3 ng/ml), IL-23 (30ng/ml) and anti-IL4 and anti-IFN-γ for the generation of TH17 cells. Mouse IL-27 (34-8271) was from eBioscience. Mouse IL-4, IL-6, IL-12, IL-23, anti-IFN-γ and anti-IL-2 (404-ML-010, 406-ML-025, 419-ML-050, 1887-ML-010, MAB485 and MAB702, respectively) were from R&D Systems; TGF-β1 (130-095-067) was from Miltenyi Biotec. ATP (A3377, Sigma-Aldrich) was added to the cultures at the indicated concentration. The P2X7R receptor antagonist A438079 (2976, Tocris bioscience) and the P2X4R receptor antagonist 5-BDBD (3579, Tocris bioscience) were used at a final concentration of 25 µM and 1.5 mM respectively. Cells were subjected to hypoxia (1% O2) in a GasPak Plus anaerobic chamber (BD Biosciences). For human Tr1 cells, FACS sorted naive CD4+ T cells (CD4+CD62L+ CD45RO−) were cultured in 96-well round bottom plates at 105 cells/well in serum-free X-Vivo 15 medium (04-418Q; Lonza) and stimulated with plate-bound anti-CD3 (2 µg/ml; 555336; BD Biosciences) and soluble anti-CD28 (2 µg/ml; 555725; BD Biosciences), in the presence of recombinant human IL-27 (100 ng/ml) and IL-2 (100 U/ml) as described24. Human IL-27 (2526-IL-010) and IL-2 (354043) were from R&D and BD biosciences respectively. Purified anti-human CD39 blocking antibody (328202) was purchased from Biolegend.
Flow Cytometry Staining and Acquisition
Antibodies for flow cytometry were purchased from eBioscience or BD Pharmingen and used as recommended by the manufacturers. Cells were then analyzed on a LSRII or Macsquant flow cytometer (BD Biosciences and Miltenyi Biotec respectively).
Proliferation assays
Proliferation assays were performed as described34, using naive CD4+ T cells labeled with 10 µM of CellTrace Violet solution (C34557, from Molecular Probes, USA). Data were acquired on a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR, USA).
Measurement of cytokines
Secreted cytokines were measured after 48h by ELISA as described34,49. For intracellular cytokine staining, cells were cultured and restimulated for 4h at 37C in culture medium containing PMA (500 ng/ml; Sigma, USA), ionomycin (500 ng/ml; Sigma) and monensin (GolgiStop; 2 µl/ml; BD Biosciences, USA). After staining for surface markers, cells were fixed and permeabilized as suggested by the manufacturer (eBiosciences, USA).
Metabolite Profiling
Lipid extracts were analyzed using a Nexera X2 U-HPLC (Shimadzu, Marlborough, MA) coupled to an Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Extracts (10µL) were injected onto a ACQUITY UPLC BEH C8 column (1.7µm, 2.1 × 100 mm; Waters; Milford, MA). The column was initially eluted isocratically at a flow rate of 450 µL/min with 80% mobile phase A (95:5:0.1 vol/vol/vol 10mM ammonium acetate/methanol/formic acid) for 1 minute followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 minutes, a linear gradient to 100% mobile phase B over 7 minutes, then 3 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization in the positive ion mode (source voltage was 3kV, source temperature was 300°C, sheath gas was 50.0, auxillary gas was 15) using full scan analysis over m/z 200-1100 and at 70,000 resolution. Hydrophilic interaction liquid chromatography (HILIC) analyses of water soluble metabolites were conducted in the positive ion mode using a Nexera X2 U-HPLC (Shimadzu, Marlborough, MA)-Q Exactive orbitrap (Thermo Fisher Scientific; Waltham, MA) LC-MS instrument. Cell extracts (10µL) were diluted using 90 µL of 74.9:24.9:0.2 v/v/v acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (valine-d8, Isotec; and phenylalanine-d8, Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9,000 × g, 4 °C) and the supernatants were injected directly onto a 150 × 2 mm Atlantis HILIC column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 250 µL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 min followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. The electrospray ionization voltage was 3.5 kV and data were acquired using full scan analysis over m/z 70-800 at 70,000 resolution. HILIC analyses of water soluble metabolites in the negative ion mode were done using a Nexera X2 U-HPLC (Shimadzu, Marlborough, MA) coupled to a Q Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Cell extracts (10µL) were directly injected onto a 150 × 2.0 mm Luna NH2 column (Phenomenex; Torrance, CA) that was eluted at a flow rate of 400 µL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide in water) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol) followed by a 10 min linear gradient to 100% mobile phase A. MS full scan data were acquired over m/z 70-750. Instrument settings were: source voltage −3.0 kV, source temperature 325°C, capillary temperature 350°C, sheath gas 55, auxiliary gas 10, and resolution 70,000. LC-MS data were processed and visually inspected using TraceFinder 3.1 software (Thermo Fisher Scientific; Waltham, MA) LC-MS.
