Abstract
Chromosomal abnormalities, including microdeletions and microduplications, have long been associated with abnormal developmental outcomes. Early discoveries relied on a common clinical presentation and the ability to detect chromosomal abnormalities by standard karyotype analysis or specific assays such as fluorescence in situ hybridization. Over the past decade, the development of novel genomic technologies has allowed more comprehensive, unbiased discovery of microdeletions and microduplications throughout the human genome. The ability to quickly interrogate large cohorts using chromosome microarrays and, more recently, next-generation sequencing has led to the rapid discovery of novel microdeletions and microduplications associated with disease, including very rare but clinically significant rearrangements. In addition, the observation that some microdeletions are associated with risk for several neurodevelopmental disorders contributes to our understanding of shared genetic susceptibility for such disorders. Here, we review current knowledge of microdeletion/duplication syndromes, with a particular focus on recurrent rearrangement syndromes.
Keywords: developmental delay, intellectual disability, copy-number variation, recurrent rearrangement, nonallelic homologous recombination, microarray
INTRODUCTION
Chromosomal microdeletions and microduplications have been associated with syndromic forms of intellectual disability (ID) and developmental delay (DD) since the 1980s. Classic examples include the 15q11–q13 deletion associated with Prader–Willi and Angelman syndromes (24), the 17p11 deletion associated with Smith–Magenis syndrome (29), the 7q11 deletion associated with Williams–Beuren syndrome (131), and the 22q11 deletions associated with velocardiofacial syndrome (45). Each of these disorders was first described based on a series of patients who shared a recognizable collection of clinical features. The clinical description of each syndrome was followed later by the discovery of the molecular basis for the syndrome—a “phenotype-first” discovery. Improved cytogenetic techniques, including high-resolution karyotype analysis and fluorescence in situ hybridization (FISH), facilitated diagnostic testing for the deletion associated with each suspected diagnosis. For example, if a physician suspected that a patient had Prader-Willi syndrome, a FISH test could be performed to determine whether the causative 15q11–q13 deletion was present.
Chromosomal microdeletions and microduplications make up a fraction of copy-number variants (CNVs). CNVs are defined as either the gain or loss of a stretch of DNA as compared with the reference human genome; they may range in size from a kilobase to several megabases or even an entire chromosome (trisomies and monosomies). CNVs can involve multiple, one, or no genes, and although some CNVs cause disease, many others remain benign variants within the population (68, 71, 103, 140, 151).
There are two major classes of CNVs: recurrent and nonrecurrent. Recurrent CNVs generally arise by nonallelic homologous recombination (NAHR) during meiosis, with breakpoints in the large duplicated blocks of sequence flanking the CNV event (Figure 1). Because the breakpoints cluster within defined regions, the extent of recurrent CNVs is essentially identical even in unrelated individuals (101). In contrast, nonrecurrent CNVs have breakpoints that generally lie within unique sequence and do not result from a predisposing genomic architecture. Nonrecurrent CNVs can arise by several different mechanisms, including nonhomologous end joining and fork stalling and template switching (FoSTeS) (25, 64, 194). As a result, although two unrelated individuals may have overlapping nonrecurrent CNVs, they are unlikely to share the same breakpoints.
Figure 1.
Nonallelic homologous recombination (NAHR), the primary mechanism underlying the generation of recurrent copy-number variants (CNVs). Segmental duplications, also termed low-copy repeats, represent regions of extended sequence identity that can provide a substrate for NAHR-mediated chromosomal rearrangements. In this schematic, two large segmental duplications (blue arrows) with high sequence similarity flank a region containing genes a, b, and c. Following misalignment of the homologs, these duplications facilitate NAHR during meiosis by assisting an illegitimate crossing-over event between paralogous, rather than allelic, segmental duplications. This results in two reciprocal products: one chromosome carrying a duplication of the intervening region and an additional copy of genes a, b, and c, and a second chromosome carrying a deletion of this same region. Such rearrangements are common causes of many recurrent genomic disorders characterized by reciprocal rearrangements of specific chromosomal regions (see Table 1).
Over the past 10 years, there have been incredible advances in high-throughput genomic technologies. It is now possible to rapidly interrogate the entire genome for CNVs, providing vastly improved screening methods for the detection of microdeletions and microduplications compared with the technology available a decade ago (183). Array comparative genomic hybridization (aCGH) was introduced in 2003 and was initially used to characterize the many large somatic rearrangements that often occur in cancer (182). aCGH is a comparative assay in which two samples are differentially labeled with fluorescent dyes. Cohybridizing these labeled genomes to an array comprising probes spaced throughout the genome allows for the detection of relative differences in copy number between the two samples. Although not originally designed for detecting CNVs, single-nucleotide polymorphism (SNP) genotyping arrays can also be used to detect copynumber changes by combining measures of fluorescence intensity and allelic ratios produced by the array (34). Throughout this review, we refer to aCGH and SNP genotyping arrays as chromosome microarrays. Although the resolution of these methods is largely dependent on probe density, modern arrays comprising hundreds of thousands or millions of probes can detect CNVs that are several orders of magnitude smaller than those visible by standard karyotype analysis. Chromosome microarrays are now widely used in both research and diagnostic settings.
Soon after their introduction, microarrays were used to profile the CNV patterns of normal individuals. The results of these studies were unexpected, revealing that the genomes of two healthy people could differ in DNA content by many millions of base pairs (140, 151, 156). Shortly thereafter, similar studies were initiated to also identify CNVs in individuals with genetic syndromes. The first large cohorts to be studied were individuals with ID (41, 88, 145, 152, 155, 159), autism spectrum disorder (ASD) (30, 92, 108, 150, 173, 187), or congenital anomalies (54, 86, 111, 141). More recently, studies of CNVs have been extended to more common phenotypes that are not traditionally considered to be chromosomal disorders, including schizophrenia (11, 82, 127, 168, 170, 184), epilepsy (39, 43, 66), amyotrophic lateral sclerosis (161), autoimmune diseases (38), and craniosynostosis (73). Through comparisons of the frequency of a given CNV in cases and controls, disease-related CNVs have been rapidly identified for each of these conditions.
The size of CNV detected by chromosome microarray technologies depends largely on the probe density, which is determined by the spacing of probes on the array. Although probe density continues to increase, the smallest CNVs detected by these methods are generally on the order of ~50 kb or larger. However, recent major advances in sequencing technologies are providing novel insights into CNVs, particularly those that have not been resolvable by chromosome microarray methods. Next-generation sequencing or massively parallel sequencing (MPS) involves highly parallelized sequencing of millions of short DNA fragments from the genome. Indeed, an entire genome can be sequenced using MPS in a matter of days. By taking advantage of these technologies, researchers have developed several approaches that enable the identification of CNVs from MPS data (192, 193, 196); these approaches have facilitated the routine genome-wide discovery of much smaller CNVs, which have not been comprehensively assayed using current technologies. Whole-genome sequencing using MPS has the potential to provide a truly unbiased assay that can identify CNVs ranging in size from a single base pair to entire chromosomes, simultaneously with a complete assessment of single-nucleotide sequence changes.
The use of these technologies has had a significant impact on the field of human genetics. Indeed, as we discuss below, many “syndromes” that are difficult to recognize solely by clinical features have now been identified and defined purely by the nature of a shared genomic rearrangement; this “genotype-first” approach to clinical diagnosis is now routine. Here, we review the discovery, genetic architecture, and evolution and characterization of novel microdeletion and microduplication syndromes. We also discuss approaches to understanding the clinical variability associated with many novel deletions and duplications and mechanisms of CNV formation. Finally, we address the clinical implications that these and other new technologies have for clinical practice. Although many of the principles of our discussion are relevant to both recurrent and nonrecurrent events, we place particular emphasis on the truly recurrent genomic disorders that are catalyzed by the local genome architecture and arise via NAHR.
HOT SPOTS OF RECURRENT REARRANGEMENTS
The term genomic disorder was first applied to diseases for which the primary underlying cause involved rearrangements of specific chromosomal regions (104). From early studies of such rearrangements, it became clear that these CNVs were recurrent, and this recurrence was strongly related to the local genome architecture. Most cases were de novo and represented reciprocal deletion and duplication of a specific chromosomal interval impacting one or more dosage-sensitive genes (104). Seminal examples are the 17p12 duplications and deletions, which result in the development of Charcot–Marie–Tooth disease type 1A (CMT1A) and hereditary neuropathy with liability to pressure palsies (HNPP), respectively (27, 105), owing to contrasting dosage effects of the PMP22 gene (26). These recurrent, reciprocal, disease-causing CNVs were two of the first genomic disorders described. More examples soon followed, including Prader–Willi and Angelman syndromes (15q11–q13) (4) and Smith–Magenis syndrome (17p11.2) (29). The recurrent CNVs causing each disorder were shown to involve NAHR-mediated rearrangements in regions of the genome exhibiting local architectures characterized by repetitive DNA features, termed segmental duplications (SDs) or low-copy repeats (LCRs) (104, 166) (Figure 1).
Upon noting that recurrent genomic disorders are catalyzed by the presence of pairs of large, highly identical flanking repeats, Eichler and colleagues (9) generated a genome-wide map of more than 8,000 SDs. Analysis of this SD map identified 169 regions of the human genome that were predicted to be potential rearrangement hot spots because of the presence of large blocks of SDs with >95% sequence similarity that were separated by 50 kb–10 Mb of intervening sequence. Interestingly, 24 of these regions had already been linked to recurrent genetic diseases (9). Testing of these hot-spot regions in a population of normal individuals using a targeted array revealed that CNVs were significantly more common within these predicted rearrangement hot spots compared with the genome average (156). However, for many of these regions, CNVs were apparently not observed in this normal population, prompting a strategy to target such regions in human disease patients. Indeed, the initial use of this targeted microarray method in patient cohorts with ID/DD and various congenital anomalies proved fruitful in the discovery of novel pathogenic recurrent rearrangements (111, 155). For example, by screening 290 patients with ID/DD for which underlying genetic causes had not previously been found, Sharp et al. (155) identified 16 individuals carrying large submicroscopic deletions and duplications that were associated with their disease. Localization of the breakpoints in each of these patients to flanking clusters of SDs defined five disease-associated NAHR hot spots, located at 1q21.1, 15q13, 15q24, 17q12, and 17q21.31. Subsequent studies have replicated all of these loci as associated with recurrent genomic disorders in a variety of cohorts of patients with ID/DD, and several of them are now associated with clinically recognizable syndromes (88, 115, 176).
