Abstract
The carbapenem-hydrolyzing class D β-lactamases OXA-23 and OXA-24/40 have emerged world-wide as causative agents for β-lactam antibiotic resistance in Acinetobacter species. Many variants of these enzymes have appeared clinically, including OXA-160 and OXA-225, which both contain a P→S substitution at homologous positions in the OXA-24/40 and OXA-23 backgrounds respectively. We purified OXA-160 and OXA-225 and used steady-state kinetic analysis to compare the substrate profiles of these variants to their parental enzymes, OXA-24/40 and OXA-23. OXA-160 and OXA-225 possess greatly enhanced hydrolytic activities against aztreonam, ceftazidime, cefotaxime and ceftriaxone when compared to OXA-24/40 and OXA-23. These enhanced activities are the result of much lower Km values, suggesting that the P→S substitution enhances the binding affinity of these drugs. We have determined the structures of the acylated forms of OXA-160 (with ceftazidime and aztreonam) and OXA-225 (ceftazidime). These structures show that the R1 oxyimino side-chain of these drugs occupies a space near the β5-β6 loop and the omega loop of the enzymes. The P→S substitution found in OXA-160 and OXA-225 results in a deviation of the β5-β6 loop, relieving the steric clash with the R1 side-chain carboxypropyl group of aztreonam and ceftazidime. These results reveal worrying trends in the enhancement of substrate spectrum of class D β-lactamases, but may also provide a map for β-lactam improvement.
The number of class D β-lactamases has exploded in the last ten years, with over 420 unique sequences identified.1 Most of the growth has occurred amongst the five subfamilies of carbapenemases categorized by their prototypes OXA-23, OXA-24/40, OXA-48, OXA-51 and OXA-58.2 In many cases, the presence of one of these enzymes (or variants containing a small number of substitutions) leads to increased levels of carbapenem resistance in their bacterial hosts, the vast majority of which are members of the clinically challenging Gram-negative rod genus Acinetobacter.
Our understanding of how these enzymes achieve specificity and catalytic hydrolysis for a variety of β-lactam substrates has been greatly enhanced by a large number of structural studies. Early studies of OXA-10 and OXA-1 established that class D β-lactamases have an unusual carboxylated lysine that acts as a general base in a serine nucleophile-based covalent catalysis mechanism.3, 4 Later studies on OXA-24/40 and OXA-23 showed that a hydrophobic bridge across the top of the active site is important for tight carbapenem binding in those enzymes,5–7 but structural analysis of OXA-48 and OXA-58 demonstrated that this bridge is not universal among carbapenemases.8, 9 Two surface loops have emerged as key determinants of substrate specificity for the class D β-lactamase family. The omega loop, named after a homologous structure from class A β-lactamases, contains a highly conserved tryptophan/leucine motif (W167/L168 in OXA-24/40). The tryptophan stabilizes the carbamate general base (carboxylysine 84), and the leucine has been implicated in modulating carbapenem hydrolysis.6, 8 The second loop, sometimes referred to as the β5-β6 loop, is positioned very close to the omega loop. This structure contributes one of the “bridge” residues (M223 in OXA-24/40), and is thought to play a key role in the ability of these enzymes to deacylate carbapenems.10
Until very recently, it has been noted that these class D carbapenemases contribute little to resistance against advanced generation cephalosporins such as ceftazidime and cefotaxime, or to the monobactam aztreonam (Figure 1).11 This view has begun to change with the discovery of carbapenem-hydrolyzing class D β-lactamase (CHDL) variants that have mutated to gain activity against cephalosporins (e.g. OXA-163, OXA-146).12, 13 These examples suggest that class D β-lactamases are evolving to widen their specificity, and may portend further difficulty in treating A. baumannii infections. As confirmation of this evolutionary trend, we report here that two clinical variants containing proline→serine substitutions at sequentially homologous positions in either the OXA-23 or OXA-24/40 background possess broad spectrum hydrolytic activity against penicillins, carbapenems, advanced generation cephalosporins and the monobactam aztreonam. These two variants (OXA-160, genbank accession ADB28891.1 and OXA-225, genbank accession AEP43731.1) were both isolated from clinical carbapenem-resistant A. baumannii strains. We have determined the structure of both variants with several ligands bound as acyl-intermediates, and describe a rationale for how the P→S mutation causes increased activity.
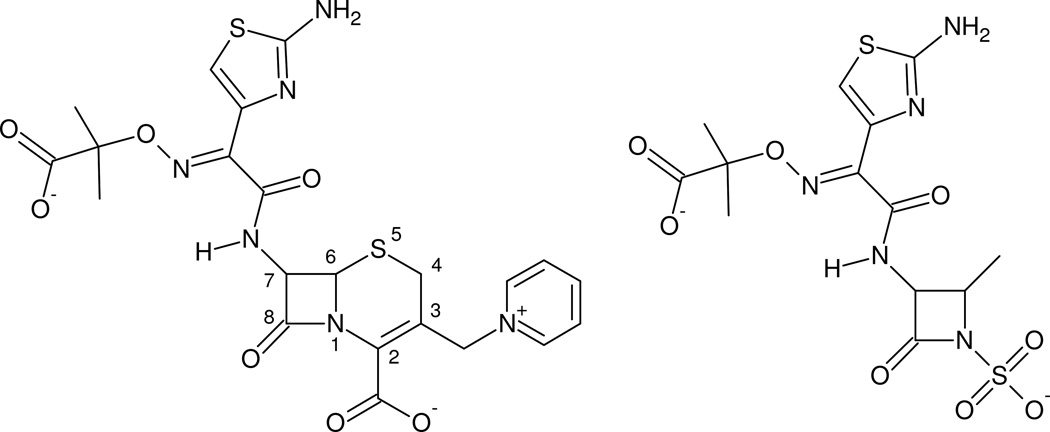
Figure 1. The structures of ceftazidime and aztreonam.
The structures of the two ligands used to produce acyl-intermediate structures in this study are shown. While ceftazidime (left) is a cephalosporin and aztreonam (right) is a monobactam, these drugs share an identical bulky oxyimino R1 side-chain connected the β-lactam ring.
Materials and Methods
Mutagenesis and protein purification
Genes for the mature proteolyzed form of OXA-23 and OXA-24/40 have previously been cloned into pET24a.7, 13 Mutations (P225S or P225S/K82D in OXA-23 and P227S or P227S/V130D in OXA-24/40) were introduced using the polymerase chain reaction (PCR) overlap extension method,14 and all substitutions were confirmed by Sanger dideoxy sequencing. Introduction of the mutated plasmids into BL21(DE3) Escherichia coli cells, culture growth and protein expression were as previously described.13 Harvested cell pellets from 1–2 L of growth were frozen overnight at −20°C, and then resuspended on ice in 20 ml 50 mM NaH2PO4, 1 mM EDTA pH 7.0 supplemented with 100 µl Halt Protease Inhibitor Cocktail (ThermoScientific). The cells were lysed with the addition of 1 mg/ml lysozyme (Sigma), and chromosomal DNA was eliminated by the addition of 5 µg/ml DNAse I (Sigma) and 5 mM MgCl2. The lysate was clarified by centrifugation (Sorvall SS34 rotor, 31500 × g, 4°C, 30 minutes), and dialyzed against 4 L 5 mM NaH2PO4, pH 5.8 overnight at 4°C. The retentate was applied to a 1.5 × 3.0 cm column of carboxy-methyl cellulose 23 (Whatman, UK) previously equilibrated with dialysis buffer. After washing with dialysis buffer, the protein was eluted with a 150 ml linear gradient from 5 mM NaH2PO4, pH 5.8 to 50 mM NaH2PO4, pH 7.0. Purified protein samples (> 95% by Coomassie staining) for kinetics were snap frozen in liquid nitrogen. Protein for crystallization was concentrated to 10–20 mg/ml using a Centricon centrifugal ultrafiltration device (10 kDa molecular weight cutoff; Amicon), and used fresh. The concentration of all protein samples was determined by measuring the absorbance at 280 nm, and using an extinction coefficient calculated by the method of Gill and von Hippel.15
Steady state kinetic analysis
Kinetic parameters were determined by the addition of aliquots of purified enzyme preparations to various concentrations of substrate in 50 mM NaH2PO4, 25 mM NaHCO3, pH 7.4 at 25°C. Changes in absorbance as a function of time were converted to velocity (µM/s) using the following Δε values (M−1 cm−1): ampicillin, −900 (λ=235 nm); doripenem, −11460 (λ=297 nm); imipenem, −9000 (λ=300 nm); cefotaxime, −7500 (λ=260 nm); ceftriaxone, −7800 (λ=255 nm); ceftazidime, −6900 (λ=260 nm); aztreonam, −700 (λ=297 nm). Initial rates from at least three trials were averaged and plotted as a function of substrate concentration. Km and Kcat values were determined from non-linear regression of the data to the Michaelis-Menten-Henri equation using the Microsoft Excel SDAS module. For Km values that were too low to be measured accurately by this method, the substrate was used as a competitive inhibitor with ampicillin as a reporter substrate. In this case, Ks values (instead of km values) were calculated from the concentration of substrate that generated 50% of uninhibited velocity (IC50) using the Cheng-Prusoff equation.16 For Km values that were very high (> 500 µM), cuvettes with shorter pathlengths were used (0.2 mm or 2 mm).
