Abstract
Functional primordial proteins presumably originated from random sequences, but it is not known how frequently functional, or even folded, proteins occur in collections of random sequences. Here we have used in vitro selection of messenger RNA displayed proteins, in which each protein is covalently linked through its carboxy terminus to the 3′ end of its encoding mRNA1, to sample a large number of distinct random sequences. Starting from a library of 6 × 1012 proteins each containing 80 contiguous random amino acids, we selected functional proteins by enriching for those that bind to ATP. This selection yielded four new ATP-binding proteins that appear to be unrelated to each other or to anything found in the current databases of biological proteins. The frequency of occurrence of functional proteins in random-sequence libraries appears to be similar to that observed for equivalent RNA libraries2,3.
The frequency of occurrence of functional proteins in collections of random sequences is an important constraint on models of the evolution of biological proteins. Here we have experimentally determined this frequency by isolating proteins with a specific function from a large random-sequence library of known size. We selected for proteins that could bind a small molecule target with high affinity and specificity as a way of identifying amino-acid sequences that could form a three-dimensional folded state with a well-defined binding site and therefore exhibit an arbitrary specific function. ATP was chosen as the target for binding to allow comparison with known biological ATP-binding motifs and also with previous selections using random-sequence RNA libraries2,3.
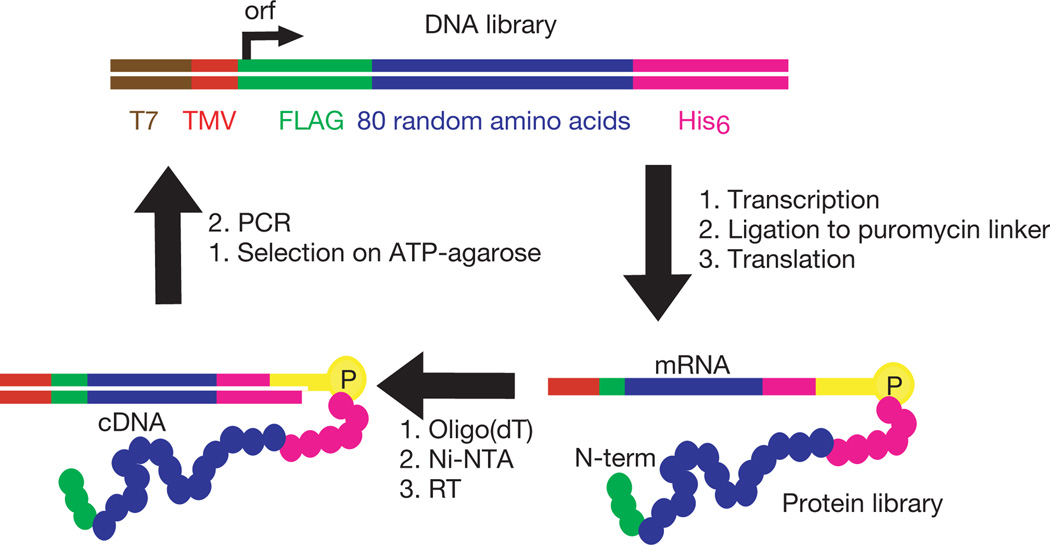
Because protein sequences with specific functions are expected to be quite rare in protein sequence space, we prepared a DNA library of 4 × 1014 independently generated random sequences. This DNA library was specifically constructed to avoid stop codons and frameshift mutations4, and was designed for use in mRNA display1 selections. This DNA library was then used to generate 6 × 1012 purified non-redundant random proteins that were used as the input into the first selection step. These proteins contain a contiguous stretch of random amino acids 80 residues in length, long enough to form known protein domains. Unlike other libraries that have been used in protein selections, this random region is not part of a larger structure that would otherwise tend to constrain or bias the conformation of the resulting proteins. This library randomly samples the whole of sequence space, rather than the vicinity of a known protein. The random region of each library member is flanked by short invariant sequences encoding affinity tags for purification (Fig. 1).