The widespread application of microarray technologies to additional series of patients with ID/DD has led to a renaissance in the understanding of the chromosomal basis of human disease. Since 2006, more than 20 recurrent microdeletion/duplication syndromes have been identified for ID/DD; these are listed with associated references in Table 1. A central conclusion drawn from these newly described syndromes is that, in each case, a series of patients were initially classified based on the characterization of common overlapping genetic lesions rather than on constellations of clinical features, reflecting a transition in the field from the phenotype-first approach to the genotype-first approach (113). Importantly, in such instances, the identification of a group of patients with shared genomic rearrangements can allow for more definitive characterization of clinical symptoms and lead to improved patient diagnoses and management.
Table 1. Known recurrent microdeletions and microduplications.
| Chromosomal location |
Rearrangement | Associated syndromes and phenotypes |
IDa | ASDa | Epilepsya | Scza | Associated inversion | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| 1q21.1 | Deletion | TAR syndrome | 87 | |||||
| 1q21.1 | Deletion/duplication | Variable ID, cataracts, MCA (deletion); autism (duplication); heart defects (both) |
✓ | ✓ | ✓ | Unknown relationship to deletion/ duplication risk |
22, 63, 116, 181 |
|
| 1q22 | Deletion | Gaucher disease, lysosomal storage disorder |
197 | |||||
| 2q11.2b | Deletion/duplication | DD, hypotonia, macrocephaly (duplication); ID, congenital heart defect (deletion) |
144 | |||||
| 2q13 | Deletion | Familial juvenile nephronophthisis | 148 | |||||
| 2q21.1 | Deletion | ID/DD, ADHD, epilepsy, behavioral abnormalities |
✓ | ✓ | ✓ | ✓ | 42 | |
| 3q29 | Deletion/duplication | Dysmorphic features, microcephaly (deletion); variable ID (both) |
✓ | ✓ | Unknown relationship to deletion/ duplication risk |
189 | ||
| 4q35 | Deletion | Facioscapulohumeral muscular dystrophy |
55, 191 | |||||
| 5q35 | Deletion/duplication | Sotos syndrome (deletion); 5q35 duplication syndrome (duplication) |
Prerequisite for deletion/duplication |
174 | ||||
| 6p21.32 | Deletion | Congenital adrenal hyperplasia III | 96 | |||||
| 7q11.23 | Deletion/duplication | Williams–Beuren syndrome (deletion); 7q11 duplication syndrome (duplication) |
✓ | Increased risk for deletion/duplication |
146 | |||
| 7q11.23 distal | Deletion/duplication | Infantile spasms, ID | ✓ | ✓ | 139 | |||
| 8p23.1 | Deletion/duplication | ID/DD, congenital heart defects, microcephaly, behavior problems |
Prerequisite for deletion/duplication |
31 | ||||
| 8q24.3 | Duplication | Glucocorticoid-remediable aldosteronism |
100 | |||||
| 10q11.21– q11.23 |
Deletion/duplication | ASD, seizures, MCA, immunodeficiency, encephalopathy, hypotonia (duplication); ID/DD, dysmorphic features (both) |
✓ | ✓ | 165 | |||
| 10q22–q23 | Deletion/duplication | Infantile juvenile polyposis, ID, behavioral abnormalities |
✓ | 10, 118 | ||||
| 12q14.2 | Deletion | Globozoospermia | 51 | |||||
| 15q11.2 | Deletion/duplication | Variable presentation of ID, behavioral abnormalities, epilepsy |
✓ | ✓ | ✓ | ✓ |
83, 114, 168, 178 |
|
| 15q11–q13 | Deletion/duplication | Prader–Willi and Angelman syndromes |
✓ | Increased risk for deletion/duplication |
124 | |||
| 15q13.3 | Deletion/duplication | Variable ID, ASD, epilepsy | ✓ | ✓ | ✓ | ✓ | Unknown relationship to deletion/ duplication risk |
66, 120, 157, 177 |
| 15q24 | Deletion/duplication | ID, ASD, dysmorphic features, MCA |
✓ | ✓ | Unknown relationship to deletion/ duplication risk |
115, 155 | ||
| 15q25.2 | Deletion | Congenital diaphragmatic hernia, ID, ASD |
✓ | ✓ | 130, 186 | |||
| 16p11.2 | Deletion/duplication | Early-onset obesity/underweight | ✓ | ✓ | ✓ | ✓ | 17, 187 | |
| 16p12.1 | Deletion/duplication | Variable DD, neuropsychiatric disorders |
✓ | Prerequisite for deletion/duplication |
60 | |||
| 16p13.11 | Deletion/duplication | Variable ID, ADHD, epilepsy, schizophrenia |
✓ | ✓ | ✓ | ✓ |
39, 65, 69, 114 |
|
| 17p11.2 | Deletion/duplication | Smith–Magenis syndrome (deletion); Potocki–Lupski syndrome (duplication) |
29 | |||||
| 17p11.2–p12 | Deletion/duplication | HNPP (deletion); CMT1A (duplication) |
26, 27, 105 | |||||
| 17q11.2 | Deletion/duplication | Neurofibromatosis type 1 | 44 | |||||
| 17q12 | Deletion/duplication | Renal cysts and diabetes | ✓ | ✓ | ✓ | ✓ | Unknown relationship to deletion/ duplication risk |
111, 123 |
| 17q21.31 | Deletion/duplication | 17q21 microdeletion syndrome (deletion) |
Prerequisite for deletion/duplication |
88, 155 | ||||
| 22q11.2 | Deletion/duplication | VCFS, DiGeorge syndrome, schizophrenia (deletion); 22q11 duplication (duplication) |
✓ | ✓ | 82, 153 | |||
| 22q11.2 distal | Deletion/duplication | Growth delay, ID, MCA (deletion); variable ID (duplication) |
✓ | 21 | ||||
| 22q11.2–q12 | Deletion | Cat eye syndrome | 110 | |||||
| Xp22.31 | Deletion | X-linked ichthyosis | 180 | |||||
| Xq28 | Deletion | Incontinentia pigmenti | 8 | |||||
| Xq28 | Deletion | Red-green color blindness | 126 | |||||
| Xq28 | Deletion/duplication | ID, ASD, dysmorphic features | ✓ | ✓ | 47 | |||
| Yq11.23 | Deletion | Azoospermia | 160 | |||||
| der(8)t(8;12) (p23.1;p13.31) |
Translocation | Obesity, DD, ID, seizures, macrocephaly, eczema, dysmorphic features |
✓ | ✓ | Unknown relationship to translocation risk |
62, 129 | ||
| der(4)t(4;11) (p16.2;p15.4) |
Translocation | Features of Wolf–Hirschhorn syndrome, Beckwith–Wiedemann syndrome, and Russell–Silver syndrome |
129 | |||||
| der(X)t(X;Y) (p22.33;p11.2) |
Translocation | 46,XX maleness | Prerequisite for translocation |
125 | ||||
| t(17;22)(q11;q11) | Translocation | Offspring at risk of aneuploidy due to malsegregation of the t(17;22) |
94 | |||||
| t(11;22)(q23;q11) | Translocation | Offspring at risk of aneuploidy due to malsegregation of the t(11;22) |
46 | |||||
| Xq28 | Inversion | Hemophilia A/factor VIII deficiency |
Pathogenic inversion | 95 | ||||
| Xq28 | Inversion | Hunter syndrome/ mucopolysaccharidosis II |
Pathogenic inversion | 19 |
Abbreviations: ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; CMT1A, Charcot–Marie–Tooth disease type 1A; DD, developmental delay; HNPP, hereditary neuropathy with liability to pressure palsies; ID, intellectual disability; MCA, multiple congenital anomalies; Scz, schizophrenia; TAR, thrombocytopenia absent radius; VCFS, velocardiofacial syndrome.
Nonsyndromic forms.
One patient each reported for deletion and duplication.
An illustrative example of the power of this approach is the 15q24 microdeletion syndrome, which was first described in 2006 and then confirmed as a site of recurrent rearrangement in 2007 (155, 158). Carriers of this syndrome may present with a spectrum of clinical features that include DD, mild to moderate ID, dysmorphic facial features, physical abnormalities, neurological symptoms, and recurrent infections as well as psychiatric traits including behavioral problems, hyperactivity, and attention deficit hyperactivity disorder (106). There have now been some 35 patients characterized with this syndrome, each carrying highly penetrant deletions that range from 0.26 to 3.75 Mb in size (5, 48, 49, 86, 115, 179). The majority of these patients’ characterized deletions (24 of 32) have breakpoints that map to five clusters of highly identical SDs within 15q24, implicating NAHR as the underlying mutational mechanism. Assessments of clinical presentations in these patients have revealed that, whereas DD and distinct facial features occur in nearly all cases, presentations of other associated traits tend to be more variable between patients (106, 115). Genotype–phenotype studies comparing the clinical features of patients with overlapping 15q24 deletions provide important insight into potentially disease-relevant genes (115). For example, 29 of the 32 fully characterized 15q24 deletions span a critical 1.2-Mb region (5, 48, 49, 86, 115, 179), strongly suggesting that this interval contains the dosage-sensitive element(s) underlying the cardinal features of the disorder. In contrast, the 3 patients with atypical deletions mapping outside of or only partially overlapping this 1.2-Mb critical region lacked certain core features seen in most other patients, such as cardiac abnormalities and seizures (115).
