Abstract
During development and regeneration, tissues emerge from coordinated sequences of stem cell renewal, specialization, and assembly that are orchestrated by cascades of regulatory factors. This complex in vivo milieu, while necessary to fully recapitulate biology and to properly engineer progenitor cells, is difficult to replicate in vitro. We are just starting to fully realize the importance of the entire context of cell microenvironment—the other cells, three-dimensional matrix, molecular and physical signals. Bioengineered environments that combine tissue-specific transport and signaling are critical to study cellular responses at biologically relevant scales and in settings predictive of human condition. We therefore developed microbioreactors that couple the application of fast dynamic changes in environmental signals with versatile, high-throughput operation and imaging capability. Our base device is a microfluidic platform with an array of microwells containing cells or tissue constructs that are exposed to stable concentration gradients. Mathematical modeling of flow and mass transport can predict the shape of these gradients and the kinetic changes in local concentrations. A single platform, the size of a microscope slide, contains up to 120 biological samples. As an example of application, we describe studies of cell fate specification and mesodermal lineage commitment in human embryonic stem cells and induced pluripotent stem cells. The embryoid bodies formed from these cells were subjected to single and multiple concentration gradients of Wnt3a, Activin A, bone morphogenic protein 4 (BMP4), and their inhibitors, and the gene expression profiles were correlated to the concentration gradients of morphogens to identify the exact conditions for mesodermal differentiation.
Keywords: Human stem cells, cardiac differentiation, microscale platforms, flow, transport, gradients
Introduction
Major advances in stem cell biology over the last decade have focused attention to the importance of the microenvironment in determining cell fate.1,2 During development and regeneration, tissues emerge from coordinated sequences of stem cell renewal, specialization, and assembly that are orchestrated by molecular and physical regulatory factors.3,4 The complexity of spatial and temporal changes of these factors in their native in vivo milieu cannot be replicated using standard in vitro methods. Tissue engineering, originally developed to meet the need for regenerating human tissues lost to injury or disease, is now becoming increasingly focused on biologically inspired culture systems.
It has been argued that we need a new generation of three-dimensional (3D) culture systems that would be “something between a Petri dish and a mouse”5 to more authentically represent the cell environment in a living organism. In order to unlock the full potential of stem cells, we need to reconstruct at least some aspects of the dynamic environments associated with their renewal, differentiation, and assembly in native tissues.6 In response to this need, tissue engineering is shifting its focus to highly controllable microtissue systems for fundamental stem cell research, study of disease, and drug screening.7–10
Tight control of transport and signaling in bioengineered cell niches would allow us to decode physiological cell responses.11 New microfluidic platforms accommodating large numbers of small-size tissues, often described as “organs on a chip” models, are increasingly predictive of human physiology in health and disease.2,11–15
Microscale platforms bring significant advantages to biological and medical research, largely due to the small transport distances, small volumes being handled, and the ability to introduce and measure fast dynamic changes in cellular responses. These features allow precise control and fine tuning of variables in a large parameter space. On a small scale, transport phenomena are more easily predicted and mathematically described and are amenable to computational modeling16–18 that in turn provides rational approaches to the optimization of culture conditions. Most importantly, working on biologically relevant scales —in space and time—enables real-time insights into cellular responses.
Several research groups have reported the effects of substrate stiffness19–24 and other physical factors such as mechanical forces25,26 on stem cells and engineered tissues. Microscale platforms were developed to simultaneously probe the roles of biochemical and biophysical factors on stem cells cultured in hydrogels with tunable stiffness and functionalized with combinations of proteins.27
The development of in vitro models of multiorgan systems is one of the most promising microscale applications. Human “organs-on-a-chip” devices, capturing a more “holistic” behavior of human tissues, would greatly improve the current standards for screening of drug efficacy and toxicity.28–30 Highly meritorious studies of this kind include “Gut-on-a-Chip” microfluidic platforms that recapitulated some aspects of normal intestinal physiology,31 microscale human liver constructs that exhibited species-specific drug responses,32,33 and a lung chip microdevice that replicated complex organ-level responses.34 Since vasculature would be needed to connect the individual “organs,” microvascular 3D networks have been engineered to connect the individual organs-on-a-chip. Such vessels were also shown to respond to inflammatory signals with a switch from a non-thrombotic to a prothrombotic state.35
In addition, continuous efforts are being devoted to interfacing microscale platforms with the automated high-throughput analysis systems. Systems such as the Fluidigm Single-Cell Gene Expression platforms (Fluidigm Corp, South San Francisco, CA 94080) can greatly improve the quality of the output data and overcome difficulties of analyzing small amounts of samples using standard techniques.36
In this mini-review, we discuss the benefits of studying biological processes on a small scale, where fast mass transport allows full expression of biological kinetics, and where perturbations in the cellular environment can be precisely introduced and measured. Exactly due to these features, the microscale technologies enable studies of physiological and medical questions in a way truly representative of human physiology. We then present a case study of early mesoendodermal differentiation of human embryonic stem cell (hESC) and induced pluripotent stem cell (iPSC) to illustrate the utility of microscale technologies in optimizing protocols for the derivation of human cardiac cells and their progenitors.