Figure 1.
In vitro selection and amplification of mRNA-displayed proteins. The DNA library encodes proteins with 80 contiguous random amino acids4. Transcription, splinted ligation to a 3′-puromycin oligonucleotide, translation, high-salt incubation, purification and reverse transcription (RT) yielded 6 × 1012 independent mRNA-displayed proteins. This library was incubated with an ATP-agarose affinity matrix. Unbound material was washed away with selection buffer at 4 °C, and bound material was collected by incubation with the same buffer containing 5 mM ATP at 4 °C. Eluted fractions were combined, amplified by PCR, and used as the input for the next round of in vitro selection and amplification.
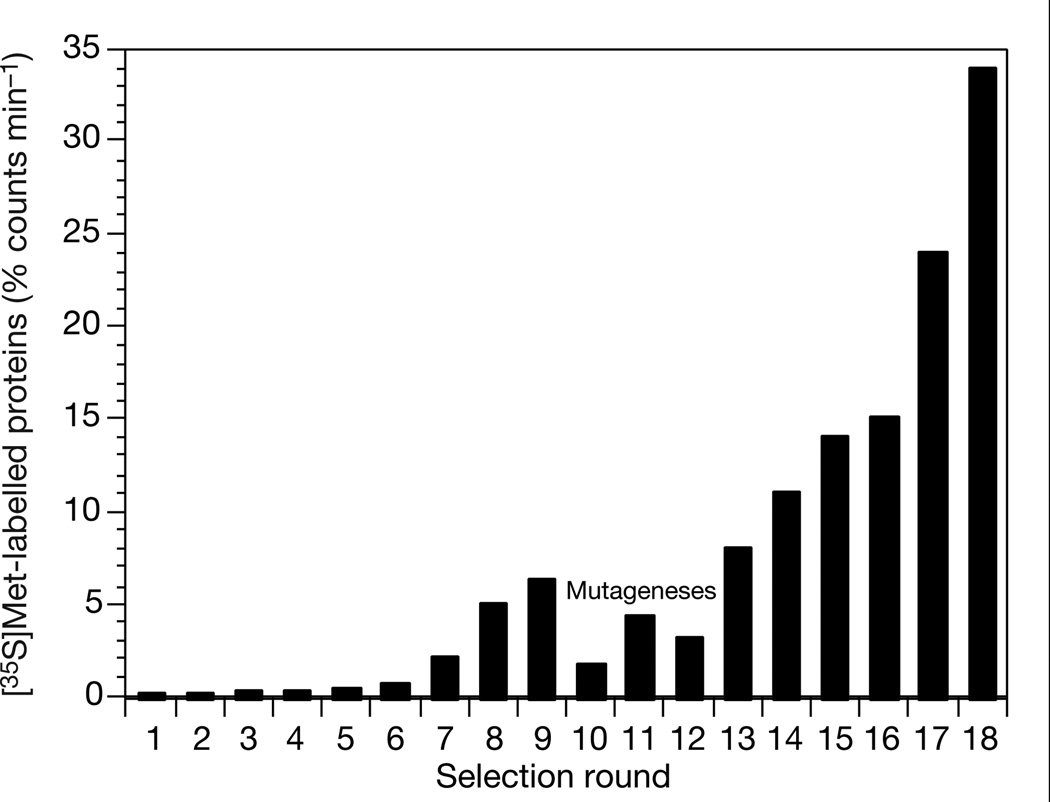
Successive rounds of in vitro selection and amplification were performed starting with this random-sequence library. In each round the mRNA-displayed proteins were incubated with immobilized ATP, washed and eluted with free ATP. The eluted fractions were collected and amplified by polymerase chain reaction (PCR); this DNA was then used to generate a new library of mRNA-displayed proteins, enriched in sequences that bind ATP, for input into the next round of selection (Fig. 1). After eight rounds, the fraction of mRNA-displayed proteins eluting with ATP had risen from 0.1 to 6.2% (Fig. 2). We cloned and sequenced 24 individual library members, which showed that the population was now dominated by 4 families of ATP-binding proteins (Fig. 3a). These families show no sequence relationship to each other or to any known biological protein. The members of each family are closely related, indicating that each family is descended from a single ancestral molecule, which was one of the original random sequences.