Crystallization
Crystals of OXA-160 V130D were prepared by hanging drop vapor diffusion using well buffer composed of 0.1 M HEPES sodium, 2% v/v polyethylene glycol 400, 2.0 M ammonium sulfate, pH 7.5. Purified protein (3.0 mg/ml) was combined with well buffer in a ratio of 3:1 (total drop volume 8 µl), and crystals formed within 1–2 days at room temperature.17 Crystals of OXA-225 K82D were grown starting with 14.0 mg/ml protein using a well buffer composed of 100 mM NaH2PO4, 100 mM citric acid, 200 mM NaCl, 20% PEG 8000, pH 4.2 with a ratio of 1:1 for protein to well buffer (total drop volume 8 µl). OXA-160 V130D crystals were soaked in either 25 mM ceftazidime (Sigma) or 25 mM aztreonam (Sigma) in well buffer with 5% sucrose added as a cryoprotectant for ~ 60 minutes and flash-cooled in liquid nitrogen. Crystals of OXA-225 K82D were soaked in 25 mM ceftazidime in well buffer (no additional cryoprotectant) for 40 minutes before flash-cooling.
Structure determination
Diffraction data for all three crystals was collected at LS-CAT at the Advanced Photon Source (Argonne, IL) at 100 K using a MarCCD detector. Reflections were indexed, integrated and scaled using autoPROC.18 The OXA-160 V130D structures were determined initially by molecular replacement with Phaser19 using the model of OXA-24/40 K84D (PDB 3PAE with all ligand and water molecules removed) as the initial phasing model. OXA-225 K82D was determined similarly using the model of OXA-23 (PDB 4K0X) for molecular replacement. Refinement and electron density map calculations were carried out with REFMAC520 in the CCP4 program suite.21 Manual rebuilding of the model was accomplished with Coot.22
Thermal denaturation and stability
For thermal denaturation, protein samples were monitored for helical content by circular dichroism (CD) in the far-UV region at 210 and 220 nm. CD experiments were done with a Jasco J-815 spectrometer (Easton, MD) with a Peltier-effect temperature controller. Quartz cells with a 0.1 cm path-length and 200 µl volume (Hellma) were used for all experiments. The behavior was identical at the two different wavelengths, suggesting a two-state thermodynamic event. The enzyme concentrations were 10 µM in 50 mM NaH2PO4, 25 mM NaHCO3 pH 7.4. Thermal melting was performed between 20–70°C with a heating rate of 2°C/min. The thermodynamic stability of OXA enzymes and variants was determined from equilibrium unfolding curves. Raw equilibrium denaturation data were normalized to the fraction of denatured protein (fU). With the assumption of a reversible two-state transition (N ↔ U), equilibrium constants (Keq) at any given temperature were calculated from equation 1:
| equation 1 |
Melting temperature (Tm) and Van’t Hoff enthalpy (ΔHVH) for the thermal denaturation were determined from the free energy using equation 2 and equation 3.
| equation 2 |
| equation 3 |
From the Van’t Hoff plot (lnKeq vs. 1000/T), ΔHVH determined as the slope × (−R), and ΔS was the intercept × R. With the assumption that the variation in enthalpy and entropy are temperature independent, we determined the melting temperature (Tm) at the midpoint of equilibrium folding.
Molecular Dynamics simulations
OXA-24/40 and its P227S variant (OXA-160) were constructed for two parallel simulations using the published OXA-24/40 structure (PDB entry 3PAE), MMTSB23 and CHARMM24 script libraries, and VMD package.25 For the mutant protein, P227 was replaced with serine using an MMTSB mutate script. OXA enzymes possess a characteristic PASTFK family motif (79–84 in OXA-24/40) that contains both the catalytic serine residue and the lysine which is modified by carboxylation to form the general base.26 Force-field parameters for the carboxylated lysine are provided in the Supplementary Material. Both proteins were then solvated with TIP3P27 water molecules with the box dimensions of 90Å × 70Å × 65Å. The CHARMM27 protein force field24 and the CHARMM version 35b2 simulation package were used. Both systems were geometry optimized using the Steepest Descent algorithm for 500 steps, and then using the Adopted Basis Newton-Raphson (ABNR) method until the gradient threshold of 0.01 kcal/mol/Å was achieved. During the minimization and MD simulations, periodic boundary conditions were applied and electrostatic interactions were calculated with the Particle Mesh Ewald method.28 A switching function was applied to calculate the van der Waals interactions with a non-bonded cutoff of 10Å. The SHAKE algorithm29 was used to constrain the bond lengths involving hydrogens. The systems were heated gradually from 10K to 300K for 30 ps (time-step 1 fs) and then simulated for 40 ps under NPT (constant number of atoms, constant pressure and temperature) to equilibrate the water molecules. A harmonic constraint of 5 kcal/mol/Å2 for all of the protein’s heavy atoms was applied throughout these steps. The systems were then further equilibrated for 1 ns in the NVT ensemble (constant number of atoms, constant volume and temperature) with harmonic constraints removed and a time-step of 2 fs. Each system was simulated for a total of 39.4 ns. To calculate electrostatic interactions, we used the Particle Mesh Ewald method and a non-bonded cutoff of 10Å with a switching function to calculate van der Waals interactions. The Cα root mean squared deviation (RMSD) profiles for wild-type and P227S are shown in Supplemental Figure S1. The conformational clusters were determined based on the Cα RMSD of the β5-β6 loop with respect to the initial frame. VMD was used to analyze the trajectories and to create images.
Results
Clinical A. baumannii strains have been the source of many β-lactamase variants that are members of the OXA-23 and OXA-24/40 subfamilies, including several that represent single residue substitutions of the parental sequences. Figure 2 shows an alignment of two such variant enzymes (OXA-160 and OXA-225) with OXA-23 and OXA-24/40. The alignment stresses that OXA-160 and OXA-225 each arose from an identical P→S substitution in the OXA-24/40 and OXA-23 backgrounds respectively (P227S in OXA-24/40 and P225S in OXA-23). A number of lines of evidence suggested to us that these substitutions could be affecting the activity of these enzymes, perhaps as positive adaptations in response to antibiotic therapies. Firstly, these two identical mutations arose in two enzymes that share only 59% identity. This makes it unlikely that the mutation arose in some common ancestor, and rather suggests a case of convergent evolution as seen many times in the class A β-lactamases (for example, the G238S substitution that has appeared in both the TEM-1 and SHV-1 families).30 Secondly, in both OXA-23 and OXA-24/40 the proline in question is found in a loop that borders the active site and has been implicated in the modulation of both substrate selectivity and catalytic activity for diverse classes of antibiotics.10, 12, 13, 17 De Luca et al noted the presence of this proline in a sequence motif typically associated with class D carbapenemases.10 Lastly, the study that identified OXA-160 (i.e. OXA-24/40 P227S) from an A. baumannii strain recovered from a transplanted lung noted that the strain producing this enzyme had elevated levels of resistance to aztreonam compared to strains containing OXA-24/40.31
Figure 2. Alignment of OXA-23, OXA-24/40 and two clinical variants containing a single P→S substitution.
The protein sequences for OXA-24/40, OXA-160, OXA-23 and OXA-225 were aligned using ClustalW. Identical residues between the two subfamilies are indicated by vertical hashes, and the site of the P→S substitution is highlighted in red (P227S in OXA-24/40; P225S in OXA-23). The site of proteolytic processing to generate the mature secreted protein is shown by a vertical green line.
To test this hypothesis, we engineered these clinical variants by introducing P227S into OXA-24/40 and P225S into OXA-23, purified the proteins to homogeneity and analyzed their substrate specificity profiles using steady-state kinetic analysis (Table 1). In both the OXA-24/40 and OXA-23 backgrounds, the P→S substitution resulted in gains of overall enzyme hydrolysis efficiency against a wide variety of substrates as measured by kcat/Km. For ampicillin, the gains are modest (~3-fold for OXA-160 and ~2-fold for OXA-225). For other substrates, the effects are much more dramatic. Of particular note is that while OXA-24/40 and OXA-23 show no activity against the extended spectrum cephalosporin ceftazidime, OXA-160 and OXA-225 both register activities against that drug (though with high Km values). A similar gain of function is observed for OXA-160 and OXA-225 with regard to the monobactam aztreonam, with the latter registering a > 500-fold increase in kcat/Km for that drug compared to OXA-23. In the case of activity against cefotaxime and ceftriaxone, the kcat/Km values increase to levels (ranging from 0.0025 – 0.110 µM−1·s−1) that are commensurate with those observed for other class D β-lactamases that confer clinical resistance levels against advanced generation cephalosporins (e.g. OXA-14, OXA-163).12, 32 In summary, for all non-carbapenem substrates tested with both the parent enzyme and variant, the P→S substitution led to a reduced Km. For carbapenems, the P→S mutation yields only slightly altered levels of activity. For instance, the conversion of OXA-24/40 to OXA-160 leads to a 40% increase in kcat/Km for doripenem, but a 10% decrease against imipenem. One possible interpretation of these results is that the mutation causes a general increase in the affinity for a broad array of non-carbapenem β-lactam substrates without any major loss of activity toward carbapenems. We should note that the use of Km as an indication of binding affinity in this way depends on the assumption that the acylation rate is much slower than the substrate dissociation rate, which has not yet been demonstrated for these β-lactamases. Even so, the mutations lead to increased turnover rates against many substrates, in some cases when the “parental” enzymes yield no detectable activity at all. Whether through reduced Km values, increased turnover rates, or both effects together, the P→S mutations have the potential to yield higher rates of antibiotic hydrolysis, particularly at low drug concentrations where increased affinity will be a major advantage.
Table 1.