Biomimetic design principles
Transport phenomena
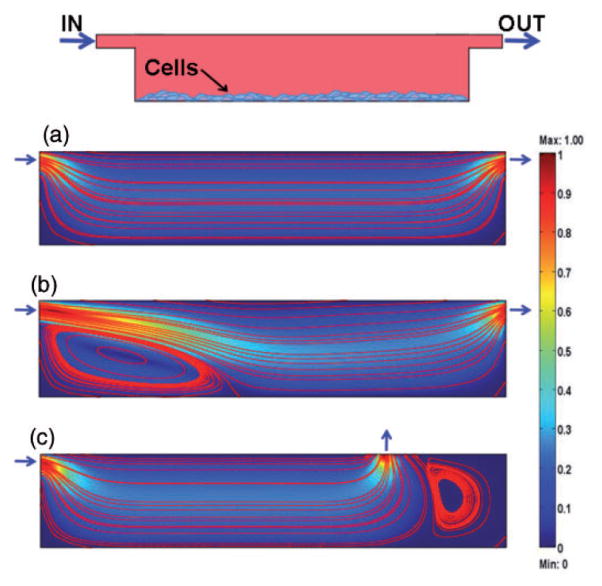
When scaling down to sub/millimeter characteristic lengths in a cell culture system, we shift from turbulent and intrinsically “chaotic” transport phenomena to more predictable and controllable molecular phenomena. At small scales intrinsic to microfluidic systems and microbioreactors, fluid flows are laminar (with low Reynolds numbers, Re<100), and molecular diffusion becomes a dominant mechanism of mass transport, allowing generation of well-defined concentration patterns. Laminar fluid flow can be utilized to maintain steady-state concentrations in cultured tissues, mimicking in vivo homeostasis much more closely than any standard culture system. Alternatively, microfluidic flows enable precise introduction of signals—such as changes in cytokines, oxygen, or pH—to replicate the physiological or pathological events of interest. In both cases, orderly behavior of microfluidic systems forms a basis for control over transport phenomena, and the measurement and modulation of multiple environmental factors of interest (Figure 1).
Figure 1.
Mathematical modeling of flow profiles. A model microwell was designed and different flow regimes and geometries of the inlets and outlets were tested for the cells adhering to the bottom surface (numerical simulations by Comsol Multiphysics). The heat map represents the dimensionless velocity of fluids with respect to the inflow velocity. Streamlines allow visualization of the patterns followed by fluids. (a) Perfectly laminar and orderly flow with low levels of shear stress experienced by the cells generated at low flow velocities. (b) Higher velocities lead to increases in shear stress and the formation of stagnant areas within the culture chamber. (c) Poorly positioned inlets and outlets result in the formation of stagnant areas even at the proper flow rates. (A color version of this figure is available in the online journal.)
In contrast, cell culture operations in well plates involve periodic changes of relatively large amounts of culture media. This simple operation results in the complete disruption of the cell niche, with stepwise changes in the levels of nutrients and depletion of cell-secreted factors. After each medium change, the properties of the system and the composition of the soluble cell microenvironment are averaged at a new level that is constant throughout the plate. The metabolic activity of the cells then starts again the accumulation of secreted factors and depletion of nutrients. The system constantly evolves toward a new equilibrium state, with diffusion alone dominating the transport of nutrients into the cells and cell-secreted factors into the medium. These sequences of events are certainly quite different from the in vivo situation where the micro-environmental conditions are finely regulated and where spatial and temporal variations in levels of multiple factors occur in an orderly and tightly controlled fashion.
The favorable flow and transport characteristics of microbioreactor systems allow maintenance of the composition of the cell niche and provision of molecular and mechanical signals in a more physiological fashion. By working at the steady state, one can keep a defined set of properties constant (i.e. concentration levels of selected factors) and ensure availability of metabolites to the cultured cells. At the same time, it is possible to introduce precise perturbations triggering desired cellular programs. Design rules and guidelines for the choice of operating parameters, based on the analysis of characteristic times and scales of transport and reaction are gradually being established.16 For example, it has been argued that scaling of the “micro-human” and “milli-human” systems for physiological and pharmacological studies will require work with small volumes of fluid in order to reach the time constants for signaling effects being studied.19
Representative results of the mathematical modeling of flow profiles in a model microwell within a microfluidic platform are shown in Figure 1. The dimensions of culture microwells are 5 mm × 1 mm (length and height, respectively), with inlet and outlet in the form of 200 μm high channels. The heat map correlates to the relative velocity of medium within the chamber with respect to the inflow velocity. Streamlines allow visualization of the patterns followed by fluids. The top panel in Figure 1 shows a schematic of the model chamber, filled with culture medium, and with inlet and outlet channels positioned as in Panel A and Panel B. The cells are adhering to the bottom surface. The example in Panel A refers to the case of low flow velocity, corresponding to laminar flow, exposing the cells to low levels of shear stress. In Panel B, the inflow velocity is two orders of magnitude higher and, although streamlines remain parallel to each other, shear stress values dramatically increase at the cell surfaces, and a stagnant area forms near the inlet port. Panel C exemplifies the case of improperly positioned inlets and outlets. Although the correct inflow velocities were set, stagnant areas form in the corner regions so that culture medium cannot reach all cultured cells, and the targeted environmental composition is not maintained.