Genome-wide screens in larger and more comprehensive collections of patients and normal controls have aided in the continued discovery of novel syndromes in ID/DD. This is particularly relevant for very rare disorders, which require large sample sizes to detect recurrence, and for those that show incomplete penetrance and/or variable expressivity (discussed in detail below). Based on an analysis of 15,767 patients with a general diagnosis of ID/DD and 8,329 normal individuals, Cooper et al. (33) recently identified 14 novel loci showing a significant enrichment of CNVs in disease cases compared with controls, with overlapping CNVs of each locus observed in multiple patients. In addition, 3 of these loci had previously been described in single case reports, indicating that these regions are likely associated with genuine syndromes that warrant further investigation and clinical classification. The relatively low frequency of these events also illustrates the increased power of surveys of larger sample sizes in the identification of novel rare pathogenic CNVs. In addition to the 14 putative pathogenic loci, another 1,400 of the 15,767 patients were found to have rearrangements in one of 45 chromosomal regions previously determined to be causative in other known genomic syndromes. Thus, from their data, the authors were able to make comprehensive estimates of prevalence and penetrance for a significant majority of known microdeletion/duplication syndromes, with an estimate that ~14% of ID/DD in their cohort was attributable to pathogenic CNVs greater than 400 kb in size. Notably, partitioning data based on CNV size and phenotypic characteristics revealed that odds ratios increased with the size of pathogenic events, and the burden of larger CNVs was greater in patients with more severe developmental phenotypes (33).
Within the past five years, genome-wide CNV studies have also been conducted in patients affected by a variety of other phenotypes. Among these are both traits and disorders that have traditionally been associated with ID/DD, such as congenital anomalies (54, 111), epilepsy (39, 66, 114, 117), and autism (14, 61, 97, 108, 133, 146, 187), as well as other complex neuropsychiatric disorders, such as schizophrenia (70, 83, 123, 124, 170, 184) and attention deficit hyperactivity disorder (50, 190). Not surprisingly, results from these studies have indicated that the impact of large regions of aneuploidy resulting from CNVs extends beyond the scope of ID/DD. In addition, particularly for neurological disorders, strong evidence has emerged in support of more prominent roles for rare and de novo events over more common CNVs.
In autism and schizophrenia, for example, both family-based genome-wide studies and those conducted in case-control cohorts have reported an increased CNV burden in affected individuals. Comparing the rates of CNV formation in 195 simplex and multiplex families with those of 99 control families, Sebat et al. (150) identified a significant enrichment for the occurrence of de novo CNVs in cases, a finding that has since been confirmed by additional studies (97, 108, 133, 146). A recent study in schizophrenia also observed an increase of de novo microdeletions and microduplications in some cases (5.1%) compared with controls (2.2%), and, similar to trends reported in autism (150), the frequency of de novo CNVs was greater in cases with a family history of disease compared with those without such a history (83).
With respect to rare CNVs (population prevalence <0.1%), using a case–control design, Pinto et al. (133) reported a 1.19-fold increase in the number of genic CNVs in autistic cases compared with controls. Notably, a 1.26-fold increase was observed when only deletions were considered, an observation consistent with an idea commonly believed in population studies, namely that deletions are more likely to result in negative phenotypic consequences than are duplications (32, 81). Similar trends have been noted for schizophrenia using case–control cohorts. For example, a survey of >3,000 individuals found that large (size > 100 kb) and rare (frequency < 0.1%) CNVs were enriched 1.15-fold in cases compared with controls. In addition, CNV burden varied depending on event type, size, and frequency and the number of genes included; the greatest difference between cases and controls was observed for deletions greater than 500 kb in size (70).
Enrichments of rare CNVs, both inherited and de novo, have also been observed in epilepsy. For example, Mefford et al. (114) detected rare CNVs in 46 out of 517 epilepsy cases, none of which were identified in a control cohort of 2,493 individuals. In addition, deletions comprised the majority (69%) of the 51 characterized events (114), also consistent with results noted above for autism (133). Thus, CNVs have a strong influence on risk for a wide variety of neurodevelopmental disorders.
The ability to assay large cohorts with different phenotypes has also been important for understanding the diverse risk profile associated with some recurrent CNVs. Collectively, findings from CNV studies conducted in the diseases noted above, including in ID/DD, have revealed that in many instances pathogenic CNVs occurring within the same microdeletion/duplication hot spot are associated with several different disorders. For example, of the five regions initially characterized by Sharp et al. (155), rearrangements in 1q21.1, 15q13, 15q24, and 17q12 have now also been identified in patients exhibiting various congenital defects, autism, schizophrenia, or epilepsy (39, 54, 66, 70, 123, 150, 168), suggesting that the underlying causes of these disorders may in some cases involve common developmental pathways.
DEFINING THE PATHOGENICITY OF COPY-NUMBER VARIANTS
Many criteria can be used to help interpret the clinical relevance of a CNV, including inheritance, size, type, and gene content (119). The inheritance pattern of a CNV, when accompanied by clinical and family history information, can be useful. De novo CNVs are more likely than inherited CNVs to be pathogenic, especially for severe disorders, though there are certainly exceptions. Conversely, as illustrated by the example of the 1q21.1 syndrome and others, inherited CNVs may cause a range of severity and presentation of neurodevelopmental disorders and can clearly be pathogenic, even when present in a phenotypically normal parent (22, 70, 112).
In the clinical setting, it may at times be difficult to test both parents for financial or social reasons, so other criteria must be considered. CNV size and type (deletion or duplication) are often used as guides to interpret pathogenicity. Large CNVs are more likely than small CNVs to cause disease (41). In part, this is because larger CNVs generally encompass more genes, with a concomitant increase in the probability of altering a dosage-sensitive element. Likewise, deletions result in haploinsufficiency, the consequences of which are known for some genes. Duplications are more difficult to interpret, and in the clinic, a larger minimum size threshold is often employed for duplications than for deletions. Gene content is also a consideration. CNVs that contain many genes or known disease genes are more likely to be pathogenic than those that contain few genes or genes of uncertain function. CNVs within “gene deserts” are particularly difficult to interpret, though as we learn more about regulatory regions within noncoding DNA (52), interpretation will become easier.
All of these criteria are probabilistic in nature, and there are documented instances in which, for example, small noncoding duplications cause genetic syndromes (37). Despite these exceptions, with careful consideration, likely determinations of pathogenicity can be made in most cases.
CLINICAL TESTING FOR COPY-NUMBER VARIANTS
Since the introduction of chromosome microarrays, several large studies have addressed the overall importance of copy-number changes in the diagnostic workups for DD, ID, and autism (119, 145, 152). Today, there is no question that chromosome microarrays have a far higher diagnostic yield than the standard karyotype, indicating that microarray analysis should replace karyotyping as the method of choice for aneuploidy screening. In 2010, a review of 33 published studies involving 21,698 patients with DD, congenital anomalies, or autism who were tested for CNVs with chromosome microarrays found a diagnostic yield of 15–20% across all studies compared with ~3% for the standard G-banded karyotype (119). Other studies of large patient cohorts screened by microarray have similarly concluded that ~14% of DD cases can be explained by a detectable CNV (33).
Importantly, in addition to providing a diagnosis, chromosome microarray testing can lead to changes in medical management recommendations. In a retrospective review of 1,792 patients with ID/DD, ASD, and/or congenital anomalies who had chromosome microarray testing, individuals who had a positive diagnosis had a higher rate of recommendation for clinical action than those who had an uncertain result (54% versus 34%, p = 0.01) (35). More recently, Riggs et al. (142) identified at least 146 disorders that can be diagnosed by chromosome microarray testing and that have published literature supporting specific clinical management implications. Approximately 7% of cases that undergo chromosome microarray testing are diagnosed with one of these conditions. The authors concluded that chromosome microarray testing impacts clinical management at a rate similar to that of other genetic tests for which insurance coverage is often more readily approved. In summary, undiagnosed individuals with ID, DD, ASD, or congenital anomalies should be referred for chromosome microarray testing, which may lead to diagnosis and management recommendations.
VARIABLE EXPRESSIVITY OF MICRODELETION AND MICRODUPLICATION SYNDROMES
Comparison of CNV findings across studies reveals several recurrent rearrangements that are associated with a wide range and severity of phenotypes (113) (Table 1). Rearrangements that fall into this category include deletions and duplications of 1q21.1, 16p11.2, 16p13.11, and 15q13.3. For most of these regions, recurrent rearrangements have been associated with risk for some or all of several neurodevelopmental disorders, including ID, ASD, epilepsy, attention deficit hyperactivity disorder, and schizophrenia. Not only is each of these disorders distinct in its presentation, but the severity of each phenotype associated with these rearrangements can also vary significantly.
The factors underlying such extreme clinical variability are still poorly understood. Several possible explanations for different clinical presentations in carriers of the same deletion or duplication have been proposed (154). Differences in genetic background—that is, the milieu of sequence and CNVs present within the genomes of specific individuals—could contribute, and there is now evidence showing that both common and rare variants can play a role in modifying phenotypic outcome. Epigenetic differences are another possible factor, and phenomena such as imprinting may also play a role. Finally, environmental or sporadic effects might interact to alter the risk of abnormalities associated with an aneuploidy event.
One way in which the genetic basis of variable phenotypes in patients with the same recurrent rearrangement is explored is by looking for sequence variants in candidate genes within the deleted region. This is a reasonable strategy based on the hypothesis that a “second hit” in a gene that is already reduced to a single copy following a deletion event on one homolog may worsen the phenotype in a carrier. An example comes from the recurrent 22q11 microdeletion, which is associated with several variable phenotypes, including schizophrenia. Exploration of genetic variants within the COMT gene identified a functional polymorphism, the presence of which appears to increase the risk of schizophrenia in carriers of the 22q11 deletion (13). COMT encodes catechol-O-methyltransferase, a reasonable candidate for the source of the schizophrenia phenotype. Similarly, mutations in SNAP29 are responsible for atypical phenotypes in some patients with 22q11 deletions (109).
Targeted studies have also been performed to explore phenotypic variability in some of the recently described microdeletion syndromes, such as in the 1q21.1 deletion syndrome, which is associated with a wide range of phenotypic severity. To date, there is no evidence that differences in deletion breakpoints, in the methylation of the 1q21 region, or in the coding sequence of at least two genes within the region are responsible for phenotypic differences (116). There is also no evidence for the role of additional pathogenic CNVs in this small set of families (63). Expression studies using lymphoblast cell lines in a subset of 1q21 patients showed that most of the unique genes within the 1q21 region (PRKAB2, CHD1L, BCL9, ACP6, GPR89A, and PDIA3P) correlate with the copy number of the region, but there were no differences among affected individuals to explain the variable phenotypes (63). Similarly, deletions of 16p13.11 are associated with ID, ASD, schizophrenia, and epilepsy. Again, expression studies showed that expression levels were decreased for genes within the region in seven patients with the 16p13.11 deletion and epilepsy, with no significant differences among patients. Sequence analysis of all genes within the 16p13.11 deletion region resulted in no evidence for secondary mutations within that region (65).