qPCR
RNA was extracted with RNAeasy columns (Qiagen, USA), cDNA was prepared following the manufacturer’s instructions (Applied Biosystems, USA) and used as template either for real-time PCR or RT2Profiler PCR Array (330231 PAMM-006ZA, Qiagen, USA). All the primers and probes were provided by Applied Biosystems, and were used on the ViiA™ 7 Real-Time PCR System (Applied Biosystems). Expression was normalized to the expression of Gapdh. Primers-probe mixtures were purchased from Applied Biosystems (USA): Il10 (Mm0043614_m1), Entpd1 (Mm00515447_m1), AHR (Mm01291777_m1), Maf (Mm02581355_s1), Foxp3 (Mm00475156_m1), Gzmb (Mm00442834_m1), Hif1a (Mm00468872_m1), Arnt (Mm00507836_m1), Eno1 (Mm01619597_g1), Ldha (Mm01612132_g1), Hk2 (Mm00443385_m1), Slc2a1 (Mm00441480_m1), Egln1 (Mm01198376_m1), Egln2 (Mm00459770_m1), Egln3 (Mm00519067_m1), Hif1an (Mm00472200_m1) and Gapdh (Mm99999915_g1).
Immunoblot analysis
Cells were lysed with Cell Lysis buffer (1X) supplemented with protease inhibitor cocktail (Cell Signaling, USA). Total cell lysates (10 µg) were resolved on 4–12% Bis-Tris Nupage gels (Invitrogen, USA) and transferred onto PVDF membranes (Millipore). The following primary antibodies were used: Anti-AHR (BML-SA210, 1/2000) from Enzo Life Sciences; anti-HIF1-α(10006421, 1/500) from Cayman Chemical and anti-GAPDH (#2111, 1/10000) from Cell Signaling Technology, USA; anti-Ubiquitin (ab7780, 1/1000), anti-HIF-1beta (ab2, 1/2000) and anti-beta actin (ab20272, 1/10000) from Abcam, USA. The Immunoblot was developed using SuperSignal West Femto Maximum Sensitivity Substrate, as described49 and instructed by the manufacturer (Pierce).
Immunoprecipitation
Cells were lysed with Cell Lysis buffer (1X) supplemented with protease inhibitor cocktail (Cell Signaling, USA). Five µg of antibody was prebound for a minimum of 4 h to protein A and protein G Dynal magnetic beads (Invitrogen, USA) and washed 3 times with ice-cold PBS plus 5% BSA, and then added to 400 µg of cell lysates and immunoprecipitated overnight. The magnetic bead-protein complexes were washed 3 times in RIPA buffer (Thermo Scientific, USA), the immunoprecipitates were re-suspended in 4X loading buffer and analyzed by immunoblot as described above.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% paraformaldehyde and lysed with 0.35 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) containing 1X protease inhibitor cocktail (Roche Molecular Biochemicals, USA). Chromatin was sheared by sonication and supernatants were collected after centrifugation and diluted in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). Five µg of antibody was prebound for a minimum of 4 h to protein A and protein G Dynal magnetic beads (Invitrogen, USA) and washed three times with ice-cold PBS plus 5% BSA, and then added to the diluted chromatin and immunoprecipitated overnight. The magnetic bead-chromatin complexes were then washed 3 times in