When designing a microfluidic platform, one must always be aware of the nonidealities of the system, especially when 2D simulations are used to model 3D fluid flow and transport. For example, common simplifying assumptions such as ignoring the role of the channels walls in shaping the velocity and concentration profiles can lead to significant deviations of the predicted from the actual behavior.18,37
Physical signals
Differentiation, migration, proliferation, and functional assembly of many cell types are regulated by physical signals: hydrodynamic, mechanical, and electrical. Since standard culture dishes do not allow for the application and control of physical signals, the studied biological phenomena often undergo nonphysiological changes that poorly replicate the in vivo events.
Control of mechanical forces—including hydrodynamic shear—is an aspect of paramount importance in the design of bioreactors and microbioreactors systems. Accurate positioning of the inlets and outlets, flow channels, and culture chambers is the first determinant of the flow patterns through the platform. Geometric design of the microwells and the changes in flow regimes in the wells and channels allow modulation of forces acting on the cultured cells. An “ideal” microbioreactor platform would be modular and versatile, allowing robust testing of various combinations of conditions. For example, we developed a microbioreactor array that, by simply varying the relative positions of the microfluidic and culture chambers layers, creates culture environments with or without hydrodynamic shear.38 We used this system for differentiation studies of hESCs into vascular lineages and established quantitative correlations between the expression of smooth muscle actin and cell density for three different flow configurations.
Complex biological stimulations
When reducing the characteristic dimensions of a system to the microscale, transport distances are shortened and time constants are reduced. This is essential for studying biological events, which, in traditional cell and tissue culture systems, are often masked by the slow kinetics of physical phenomena. As the overall rate of a process is always limited by the slowest step in the series, the biological processes can only be studied in systems providing kinetics of mass transport that are faster than the biological kinetics being measured. This requirement can only be met on a small scale.
Small dimensions also translate into small working volumes, making it easy to introduce precise perturbations of input parameters and rapidly establish a new steady state. Developmental and functional processes in vivo involve integration of the signaling events at multiple scales, in space and time. Several morphogens (i.e. Wnts, BMPs, fibroblast growth factor) are known to elicit cell responses in a concentration-dependent manner, by responding to gradients that are acting both over the short and long ranges.39–41 Being able to replicate complex patterns of stimulation is a fundamental component of recreating in vitro the cellular environments similar to those encountered in vivo. Since the shape of concentration gradients is mostly determined by the diffusion coefficient of a particular species of interest (which is in turn determined by the molecular weight of the species) and the flow regime, microbioreactors are becoming the tools of choice for controlling the concentration gradients around the cells.8
Throughput
Microscaling can also increase the throughput of the systems and the number of the experimental variables. Small-scale platforms are capable of running multiple samples and can be used for parallelized studies when screening a large matrix of experimental variables. Besides increasing the density of the output data and thereby the statistical significance of the experiments, this approach also leads to a reduction of the batch-to-batch variability in the reagents and cells. When searching for small changes in cell behavior, the actual differences in the output measures can be masked by the intrinsic variability of the cell samples. Since microbioreactor platforms require small numbers of cells per condition, researchers can use cells from a single passage (if not from a single plate) thus further reducing inter-sample variability. Similar observations can be made with regards to the culture medium, as the use of small amounts of culture media helps avoiding the nonuniformities typical of larger media volumes. Moreover, the automation of most operations removes the operator dependency, further reducing the sources of unpredictable errors.
Case study: Activation of mesodermal and mesoendodermal programs in hESC and iPSC by space-resolved gradients of molecular factors
Numerous studies have shown that it is possible to generate cardiomyocytes (CM) from mouse42 and human Embryonic Stem Cells43 (hESC) and induced Pluripotent Stem Cells44 (iPS). The most efficient and reproducible protocols to date are those that replicate the signaling pathways regulating lineage commitment in the embryo. The earliest stages of cardiovascular development were mapped in ES cell cultures by identifying a cardiovascular progenitor that has capacity to generate cardiac and vascular progeny.42,44 The expression of fetal liver kinase 1 in mouse and kinase insert domain receptor (KDR) in human can be used to enrich differentiating cells for cardiac specified mesoderm.45 When isolated from embryoid bodies (EBs) and cultured in monolayers, these cardiac progenitors generate contracting CMs,42,44 and progress through developmental stages involved in the establishment of the cardiovascular lineages in vivo, which in turn require presentation of specific cytokines.