Distinct from the more distal 1q21.1 deletions mentioned above, deletions of proximal 1q21.1 were recently identified in patients with thrombocytopenia absent radius (TAR) syndrome (87). Interestingly, individuals with TAR syndrome may carry either an inherited or de novo deletion, and in many cases an unaffected parent also carries the deletion. Because parent-to-child transmission is rare, the disorder was thought to be due to either autosomal recessive or biallelic inheritance. To look for potential second hits that could act as modifiers of penetrance in 1q21.1 deletion carriers, Albers et al. (3) performed exome sequencing in five affected deletion carriers. In four of the five cases, they identified a low-frequency SNP within the 5′ untranslated region of RBM8A, a gene within the TAR deletion region, with an intronic variant identified in the fifth case. Functional assays showed that both variants significantly reduced RMB8A promoter activity, indicating that TAR syndrome results from an insufficiency of RMB8A. Investigation of 25 additional patients with TAR syndrome who inherited a 1q21.1 deletion from a normal parent revealed that a second RBM8A SNP was inherited from the other parent. This confirmed that compound inheritance of a deletion together with a low-frequency noncoding SNP in RBM8A causes TAR syndrome, likely by reducing protein levels below a critical threshold in certain tissues.
Taking advantage of the fact that microarrays provide data on genome-wide CNVs, Girirajan et al. (59) investigated the hypothesis that the co-occurrence of additional genetic variants can contribute to the phenotypic outcome of genomic disorders. In 2,312 children with a disease-associated CNV, 10% were found to carry at least one additional large CNV elsewhere in their genome. Interestingly, these second hits were more likely to occur in patients who harbored a CNV that is associated with a range of neurodevelopmental disorders, including 1q21.1 deletions and duplications. This study therefore suggests that the overall mutational burden, detected here in the form of additional CNVs elsewhere in the genome, likely influences the phenotypic outcome and penetrance of microdeletion/duplication events.
Exome sequencing data taken from patients with ASD also suggest that multiple mutations may affect phenotypic outcome. Moreover, O’Roak et al. (128) observed that patients carrying both a rare or de novo CNV and at least one other de novo damaging point mutation scored significantly lower in nonverbal IQ than did individuals with no events. In previously published series of patients with 1q21 rearrangements, it was already clear that 10–15% of cases have additional CNVs. However, a comprehensive analysis of second hits in a large series of patients has not yet been performed.
Studies of mice with haploinsufficiency for genes with roles in vertebral development showed a strong interaction between the penetrance of the accompanying scoliosis phenotype and maternal environmental factors. Using a mouse model, Sparrow et al. (163) showed that exposure of the developing embryo to hypoxic conditions for even a few hours—a relatively common event that can occur during pregnancy—resulted in a significant increase in the rate of scoliosis in the offspring via a disruption of FGF signaling (163). Although scoliosis is not a common finding in most genomic disorders, this study demonstrates an important principle: The penetrance of genetic mutations can be modified by interaction with common environmental factors, potentially explaining the phenotypic heterogeneity associated with many recurrent microdeletion/duplication syndromes.
MECHANISMS OF RECURRENT COPY-NUMBER VARIANTS
The mechanisms underlying the generation of structural variations in mammalian genomes can be broadly divided into two main categories. In the vast majority of cases, truly recurrent CNVs (i.e., those in which the breakpoints in unrelated individuals recur de novo in local hot-spot regions) are now known to occur as a result of recombination-based mechanisms, specifically NAHR. Here, deletion, duplication, inversion, or translocation events are generally mediated by large blocks of flanking homologous sequences that typically share 95–99% sequence identity of over tens to hundreds of kilobases, allowing illegitimate recombination to occur. NAHR appears to occur overwhelmingly during meiosis, although in some rare cases mitotic NAHR rearrangements are observed, potentially leading to somatic mosaicism (77, 99, 199). In contrast, sporadic or nonrecurrent CNVs (i.e., those in which, even where there may be a common overlapping region of aneuploidy between two unrelated carriers, different breakpoints are observed in each instance) result from errors during the repair of double-strand chromosomal breaks and are mediated by non-homology-based mechanisms. Several such mechanisms have been proposed, including nonhomologous end joining, FoSTeS, and microhomology-mediated break-induced replication (64). Studies of the rates of de novo rearrangement in sperm at several genomic disorder loci have revealed marked variation in the frequency of NAHR at different loci (175) and have shown that rates of NAHR can vary among different individuals at any one locus (16, 122). For a given microdeletion/duplication syndrome, the rate of formation of de novo rearrangements by NAHR is directly related to both the length and level of sequence identity between the flanking SDs that catalyze the rearrangement, and this rate is inversely related to their separation (102).
In addition to these fundamental mechanisms, studies of commonly identified CNVs have provided additional insights into some of the factors contributing to the generation of structural rearrangements. For example, analysis of the breakpoint regions of thousands of CNVs identified in the normal population has recognized enrichments for secondary structures such as G-quadruplexes and cruciforms, recombination motifs, Alu signal recognition particle motifs, and microsatellites (32). These observations indicate that local sequence features contribute toward CNV formation, and the generation of even nonrecurrent CNVs is not an entirely random process. Indeed, a small number of recurrent rearrangement syndromes are thought to be attributable to the presence of breakpoint motifs that form inherently unstable secondary structures. Two recurrent translocations, t(11;22)(q23;q11) and t(17;22)(q11;q11), have been shown to be catalyzed by large AT-rich repeats that are thought to form cruciform structures that mediate double-strand breaks and subsequent translocation (46, 93, 94). Similarly, Béna et al. (15) described a recurrent microdeletion of 14q32.2 that is catalyzed by large (TGG)n tandem repeats that are predicted to have extremely strong secondary structure (15).
In addition to these sequence-based features, it is now recognized that the relative stage at which a genomic region replicates its DNA during cell division influences the formation of CNVs. By comparing different classes of CNV with genome-wide maps of replication timing, Koren et al. (89) observed that human CNVs generated by NAHR-mediated events were enriched >4-fold in early-replicating regions, whereas events attributable to nonhomologous end joining were enriched ~2-fold in late-replicating regions. Data from the study of recurrent translocations that occur somatically in cancer have shed light on additional genomic features that can influence rates of chromosomal rearrangement. Burrow et al. (23) showed that the majority of such translocations involve regions known to be common fragile sites, representing regions of frequent chromosomal breakage. Furthermore, spatial organization within the nucleus also appears to influence rates of mitotic rearrangement; genomic regions that show stronger interactions undergo elevated rates of translocation (53, 195).
There are several examples of structural variations at genomic disorder loci that act to modify the rate at which subsequent deletions/duplications occur. Cuscó et al. (36) described a deletion variant in the flanking SDs that increases susceptibility to the recurrent 1.5-Mb deletion underlying Williams–Beuren syndrome (36). Many genomic disorder loci are also now known to be sites of common inversion in the general population (Table 1). A theme common to most such cases is that, although the inversions themselves are apparently benign variants, one specific orientation predisposes the region to further rearrangement, presumably creating a local architecture that increases the probability of NAHR by aligning SDs into a direct orientation (6, 7, 57, 88). In some cases, such as at 7q11.23 and 15q11–q13, the presence of an inversion alters the frequency of subsequent rearrangement, whereas for other loci the presence of an inversion appears to be a prerequisite for deletion/duplication (154). For example, 17q21.31 microdeletions (88, 155, 159) are strongly associated with a haplotype-specific common inversion polymorphism (88). A detailed screening of 22 patients with 17q21.31 microdeletions showed that all these deletions arose in a parent who was a carrier of the inversion in either a homozygous or heterozygous form (88). Interestingly, this inversion haplotype occurs at a relatively high frequency in Europeans (~20%) compared with Asians (1%) and Africans (6%) (167), and, not surprisingly, the 17q21.31 deletion syndrome occurs at a much higher frequency in European populations. In fact, an estimated 97% of all reported cases are in individuals of European descent (33). For several other genomic disorder loci, although common inversions have been recognized, whether these have any effect on the rate of subsequent deletion/duplication in carrier individuals is not known. Sharp (154) also previously proposed an alternative mechanism in which the presence of a heterozygous inversion at a locus could be mutagenic by suppressing meiotic synapsis, leading to the formation of an unstable asynaptic bubble. Although this hypothesis has not been formally tested, detailed sequencing studies of several hundred CNVs have since revealed that ~5% of deletions do indeed have inverted sequences around their breakpoints (32). Although this observation is also compatible with some CNVs occurring via more complex underlying rearrangements involving simultaneous deletion and inversion of a region, one interpretation of these data is that these inversions were preexisting and rendered the local regions prone to further rearrangements.
In addition to these local variants, trans-acting factors have been shown to influence rates of CNV formation. The best described of these is PRDM9, a zinc finger protein that specifies the location of meiotic crossovers in mammals (12). PRDM9 contains a zinc finger repeat that codes for its DNA-binding domain, and this repeat is highly polymorphic in humans. Different PRDM9 alleles are associated with significantly altered patterns of meiotic recombination across the genome, and Berg et al. (16) showed that an individual’s PRDM9 genotype can alter the usage of a pair of recombination hot spots in 17p11.2, leading to interindividual variation in the risk of producing offspring with CMT1A or HNPP. Studies of Williams–Beuren syndrome trios have also shown a significant increase in the frequency of uncommon PRDM9 alleles in parents who transmit de novo 7q11.23 deletions to their offspring, suggesting that allelic variation in PRDM9 is associated with alterations in the rate of NAHR at this locus (20).
Even though these known factors can result in modest increases in the frequency of certain genomic disorders in some families, it is generally presumed that all cases of pathogenic de novo microdeletion/duplication result from sporadic NAHR events that are mediated purely by the underlying genomic architecture. However, could it be that some cases of de novo microdeletion are not due simply to chance rearrangement? Specifically, what if some individuals are genetically predisposed to have children with genomic disorders? Relevant to this question are studies showing that, in yeast, there are numerous genes that act to suppress both spontaneous chromosomal rearrangements and recurrent rearrangements attributable to NAHR. Many of these CNV suppressor genes are involved in pathways such as double-strand break repair, DNA replication stress checkpoint signaling, mismatch repair, and chromatin modification. Interestingly, mutation of these genes can lead to massively elevated rates of spontaneous chromosomal rearrangement in the yeast genome that can be up to 1,000-fold higher than background mutation rates (28, 138, 162). It therefore seems plausible that if defects in similar pathways in humans were also associated with increased rates of meiotic rearrangement, then the offspring of any such individuals might have a significantly increased probability of inheriting a de novo aneuploidy event. We suggest that this represents an interesting hypothesis that warrants future exploration.