RIPA buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl) followed by 2 times with TE buffer. Immunoprecipitated chromatin was then extracted with 1% SDS, 0.1 M NaHCO3 and heated at 65°C for at least 6 h to reverse the paraformaldehyde cross-linking. DNA fragments were purified with a QIAquick DNA purification Kit (Qiagen, USA) and analyzed using SYBR green real time PCR (Takara Bio Inc., USA). We used the following antibodies for ChIP: Anti-AHR (BML-SA210, Enzo Life Sciences, USA), anti-STAT3 (Cat. #9132, Cell Signaling Technology, Inc., USA), anti-HIF-1 (10006421) from Cayman Chemical USA), The following primer pairs were used: Entpd1:STAT3 (SRE): for: 5’-GCTGGGCTTTAGAGACTTGTGGGC-3’, rev: 5’-ACCCATGCAAATGGTTTGGGCA-3’; AHR:STAT3 (SRE): for: 5’-GGGTCTCATTCTGCAGACAAC-3’, rev: 5’-TCAATTTCCCAGCGATTCTC-3’; Entpd1:AHR (XRE-1): for: 5’-CTTACACCGTCCTCCCTGAG-3’. rev: 5’-GCCAGCTGTGAAATGACAAA-3’; Entpd1:AHR (XRE-2): for: 5’-CCTGGGCCTCTCACTCTCTT-3’, rev: 5’-GAGTGCTGCGCAGGTGTA-3’; Entpd1:AHR (XRE-3): for: 5’-AAGGAGGTGGACACAACCAG-3’, rev: 5’-TGAATAAATGTGTGCAGAAGGA-3’; Il10:AHR (XRE): for: 5’-GGTGACTTCCGAGTCAGCAA-3’. rev: 5’-ACAGCTATTTTTAGTATGGGCTACC-3’; Il21:AHR (XRE): for: 5’-CCTGCCATAACACCTGCTTT-3’, rev:5’-TCCAGAGAACCAAATAACCCTA-3’; Eno1:AHR (XRE-1): for: 5’-GAGCGCCACACCCTAGTAGA-3’, rev: 5’-GAATATTCCACAGCAGAGATGG-3’; Eno1:AHR (XRE-2): for: 5’-ATTGGTGATATGAATGTTTTTCCTC-3’. rev: 5’-CAAGAAACAGAGTGGGTGCAT-3’; Ldha:AHR (XRE-1): for: 5’-CCTGCCAGTCTTCTGGATCT-3’, rev: 5’-GACCCCAAGGCATAGCAAC-3’; Ldha:AHR (XRE-2): for: 5’-TAAGAGGCTCGCTCCTGGT-3’, rev: 5’-GGTGCCTCTACTTCCCACCT-3’; Hk2:AHR (XRE-1): for: 5’-CAGCTACCCAGACTGGTTCC-3’, rev: 5’-CAACCAAAACCAAGTGCAGA-3’; Hk2:AHR (XRE-2): for: 5’-TCCCGTCGCTAACTTCACTC-3’. rev: 5’-AGGCTACCCGGAGTTGTTCT-3’; Slc2a1:AHR (XRE-1): for: 5’-CTCCCCCAAGAGCAGAGG-3’, rev: 5’-GCTGCGCTCGAAGCTCTA-3’; Slc2a1:AHR (XRE-2): for: 5’-CACACACACACACACACACACT-3’, rev: 5’-TGTTGCTAGCTTCAGGGAGTC-3’; Egln1:AHR (XRE-1): for: 5’-AGAATCTGCCAATAGTGTCTGC-3’, rev: 5’-AGTTGGCAAAGAGGGCTCAC-3’; Egln1:AHR (XRE-2): for: 5’-CTCATAGAGCTTGGCACGGT-3’. rev: 5’-ACAAATCACACGTGGAACCG-3’; Egln2:AHR (XRE-1): for: 5’-CGGCTAGACTCCAAGGACTC-3’, rev: 5’-ATGAAGACACTGCTGCCATG-3’; Egln2:AHR (XRE-2): for: 5’-TAGATGACAGACTGGGCCAC-3’, rev: 5’-TGTCTCCCTATCACCTTCCTC-3’; Egln3:AHR (XRE-1): for: 5’-CAGCTGCTTGACTCGCTC-3’, rev: 5’-GAAGATCGCCCTGGAGTACA-3’; Egln3:AHR (XRE-2): for: 5’-ACTTTAGCTCCAGAGGCGG-3’. rev: 5’-GGTGGCTCTGCATCTCCA-3’; Hif1an:AHR (XRE-1): for: 5’-GGCCGTCCCTAGAGTAGAGA-3’, rev: 5’-TGGCTCAGACGTGGGATG-3’; Hif1an:AHR (XRE-2): for: 5’-CTTCAGGACAGCTTTACCCA-3’, rev: 5’-TGCTGCCTCCCTATGTTCAA-3’; Eno1:HIF (HRE-1): for: 5’-GAGACAAGGTGCCCTTTTGA-3’, rev: 5’-AAACTCCTTGGCCTGTGGT-3’; Eno1:HIF (HRE-2): for: 5’-ACGTTTGCTGCCTAGCCTAA-3’. rev: 5’-GCATCCCTAGCGCTACACTT-3’; Ldha:HIF (HRE-1): for: 5’-CCTTTTGGGGTTCATCAAGA-3’, rev: 5’-CTCACCAGGAAGGCTCTCAC-3’; Ldha:HIF (HRE-2): for: 5’-CTCTGCACCCTAGACGGAAG-3’, rev: 5’-AGCCTTGTGAGGTCTCTGCT-3’; Hk2:HIF (HRE-1): for: 5’-GACTTTCCCAGCTACCCAGA-3’, rev: 5’-GACCAAGTTGCGTGCTCTC-3’; Hk2:HIF (HRE-2): for: 5’-AGCCAATCAGCGCCTAGAG-3’. rev: 5’-ACCCGAAGCTGAGCCTGAC-3’; Slc2a1:HIF (HRE-1): for: 5’-GCGGCGGTCCTATAAAAAG-3’, rev: 5’-GCTGCGCTCGAAGCTCTA-3’; Slc2a1:HIF (HRE-2): for: 5’-AGCATCCAGAGCCAGACTGT-3’, rev: 5’-GCGGAGCTGCCTTTTTATAG -3’.