The combination of Activin A and BMP4 on days 1–4 of EB differentiation induces a primitive-streak-like population and leads to mesoderm development. Subsequent application of Wnt inhibitor Dickkopf-related protein 1 (Dkk1) and KDR ligand vascular endothelial growth factor significantly enhances the differentiation of KDR+ progenitors into CMs,42,44 while basic fibroblast growth factor supports continued expansion. The concentration levels and times of application and withdrawal of morphogens and their inhibitors involved in early cardiac differentiation are critical for the cell differentiation outcomes,41 and the optimal values of these parameters change with cell type (hESC or hiPSC), line, and culture setting (e.g. EBs or cell monolayers). We show here that a microscale system, specifically developed for this purpose, enables detailed studies of cardiac differentiation protocols used for hESC and iPSC.
The basic design of the microbioreactor system is shown in Figure 2 and is described in detail in Cimetta et al. 12 Briefly, our platform combines a matrix of conical microwells and a network of microfluidic channels connected to the two inlets and two outlets. The characteristic dimensions and the numbers of microwells can be easily varied, and the corresponding molds are rapidly prototyped, using 3D printing. In the specific configuration used in previous human stem cell differentiation experiments, the microwells were sized to accommodate spherical cell aggregates with diameters ranging from 300 to 400 μm.
Figure 2.
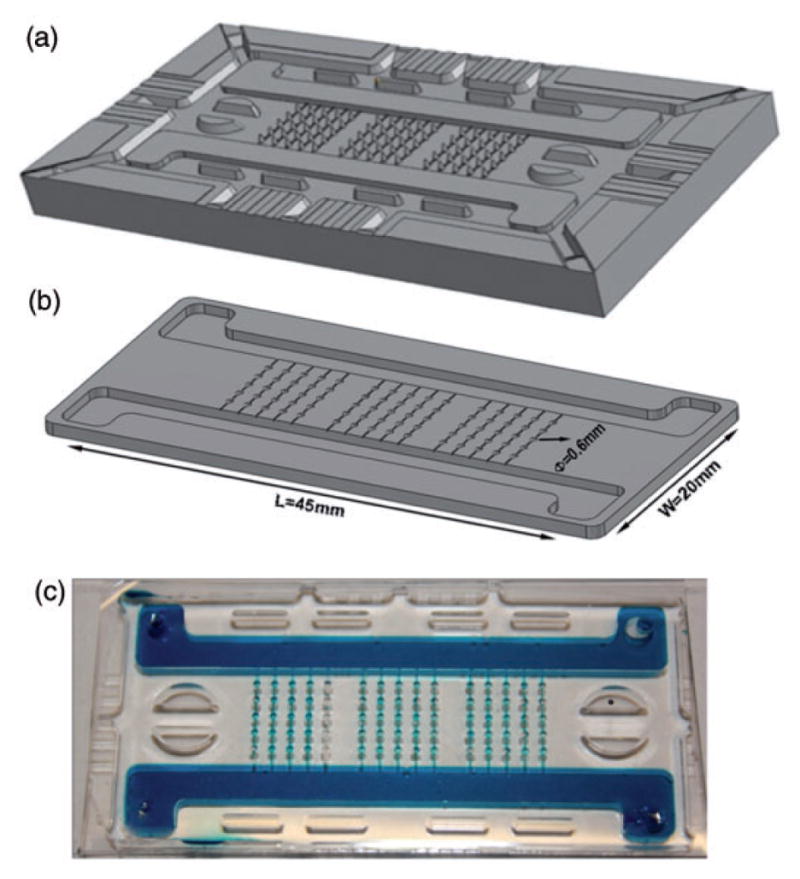
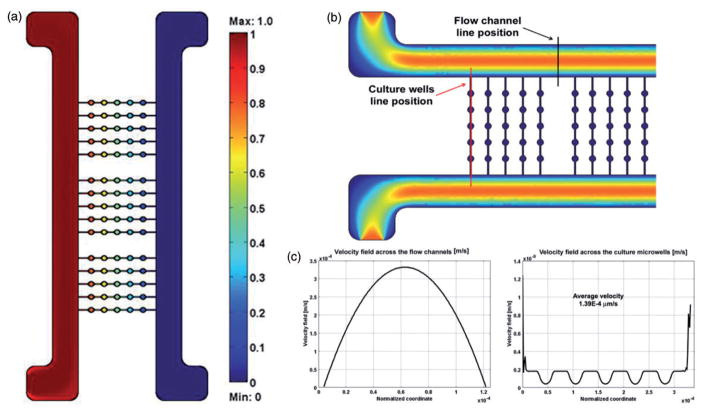
Microscale platform for studies of human stem cells. The platform was designed using a 3D CAD software with the following design criteria: (i) Capability of generating multiple concentration gradients in a no-shear environment, (ii) 3D culture of EBs, (iii) high-throughput, (iv) compatibility with online imaging, (v) easy retrieval of the EBs for additional analyses. The platform comprises a matrix of conical microwells accommodating EBs while shielding them from shear forces. The integrated microfluidic platform generates concentration gradients across the rows of microwells. The overall dimensions of the device are comparable to a standard microscope slide. Multiple prototypes with varying numbers of microwells were made; a prototype with five microwells per row is shown. (a) From the CAD file a mold was produced via Stereolithography Rapid Prototyping. (b) The mold could be used indefinitely to produce replicates of the microbioreactor via replica molding in poly(dimethylsiloxane). (c) A dye-tracer fluid fills the device, for visualization of the channels and microwells. (A color version of this figure is available in the online journal.)