THE IMPACT OF TECHNOLOGY
Major strides in patient screening and diagnosis have been made over the past decade with the advent of genome-wide technologies (113). Chromosome microarray technologies were rapidly introduced into the clinical diagnostic laboratory after they appeared in the research arena. Although aCGH arrays were the first chromosome microarrays used in the clinic, within a short period of time, SNP microarrays were also utilized. Both platforms are now routinely used for clinical diagnosis, though there are a wide variety of specific array designs that have evolved over the past several years. Notably, early aCGH testing used targeted arrays with a higher density of probes in genomic regions known to be important for disease (e.g., 15q11–q13 for Prader–Willi and Angelman syndromes and subtelomeric regions). These were generally combined with backbone probes spaced evenly, but at lower density, throughout the genome to facilitate the detection of sporadic large deletions and duplications. As a result, the majority of CNVs that were detected by these platforms were pathogenic, because they were either within clinically relevant regions or large in size. With the evolution of the technology, most chromosome microarrays in use now have a large number of probes (often greater than 1 million) that tile across the human genome, allowing the detection of very small CNVs (size < 10 kb). These dense genome-wide arrays as well as more refined targeted arrays have greatly enhanced the ability to detect CNVs.
For example, Girirajan et al. (58) recently conducted a large CNV association study in a cohort of 2,588 ASD cases using a custom targeted array with a probe density of one probe per 50–1,000 base pairs. Building on a previous strategy that targeted regions of the genome flanked by pairs of large SDs, probes on this array were preferentially placed in 1,367 gene-containing rearrangement hot spots, including 1,247 regions characterized by shorter flanking repetitive sequences, such as Alu and other repeats and short SDs with >100 base pairs of identical sequence. Approximately 10% of these regions contained CNVs, 86% of which were inherited. Although none of these variants were found to be significantly enriched in ASD cases compared with controls, several showed evidence of shared breakpoints between unrelated individuals, indicating that there are potentially many novel recurrent microdeletion/duplication regions in the genome that are below the resolution of older microarray platforms.
Adding to the wealth of data generated by array-based methods, the more recent development and application of novel sequencing methods and associated analysis pipelines have continued to pave the way for improvements in the genetic detection and characterization of causative variants underlying microdeletion/duplication syndromes and related disorders. A key prerequisite for building technologies for variant detection is the availability of a reliable reference genome, which can affect both the development and interpretation of genome-wide assays (143); this is particularly true for structurally complex regions of the genome for which complete assemblies are lacking. Indeed, in many instances, pathogenic CNVs map to regions with known assembly gaps (72). This notion has motivated attempts in the field to construct more comprehensive maps of structural variation and improve existing reference assemblies, both of which have contributed to significant advances in CNV detection and characterization. An important contribution toward this aim evolved from the development of methods that enable the discovery of structural variants from the end sequencing and mapping of large-insert clones (78-80, 176). The power of this approach is that, following the identification of clones harboring structural variants, these loci can be fully sequenced and assembled to provide base-pair characterization of novel sequence not yet represented in the genome reference assembly.
Additional improvements to maps of structural variation have come with the increased use of next-generation sequencing technologies (e.g., Illumina, SOLiD, and 454 sequencing). Owing to the relatively low cost of these methods, at least when compared with Sanger sequencing, an unprecedented amount of genome sequence data has been generated in the past few years, including high-coverage assemblies of individual genomes (84, 149, 185, 188). The 1000 Genomes Project, one of the largest and most comprehensive sequencing efforts currently under way, is tasked with cataloging standing genome-wide genetic variation in a broad range of human populations (1, 2, 121). These large sequencing projects have sparked parallel efforts to develop effective analytical methods that utilize different aspects of these data for the discovery and characterization of structural genome variants, including not only copy-number changes that are detectable by microarrays but also balanced events such as inversions and translocations that have been invisible to array-based methods. These approaches include de novo sequence assembly, paired-end read mapping, split-read mapping, and read-depth analysis (see Figure 2). Not unexpectedly, family-based and population-scale screens using these methods have resulted in descriptions of thousands of novel structural variants (1, 2, 67, 79, 121, 172, 193, 194, 196). In addition, the directed application of these approaches has already proven useful in the context of microdeletion/duplication syndromes. For example, recent efforts in the 17q21.31 microdeletion region utilized a suite of methods—including custom microarrays, SNP imputation, large-insert clone mapping/sequencing/assembly, and next-generation sequencing analysis—to construct better haplotype maps of the locus, allowing refined characterization of breakpoints in microdeletion carriers and yielding insights into their underlying mechanisms, the genes impacted, and the local architectures that predispose to the occurrence of these deletions (18, 72, 169).
Figure 2.
Analysis methods for the identification and characterization of genomic structural variants from massively parallel sequencing data. (a) Libraries are constructed from individual DNA samples, and the ends of the cloned DNA inserts/adapter-ligated fragments are sequenced using either traditional Sanger sequencing or next-generation methods. End sequences can then be mapped to the human reference genome, providing mapping data that can be used to infer the position, size, and type of structural variants carried by the individual used to generate the sequencing library with respect to the reference assembly. Several commonly used methods include paired-end read mapping, split-read mapping, and read-depth analysis (panels b, c, and d, respectively). (b) In paired-end read mapping, paired reads derived from a single DNA molecule that show relative discordancy in their mapping (in terms of relative separation or orientation of the read pairs) allow the detection of deletions, insertions, and inversions. The maximum size and resolution of the events detected are dictated by the size and distribution of the library that is sequenced. If appropriate mapping parameters are used, paired-end reads located on separate chromosomes can also be used to infer translocation events (not shown). (c) Split-read mapping is used to identify instances in which portions of a single read align discontiguously to the reference genome; this approach is useful for directly mapping and characterizing the exact breakpoints of insertion, deletion, and inversion events. The power of this approach is dictated largely by read length. (d) Read-depth analysis utilizes the relative sequencing coverage across the genome from a population of individuals to identify regions with significantly altered read depth among individuals, highlighting loci that harbor deletions or duplications. The resolution of this method is strongly influenced by depth of coverage. Importantly, both paired-end and split-read mapping methods often perform poorly where the breakpoints are embedded in segmental duplications owing to the potential for mismapping of reads in paralogous sequences. In such cases, the read-depth approach is often more tractable. Targeted sequencing and assembly in regions harboring novel structural events is another powerful method in structurally complex regions (not shown); de novo assemblies can allow for the delineation of complex event breakpoints as well as complete descriptions at nucleotide resolution of insertion and duplication sequences not represented in the reference genome.
Methods and analysis pipelines centered on gleaning CNV information from whole-exome sequencing data are also beginning to emerge (56, 91, 98, 134, 147). Exome-based CNV detection methods build on read-depth approaches, such as those mentioned above for whole-genome data. Comparisons made between these methods have revealed some notable discrepancies with respect to both the total number and rate of de novo calls made (56) and in some cases have revealed relatively low concordance rates with high-confidence calls from microarrays, thus indicating that further technological developments are needed in this area (40). Nonetheless, several methods have been shown to have reasonable sensitivity (>75%) when there is sufficient overlap between array probes and exome capture targets (40, 56, 91), and similar results have been observed using whole-genome sequencing calls for comparison (91). Importantly, these exome-based methods are also able to outperform SNP arrays for the detection of smaller CNVs (90, 91). These methods have already been applied to two ASD cohorts (90, 136), demonstrating their power in the research laboratory.
Taken together, the advent and application of exome and whole-genome sequencing highlight the obvious potential benefits to clinical medicine, offering potentially novel approaches complementary to commonly used karyotyping or chromosome array–based methods. The strength of these methods is that they allow parallel assessments of a broad range of variants, from single-nucleotide mutations to large structural variants, including smaller CNVs and translocations that are often missed by standard chromosome arrays. Of course, as with array-based technologies, MPS approaches also come with considerations, and certain methods carry advantages or disadvantages depending on the exact question at hand. For example, exome sequencing provides targeted data for only coding portions of the genome at an increased depth of coverage and at lower cost, enabling researchers to interrogate larger cohorts with increased power. However, the obvious downfall is that, in contrast to whole-genome sequencing, much of the genome is left uninterrogated, and the nature of the data in most cases does not allow for the specific ascertainment of variant characteristics, such as total size or event breakpoint analysis. Thus, future prospects, especially as sequencing costs decrease, may well place more emphasis on whole-genome methods for patient screening.
The utility of whole-genome next-generation sequencing for the detection of lesions underlying genomic disorders is already being demonstrated, including in the introduction of novel noninvasive methods for prenatal screening (74, 85, 132, 164). For example, an approach developed by Srinivasan et al. (164) using cell-free maternal plasma DNA was able to identify a variety of pathogenic lesions (deletions, duplications, and translocations), including a 300-kb microdeletion.
THE EVOLUTIONARY HISTORY OF RECURRENT GENOMIC DISORDERS
Comparative studies of different human populations and closely related primate species have revealed that several loci associated with genomic disorders show evidence of unusual evolutionary pressures. In particular, there are several gene families that are known to have undergone rapid expansion and coding sequence evolution during recent primate evolution and are also associated with recurrent genomic disorders in humans.
Detailed genome-wide analysis of the evolutionary history of SDs shows that many appear to have proliferated in recent human and primate evolution, at the time of the African great ape ancestor, via expansion of key sequences termed core duplicons (75, 107). In many cases, these core duplicons contain transcriptionally active fragments that presumably drive the proliferation of these sequences, and the coding regions of some of these transcripts also show evidence for positive selection driving rapid evolution at the amino acid level (76). Duplication of these cores can often include flanking sequences that hitchhike with them, producing chromosomal regions containing complex mosaics of SD clusters comprising multiple copies of a particular core duplicon (75). This creates a particular pattern in which older duplications are surrounded by younger duplications (107).