Plasmids
The dual reporter constructs expressing Gaussia luciferase under the murine Eno1, Ldha, Hk2 and Slc2a1 promoters were from GeneCopoeia, Inc; the constructs for Il10 and Il21, Entpd1 were previously described20,34; the stabilized Hif1a encoding construct was previously described6. The vector coding for STAT3C was provided by Dr. David Frank (Dana-Farber Cancer Institute, Boston, MA, USA).
Transfection and luciferase assays
HEK293 cells grown in DMEM supplemented with 10% fetal bovine serum were transfected using the FuGENE HD (Roche, USA) and 2 µg of each plasmids according the manufacturer’s instructions. Luciferase activity was analyzed 48 h after transfection using a dual luciferase assay kit (Promega, USA).
Retroviral infection
Naive CD62LhiCD25−CD44loCD4+ T cells were transduced with Cre-retrovirus as described51 after activation with plate-bound anti-CD3 and anti-CD28.
Glucose uptake assay
Glucose uptake was measured following 15 minutes of incubation of T cells with 2NBDG (Life Technologies, USA) as described52. Briefly, Naïve T cells were differentiated under Th0, Tr1, iTreg and Th17 polarizing condition and after 48 h cells were washed twice and T-cell media was replaced by a glucose free T-cell media. 2 NBDG (Life Technologies, USA) was added at a final concentration of 150 µg/ml for 15 min and cells analyzed by flow cytometry.
Free ATP measurement and lactate assay
Tr1 cells were differentiated as described above for 48 h. Cells were then washed 2 times with phenol red-free RPMI 1640 (Gibco, USA) and serum starved for 24 h. Cell-free medium was then analyzed for ATP using ENTILEN rLuciferase/Luciferin Reagent (Promega, USA). Bioluminescent activity was measured using an Infinite 200 Pro luminometer (Tecan, USA). Lactate levels in cell free medium were measured using the Lactate Colorimetric Assay Kit II (Sigma, USA).
Ectonucleotidase enzymatic activity analysis
Thin Layer Chromatography (TLC) was performed as described 27,53 with slight modifications. Specifically, 1×105 naive, TH0, Tr1 or TH17 cells were incubated with 2 mCi/ml [C14]ADP (GE Healthcare Life Sciences, USA) in 10 mM Ca2+ and 5 mM Mg2+. Five ml aliquots, collected at 1.5, 3, and 6 min, were analyzed for the presence of [C14]ADP hydrolysis products by TLC and applied onto silica gel matrix plates (Sigma-Aldrich, USA). [C14]ADP and radiolabeled derivatives were separated using a solvent mixture as previously described23,27. Adenosine uptake and deamination was blocked with 10 mM of dipyridamole. [C14]ADP, [C14]AMP and [C14]ADO were incubated in PBS served as standards.