Cell culture medium flows through the two side channels that are much larger than the microchannels between the individual wells, so that there is no convective flow in the microchannels. Each of the two streams can be supplemented with specific morphogens or inhibitors at desired concentrations that are different between the two channels. Gradients of factors are established across the cell culture field (perpendicular to the flow channels) by molecular diffusion between the neighboring microwells, through microchannels connecting each row of wells. The parallel rows of wells are fully independent from each other. The flow in the large channels is large enough to effectively eliminate longitudinal gradients. This arrangement of microwells allows formation of concentration gradients across the rows of microwells (left to right and/or right to left) in a culture environment protected from hydrodynamic shear (Figure 2). As a result, each biological sample is exposed to a different microenvironment, with separate rows serving as replicates (Figure 3).
Figure 3.
Mathematical modeling of flow and mass transport. (a) Color heat map correlates to the dimensionless concentration of metabolites relative to the initial values. The concentration gradient is established from left to right across the rows of microwells. Parallel rows are replicates of the same conditions. (b) Color heat map represents the velocity. The position of two lines for further analysis is highlighted. (c) The parabolic velocity profiles across the two flow channels (left) and above the microwells (right). (A color version of this figure is available in the online journal.)
The assembled platform is optically transparent in the cell culture area, allowing online monitoring of the cultured samples. The base configuration is capable of generating up to eight different conditions (composition across the linear concentration gradient) with 15 replicates for each condition, totaling 120 data points (Figure 2). After seeding the EBs into the wells, the open-well configuration is reversibly sealed using permeable membranes and a clamping system, by following a very simple protocol.
In our published work, we used hESC and hiPSc and studied their early mesodermal and mesoendodermal differentiation. Based on the hypothesis that the application of concentration gradients of soluble factors would provide more reliable in vitro models of development, we used this microbioreactor to expose EBs to controlled environments with compositions changing in a graded fashion. We selected a set of known activators of the mesodermal/ mesoendodermal programs: Wnt3a, Activin A, BMP4, Dkk1, and SB-431542 (Activin A inhibitor).
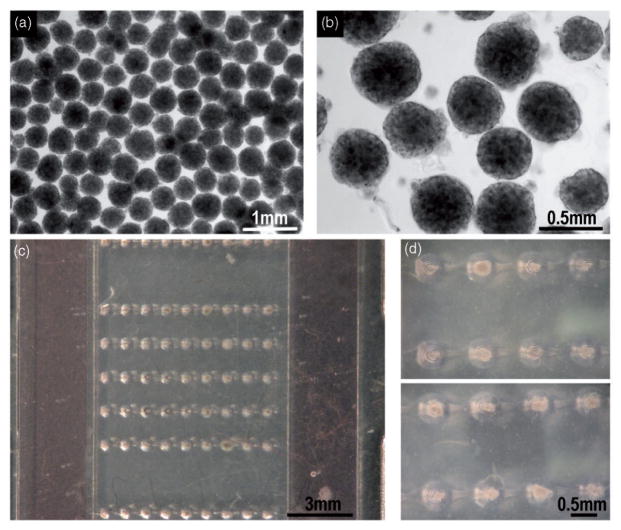
Large numbers of EBs were formed starting from the same cell passage and the same culture plate. The size of EBs is important, as it determines the concentrations of factors (through diffusional distances).46 Uniformly sized EBs were formed in AggreWell® plates from single cell suspensions (1000–1200 EBs are formed in one well of a 6-well AggreWell® plate). An aliquot of suspension of EBs is pipetted onto the microbioreactor and the EBs are docked by gravity settling into the conical wells (one EB per well, ~100% yield), by placing the platform onto an orbital shaker (Figure 4, hESC-derived EBs).
Figure 4.
The formation and seeding of EBs made of hESCs and iPSCs. (a, b) Uniformly sized EBs were formed in using Aggrewell® plates. Images refer to hESC-derived EBs. Magnifications: 4× and 10×, respectively. (c) An entire matrix composed by five rows of microwells filled with individual EBs can be visualized. Panel d: two magnifications of selected microwells. Again, a single EB fills each microwell and settles in a shear-protected environment. (A color version of this figure is available in the online journal.)
The assembled platform was connected to the syringe pump infusing culture medium through the two inlets. Each syringe could be filled with medium enriched with different cocktails of factors, thus broadening the spectrum of conditions to be tested.
The platform also allowed online monitoring of cell differentiation by using reporter cell lines (i.e. in Cimetta et al. 12 we used H1-BAR-Venus: H1 hESC with BAR, a β-catenin activated reporter, see Figure 4). Images of the fluorescing cells could be taken using a microscope and then analyzed via image analysis tools, resulting in semi-quantitative data.12
hESC and iPSc in the microbioreactors were exposed to a range of mesodermal-inducing environments. The concentration levels of soluble factors were systematically varied, by using the gradient feature of the system, and the effects of simultaneous activation and inhibition of selected pathways were analyzed by quantitative Reverse transcription polymerase chain reaction (RT-PCR) of mesodermal genes.