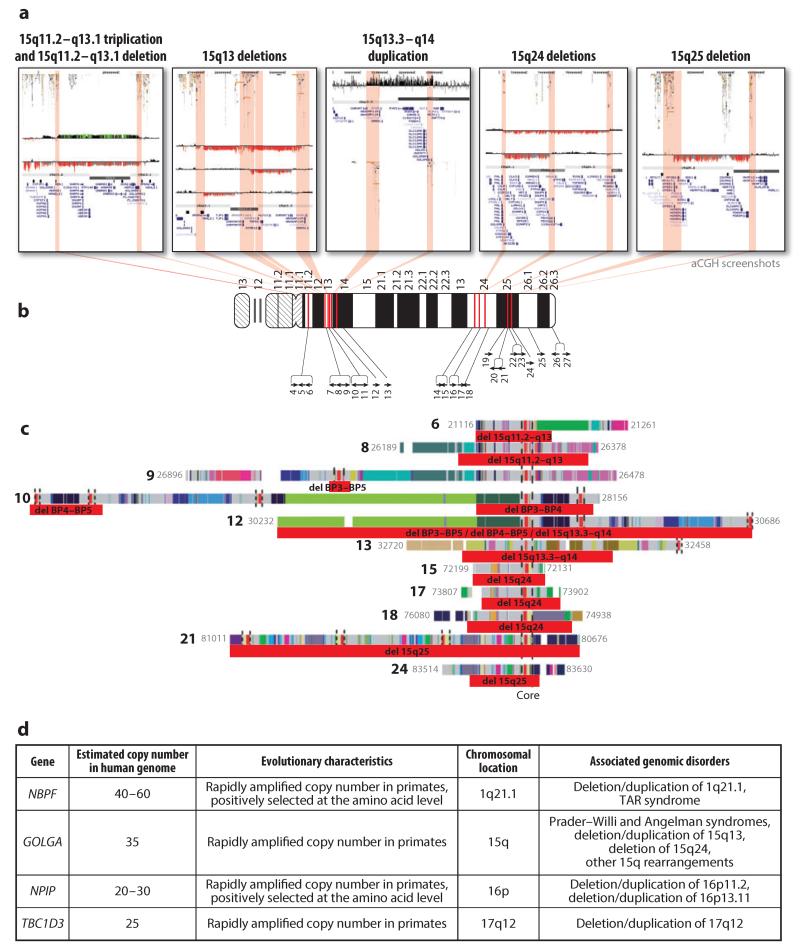
Intriguingly, there are several known instances where these core duplicons are located precisely at the breakpoints of recurrent genomic disorders (Figure 3). These include gene families such as NBPF, which shows the most extreme increase in human-specific copy number of any gene identified to date (135), and NPIP, which both has expanded in copy number specifically in the great ape lineage and shows one of the highest levels of amino acid replacement of any human gene (76). Perhaps the most striking example of this phenomenon is provided by the GOLGA gene family, which contains several dozen copies scattered over human chromosome 15. Microarray studies of many different chromosome 15 rearrangements, including several recurrent microdeletion/duplication syndromes and more complex rearrangements such as inverted duplications and triplications of chromosome 15, have shown that the breakpoints of all of these appear to coincide precisely with the location of a duplication family containing the GOLGA gene (Figure 3). The recent proliferation of these highly duplicated gene families therefore seems to have been the key driver in creating local architectures that predispose our genome to recurrent pathogenic rearrangements. Intriguingly, although their evolutionary history strongly suggests that all of these genes have undergone strong positive selection in the primate lineage, and therefore presumably have important phenotypic consequences, almost nothing is known about their function. The high frequency of genomic disorders observed in humans can be regarded as a negative side effect resulting from the rapid expansion of these paralogous gene families.
Figure 3.
Gene families that have undergone rapid expansion during recent primate evolution that are associated with several recurrent genomic disorders in humans. (a) Studies of multiple chromosome 15 rearrangements by array comparative genomic hybridization (aCGH) show that the breakpoints of these rearrangements coincide with the location of a duplication family containing the GOLGA gene. These include both recognized recurrent genomic disorders and rarer rearrangements, including (left to right) a triplication of 15q11.2–q13.1 (157), a deletion of 15q11.2–q13.1 associated with Angelman syndrome (157), BP3–BP5 deletions of 15q13, a duplication of 15q13.3–q14 associated with epilepsy, deletions of 15q24 (158), and a deletion of 15q25 associated with congenital diaphragmatic hernia (111). In each image, the locations of duplication blocks containing the GOLGA gene (75) are indicated by red shaded regions. Tracks show segmental duplications, cytogenetic bands, assembly gaps, and RefSeq genes. (b) This panel shows the locations of 27 assembled GOLGA-containing duplication blocks on chromosome 15. Each block is indicated by a black arrow (75), numbered according to its genomic location along the chromosome. Those that coincide with the breakpoints of deletion/duplication events are highlighted (red bars). (c) GOLGA duplication blocks coincide with sites of rearrangement breakpoints on chromosome 15. Red bars below each duplication block indicate the interval in which rearrangement breakpoints occur. Although the presence of structural polymorphisms and cross-hybridization between paralogous sequences makes it difficult to precisely determine the breakpoints by aCGH, in every case data showed that the intervals in which the breakpoints occur overlap a GOLGA core. Blocks are numbered, with colored bars denoting the ancestral chromosomal origin of each subelement (75). The core element, which contains the GOLGA gene and is shared by all duplication blocks, is highlighted by vertical dashed lines. Note that some blocks contain multiple GOLGA sequences. (d) At least four different gene families, all of which have undergone rapid amplification of copy number in the past 20–30 million years of primate evolution, are associated with the breakpoints of recurrent genomic disorders in the human genome. This includes NBPF, which shows the most extreme increase in human-specific copy number of any gene identified to date (135), and NPIP, which both has expanded in copy number specifically in the great ape lineage and shows one of the highest levels of amino acid replacement of any human gene (76). Abbreviation: TAR, thrombocytopenia absent radius. Panels a–c have been adapted in part from a figure first published by the Nature Publishing Group (157).
A particularly good example for recurrence of architectures is the 17q21.31 region, which contains an alternative haplotype in humans that includes a large inversion encompassing the recurrent microdeletion region. The inverted H2 haplotype occurs almost exclusively in northern European populations, and population analysis suggests that it has undergone recent positive selection, potentially owing to an association with increased fecundity in carrier individuals (167). Detailed analysis showed that the inverted H2 haplotype represents the ancestral configuration, but, remarkably, this region is highly mutagenic and has undergone multiple inversions at different times during the past ~10 million years, such that it is now polymorphic in both humans and chimpanzees (198).
Despite these rapid changes in the SD architecture of primate genomes, consistent with theoretical predictions, it has recently been demonstrated that genomic disorders are not unique to humans. The advent of whole-genome studies of primates has for the first time identified a chimpanzee with a microdeletion syndrome. Susie, a female chimpanzee from the Biomedical Primate Research Centre, showed abnormal behavioral and phenotype characteristics but had never been formally diagnosed with a specific disease. However, a screen for CNVs in a large panel of 80 great apes (137) found a 1.7-Mb deletion of 17p11.2 (including the RAI1 gene) present in Susie, corresponding to the region responsible for Smith–Magenis syndrome in humans. This syndrome is characterized by some of the same phenotypes observed in Susie, including aggressiveness, obesity, and renal problems. Surprisingly, the chimpanzee presented a more complex architecture in the region and different deletion breakpoints compared with the human counterpart, suggesting that different duplication blocks that have expanded exclusively in the Pan lineage likely catalyze NAHR at this locus (171).