Figure 2.
Proportion of the mRNA-displayed protein library bound to immobilized ATP and subsequently eluted with free ATP, as a function of selection round. The inputs into rounds 10–12 were subjected to mutagenic PCR amplification with an average mutagenesis rate of 3.7% at the amino-acid level per amplification.
Figure 3.
Sequences of selected ATP-binding proteins. a, Consensus sequences of the four selected protein families at round 8 (A, B, C, D), before mutagenesis. Flanking constant region residues are indicated by dashes. These protein sequences are unrelated to each other or to any known biological protein. b, The consensus sequence of the single remaining protein family at round 18 (18predom). Four predominating substitutions (>39/56 clones) and sixteen other selectively enriched (>4/56 clones) substitutions are also indicated (18select). Clone 18–19 had the lowest Kd for ATP (100 nM) of those assayed. c, Deletion analysis of clone 18–19 using DNA-tagged proteins5. Purified constructs were incubated with ATP-agarose, washed and eluted with free ATP. The fraction present in the elution phase is shown, normalized with respect to the full-length DNA-tagged protein. Invariant residues are indicated by dashes. d, Deletion analysis of clone 18–19 using MBP fusion proteins. Successive deletion constructs of clone 18–19 were generated as MBP fusion proteins, then cleaved from the MBP with thrombin. Apparent Kds were determined by displacement equilibrium filtration14. Invariant residues are indicated by dashes.
Single representatives of each of these protein families (round 8) were chosen for further study. Only 5–15% of the mRNA-displayed protein prepared from each of these clones binds to immobilized ATP and then elutes with free ATP under selection conditions, consistent with the 6.2% binding and elution with ATP for the library as a whole. One possible explanation for this low level of ATP-binding is conformational heterogeneity, possibly reflecting inefficient folding of these primordial protein sequences.
In an effort to increase the proportion of these proteins that fold into an ATP-binding conformation, we mutagenized the library and carried out further rounds of in vitro selection and amplification. Three consecutive rounds with mutagenic PCR amplification were performed with an average mutagenic rate of 3.7% per amino acid for each round. After six subsequent rounds of in vitro selection and amplification without mutagenesis, the proportion of the library of mRNA-displayed proteins that bound and eluted with ATP rose to 34% (round 18) (Fig. 2). At this point the library was entirely composed (56/56 clones sequenced) of the descendents of one of the four originally selected protein families (family B) (Fig. 3b).
Comparing the round 18 sequences with the ancestral sequence showed that four amino-acid substitutions had become predominant in the selected population (present more than 39 times in 56 sequences, Fig. 3b), and that 16 other substitutions had also been selectively enriched (present more than 4 times in 56 sequences, Fig. 3b). In addition, each clone contained a variable number of other substitutions. The selectively enriched substitutions are distributed over the 62 amino-terminal amino acids of the original 80-amino-acid random region, suggesting that amino acids throughout this region are contributing to the formation of a folded structure, at least in the complex with ATP. The substitutions in each of the assayed clones improve ATP-binding relative to the ancestral sequence.
Eight individual proteins from the final round of in vitro selection (round 18) were chosen randomly for further study. As free proteins, the proportion that bound to the column and then eluted with ATP varied from 5% to 40%. The four proteins that bound and eluted to the greatest extent were expressed in Escherichia coli as C-terminal fusion proteins of maltose binding protein (MBP), as the solubilities of the free proteins were too low to permit full characterization. Purification on an amylose column and subsequently on a Ni-NTA column under denaturing conditions yielded several milligrams of each as highly pure fusion proteins.