As an example, in Figure 5 we report results from the exposure of hiPSC-derived EBs to the opposing gradients of Activin A and BMP4 and either SB-431542 (Activin A inhibitor) or Dkk1 (canonical Wnt signaling inhibitor). To this end, Activin A and BMP4 were supplemented (one or both) to culture medium introduced through one flow channel, and SB-431542 and Dkk1 were introduced (one or both) through the other flow channel.
Figure 5.
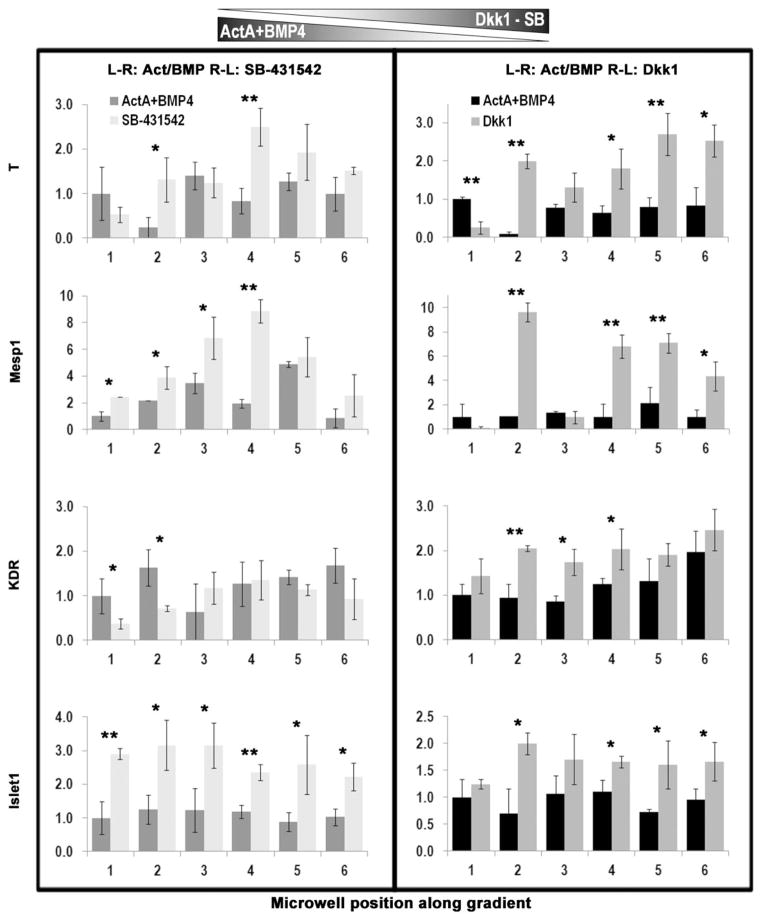
Expression of mesodermal genes after exposure to multiple, opposing gradients of ActivinA/BMP4 and SB-431542 or Dkk1. We quantified the expression levels of representative mesodermal and mesendodermal genes via qPCR. Data were obtained from ΔCt values and normalized to GAPDH. For each assay, 3–5 EBs per condition were pooled, from microwells in one row of the platform, corresponding to one single condition. Experiments without opposing inhibitors served as controls. hiPSC-derived EBs were subjected to a stable gradient of the chosen morphogens for 24 h between day 3 and day 4 after induction. Left inlet: Activin A (9 ng/ mL), BMP4 (13 ng/mL). Right inlet: SB-431542 (5 μM) for results in the left box, and Dkk1 (150 ng/mL) for results in the right box. Statistics were measured between control experiment and the corresponding inhibitor-based gradient for each microwell. *p <0.05, **p <0.005
Histograms show the quantification of gene expression profiles following inhibition of Activin signaling via SB counter-gradient (Figure 5, left) and inhibition of Wnt3a signaling via Dkk1 counter-gradient (Figure 5, right). Bars in the histograms are labeled by the position of the microwell across the gradient (1–6) that they represent. We measured the most dramatic differences in two early genes: T (T Brachyury) and Mesp1 (a key regulator of cardiovascular lineage commitment). The inhibition of Activin A resulted in increased expression levels of both genes, suggesting that cells might also endogenously produce Activin A and thus bring its local concentration to suboptimally high levels, such that partial inhibition might be required to optimally induce early mesodermal/mesoendodermal specification. Wnt inhibition resulted in an almost linear correlation, with differentiation progressing in direct proportionality for T and KDR, and inverse proportionality for Mesp1. The cells were differentially activated by the local microenvironment in response to concentration gradients and significant differences were observed in the early versus later genes expression profiles.