Steady-state kinetic analysis
| Km or Ks (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) | |
|---|---|---|---|
| OXA-23a | |||
| Ampicillin | 82 ± 9 | 460 ± 10 | 5.7 ± 0.6 |
| Imipenem | 0.20 ± 0.02b | 0.49 ± 0.01 | 2.2 ± 0.2 |
| Doripenem | 0.018 ± 0.002b | 0.028 ± 0.003 | 1.6 ± 0.3 |
| Cefotaxime | 340 ± 30 | 5.5 ± 0.1 | 0.016 ± 0.002 |
| Ceftriaxone | 5.4 ± 0.5 | 0.021 ± 0.001 | 0.0039 ± 0.0004 |
| Ceftazidime | NA | <0.02 | NA |
| Aztreonam | 2,400 ± 100 | 0.24 ± 0.01 | 0.00010 ± 0.00001 |
| OXA-24/40a | |||
| Ampicillin | 180 ± 20 | 480 ± 20 | 2.6 ± 0.3 |
| Imipenem | 0.68 ± 0.09b | 2.1 ± 0.1 | 3.1 ± 0.4 |
| Doripenem | 0.024 ± 0.003b | 0.074 ± 0.001 | 3.1 ± 0.4 |
| Cefotaxime | 750 ± 70 | 0.38 ± 0.01 | 0.00051 ± 0.00005 |
| Ceftriaxone | 110 ± 10 | 0.035 ± 0.001 | 0.00031 ± 0.00004 |
| Ceftazidime | NA | <0.02 | NA |
| Aztreonam | >1,500 | >0.3 | ------ |
| OXA-225 | |||
| Ampicillin | 12 ± 1 | 150 ± 10 | 12 ± 1 |
| Imipenem | 0.20 ± 0.02b | 0.39 ± 0.01 | 1.9 ± 0.2 |
| Doripenem | 0.027 ± 0.005b | 0.026 ± 0.002 | 1.0 ± 0.2 |
| Cefotaxime | 100 ± 20 | 3.2 ± 0.2 | 0.031 ± 0.006 |
| Ceftriaxone | 0.48 ± 0.05b | 0.051 ± 0.002 | 0.11 ± 0.01 |
| Ceftazidime | 1,400 ± 200 | 1.4 ± 0.1 | 0.0010 ± 0.0002 |
| Aztreonam | 13 ± 2 | 0.79 ± 0.03 | 0.061 ± 0.01 |
| OXA-160 | |||
| Ampicillin | 13 ± 1 | 93 ± 2 | 7.5 ± 0.9 |
| Imipenem | 0.043 ± 0.005b | 0.12 ± 0.01 | 2.8 ± 0.4 |
| Doripenem | 0.014 ± 0.003b | 0.064 ± 0.001 | 4.4 ± 1 |
| Cefotaxime | 170 ± 5 | 0.41 ± 0.01 | 0.0025 ± 0.0008 |
| Ceftriaxone | 14 ± 0.8 | 0.24 ± 0.01 | 0.017 ± 0.001 |
| Ceftazidime | >3,000 | >0.80 | ------ |
| Aztreonam | 170 ± 40 | 2.7 ± 0.2 | 0.016 ± 0.004 |
OXA-24/40 and OXA-23 wild-type values were previously reported in Kaitany et al.13
KS values determined by competition kinetics with ampicillin as a reporter substrate.
Residue P227 (P225 in OXA-23) is located in the β5-β6 loop (residues 220–228 in OXA-24/40)*, which as noted before, borders the active site and has been shown to affect the binding of all classes of β-lactam drugs.10, 12, 13, 17 The proline, however, resides near the point where the loop reenters the enzyme core as strand β6, and is thus unlikely to make any direct contacts with substrates. This, and the fact that proline’s unique structural properties would be altered upon conversion to serine, suggested to us that the mutation may lead to extensive structural rearrangements of the β5-β6 loop and thus affect substrate binding indirectly. To explore this hypothesis, we sought to determine the crystal structures of OXA-225 and OXA-160 with relevant ligands bound. We were able to crystallize these variants under the same conditions that had been successful with the parent enzymes OXA-24/4017 and OXA-23.13 Although these crystals yielded protein structures at high resolution, all attempts to capture ligands bound as acylintermediates failed. We then introduced mutations that are known to diminish or eliminate deacylation of substrates,33, 34 and have been used successfully in the past to capture acyl-intermediates in class D enzymes.7, 17 We used site-directed mutagenesis to introduce a V130D substitution into the blaOXA-160 gene and used it to express and purify the variant enzyme. The protein was then crystallized using hanging drop vapor diffusion, and the crystals that were produced were identical in size and shape to OXA-24/40 crystals prepared in the same manner.7, 17 After soaking these crystals with various ligands and collecting X-ray diffraction data, we were able to determine the structure of OXA-160 V130D with ceftazidime (2.28 Å; PDB 4X55) and aztreonam (2.30 Å; PDB 4X53) bound in the active site (Table 2). Similarly, we introduced a K82D substitution into blaOXA-225. After expression and purification, we crystallized the enzyme and again, the crystals matched those produced previously for OXA-23.13 Thus neither the P→S mutation or the deacylation-deficiency substitutions affected our ability to produce diffraction-quality crystals in either protein. For OXA-225 K82D, we soaked ceftazidime into the crystals and produced a structure with that drug bound in the active site (1.94 Å; PDB 4X55). The quality of the final models was analyzed with MolProbity.35 For OXA-160 V130D bound to aztreonam, 97% of residues were in the favored region with no Ramachandran outliers, and for OXA-160 V130D with ceftazidime bound 98% of residues were in the favored region with one outlier (V225). For OXA-225 K82D, 95% of the residues were in the favored region, with three outliers of the Ramachandran plot (L224, S257, A58).
Table 2.
Structure Parameters
| OXA-23 P225S K82D ceftazidime (PDB 4X55) |
OXA-24 P227S V130D aztreonam (PDB 4X53) |
OXA-24 P227S V130D ceftazidime (PDB 4X56) |
|
|---|---|---|---|
| cell constants (Å;0) | a = 98.78, b = 143.86, c= 44.18, α=β=γ=90 |
a = b = 102.5 c= 87.2 α=β=γ=90 |
a = b = 102.3 c= 87.2 α=β=γ=90 |
| space group | P 21 21 2 | P 41 21 2 | P 41 21 2 |
| resolution (Å) | 1.94 (1.99 – 1.94)a |
2.30 (2.30 – 2.36)a |
2.28 (2.34–2.28)a |
| No. of unique reflections | 47536 | 21202 | 20839 |
| total no. of reflections | 342979 | 169442 | 174691 |
| Rmerge (%) | 7.0 (87.2) | 7.7 (61.4) | 7.7 (62.7) |
| completeness (%)b | 100.0 (100.0)b | 100.0 (100.0)b | 95.7 (97.6) b |
| (I)/(σI) a | 15.2 (2.4) a | 21.0 (3.5) a | 21.9 (3.5) a |
| resolution range for refinement (Å) | 30.0 – 1.94 | 72.4 – 2.30 | 75.0 – 2.28 |
| No. of protein residues | 475 | 243 | 244 |
| no. of water molecules | 193 | 96 | 112 |
| rmsd for bond lengths (Å) | 0.013 | 0.015 | 0.016 |
| rmsd for bond angles (0) | 1.66 | 1.95 | 1.85 |
| R-factor (%) | 0.214 | 0.192 | 0.181 |
| Rfree (%)c | 0.261 | 0.228 | 0.213 |
| average B-factor (Å2), protein atoms | 43.5 (monomer A) 45.0 (monomer B) |
36.4 | 36.2 |
| average B-factor (Å2), ligand atoms | 59.1 | 56.1 | 48.9 |
| average B-factor (Å2), water molecules | 44.3 | 31.2 | 38.0 |
Values in parentheses are for the highest resolution shell.
Fraction of theoretically possible reflections observed.
Rfree was calculated with 5% of reflections set aside randomly.
Ligand Structures
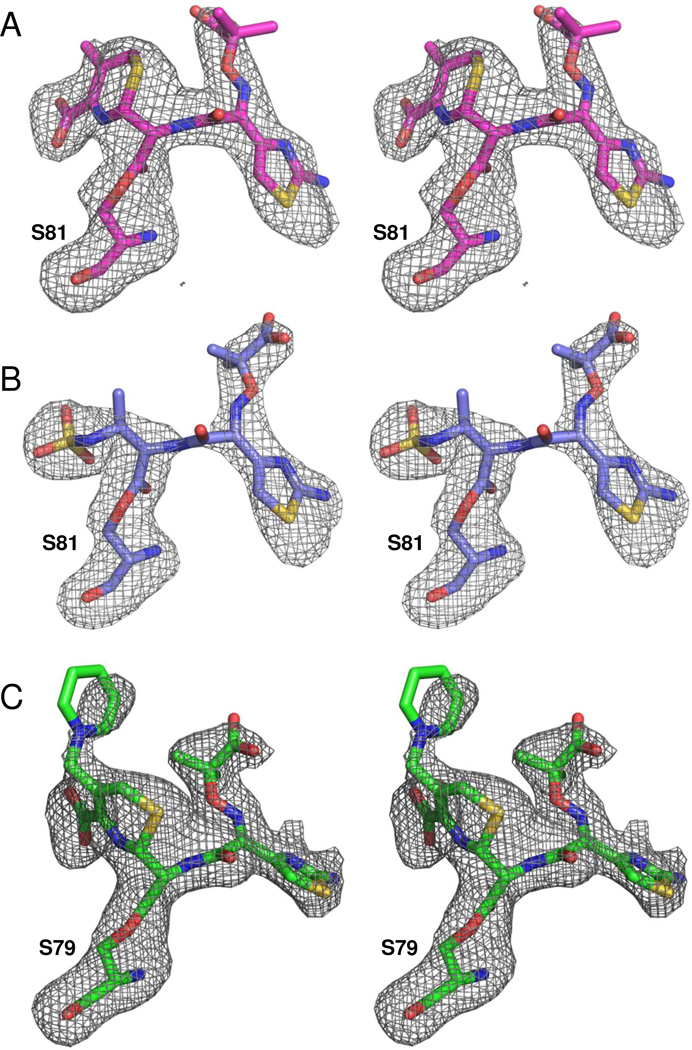
Figure 3 shows that for both OXA-160 structures, significant Fo-Fc electron density (contoured at 3.0 σ) is present in the active site (Figure 3a and b). In each case, the density extends from the γ-hydroxyl oxygen atom of S81 to the drug’s β-carbonyl carbon, indicating that the ligands are bound as acyl-intermediates. We were able to successfully model in the full drug structures in each case, with the exception of the R2 leaving group for ceftazidime. The R2 leaving group is expected to be eliminated as part of the molecular rearrangement that occurs upon breaking of the β-lactam bond (Supplemental Figure S2). For both structures, the carbonyl oxygen of the acyl moiety is bound in the oxyanion hole formed by the main-chain amide nitrogens of S81 (2.8 Å in OXA-160/aztreonam) and W221 (2.9 Å). The dihydrothiazine ring of ceftazidime is anchored to the active site by interactions between the C2 carboxylate group and R261, S219, K217 and S128, much as has been observed for doripenem and oxacillin.7, 17 In the case of aztreonam, the sulfate group that mimics the carboxylate occupies the analogous space, with an additional salt bridge to K218 (3.6 Å). An alignment of the aztreonam-acylated structures of OXA-160 (PDB: 4X53, this study), Toho-1 (PDB: 2ZQC) and the β-lactamase from Citrobacter freundii (PDB: 1FR6) shows the sulfate group aligns very closely, though the stabilizing interactions differ (data not shown).36, 37
Figure 3. Ligand electron density.