FUTURE PERSPECTIVES
Chromosome microarray testing is a first-line test in the clinic for the diagnosis of patients with ID/DD, ASD, or multiple anomalies, and it has largely replaced traditional karyotype analysis (119). As costs continue to fall and analytical methods evolve, MPS technologies are now poised to replace chromosome microarrays in the near future. Indeed, sequencing-based approaches are already being used in the clinic for noninvasive prenatal diagnosis of fetal aneuploidy (74, 85, 132, 164). Over the past decade, the introduction of first microarrays (40) and then exome sequencing (90, 136) led to marked increases in the proportion of cases of presumptive genetic disease for which an underlying pathogenic mutation can be identified. However, given (a) the limited resolution of many array platforms and (b) that exome sequencing effectively assays only a small fraction of the genome, we anticipate that the introduction of whole-genome sequencing in the clinic will lead to further improvements in patient diagnosis.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01DA033660, R01HG006696, and R03HD073731, March of Dimes grant 6-FY13-92, and Alzheimer’s Association grant 2012ALZNIRG69983 to A.J.S.; by National Institutes of Health grant R01NS069605 and a Burroughs Wellcome Fund Career Award for Medical Scientists to H.C.M.; and by European Research Council Starting Grant 260372 and Spanish Government Grants BFU2011-28549 to T.M.-B.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.1000 Genomes Proj. Consort. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.1000 Genomes Proj. Consort. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 2012;44:435–39. doi: 10.1038/ng.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am. J. Hum. Genet. 1999;65:370–86. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrieux J, Dubourg C, Rio M, Attie-Bitach T, Delaby E, et al. Genotype-phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am. J. Med. Genet. A. 2009;149A:2813–19. doi: 10.1002/ajmg.a.33097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat. Genet. 2010;42:745–50. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonacci F, Kidd JM, Marques-Bonet T, Ventura M, Siswara P, et al. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 2009;18:2555–66. doi: 10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aradhya S, Woffendin H, Jakins T, Bardaro T, Esposito T, et al. A recurrent deletion in the ubiquitously expressed NEMO (IKK-γ) gene accounts for the vast majority of incontinentia pigmenti mutations. Hum. Mol. Genet. 2001;10:2171–79. doi: 10.1093/hmg/10.19.2171. [DOI] [PubMed] [Google Scholar]
- 9.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 10.Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, et al. Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am. J. Hum. Genet. 2007;80:938–47. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW, et al. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum. Mol. Genet. 2008;17:4045–53. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bearden CE, Jawad AF, Lynch DR, Sokol S, Kanes SJ, et al. Effects of a functional COMT polymorphism on prefrontal cognitive function in patients with 22q11.2 deletion syndrome. Am. J. Psychiatry. 2004;161:1700–2. doi: 10.1176/appi.ajp.161.9.1700. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J. Med. Genet. 2009;46:382–88. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Béna F, Gimelli S, Migliavacca E, Brun-Druc N, Buiting K, et al. A recurrent 14q32.2 microdeletion mediated by expanded TGG repeats. Hum. Med. Genet. 2010;19:1967–73. doi: 10.1093/hmg/ddq075. [DOI] [PubMed] [Google Scholar]
- 16.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–63. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. 2012;44:881–85. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondeson ML, Dahl N, Malmgren H, Kleijer WJ, Tönnesen T, Carlberg BM, et al. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum. Med. Genet. 1995;4:615–21. doi: 10.1093/hmg/4.4.615. [DOI] [PubMed] [Google Scholar]
- 20.Borel C, Cheung F, Stewart H, Koolen DA, Phillips C, et al. Evaluation of PRDM9 variation as a risk factor for recurrent genomic disorders and chromosomal non-disjunction. Hum. Genet. 2012;131:1519–24. doi: 10.1007/s00439-012-1180-4. [DOI] [PubMed] [Google Scholar]
- 21.Breckpot J, Thienpont B, Bauters M, Tranchevent LC, Gewillig M, et al. Congenital heart defects in a novel recurrent 22q11.2 deletion harboring the genes CRKL and MAPK1. Am. J. Med. Genet. A. 2012;158A:574–80. doi: 10.1002/ajmg.a.35217. [DOI] [PubMed] [Google Scholar]
- 22.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–71. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrow AA, Williams LE, Pierce LC, Wang YH. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. doi: 10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler MG, Meaney FJ, Palmer CG. Clinical and cytogenetic survey of 39 individuals with Prader-Labhart-Willi syndrome. Am. J. Med. Genet. 1986;23:793–809. doi: 10.1002/ajmg.1320230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, et al. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 2013;45:1319–26. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chance PF. Inherited focal, episodic neuropathies: hereditary neuropathy with liability to pressure palsies and hereditary neuralgic amyotrophy. Neuromol. Med. 2006;8:159–74. doi: 10.1385/NMM:8:1:159. [DOI] [PubMed] [Google Scholar]
- 27.Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–51. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 29.Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, et al. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 1997;17:154–63. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 30.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol. Psychiatry. 2008;63:1111–17. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciccone R, Mattina T, Giorda R, Bonaglia MC, Rocchi M, et al. Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J. Med. Genet. 2006;43:e19. doi: 10.1136/jmg.2005.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010;42:385–91. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper GM, Mefford HC. Detection of copy number variation using SNP genotyping. Methods Mol. Biol. 2011;767:243–52. doi: 10.1007/978-1-61779-201-4_18. [DOI] [PubMed] [Google Scholar]
- 35.Coulter ME, Miller DT, Harris DJ, Hawley P, Picker J, et al. Chromosomal microarray testing influences medical management. Genet. Med. 2011;13:770–76. doi: 10.1097/GIM.0b013e31821dd54a. [DOI] [PubMed] [Google Scholar]
- 36.Cuscó I, Corominas R, Bayés M, Flores R, Rivera-Brugués N, et al. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams-Beuren syndrome deletion. Genome Res. 2008;18:683–94. doi: 10.1101/gr.073197.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dathe K, Kjaer KW, Brehm A, Meinecke P, Nürnberg P, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 2009;84:483–92. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Cid R, Riveira-Munoz E, Zeeuwen PLJM, Robarge J, Liao W, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 2009;41:211–15. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Ligt J, Boone PM, Pfundt R, Vissers LE, Richmond T, et al. Detection of clinically relevant copy number variants with whole-exome sequencing. Hum. Mutat. 2013;34:1439–48. doi: 10.1002/humu.22387. [DOI] [PubMed] [Google Scholar]
- 41.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–16. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dharmadhikari AV, Kang SH, Szafranski P, Person RE, Sampath S, et al. Small rare recurrent deletions and reciprocal duplications in 2q21.1, including brain-specific ARHGEF4 and GPR148. Hum. Med. Genet. 2012;21:3345–55. doi: 10.1093/hmg/dds166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dibbens LM, Mullen S, Helbig I, Mefford HC, Bayly MA, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum. Mol. Genet. 2009;18:3626–31. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K, et al. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum. Med. Genet. 2000;9:35–46. doi: 10.1093/hmg/9.1.35. [DOI] [PubMed] [Google Scholar]
- 45.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–67. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 46.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, et al. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Hattab AW, Fang P, Jin W, Hughes JR, Gibson JB, et al. Int22h-1/int22h-2-mediated Xq28 rearrangements: intellectual disability associated with duplications and in utero male lethality with deletions. J. Med. Genet. 2011;48:840–50. doi: 10.1136/jmedgenet-2011-100125. [DOI] [PubMed] [Google Scholar]
- 48.El-Hattab AW, Smolarek TA, Walker ME, Schorry EK, Immken LL, et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum. Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El-Hattab AW, Zhang F, Maxim R, Christensen KM, Ward JC, et al. Deletion and duplication of 15q24: molecular mechanisms and potential modification by additional copy number variants. Genet. Med. 2010;12:573–86. doi: 10.1097/GIM.0b013e3181eb9b4a. [DOI] [PubMed] [Google Scholar]
- 50.Elia J, Gai X, Xie HM, Perin JC, Geiger E, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–46. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elinati E, Kuentz P, Redin C, Jaber S, Vanden Meerschaut F, et al. Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum. Med. Genet. 2012;21:3695–702. doi: 10.1093/hmg/dds200. [DOI] [PubMed] [Google Scholar]
- 52.ENCODE Proj. Consort. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engreitz JM, Agarwala V, Mirny LA. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS ONE. 2012;7:e44196. doi: 10.1371/journal.pone.0044196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erdogan F, Larsen LA, Zhang L, Tumer Z, Tommerup N, et al. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J. Med. Genet. 2008;45:704–9. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- 55.Fisher J, Upadhyaya M. Molecular genetics of facioscapulohumeral muscular dystrophy (FSHD) Neuromuscul. Disord. 1997;7:55–62. doi: 10.1016/s0960-8966(96)00400-2. [DOI] [PubMed] [Google Scholar]
- 56.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am. J. Hum. Genet. 2012;91:597–60. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Med. Genet. 2003;12:849–58. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- 58.Girirajan S, Dennis MY, Baker C, Malig M, Coe BP, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013;92:221–37. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–31. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldlust IS, Hermetz KE, Catalano LM, Barfield RT, Cozad R, et al. Mouse model implicates GNB3 duplication in a childhood obesity syndrome. Proc. Natl. Acad. Sci. USA. 2013;110:14990–94. doi: 10.1073/pnas.1305999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, et al. Understanding the impact of 1q21.1 copy number variant. Orphanet J. Rare Dis. 2011;6:54. doi: 10.1186/1750-1172-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am. J. Hum. Genet. 2010;86:707–18. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–62. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hormozdiari F, Hajirasouliha I, McPherson A, Eichler EE, Sahinalp SC, et al. Simultaneous structural variation discovery among multiple paired-end sequenced genomes. Genome Res. 2011;21:2203–12. doi: 10.1101/gr.120501.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 69.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol. Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Int. Schizophr. Consort. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Itsara A, Cooper GM, Baker C, Girirajan S, Li J, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Itsara A, Vissers LE, Steinberg KM, Meyer KJ, Zody MC, et al. Resolving the breakpoints of the 17q21.31 microdeletion syndrome with next-generation sequencing. Am. J. Hum. Genet. 2012;90:599–613. doi: 10.1016/j.ajhg.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jehee FS, Krepischi-Santos AC, Rocha KM, Cavalcanti DP, Kim CA, et al. High frequency of submicroscopic chromosomal imbalances in patients with syndromic craniosynostosis detected by a combined approach of microsatellite segregation analysis, multiplex ligation-dependent probe amplification and array-based comparative genome hybridisation. J. Med. Genet. 2008;45:447–50. doi: 10.1136/jmg.2007.057042. [DOI] [PubMed] [Google Scholar]
- 74.Jensen TJ, Dzakula Z, Deciu C, van den Boom D, Ehrich M, et al. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin. Chem. 2012;58:1148–51. doi: 10.1373/clinchem.2011.180794. [DOI] [PubMed] [Google Scholar]
- 75.Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–68. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 76.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–19. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 77.Juyal RC, Kuwano A, Kondo I, Zara F, Baldini A, et al. Mosaicism for del(17)(p11.2p11.2) underlying the Smith-Magenis syndrome. Am. J. Med. Genet. 1996;66:193–96. doi: 10.1002/(SICI)1096-8628(19961211)66:2<193::AID-AJMG13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 78.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–47. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kidd JM, Sampas N, Antonacci F, Graves T, Fulton R, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat. Methods. 2010;7:365–71. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiezun A, Pulit SL, Francioli LC, van Dijk F, Swertz M, et al. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 2013;9:e1003301. doi: 10.1371/journal.pgen.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Med. Genet. 2009;18:1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry. 2012;17:142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, et al. Noninvasive whole-genome sequencing of a human fetus. Sci. Transl. Med. 2012;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klopocki E, Graul-Neumann LM, Grieben U, Tönnies H, Ropers HH, et al. A further case of the recurrent 15q24 microdeletion syndrome, detected by array CGH. Eur. J. Pediatr. 2008;167:903–8. doi: 10.1007/s00431-007-0616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klopocki E, Schulze H, Strauss G, Ott CE, Hall J, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am. J. Hum. Genet. 2007;80:232–40. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koolen DA, Sharp AJ, Hurst JA, Firth HV, Knight SJ, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J. Med. Genet. 2008;45:710–20. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 2012;91:1033–40. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krumm N, O’Roak BJ, Karakoc E, Mohajeri K, Nelson B, et al. Transmission disequilibrium of small CNVs in simplex autism. Am. J. Hum. Genet. 2013;93:595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 93.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Med. Genet. 2001;10:2605–17. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 94.Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS, et al. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–38. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lakich D, Kazazian HH, Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat. Genet. 1993;5:236–41. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 96.Lako M, Ramsden S, Campbell RD, Strachan T. Mutation screening in British 21-hydroxylase deficiency families and development of novel microsatellite based approaches to prenatal diagnosis. J. Med. Genet. 1999;36:119–24. [PMC free article] [PubMed] [Google Scholar]