These experiments showed that linear concentration gradients elicited nonlinear cell responses in terms of mesodermal gene expression. The application of gradients can help mimic some of the traits of the signaling patterns that dictate mesoderm induction in vivo. Quantification of gene expression in the EBs following culture in the microbioreactor yielded sets of biologically meaningful results. The high-throughput mode of experimentation allowed the use of high numbers of replicates per condition (n = 15 EBs per condition) and improved statistical significance of the measured effects.
In ongoing studies, we are further exploring the roles of concentration gradients within EBs, by patterning cell behavior in an even more precise and spatially defined manner. A graded expression of fluorophores was observed in simple experiments performed by exposing EBs to opposing gradients of 4′,6-diamidino-2-phenylindole (DAPI) and Calcein, used as fluorescent dyes. When generating a left-to-right gradient with DAPI and right-to-left with Calcein, we observed that the EBs stained consistently with the double opposing gradient (Figure 6). This opens new perspectives for studies of cell patterning and polarization during differentiation processes and at the onset of disease states (i.e. tumor progression).
Figure 6.
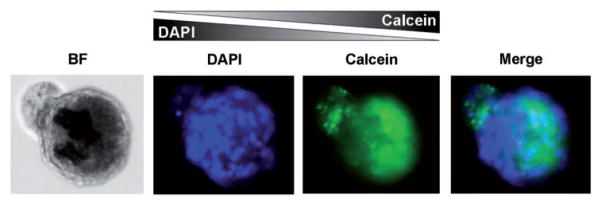
Dye tracer study of EBs subjected to opposing concentration gradients. The microbioreactor was used to generate the opposing gradients of DAPI (left to right) and Calcein (right to left) using fluorescent tracers. hESC-derived EBs stained consistently with the dye concentrations in the double opposing gradient. (A color version of this figure is available in the online journal.)
In another related study of stem cell differentiation in a microfluidic device,36 we utilized regional depletion of the Nanog transcriptional regulator through the exposure of cells to microfluidic gradients of morphogens. In this way, we established pluripotency-differentiation boundaries between Nanog expressing cells (pluripotency zone) and Nanog suppressed cells (early differentiation zone) within the same cell population, with a gradient of Nanog expression across the individual cell colonies. Notably, gene expression patterns at the pluripotency-differentiation boundaries recreated in vitro were similar to those in a developing blastocyst.
Summary
There is a clear need to study normal and pathological tissue function under conditions representative of the native in vivo milieu. Tissue engineering is responding to this need by developing microfluidic platforms with human tissues grown under biologically sound conditions and with signaling patterns representative of those in a living organism. Most advanced technologies of this kind are providing multitissue platforms allowing physiological studies of interacting tissues/organs and predictive testing of drug toxicity.
The miniaturization of culture systems further improves the throughput and accuracy of measurements conducted in microfluidic platforms, due to the fast transport of nutrients, oxygen, and regulatory molecules over short distances. However, over-complicated platforms end up being just “technology exercises” if not applied to significant biological phenomena and proven practical enough for broad use. In this sense, there should be a constant dialogue between “technology” and “biology,” i.e. bioengineers and life scientists in order to successfully coordinate the design and development of devices for biological experimentation.
As an illustration of the utility of microfluidic platforms, we described the design and use of a system supporting cultivation of stem cell EBs subjected to developmentally relevant gradients of morphogens and their inhibitors. Precise regulation of regulatory signals that normally guide cell choices between self-renewal, differentiation, differentiation, and functional assembly can greatly advance fundamental research of stem cells and their application in regenerative medicine.
Acknowledgments
The authors gratefully acknowledge funding support of NIH (grants UH2 EB17103, EB002520, HL076485), NYSCF (fellowship to EC), The Leona M. and Harry B. Helmsley Charitable Trust, and NYSTEM (grant C028119).
Footnotes
Authors contributions: Dr EC and Dr GV-N jointly wrote the manuscript. Dr GV-N approved the final version.