Fo-Fc omit maps (contoured to 3.0 σ) were calculated to show the structures of the ligand connected as an acyl-intermediate to the nucleophilic serine (S81 in OXA-160 or S79 in OXA-225). (A) OXA-160 V130D bound to ceftazidime (magenta; PDB 4X56). (B) OXA-160 V130D bound to aztreonam (blue; PDB 4X53). (C) OXA-225 K82D bound to ceftazidime (green; PDB 4X55). Figures were made with Pymol.55
Initial Fo-Fc electron density maps (contoured at 3.0 σ) of OXA-225 K82D also indicated that ceftazidime is bound as an acyl intermediate, with continuous electron density from the side-chain of S79 to the β-carbonyl of the broken lactam ring (Figure 3c). As in OXA-160, the C2 carboxylate of ceftazidime forms interactions with R259 and T217, but the rotation of the drug prevents an ionic bond to K216. Another major difference between ceftazidime bound to OXA-160 and OXA-225 involves the R2 side-chain pyridine. In the OXA-160 structure, ceftazidime has undergone the molecular rearrangement that results in the release of the pyridine, as observed in all other β-lactamase/ceftazidime complexes determined to date (e.g. 1IEL, 2ZQD, 1XKZ).38, 39 In the OXA-225 structure, the pyridine ring is still attached to the C3 exocyclic carbon of ceftazidime at 80% occupancy suggesting that the post-acylation rearrangement has not occurred for all molecules in the crystal lattice. It is likely that this difference simply reflects a serendipitous ligand soaking time, but we cannot rule out the possibility that there is some difference in the two active sites that results in a pause at this step in OXA-225.
Aztreonam/ceftazidime structure in OXA-160
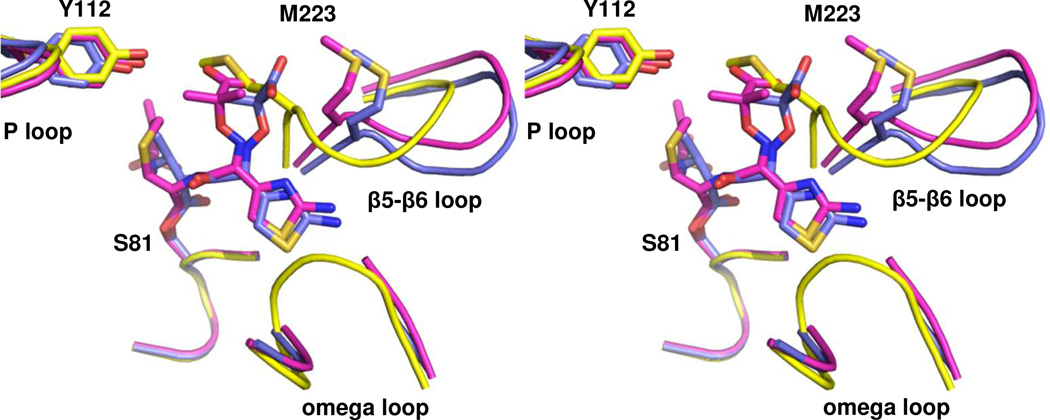
Aztreonam and ceftazidime share an identical bulky Y-shaped R1 side-chain with a thiazole ring on one branch, and a carboxypropyl-modified oxyimino group on the other (Figure 1). Not surprisingly, this group adopts a similar conformation for both drugs bound to OXA-160 V130D, as shown in an alignment of the two structures (Figure 4). Most notably, the thiazole ring occupies a narrow pocket between the omega loop and the end of β-strand β5 (in the area of G222/M223). In both cases, there is a loss of electron density for the main-chain and side-chain of L168, indicating that the thiazole ring can only be accommodated upon rearrangement of the omega loop in that region. The carboxypropyl group of both ligands occupies a similar space near the end of strand β5 (also near G222, but on the other side of the strand compared to the thiazole ring). For both drugs, the space occupied by the carboxypropyl group would clash with the position typically occupied by the main-chain and side-chain of M223, but conformational changes in that area of the protein relieve the potential clash.
Figure 4. Comparison of acyl-enzyme intermediates in the active site of OXA-160.
Stereoview of the structures of OXA-160 V130D with ceftazidime (magenta; PDB 4X56) and aztreonam (blue; PDB 4X53) bound. To generate the structural alignment, key active site residues from OXA-160 V130D/ceftazidime (S81, S128, K218 and G220) were superposed with the same residues of OXA-24/40 K84D (yellow; PDB 3PAE) using the align function of PyMOL generating a relative mean square deviation (RMSD) of 0.174 Å. This process was repeated for OXA-160 V130D/aztreonam and OXA-24/40 K84D (RMSD, 0.321 Å).
Ceftazidime structure in OXA-225
Because OXA-225 contains the same P→S mutation as OXA-160, our structure of the deacylation-deficient mutant OXA-225 K82D bound to ceftazidime gave us an opportunity to look for common elements in the binding mode of that drug to class D carbapenemases. Additionally, we have previously shown that OXA-23 binds substrates possessing bulky R1 side-chains with modestly higher affinity than OXA-24/40,13 and we note that the same trend holds for OXA-225 (from the OXA-23 subfamily) compared to OXA-160 (from the OXA-24/40 subfamily) (Table 1). We therefore sought a molecular explanation for this difference by comparing the two P→S variants with ceftazidime bound.
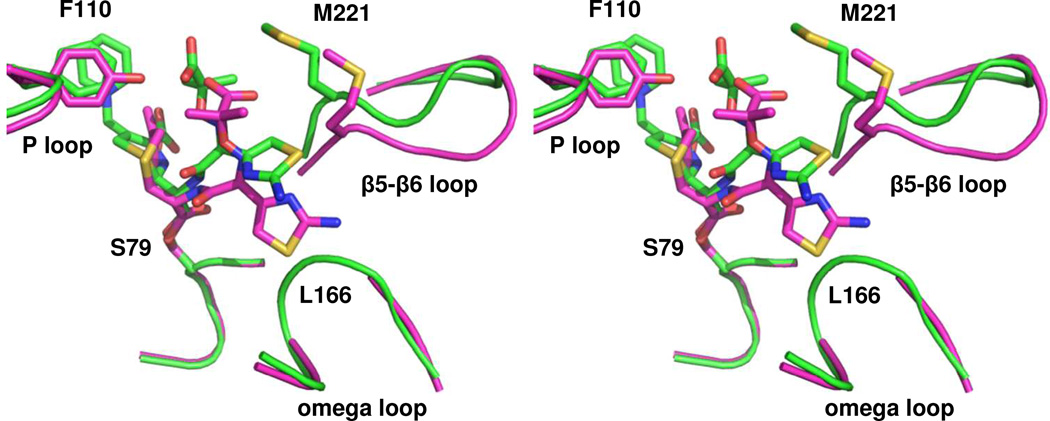
An overlay of the structure of OXA-160 V130D and OXA-225 K82D bound to ceftazidime (Figure 5) shows that while the drug binds in a roughly similar manner, there is one main difference between the ceftazidime conformation in the two variant enzymes. First, while the thiazole ring binds in the same orientation projecting toward L168, the entire R1 side-chain is rotated approximately 15° away from that residue and the rest of the omega loop. Unlike the structure of OXA-160/ceftazidime, there is significant electron density for both the main-chain and side-chain of L168 in OXA-225/ceftazidime. This suggests that the rotation of the R1 side-chain relieves the steric clash that was present in the former, and thus explains the generally tighter binding of substrates with bulky R1 side-chains for OXA-23 family members. We sought to identify which structural features of OXA-225 are responsible for the rotation of ceftazidime’s side-chain. For one thing, the main-chain and side-chain of M221 are moved back nearly 2 Å in OXA-225 compared to M223 in OXA-160. As we proposed in an earlier study, this is likely the result of the presence of two OXA-160 glycines (G222/G224) occupying positions where residues with bulkier side-chains are found in OXA-225 (A220/D222).13 The β carbons of the latter push the main-chain of the β5-β6 loop back from the omega loop and create a larger binding pocket. Indeed, a recent report showed that a G222V substitution in the OXA-24/40 subfamily member OXA-207 leads to increased activity against oxacillin, a penicillin with a bulky side-chain.40 The overlay shown in Figure 5 suggests another surprising possibility: ceftazidime is unable to rotate away from L168 in OXA-24/40 because its carboxypropyl moiety would approach too closely to the hydroxyl group of Y112, one of the so-called “bridge” residues that caps the active site. OXA-225 has a phenylalanine at this position, and thus allows ceftazidime to rotate further away from the omega loop.