EAE Induction and Tr1 transfer
EAE was induced by s.c. immunization with 200 µg of the MOG35–55 peptide (ANASPEC, USA) 34. Adoptive transfer EAE was induced as described28 with some modifications. WT or Nt5e−/− mice were immunized with 200 µg MOG35–55 in complete Freund’s adjuvant, and draining lymph nodes and spleens were collected 7 d after immunization and cultured for 48 h with MOG35–55 (20 µg/ml). Subsequently, 5x106 cells were transferred intravenously into Rag1-deficient mice. Recipient mice were further immunized with MOG as described above and on day 10 (Onset of disease), 5x105 Tr1 cells from WT or Entpd1−/− mice were administered i.v. into recipient mice. Clinical signs of EAE were assessed by investigators blind to treatment according to the following score: 0, no disease; 1, loss of tail tone; 1.5, poor righting ability; 2, hind-limb weakness; 3, hind-limb paralysis; 4, quadreparesis; and 5, moribund.
T Cell-Induced colitis
Naive (CD90.2+CD44loCD62L+IL-10:GFP–) CD4+ T cells were purified from WT or Entpd1−/−IL-10gfp mice and were then stimulated with plate bound anti-CD3 and anti-CD28 and 20 ng/ml IL-27 for 3 days. CD90.1+CD4+CD25–CD45RBhi naive T cells (7 × 105) were injected intraperitoneally (i.p.) with or without in vitro differentiated and resorted WT or Entpd1−/− Tr1 CD4+IL-10:GFP+ (1.5 × 105) into age- and sex-matched Rag1−/− mice, and weight loss was monitored.
Histological evaluation of colitis
The histologic score of colonic tissue represents the sum of non-ulcerative regions of epithelium and an infiltration score ranging from 0 to 4 as follows: epithelium: 0, normal morphology; 1, loss of goblet cells in small areas; 2, loss of goblet cells in large areas; infiltration: 0, no infiltration; 1, infiltration around crypt basis; 2, infiltration reaching the lamina muscularis mucosae; 3, extensive infiltration reaching the lamina muscularis mucosae and thickening of the mucosa with abundant oedema; 4, infiltration of the lamina submucosa.
Mathematical model
We constructed a model based on 3 nonlinear ordinary differential equations, where H = [H], A = [A] and E = [E] denote the levels of HIF1-α, AHR and HIF1-α degrading PHD, respectively. The model assumes that HIF1-α is synthesized at a constant rate kH0 and degraded by PHD proteins at a rate that depends on their amount factored by kHE. We also assume that AHR is synthesized at a constant level kA0, degraded in a HIF1-α dependent manner with a factor kHA, and promotes its own synthesis with a factor kA. Finally, we assume that PHD proteins are synthesized at a constant level kE0 and have a half-life defined by kE. In addition their AHR induced production has a maximal rate of kAE with a dissociation constant kD. Models 1 and 2 differ only by the AHR-induced production of PHD proteins (Supplementary Fig. 5b).
| Model 1 |
| Model 2 |
To solve the system of equations we fixed the parameters KH0 = 0.06, KA0 = 0.03, and also set KE = 0.12 to reflect halftime of ~6hrs for the enzymes. The same model was used for each PHD protein, however the parameters were estimated for each separately, hence we had 4 sets of model parameters. The parameters were estimated using the Levenberg-Marquardt nonlinear least-squares algorithm. The initial conditions were randomly set and the best solution in the L2 sense was chosen.
Statistical analysis
GraphPad Prism software was used for statistical analysis. For comparison of two groups, linear regression with 95% confidence interval, and unpaired, two-tailed Student’s t-test were used. One-way ANOVA test for paired data was used to determine the significance of the time-response curves. Values of P<0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants AI075285, and AI093903 from the National Institutes of Health and RG4111A1 from the National Multiple Sclerosis Society to FJQ. I.D.M. received support from an educational grant from Questcor (A219074) and by a postdoctoral fellowship (FG 2036-A1/1) from National Multiple Sclerosis Society. M.C.T. is a graduate student in the PhD program of the Federal University of Sao Paulo, and was supported by fellowship 246252/2012-0 from Ciências sem Fronteiras CNPq, Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil.