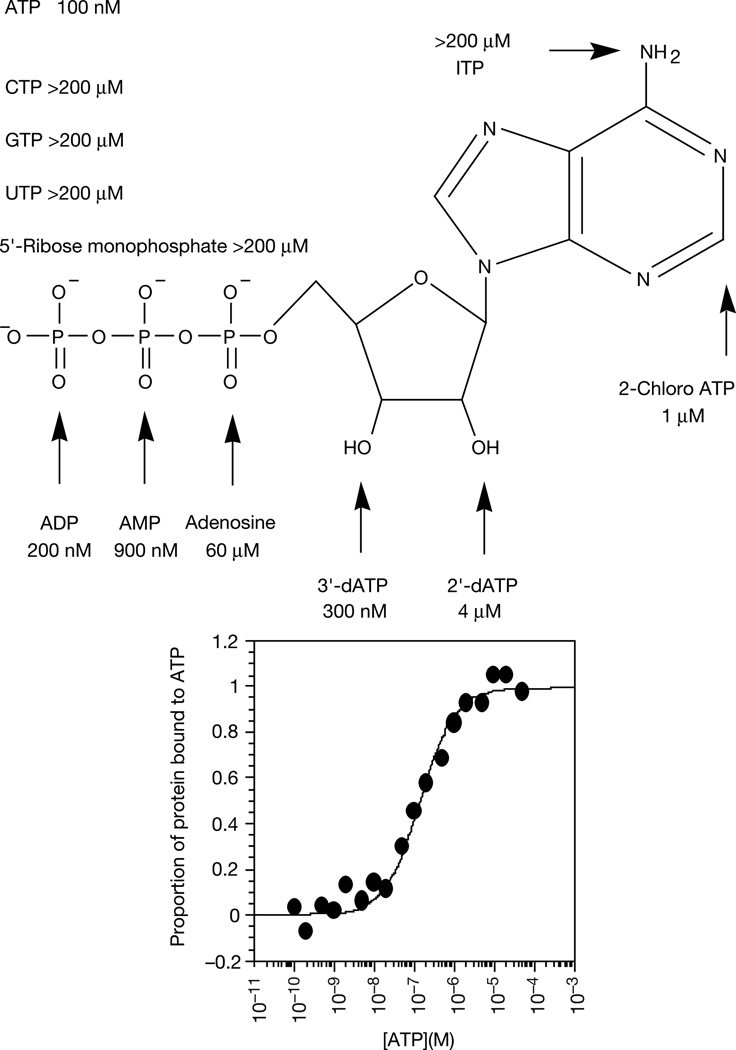
The four MBP fusion proteins have dissociation constants (Kd) for ATP that fall within the range 100 nM to 10 µM at 4°C, as determined by equilibrium dialysis with [α-32P] ATP. Of these, protein 18–19 binds most strongly, with a Kd of 100 nM for ATP at 4 °C and a 1:1 stoichiometry. Gel filtration indicates that 65% of this protein is monomeric in selection binding buffer, with the remainder as high-order aggregates running at the void volume.
Only the monomer binds ATP, and no low-order aggregates such as dimers or tetramers were observed. Competition experiments with ATP analogues show that protein 18–19 interacts with several distinct parts of the ATP molecule (Fig. 4). Removing one, two or three phosphate groups successively raises the Kd, with a particularly large increase accompanying the removal of the a-phosphate (70-fold). Removing the ribose 2′ or 3′ hydroxyl groups reduces binding by factors of 40 and 3, respectively. Binding to ITP (exocyclic amino group changed to carbonyl oxygen and N1 protonated) was undetectable using this assay (a more than 2,000-fold increase in Kd).
Figure 4.
Dissociation constants of protein–ATP (and ATP analogue) complexes as determined by displacement equilibrium filtration14 for protein 18–19 as an MBP fusion protein.