Figure 5. Comparison of the ceftazidime-bound structures of OXA-225 and OXA-160.
A stereoview of the aligned structures of OXA-225 K82D (green; PDB 4X55) and OXA-160 V130D with ceftazidime bound as an acyl-enzyme intermediate (magenta; PDB 4X56). In OXA-225, ceftazidime is able to rotate higher in the active site, relieving steric clash with the omega loop in the area of L166. Alignment was carried out as described in Figure 4.
β5-β6 loop structure
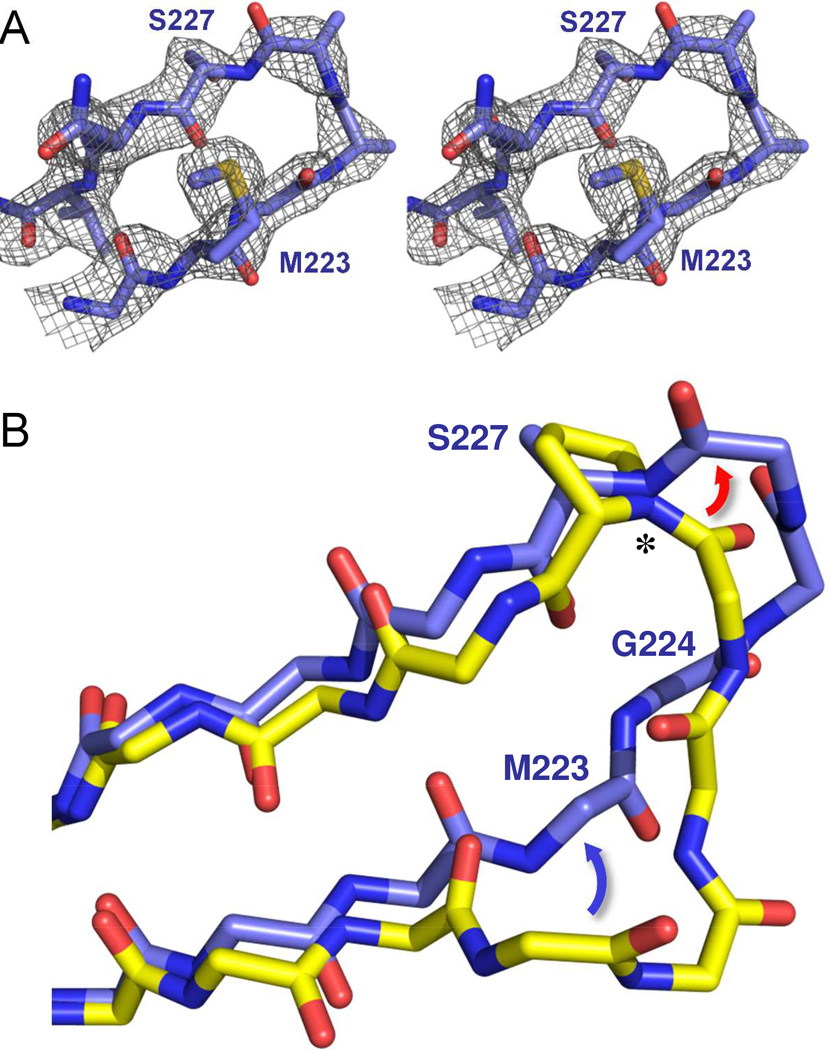
In order to understand how the P→S mutation affects ligand binding and overall protein structure, we compared the two OXA-160 structures with the structure of OXA-24/40 K84D/doripenem (PDB 3PAE)7 and OXA-225 with apo OXA-23 WT (PDB 4K0X).13 For all three structures, the largest difference with the parental enzymes occurs in the loop that contains the P→S mutation itself (residues G222-G230 in OXA-160; A220-G228 in OXA-225). In all three P→S variant structures, the loop contains relatively high B-factors, and shows evidence of multiple conformations. Still, a predominant main-chain trace is visible, as observed for the Fo-Fc loop omit map of the OXA-160 V130D/aztreonam structure (Figure 6a).
Figure 6. Structure of the β5-β6 loop in OXA-160.
(A) An Fo-Fc omit map of the β5-β6 loop (residues 222–229) contoured at 3.0 σ (PDB 4X53). (B) Effect of the P→S mutation on the trajectory of the β5-β6 loop. OXA-24/40 K84D/doripenem (3PAE, yellow) was aligned with OXA-160 V130D/aztreonam (blue; PDB 4X53) as described in Figure 4. Only the main-chain atoms are shown for clarity, with the exception of P/S227. The largest difference between OXA-24/40 and OXA-160 is in the region of G222-T226, with the area of bridge residue M223 moving away from the active site (blue arrow) and the region of T226 extending further from the surface of the enzyme (red arrow) to form a turn. The deviation is accompanied by a large change in the φ torsion angle (*) for residue 227.
In the doripenem-bound structure of OXA-24/40, the β5-β6 loop turns inward toward the active site starting at residue 222 (yellow structure; Figure 4 and Figure 6b). In the two OXA-160 structures however, the antiparallel interactions of strand β5 and β6 are extended further from the core of the protein, and are connected by a Type I β-turn at residues 224–227.41 The difference between OXA-24/40 and OXA-160 is most striking between residues 223–226, with maximal Cα displacement occurring at G224 (6.3 Å between Cα carbons). Notably, this deviation moves the main-chain and side-chain of M223 away from the area occupied by the carboxypropyl group of both aztreonam and ceftazidime. We observe essentially the same trajectory difference when comparing the same loop in OXA-225/ceftazidime and OXA-23 (not shown), although alternate conformations make it difficult to assign the turn in OXA-225 as Type I. The deviation of the loop therefore provides a simple explanation for the tighter affinity of these drugs for OXA-160 (and OXA-225) compared to OXA-24/40 (and OXA-23). The enlargement of the active site in this area is also consistent with a similar phenomenon caused by a duplicated alanine (A220/A221) in OXA-146 (PDB 4K0W), a variant that also displays strong gain-of-function binding toward bulky cephalosporins and aztreonam.13
A detailed comparison of the structure of the mutated residue (proline or serine 227) in OXA-24/40 and OXA-160 provides a simple explanation for the adaptive alteration of the β5-β6 loop’s trajectory. Figure 6b shows that, if one traces backwards through the loop (toward M223), it is precisely at the main-chain nitrogen of residue 227 (marked by an *) that the deviation is first seen. The Ramachandran φ angle (describing the torsional rotation around the bond between this nitrogen and its α carbon) is markedly different between the two structures. With its fixed ring side-chain, the proline of OXA-24/40 has a φ angle of −72°, a value that lies in the narrow acceptable band for that residue. The serine present in OXA-160 has a φ angle of −141°, a value that lies in the acceptable region for serine, but not for proline. Additionally, the fixed φ angle of proline makes it almost completely incompatible with the fourth position of a Type I β-turn, while serine is often found at that position.41 Therefore, the structural change in the area of M223 that is required for the accommodation of bulky substrates (Figure 6b, blue arrow) likely requires the movement of residues 225 and 226 (red arrow) in a manner that is simply not compatible with the torsional restrictions of proline at position 227.