- 97.Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–97. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–13. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liehr T, Rautenstrauss B, Grehl H, Bathke KD, Ekici A, et al. Mosaicism for the Charcot-Marie-Tooth disease type 1A duplication suggests somatic reversion. Hum. Genet. 1996;98:22–28. doi: 10.1007/s004390050154. [DOI] [PubMed] [Google Scholar]
- 100.Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, et al. A chimaeric 11β-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–65. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 101.Lindsay SJ, Khajavi M, Lupski JR, Hurles ME. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, et al. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 2011;89:580–88. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–90. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 105.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–32. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 106.Magoulas PL, El-Hattab AW. Chromosome 15q24 microdeletion syndrome. Orphanet J. Rare Dis. 2012;7:2. doi: 10.1186/1750-1172-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–81. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McDonald-McGinn DM, Fahiminiya S, Revil T, Nowakowska BA, Suhl J, et al. Hemizygous mutations in SNAP29 unmask autosomal recessive conditions and contribute to atypical findings in patients with 22q11.2DS. J. Med. Genet. 2013;50:80–90. doi: 10.1136/jmedgenet-2012-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, et al. Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet. Cell Genet. 1998;81:222–28. doi: 10.1159/000015035. [DOI] [PubMed] [Google Scholar]
- 111.Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mefford HC, Cooper GM, Zerr T, Smith JD, Baker C, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–85. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr. Opin. Genet. Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mefford HC, Rosenfeld JA, Shur N, Slavotinek AM, Cox VA, et al. Further clinical and molecular delineation of the 15q24 microdeletion syndrome. J. Med. Genet. 2012;49:110–18. doi: 10.1136/jmedgenet-2011-100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mefford HC, Yendle SC, Hsu C, Cook J, Geraghty E, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann. Neurol. 2011;70:974–85. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Menko FH, Kneepkens CM, de Leeuw N, Peeters EA, Van Maldergem L, et al. Variable phenotypes associated with 10q23 microdeletions involving the PTEN and BMPR1A genes. Clin. Genet. 2008;74:145–54. doi: 10.1111/j.1399-0004.2008.01026.x. [DOI] [PubMed] [Google Scholar]
- 119.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 2009;46:242–48. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Molina O, Blanco J, Vidal F. Deletions and duplications of the 15q11-q13 region in spermatozoa from Prader-Willi syndrome fathers. Mol. Hum. Reprod. 2010;16:320–28. doi: 10.1093/molehr/gaq005. [DOI] [PubMed] [Google Scholar]
- 123.Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am. J. Hum. Genet. 2010;87:618–30. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moreno-De-Luca D, Sanders SJ, Willsey AJ, Mulle JG, Lowe JK, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol. Psychiatry. 2013;18:1090–95. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakashima S, Watanabe Y, Okada J, Ono H, Nagata E, et al. Critical role of Yp inversion in PRKX/PRKY-mediated Xp;Yp translocation in a patient with 45,X testicular disorder of sex development. Endocr. J. 2013;60:1329–34. doi: 10.1507/endocrj.ej13-0334. [DOI] [PubMed] [Google Scholar]
- 126.Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS, et al. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–10. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 127.Need AC, Ge D, Weale ME, Maia J, Feng S, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Palumbo O, Palumbo P, Palladino T, Stallone R, Miroballo M, et al. An emerging phenotype of interstitial 15q25.2 microdeletions: clinical report and review. Am. J. Med. Genet. A. 2012;158A:3182–89. doi: 10.1002/ajmg.a.35631. [DOI] [PubMed] [Google Scholar]
- 131.Pérez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U, et al. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am. J. Hum. Genet. 1996;59:781–92. [PMC free article] [PubMed] [Google Scholar]
- 132.Peters D, Chu T, Yatsenko SA, Hendrix N, Hogge WA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N. Engl. J. Med. 2011;365:1847–48. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–72. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–54. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Popesco MC, MacLaren EJ, Hopkins J, Dumas L, Cox M, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–7. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 136.Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am. J. Hum. Genet. 2013;93:607–19. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–75. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–89. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ramocki MB, Bartnik M, Szafranski P, Kolodziejska KE, Xia Z, et al. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am. J. Hum. Genet. 2010;87:857–65. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, et al. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr. Res. 2008;64:358–63. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 142.Riggs ER, Wain KE, Riethmaier D, Smith-Packard B, Faucett WA, et al. Chromosomal microarray impacts clinical management. Clin. Genet. 2013;85:147–53. doi: 10.1111/cge.12107. [DOI] [PubMed] [Google Scholar]
- 143.Rosenfeld JA, Mason CE, Smith TM. Limitations of the human reference genome for personalized genomics. PLoS ONE. 2012;7:e40294. doi: 10.1371/journal.pone.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rudd MK, Keene J, Bunke B, Kaminsky EB, Adam MP, et al. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum. Med. Genet. 2009;18:2957–62. doi: 10.1093/hmg/ddp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sagoo G, Butterworth A, Sanderson S, Shaw-Smith C, Higgins J, et al. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet. Med. 2009;11:139–46. doi: 10.1097/GIM.0b013e318194ee8f. [DOI] [PubMed] [Google Scholar]
- 146.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics. 2011;27:2648–54. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saunier S, Calado J, Benessy F, Silbermann F, Heilig R, et al. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am. J. Hum. Genet. 2000;66:778–89. doi: 10.1086/302819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–47. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–49. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–28. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 152.Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, et al. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J. Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 153.Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Med. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 154.Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum. Mutat. 2009;30:135–44. doi: 10.1002/humu.20843. [DOI] [PubMed] [Google Scholar]
- 155.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 156.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, et al. Segmental duplications and copynumber variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–28. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007;16:567–72. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 159.Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–37. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 160.Shirakawa T, Fujisawa M, Kanzaki M, Okada H, Arakawa S, et al. Y chromosome (Yq11) microdeletions in idiopathic azoospermia. Int. J. Urol. 1997;4:198–201. doi: 10.1111/j.1442-2042.1997.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 161.Shoichet SA, Waibel S, Endruhn S, Sperfeld AD, Vorwerk B, et al. Identification of candidate genes for sporadic amyotrophic lateral sclerosis by array comparative genomic hybridization. Amyotroph. Lateral Scler. 2009;10:162–69. doi: 10.1080/17482960802535001. [DOI] [PubMed] [Google Scholar]
- 162.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, et al. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2004;101:9039–44. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. doi: 10.1016/j.cell.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 164.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP, et al. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am. J. Hum. Genet. 2013;92:167–76. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stankiewicz P, Kulkarni S, Dharmadhikari AV, Sampath S, Bhatt SS, et al. Recurrent deletions and reciprocal duplications of 10q11.21q11.23 including CHAT and SLC18A3 are likely mediated by complex low-copy repeats. Hum. Mutat. 2012;33:165–79. doi: 10.1002/humu.21614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 167.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 168.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–36. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 2012;44:872–80. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stone JL, O’Donovan MC, Gurling H, Kirov GK, Blackwood DH, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–82. doi: 10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–46. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–28. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tatton-Brown K, Douglas J, Coleman K, Baujat G, Chandler K, et al. Multiple mechanisms are implicated in the generation of 5q35 microdeletions in Sotos syndrome. J. Med. Genet. 2005;42:307–13. doi: 10.1136/jmg.2004.027755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 177.van Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J. Med. Genet. 2009;46:511–23. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.van der Zwaag B, Staal WG, Hochstenbach R, Poot M, Spierenburg HA, et al. A co-segregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. Am. J. Med. Genet. B. 2010;153B:960–66. doi: 10.1002/ajmg.b.31055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Esch H, Backx L, Pijkels E, Fryns JP. Congenital diaphragmatic hernia is part of the new 15q24 microdeletion syndrome. Eur. J. Med. Genet. 2009;52:153–56. doi: 10.1016/j.ejmg.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 180.Van Esch H, Hollanders K, Badisco L, Melotte C, Van Hummelen P, et al. Deletion of VCX-A due to NAHR plays a major role in the occurrence of mental retardation in patients with X-linked ichthyosis. Hum. Med. Genet. 2005;14:1795–803. doi: 10.1093/hmg/ddi186. [DOI] [PubMed] [Google Scholar]
- 181.Vassos E, Collier DA, Holden S, Patch C, Rujescu D, et al. Penetrance for copy number variants associated with schizophrenia. Hum. Med. Genet. 2010;19:3477–81. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 182.Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003;63:2872–80. [PubMed] [Google Scholar]
- 183.Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J. Med. Genet. 2010;47:289–97. doi: 10.1136/jmg.2009.072942. [DOI] [PubMed] [Google Scholar]
- 184.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 185.Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wat MJ, Enciso VB, Wiszniewski W, Resnick T, Bader P, et al. Recurrent microdeletions of 15q25.2 are associated with increased risk of congenital diaphragmatic hernia, cognitive deficits and possibly Diamond–Blackfan anaemia. J. Med. Genet. 2010;47:777–81. doi: 10.1136/jmg.2009.075903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 188.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–76. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 189.Willatt L, Cox J, Barber J, Cabanas ED, Collins A, et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am. J. Hum. Genet. 2005;77:154–60. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–8. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–34. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 192.Xi R, Hadjipanayis AG, Luquette LJ, Kim TM, Lee E, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc. Natl. Acad. Sci. USA. 2011;108:E1128–36. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J, et al. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–92. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–53. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhu M, Need AC, Han Y, Ge D, Maia JM, et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am. J. Hum. Genet. 2012;91:408–21. doi: 10.1016/j.ajhg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zimran A, Sorge J, Gross E, Kubitz M, West C, et al. A glucocerebrosidase fusion gene in Gaucher disease. Implications for the molecular anatomy, pathogenesis, and diagnosis of this disorder. J. Clin. Investig. 1990;85:219–22. doi: 10.1172/JCI114415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zori RT, Lupski JR, Heju Z, Greenberg F, Killian JM, et al. Clinical, cytogenetic, and molecular evidence for an infant with Smith-Magenis syndrome born from a mother having a mosaic 17p11.2p12 deletion. Am. J. Med. Genet. 1993;47:504–11. doi: 10.1002/ajmg.1320470414. [DOI] [PubMed] [Google Scholar]