References
- 1.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annual review of immunology. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annual review of immunology. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science (New York, N.Y.) 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nature reviews. Immunology. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annual review of immunology. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nature immunology. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JS, et al. Natural and inducible TH17 cells are regulated differently by Akt and mTOR pathways. Nature immunology. 2013;14:611–618. doi: 10.1038/ni.2607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. Journal of immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junger WG. Immune cell regulation by autocrine purinergic signalling. Nature reviews. Immunology. 2011;11:201–212. doi: 10.1038/nri2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacological reviews. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pot C, Apetoh L, Awasthi A, Kuchroo VK. Induction of regulatory Tr1 cells and inhibition of T(H)17 cells by IL-27. Seminars in immunology. 2011;23:438–445. doi: 10.1016/j.smim.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roncarolo MG, et al. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Roncarolo MG, Gregori S, Bacchetta R, Battaglia M. Tr1 cells and the counter-regulation of immunity: natural mechanisms and therapeutic applications. Current topics in microbiology and immunology. 2014;380:39–68. doi: 10.1007/978-3-662-43492-5_3. [DOI] [PubMed] [Google Scholar]
- 15.Awasthi A, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nature immunology. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 16.Stumhofer JS, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nature immunology. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald DC, et al. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nature immunology. 2007;8:1372–1379. doi: 10.1038/ni1540. [DOI] [PubMed] [Google Scholar]
- 18.Pot C, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. Journal of immunology. 2009;183:797–801. doi: 10.4049/jimmunol.0901233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spolski R, Kim HP, Zhu W, Levy DE, Leonard WJ. IL-21 mediates suppressive effects via its induction of IL-10. Journal of immunology. 2009;182:2859–2867. doi: 10.4049/jimmunol.0802978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apetoh L, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nature immunology. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi R, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nature immunology. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eltzschig HK, Sitkovsky MV, Robson SC. Purinergic signaling during inflammation. The New England journal of medicine. 2012;367:2322–2333. doi: 10.1056/NEJMra1205750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X, et al. CD39/ENTPD1 expression by CD4+Foxp3+ regulatory T cells promotes hepatic metastatic tumor growth in mice. Gastroenterology. 2010;139:1030–1040. doi: 10.1053/j.gastro.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murugaiyan G, et al. IL-27 is a key regulator of IL-10 and IL-17 production by human CD4+ T cells. Journal of immunology. 2009;183:2435–2443. doi: 10.4049/jimmunol.0900568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, et al. Induction of pathogenic T17 cells by inducible salt-sensing kinase SGK1. Nature. 2013 doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills JH, et al. CD73 is required for efficient entry of lymphocytes into the central nervous system during experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9325–9330. doi: 10.1073/pnas.0711175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintana FJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nature immunology. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagliani N, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nature medicine. 2013;19:739–746. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 31.Kamanaka M, et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–952. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 33.Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509:310–317. doi: 10.1038/nature13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mascanfroni ID, et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat Immunol. 2013;14:1054–1063. doi: 10.1038/ni.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CM, Ploia C, Anselmi F, Sarukhan A, Viola A. Adenosine triphosphate acts as a paracrine signaling molecule to reduce the motility of T cells. The EMBO journal. 2014 doi: 10.15252/embj.201386666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amoroso F, Falzoni S, Adinolfi E, Ferrari D, Di Virgilio F. The P2X7 receptor is a key modulator of aerobic glycolysis. Cell death & disease. 2012;3:e370. doi: 10.1038/cddis.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X, et al. Disordered purinergic signaling and abnormal cellular metabolism are associated with development of liver cancer in Cd39/ENTPD1 null mice. Hepatology. 2013;57:205–216. doi: 10.1002/hep.25989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annual review of pathology. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 40.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of experimental medicine. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalek RD, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. The New England journal of medicine. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraiva M, et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enjyoji K, et al. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nat Med. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- 49.Yeste A, et al. IL-21 induces IL-22 production in CD4+ T cells. Nature communications. 2014;5:3753. doi: 10.1038/ncomms4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandhi R, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nature Immunology. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quintana FJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–2111. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng L, et al. Vascular CD39/ENTPD1 directly promotes tumor cell growth by scavenging extracellular adenosine triphosphate. Neoplasia. 2011;13:206–216. doi: 10.1593/neo.101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.