Thermodynamic stability
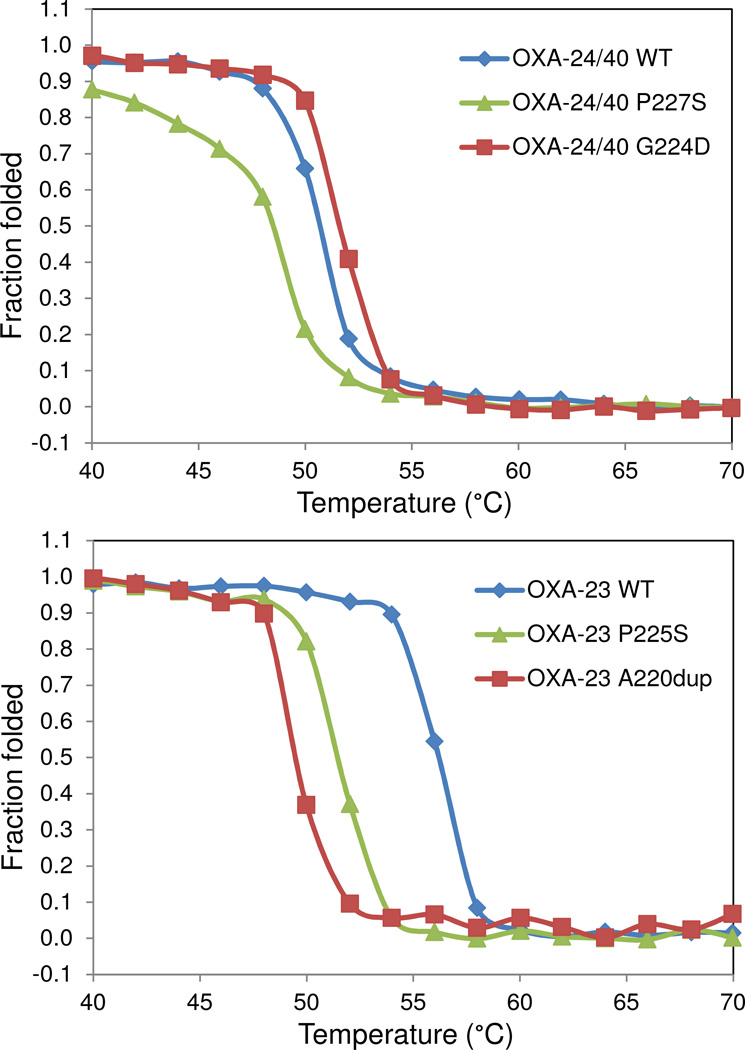
It has been observed that the advantage conferred by gain-of-function mutations in β-lactamases may be accompanied by loss of thermodynamic stability in the overall protein structure.42 In order to determine if the P→S substitutions had this effect, we used circular dichroism to determine the melting temperature of OXA-160 and OXA-225 as well as the two “parental” enzymes OXA-24/40 and OXA-23. Because the carboxylation of K84 (K82) is thought to affect protein stability, we carried out these experiments in phosphate buffer alone, or phosphate with the addition of 25 mM NaHCO3. As Figure 7 and Table 3 show, the P→S substitution in OXA-24/40 and OXA-23 did indeed lead to a destabilization of both as seen in 3–5°C lower melting temperatures for OXA-160 and OXA-225 in phosphate buffer. This destabilization effect was also observed in the presence of bicarbonate (data not shown), though as predicted, enhancing K84 carboxylation leads to overall higher melting temperatures. To further explore this result, we tested two other clinically-observed CHDL variants with substitutions in the β5-β6 loop. OXA-146 is the name given to OXA-23 with an alanine duplication (A220dup) in the β5-β6 loop, and like OXA-160 and OXA-225, this variant possesses greatly improved binding of extended spectrum cephalosporins and aztreonam.13 OXA-72 is a wide-spread clinical variant formed from a different mutation (G224D) in the same loop in OXA-24/40. Interestingly, the alanine duplication in OXA-146 leads to an even larger destabilization of OXA-23 (~ 8°C) than the P→S substitution (~ 5°C). Conversely, the G224D mutation leads to an increase in melting temperature of + 3°C, suggesting this mutant may stabilize the loop.
Figure 7. Thermal denaturation curves for OXA-23, OXA-24/40 and several of their variants.
Unfolding was monitored by slowly raising the temperature of each protein solution and monitoring by circular dichroism. The P→S variant lowered the melting temperature in both the OXA-24/40 background (top panel) and the OXA-23 background (lower panel).
Table 3.
Thermal Denaturation Parameters
| Enzyme | Tm (°C) | ΔHVH (kcal·mol−1) | ΔTm (°C)a | ΔΔG (kcal·mol−1) |
|---|---|---|---|---|
| OXA-23 | 56 ± 0.2 | 177.80 ± 44.27 | - | - |
| OXA-23 P225S (OXA-225) | 51 ± 0.1 | 155.43 ± 45.10 | − 5 ± 0.1 | − 2.70 ± 0.14b |
| OXA-23 A220dup (OXA-146) | 48 ± 0.2 | 130.37 ± 47.30 | − 8 ± 0.2 | − 4.32 ± 0.19 |
| OXA-24/40 | 49 ± 0.2 | 137.02 ± 27.58 | - | - |
| OXA-24/40 P227S (OXA-160) | 46 ± 0.2 | 99.28 ± 24.21 | − 3 ± 0.2 | − 1.27 ± 0.08 |
| OXA-24/40 G224D (OXA-72) | 52 ± 0.2 | 159.82 ± 33.54 | + 3 ± 0.2 | + 1.27 ± 0.09 |
ΔTm = Tvariant-TWT are relative to wild-type
Errors are based on Shellman equation, ΔΔG = ΔTm·ΔSWT
ΔSWTOXA-23 = 0.54 cal·mol−1·K
ΔSWTOXA-24/40 = 0.425 cal·mol−1·K
Molecular Dynamics Simulations of OXA-24/40 and OXA-160
Molecular Dynamics (MD) simulations were used to compare local and global dynamics of OXA-24/40 and OXA-160 (OXA-24/40 P227S), and to identify differences in the interactions between specific residues resulting from the P→S mutation. Substitution of proline by serine may be expected to introduce not only increased flexibility of the backbone at the end of the β5-β6 loop but also new hydrogen bond interactions, both of which may affect the loop conformation and its interactions with neighboring residues. The analysis focused on the most stable portion of the trajectory, the last 22 ns (see the root mean square deviation profiles in Supplementary Figure S1). The Cα root mean squared fluctuations (RMSF) difference between OXA-24/40 P227S (OXA-160) and OXA-24/40 reveal differences in the mean residue fluctuations of different segments of the two enzymes (Supplementary Figure S3a and b). The results indicate that the P→S mutation impacts protein dynamics not only in the direct vicinity of the mutation site but also in other more distal regions of the protein. We observe reduced fluctuations in the catalytic motif STFK (81–84), but increased fluctuations in S128-V130 and the omega loop (most notably N165, W167-V169). Fluctuations in the P loop (residues 98–119) 43 are also higher in OXA-160 compared to OXA-24/40, although a longer time-scale simulation would be required to fully sample the fluctuations of this extensive structural element. Other regions of higher fluctuations are the β6-β7 loop (residues 237–244), and the β7-α11 loop (residues 255–260). Fluctuations of R261 are slightly increased.
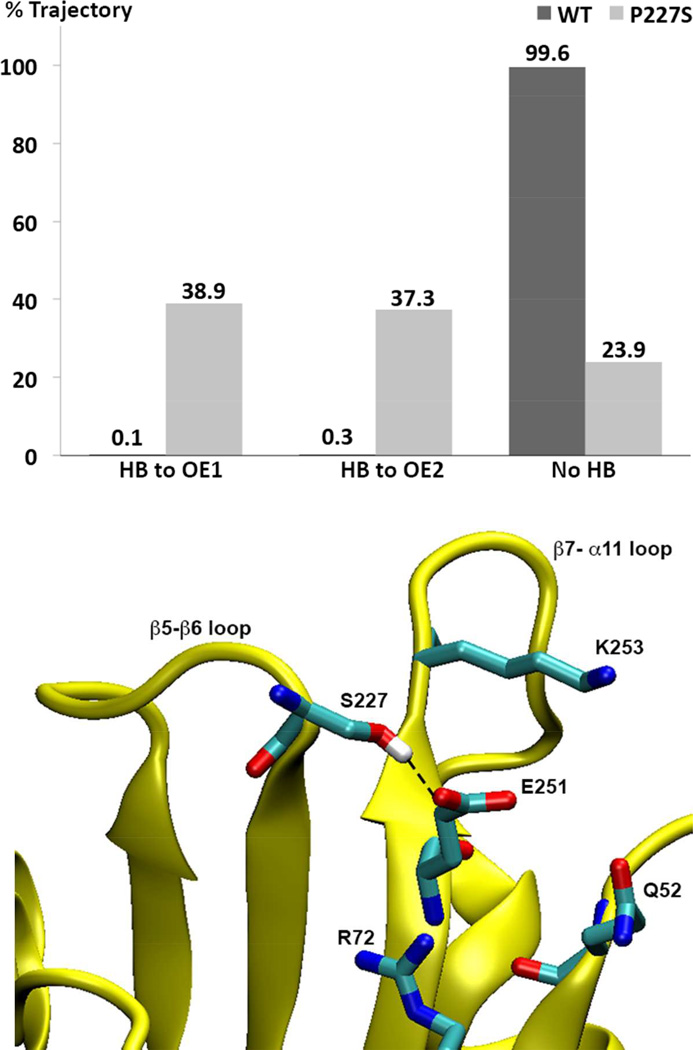
During the MD simulations of OXA-24/40, the side-chain of S227 is hydrogen bonded to either of the Oε atoms of E251 for 76% of the total productive trajectory time (see histogram in Figure 8). In contrast, the wild-type equivalent P227 Cγ atom and Oε atoms of E251 reside within 3.2 Å of each other for only 0.4% of the simulation time (the trajectory average is 4.8 Å). Further analysis of the hydrogen bonding interactions with E251 in the OXA-24/40 simulation shows that this residue interacts with K253 (33% of time), Q52 (14%) and R72 (29%). In the P→S mutant, these interactions are significantly less frequent (6%, 7%, and 3%, respectively) due to the hydrogen bond forming between E251 and S227. Thus, this new hydrogen bond affects the conformational equilibrium of the β5-β6 loop and perhaps other tertiary contacts in this region. Interestingly, the X-ray structure of OXA-160 V130D/aztreonam suggests that the side-chain hydroxyl of S227 is close enough to E251 on strand β7 to form a hydrogen bond.
Figure 8. Prevalence of hydrogen bonding between S227 and E251 in OXA-160 and OXA-24/40.
Upper panel: For OXA-160 (OXA-24/40 P227S), the hydroxyl oxygen atom of S227 remains within 3.2 Å distance from E251 Oε1 and Oε2 atoms for 38.9% and 37.3% of the time, respectively. In contrast, the corresponding atoms in P227 (Cγ) and E251 in the WT enzyme remain greater than 3.2 Å apart 99.6% of the time. Hydrogen bonding was determined over the 38 ns after equilibration based on the following geometric criteria: D-A < 3.2 Å, H-A < 2.2 Å and DHA > 120°. When hydrogen bonded, the average D-A distance in OXA-160 is 2.7 Å (with an entire trajectory average of 3.8 Å). Lower Panel: A trajectory snapshot of a conformation that displays a typical hydrogen bond between S227 and E251.