References
- 1.Kaplan D, Moon RT, Vunjak-Novakovic G. It takes a village to grow a tissue. Nat Biotechnol. 2005;23:1237–9. doi: 10.1038/nbt1005-1237. [DOI] [PubMed] [Google Scholar]
- 2.Powell K. Stem-cell niches: It’s the ecology, stupid! Nature. 2005;435:268–70. doi: 10.1038/435268a. [DOI] [PubMed] [Google Scholar]
- 3.Metallo CM, Mohr JC, Detzel CJ, de Pablo JJ, Van Wie BJ, Palecek SP. Engineering the stem cell microenvironment. Biotechnol Prog. 2007;23:18–23. doi: 10.1021/bp060350a. [DOI] [PubMed] [Google Scholar]
- 4.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–80. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S. Beyond the petri dish. Nat Biotechnol. 2004;22:151–2. doi: 10.1038/nbt0204-151. [DOI] [PubMed] [Google Scholar]
- 6.Burdick JA, Vunjak-Novakovic G. Engineered microenvironments for controlled stem cell differentiation. Tissue Eng Part A. 2009;15:205–19. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen DS, Davis MM. Molecular and functional analysis using live cell microarrays. Curr Opin Chem Biol. 2006;10:28–34. doi: 10.1016/j.cbpa.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Cimetta E, Cannizzaro C, James R, Biechele T, Moon RT, Elvassore N, Vunjak-Novakovic G. Microfluidic device generating stable concentration gradients for long term cell culture: Application to wnt3a regulation of b-catenin signaling. Lab Chip. 2010;10:3277–83. doi: 10.1039/c0lc00033g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keenan TM, Folch A. Biomolecular gradients in cell culture systems. Lab Chip. 2007;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–73. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 11.Watt FM, Hogan BLM. Out of Eden: Stem cells and their niches. Science. 2000;287:1427–30. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 12.Cimetta E, Sirabella D, Yeager K, Davidson K, Simon J, Moon RT, Vunjak-Novakovic G. Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells. Lab Chip. 2013;13:355–64. doi: 10.1039/c2lc40836h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. PNAS. 2006;103:2480–7. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–5. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 15.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 16.Cimetta E, Figallo E, Cannizzaro C, Elvassore N, Vunjak-Novakovic G. Micro-bioreactor arrays for controlling cellular environments: Design principles for human embryonic stem cell applications. Methods. 2009;47:81–9. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–6. [Google Scholar]
- 18.Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Rev Mod Phys. 2005;77:977–1016. [Google Scholar]
- 19.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 20.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler AJ, Griffin MA, Sen S, Bönnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–87. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 23.Serena E, Cimetta E, Zatti S, Zaglia T, Zagallo M, Keller G, Elvassore N. Micro-arrayed human embryonic stem cells-derived cardiomyocytes for in vitro functional assay. PLoS One. 2012;7:e48483. doi: 10.1371/journal.pone.0048483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serena E, Zatti S, Reghelin E, Pasut A, Cimetta E, Elvassore N. Soft substrates drive optimal differentiation of human healthy and dystrophic myotubes. Integr Biol. 2010;2:193–201. doi: 10.1039/b921401a. [DOI] [PubMed] [Google Scholar]
- 25.Kensah G, Gruh I, Viering J, Schumann H, Dahlmann J, Meyer H, Skvorc D, Bär A, Akhyari P, Heisterkamp A, Haverich A, Martin U. A novel miniaturized multimodal bioreactor for continuous in situ assessment of bioartificial cardiac tissue during stimulation and maturation. Tissue Eng Part C Methods. 2011;17:463–73. doi: 10.1089/ten.tec.2010.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abaci HE, Devendra R, Soman R, Drazer G, Gerecht S. Microbioreactors to manipulate oxygen tension and shear stress in the microenvironment of vascular stem and progenitor cells. Biotechnol Appl Biochem. 2012;59:97–105. doi: 10.1002/bab.1010. [DOI] [PubMed] [Google Scholar]
- 27.Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8:949–55. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- 28.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–54. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yum K, Hong SG, Healy KE, Lee LP. Physiologically relevant organs on chips. Biotechnol J. 2014;9:16–27. doi: 10.1002/biot.201300187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens KR, Ungrin MD, Schwartz RE, Ng S, Carvalho B, Christine KS, Chaturvedi RR, Li CY, Zandstra PW, Chen CS, Bhatia SN. Invert molding for scalable control of tissue microarchitecture. Nat Commun. 2013;4:1847. doi: 10.1038/ncomms2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kima HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol. 2013;5:1130. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 32.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci USA. 2007;104:5722–6. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–6. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 34.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–8. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. In vitro microvessels for the study of angiogenesis and thrombosis. PNAS. 2012;109:9342–7. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Sevilla A, Wan LQ, Lemischka IR, Vunjak-Novakovic G. Patterning pluripotency in embryonic stem cells. Stem Cells. 2013;31:1806–15. doi: 10.1002/stem.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ismagilov RF, Stroock AD, Kenis PJA, Stone HA, Whitesides G. Experimental and theoretical scaling laws for transverse diffusive broadening in two-phase laminar flows in microchannels. Appl Phys Lett. 2000;76:2376–8. [Google Scholar]
- 38.Figallo E, Cannizzaro C, Gerecht S, Burdick JA, Langer R, Elvassore N, Vunjak-Novakovic G. Micro-bioreactor array for controlling cellular microenvironments. Lab Chip. 2007;7:710–9. doi: 10.1039/b700063d. [DOI] [PubMed] [Google Scholar]
- 39.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and bmp signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Coudreuse DYM, Roel G, Betist MC, Destre O, Korswagen HC. Wnt gradient formation requires retromer function in wnt-producing cells. Science. 2006;312:921–4. doi: 10.1126/science.1124856. [DOI] [PubMed] [Google Scholar]
- 41.Jackson SA, Schiesser J, Stanley AG, Elefanty AG. Differentiating embryonic stem cells pass through ‘temporal windows’ that mark responsiveness to exogenous and paracrine mesendoderm inducing signals. PLoS One. 2010;5:e10706. doi: 10.1371/journal.pone.0010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–32. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr1 embryonic-stem-cell-derived population. Nature. 2008;453:524–8. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. PNAS. 2005;102:13170–5. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–10. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]