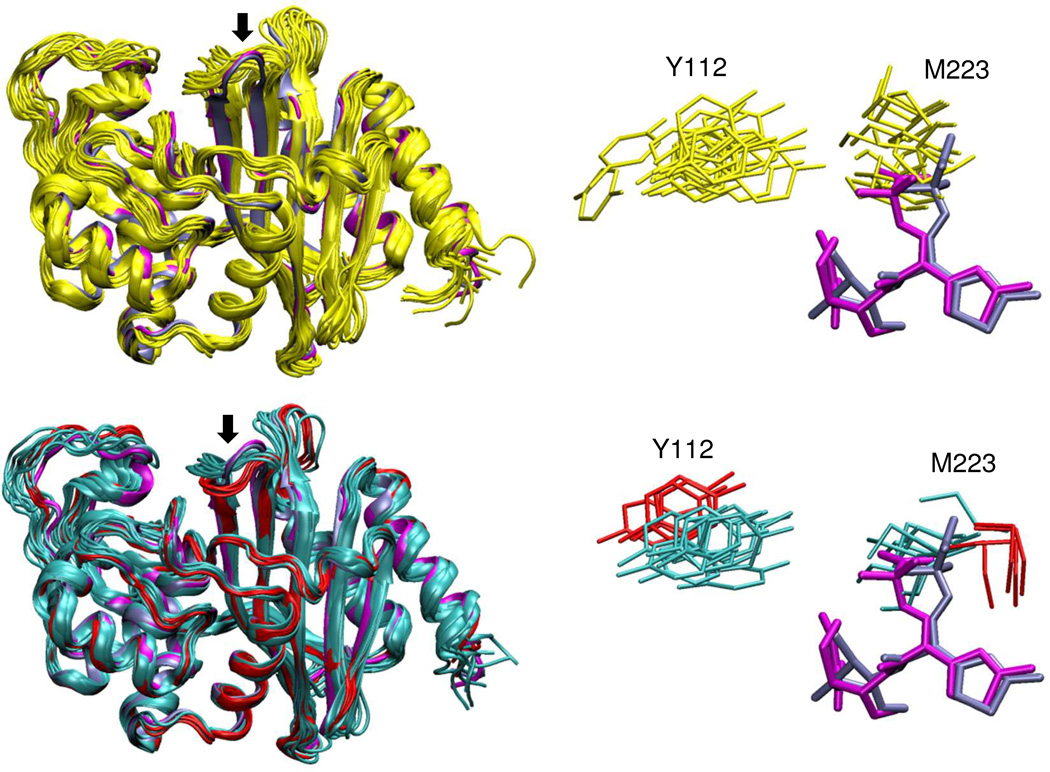
In light of the lower RMSFs of the β5-β6 loop in OXA-160, the conformational diversity of the loop in OXA-24/40 versus OXA-160 was examined. The β5-β6 loop in OXA-160 occupies two main conformational clusters (shown as cyan and red in Figure 9, lower panel). Together, these two clusters span an RMSD range of 0.3Å to 3.1Å with respect to the initial frame, which is much less diverse than that observed in OXA-24/40 (yellow; RMSD range 0.3Å to 4.6Å). As a result of this conformational redistribution in the β5-β6 loop, changes in the M223-Y112 bridge are observed (Figure 9; right panel). The bridge is much wider in the majority of OXA-160 conformers than in OXA-24/40, with average distances between Y112 and M223 increasing 14.7→16.6 Å (Cα-Cα), 5.5→7.4 Å (OH-Cε) and 5.7→7.6 Å (OH-Sδ). This widening could facilitate diffusion of the drug into the pocket and thus enhance binding. After superposing the ligands from the current OXA-160 X-ray structures (magenta, ceftazidime; blue, aztreonam) with the MD β5-β6 loop conformers, we observe steric clashes between M223 and the carboxypropyl-modified oxyimino group of ceftazidime in both proteins, but much less in OXA-160 compared to OXA-24/40 (Figure 9; right panel). Interestingly, both the widening of the gap between the bridge residues and the decrease in steric clashes with ceftazidime are more conspicuous in one of the OXA-160 β5-β6 loop clusters (Figure 9, right panel, red clusters). This cluster, which adopts a conformation closer to that observed in the two OXA-160 X-ray structures, may be the strongest contributor to the increased binding affinity suggested by our kinetic studies.
Figure 9. Comparison of conformational diversity in OXA-160 and OXA-24/40.
Left panels: representative conformers observed in molecular dynamics simulations for OXA-24/40 (yellow) and OXA-160 (two clusters shown in cyan and red). The two OXA-160 structures from this study (PDB 4X53 and 4X56, blue and magenta respectively) are included in the alignment (ligands not shown). The β5-β6 loop is marked with an arrow in both structures. Right panels: Side-chains for the bridge residues Y112 and M223 from the same representative structures. Also shown are ceftazidime (magenta) and aztreonam (blue) from the two OXA-160 X-ray structures after alignment of those structures with the simulation conformers.
We also expected that our trajectories would shed light on the role of omega loop movement and the degree to which L168 can accommodate the thiazole ring (L168 density was not observed in the X-ray structure after the ligand was soaked in). It appears, however, that a steric clash between the ligand and L168 would be present in both proteins. The omega loop in the OXA-24/40 conformers appears further away from β5-β6 loop than in OXA-160 but it shows increased fluctuations in the latter. Such increased fluctuations could promote loop widening when the ligand is present, a hypothesis that could be tested by future simulations with the ligand bound.
Discussion
The structures we report here are the first examples of class D β-lactamases with either an advanced generation cephalosporin or aztreonam bound, and they begin to clarify the answer to a fundamental question: how do enzymes of this class achieve ESBL status? The key lies in the two different areas of steric clash between the enzyme and the oxyimino side-chain that must be overcome in order to have productive binding of these bulky drugs. First, there is a strong incompatibility between the thiazole ring of the drug and the enzyme’s omega loop, particularly in the area of W167/L168. As first noted by Paetzel et al.,44 most of the ESBL variants in the OXA-10 subfamily have mutations either in or near this loop. Indeed, mutation of the tryptophan or leucine has been shown to generate enzymes that confer strong ceftazidime resistance on Pseudomonas aeruginosa.45–47 Three other common OXA-10 ESBL mutations either contact the tryptophan’s side-chain (A124T, N73S/T) or reside near it on the omega loop (G157D). Thus, one reasonable hypothesis is that these mutations destabilize the omega loop and reduce its steric clash with the drug. A similar mechanism has been observed in the class C enzyme P99, where steric clashes with the thiazole ring of ceftazidime can be relieved by a tripeptide insertion in the omega loop (creating the ESBL variant GC1).38, 48 One disadvantage of this mode of achieving ESBL status in class D enzymes however, is that mutations that perturb the omega loop in the area of W167 reduce the carboxylation of K84 through disruption of the hydrogen bond between the tryptophan’s side-chain and the carbamate general base of KCX84.49, 50 Thus gains in affinity for bulky cephalosporins in these cases may be partially offset by reduction of the catalytic turnover rate.
Alternatively, as we show here, ESBL status can be achieved by reducing the hindrance between the carboxypropyloxyimino portion of ceftazidime (or aztreonam) and the β5-β6 loop, particularly in the area of the bridge methionine. The duplication of A220 in OXA-23 to create OXA-146, for instance, moves the side-chain of M221 away from the area expected to be occupied by ceftazidime.13 As with the P→S mutations reported in this study, the duplication in OXA-146 also pushes back the main-chain of that loop. The deletion of four residues of the β5-β6 loop in OXA-48 (creating OXA-163) greatly enhances ceftazidime hydrolysis, but also reduces carbapenem turnover.12 The identification of the β5-β6 loop as a hot-spot for cephalosporinase activity may help in the discovery of other clinical mutants that display higher hydrolytic activity against cephalosporins and monobactams. OXA-72, for instance, is a wide-spread OXA-24/40 subfamily member with a G224D mutation in this loop. OXA-239 is an OXA-23 variant with three mutations: the P225S substitution reported here and two others (S109L/D222N), both of which contact the hydrophobic bridge residues. The OXA-51 subfamily of A. baumannii chromosomal carbapenemases (about which very little is known biochemically and structurally), includes a variant (OXA-173) with a substitution (P226L) at the same residue that is explored in this study.51
The fact that both OXA-160 and OXA-225 possess strong activity against cephalosporins and aztreonam suggests that the P→S mutation arose as a selective adaptation upon treatment with one or more of these drugs. OXA-160 was derived from an A. baumannii strain that was highly resistant to carbapenems, cephalosporins and aztreonam (MIC values, µg/ml: meropenem, 16; cefepime, 64; aztreonam 128).31 The strain that yielded OXA-225 was also highly resistant to the same classes of drugs (MIC values, µg/ml: meropenem, >16; ceftriaxone, 64; ceftazidime >32; personal communication, Dr. Paul Higgins). While cephalosporins and aztreonam are not typically used in the United States to treat A. baumannii infections, it is very possible that these strains could have been exposed to them during the treatment of another infection or during prophylactic treatment. For instance, the A. baumannii strain that yielded OXA-160 came from a transplant donor lung that also yielded Pseudomonas aeruginosa (personal communication, Dr. Yohei Doi), a species for which aztreonam, cefepime and ceftazidime are commonly used as treatment.
The fact that these class D β-lactamases can achieve broad spectrum activity against all classes of β-lactam antibiotics has implications for future infection control efforts. First, horizontal transfer of these genes could impart cephalosporin and/or aztreonam resistance on species for which these drugs remain a viable treatment (Pseudomonas aeruginosa, Acinetobacter spp and other non-fermentors). Second, efforts are underway to develop new monobactams (e.g. BAL30072), cephalosporins (e.g. S-649266) and cephalosporin combination therapies (e.g. ceftazidime/avibactam) for the treatment of A. baumannii and other Gram-negative microbes.52–54 The knowledge of how class D carbapenemases are evolving to broaden their substrate specificity may help direct those efforts.
Supplementary Material
Footnotes
Funding
This research was supported by National Institutes of Health grant 1R15AI082416 (D.A.L.) and R15AI094489 (R.A.P). Additional support was also provided by the Cleveland Department of Veterans Affairs, the Department of Veterans Affairs Merit Review Program 1I01BX001974, the Veterans Integrated Service Network 10 Geriatric Research, Education, and Clinical Center (VISN 10 GRECC), and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01 AI100560 and R01 AI063517 (R.A.B). This work was also supported by the National Center for Multiscale Modeling of Biological Systems (MMBioS) through National Institutes of Health grant P41GM103712 (T.W.). Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
For labeling of β strands and loops, we are following the nomenclature used by De Luca et al. for the related class D carbapenemase OXA-48. [De Luca, F., Benvenuti, M., Carboni, F., Pozzi, C., Rossolini, G. M., Mangani, S., and Docquier, J. D. (2011) Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design, Proc. Natl. Acad. Sci. U.S.A. 108, 18424–18429.]
Supporting Information
Root mean square deviation (RMSD) for the molecular dynamics simulation (Figure S1), post-acylation rearrangement of ceftazidime (Figure S2), Cα root mean squared fluctuations (RMSF) difference between OXA-24/40 and OXA-160 enzymes (Figure S3) and CHARMM force-field parameters for carboxylated lysine. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kamolvit W, Derrington P, Paterson DL, Sidjabat HE. A case of IMP-4, OXA-421, OXA-96 and CARB-2-producing Acinetobacter pittii ST119 in Australia. J. Clin. Microbiol. 2014 doi: 10.1128/JCM.02726-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans BA, Hamouda A, Amyes SG. The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 2013;19:223–238. [PubMed] [Google Scholar]
- 3.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun T, Nukaga M, Mayama K, Braswell EH, Knox JR. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5354–5359. doi: 10.1073/pnas.0607557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem. Biol. 2013;20:1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J. Mol. Biol. 2011;406:583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. Crystal structure of the OXA- 48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Smith CA, Antunes NT, Toth M, Vakulenko SB. The Crystal Structure of the Carbapenemase OXA-58 from Acinetobacter baumannii. Antimicrob. Agents and Chemother. 2014;58:2135–2143. doi: 10.1128/AAC.01983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Luca F, Benvenuti M, Carboni F, Pozzi C, Rossolini GM, Mangani S, Docquier JD. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18424–18429. doi: 10.1073/pnas.1110530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Naas T, Nordmann P. Diversity, Epidemiology, and Genetics of Class D β-lactamases. Antimicrob. Agents and Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Castanheira M, Carrër A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents and Chemother. 2011;55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, Leonard DA. Structures of the class D Carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins, and aztreonam. Antimicrob. Agents and Chemother. 2013;57:4848–4855. doi: 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 17.June CM, Vallier BC, Bonomo RA, Leonard DA, Powers RA. The structural origins of oxacillinase specificity in Class D β-lactamases. Antimicrob. Agents and Chemother. 2013;58:333–341. doi: 10.1128/AAC.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Feig M, Karanicolas J, Brooks CL. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graphics Modell. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 26.Leonard DA, Bonomo RA, Powers RA. Class D β-Lactamases: A Reappraisal after Five Decades. Acc. Chem. Res. 2013;46:2407–2415. doi: 10.1021/ar300327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 28.Darden T, York D, Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 29.Ryckaert J-P, Ciccotti G, Berendsen HJ. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 30.Hujer AM, Hujer KM, Bonomo RA. Mutagenesis of amino acid residues in the SHV-1 β-lactamase: the premier role of Gly238Ser in penicillin and cephalosporin resistance. Biochim. Biophys. Acta. 2001;1547:37–50. doi: 10.1016/s0167-4838(01)00164-9. [DOI] [PubMed] [Google Scholar]
- 31.Tian GB, Adams-Haduch JM, Bogdanovich T, Pasculle AW, Quinn JP, Wang HN, Doi Y. Identification of diverse OXA-40 group carbapenemases, including a novel variant, OXA-160, from Acinetobacter baumannii in Pennsylvania. Antimicrob. Agents and Chemother. 2010;55:429–432. doi: 10.1128/AAC.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danel F, Frère JM, Livermore DM. Evidence of dimerisation among class D β-lactamases: kinetics of OXA-14 β-lactamase. Biochim. Biophys. Acta. 2001;1546:132–142. doi: 10.1016/s0167-4838(01)00133-9. [DOI] [PubMed] [Google Scholar]
- 33.Schneider KD, Bethel CR, Distler AM, Hujer AM, Bonomo RA, Leonard DA. Mutation of the active site carboxy-lysine (K70) of OXA-1 β-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48:6136–6145. doi: 10.1021/bi900448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vercheval L, Bauvois C, di Paolo A, Borel F, Ferrer JL, Sauvage E, Matagne A, Frère JM, Charlier P, Galleni M, Kerff F. Three factors that modulate the activity of class D β-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem. J. 2010;432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- 35.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oefner C, D’Arcy A, Daly JJ, Gubernator K, Charnas RL, Heinze I, Hubschwerlen C, Winkler FK. Refined crystal structure of β-lactamase from Citrobacter freundii indicates a mechanism for β-lactam hydrolysis. Nature. 1990;343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- 37.Shimamura T, Ibuka A, Fushinobu S, Wakagi T, Ishiguro M, Ishii Y, Matsuzawa H. Acyl-intermediate structures of the extended-spectrum class A β-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J. Biol. Chem. 2002;277:46601–46608. doi: 10.1074/jbc.M207884200. [DOI] [PubMed] [Google Scholar]
- 38.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry. 2001;40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 39.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated β-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J. Am. Chem. Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 40.Cayô R, Merino M, Ruiz Del Castillo B, Cano ME, Calvo J, Bou G, Martínez-Martínez L. OXA-207, a novel OXA-24 variant with reduced catalytic efficiency against the carbapenems in Acinetobacter pittii from Spain. Antimicrob. Agents and Chemother. 2014;58:4944–4948. doi: 10.1128/AAC.02633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchinson EG, Thornton JM. A revised set of potentials for β-turn formation in proteins. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 43.Szarecka A, Lesnock KR, Ramirez-Mondragon CA, Nicholas HB, Wymore T. The Class D β-lactamase family: residues governing the maintenance and diversity of function. Protein Eng., Des. Sel. 2011;24:801–809. doi: 10.1093/protein/gzr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 45.Danel F, Hall LM, Livermore DM. Laboratory mutants of OXA-10 β-lactamase giving ceftazidime resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 1999;43:339–344. doi: 10.1093/jac/43.3.339. [DOI] [PubMed] [Google Scholar]
- 46.Fournier D, Hocquet D, Dehecq B, Cholley P, Plésiat P. Detection of a new extended-spectrum oxacillinase in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2010;65:364–365. doi: 10.1093/jac/dkp438. [DOI] [PubMed] [Google Scholar]
- 47.Hocquet D, Colomb M, Dehecq B, Belmonte O, Courvalin P, Plésiat P, Meziane-Cherif D. Ceftazidime-hydrolysing β-lactamase OXA-145 with impaired hydrolysis of penicillins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2011;66:1745–1750. doi: 10.1093/jac/dkr187. [DOI] [PubMed] [Google Scholar]
- 48.Crichlow GV, Kuzin AP, Nukaga M, Mayama K, Sawai T, Knox JR. Structure of the extended-spectrum class C β-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry. 1999;38:10256–10261. doi: 10.1021/bi9908787. [DOI] [PubMed] [Google Scholar]
- 49.Leonard DA, Hujer AM, Smith BA, Schneider KD, Bethel CR, Hujer KM, Bonomo RA. The role of OXA-1 β-lactamase Asp(66) in the stabilization of the active-site carbamate group and in substrate turnover. Biochem. J. 2008;410:455–462. doi: 10.1042/BJ20070573. [DOI] [PubMed] [Google Scholar]
- 50.Baurin S, Vercheval L, Bouillenne F, Falzone C, Brans A, Jacquamet L, Ferrer JL, Sauvage E, Dehareng D, Frère JM, Charlier P, Galleni M, Kerff F. Critical role of tryptophan 154 for the activity and stability of class D β-lactamases. Biochemistry. 2009;48:11252–11263. doi: 10.1021/bi901548c. [DOI] [PubMed] [Google Scholar]
- 51.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. Emergence and Distribution of Plasmids Bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents and Chemother. 2010;54:4575–4581. doi: 10.1128/AAC.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mushtaq S, Woodford N, Hope R, Adkin R, Livermore DM. Activity of BAL30072 alone or combined with β-lactamase inhibitors or with meropenem against carbapenem-resistant Enterobacteriaceae and non-fermenters. J. Antimicrob. Chemother. 2013;68:1601–1608. doi: 10.1093/jac/dkt050. [DOI] [PubMed] [Google Scholar]
- 53.Zhanel GG, Lawson CD, Adam H, Schweizer F, Zelenitsky S, Lagacé-Wiens PR, Denisuik A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, Karlowsky JA. Ceftazidime-avibactam: a novel cephalosporin/β-lactamase inhibitor combination. Drugs. 2013;73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]
- 54.Qin W, Panunzio M, Biondi S. β-Lactam Antibiotics Renaissance. Antibiotics. 2014;3:193–215. doi: 10.3390/antibiotics3020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrödinger . The PyMOL Molecular Graphics System, Version 1.3. 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.