We determined the minimal region of this protein required for ATP-binding by deletion analysis. For this study we used DNA-tagged proteins generated by RNase treatment of mRNA-displayed proteins, leaving the protein attached to the short DNA linker5. For each construct, we measured the fraction of protein that bound to the column and eluted with ATP (Fig. 3c). This analysis defines a core domain of 45 amino acids sufficient for ATP-binding. Using these data, a second series of constructs were generated as MBP fusion proteins and thrombin-released deletion fragment proteins were used to measure Kd values (Fig. 3d). These solution binding experiments reveal a progressive loss of affinity as the deletion extends into the 70 N-terminal amino acids of the random region, from a Kd of 100 nM for the intact protein to 5 mM for the 45-amino-acid core domain. The regions flanking the core domain might stabilize its structure, or might contribute additional distinct interactions with ATP.
These (family B) proteins contain two invariant CXXC sequences (56/56 clones sequenced) within the core domain. Also, protein 18–19 is functional in the presence of 5 mM dithiothreitol, indicating that disulphide bonds are probably not required for ATP binding. In addition, a deletion construct removing only one of these cysteines did not bind ATP (Fig. 3c). These observations suggested that family B proteins might require a coordinated metal ion to be functional. Elemental analysis by atomic absorption spectroscopy revealed the presence of one equivalent of bound zinc, and no other divalent metals. Incubation of the protein with EDTA results in a concentration-dependent loss of ATP-binding activity. Activity can be restored to protein that has been extensively dialysed in the presence of EDTA by the addition of Zn2+, but not Mg2+, ions.
We suggest that the family B proteins bind to ATP with a folded structure nucleated around, or stabilized by, a Zn2+ ion coordinated to the four invariant cysteines of the CXXC sequences. None of the other three protein families selected in this study contains this zinc-binding motif, and no known biological nucleotide-binding domain is a zinc-stabilized structure, although the glucocorticoid receptor contains a similar pair of CXXC sequences and binds to DNA in a zinc-dependent manner6. A NetBLAST7 search of the NCBI protein sequence databases shows that the most closely related known protein sequence to the 45 amino acids of the minimal functional sequence of this protein has only a 33% identity. This is not a significant homology for a sequence this short, and did not include any of the four conserved cysteines. Zinc was not added to the selection buffer, but is present at about 10 µM in mammalian blood8, from which the reticulocyte lysate used for mRNA translation is prepared. This result suggests that metal ion coordination may be one of the simplest ways of generating folded proteins while minimizing the information required to specify a functional sequence.
The frequency with which ATP-binding proteins occur in sequence space can be estimated from the observed recovery of four such proteins from a non-redundant library of 6 × 1012 random sequences. On the basis of the average behaviour of the proteins isolated before mutagenesis (Fig. 2), only about 10% of the potentially functional sequences present in the first round would be expected to generate correctly folded active proteins and thus survive to be amplified. Detailed measurements of the efficiency of each step in the selection and amplification process confirmed that there were no significant material losses that would have affected the recovery of active proteins.
We therefore estimate that roughly 1 in 1011 of all random-sequence proteins have ATP-binding activity comparable to the proteins isolated in this study. This frequency is similar to the recovery of ATP-binding RNAs from random-sequence RNA libraries (with similar Kd values)2,3. This suggests that the greater functional diversity of amino acids as compared with nucleotides does not make functional proteins more common than functional RNAs, at least with regard to binding small molecules. This may be due to the difficulty of forming a buried hydrophobic core that will fold in a consistent manner, in contrast to the ease with which nucleic acids can form compact structures stabilized by complementary base pairing. Although only a small proportion of each of the initially selected proteins folds into the active conformation, this proportion can be increased by directed evolution.
The combination of mRNA display with in vitro selection is a powerful approach to the exploration of protein sequence space. Our isolation of new functional proteins shows that it should be possible to obtain an unbiased view of the inherent diversity of all possible protein structures, and to determine whether biological proteins represent only a small subset of this diversity. Comparing the sequences of our newly evolved ATP-binding proteins with biological ATP-binding proteins has not revealed any significant similarity; structural data will also be required to reveal whether these proteins, especially the Zn2+ metalloprotein, are similar to those of any biological proteins.
In conclusion, we suggest that functional proteins are sufficiently common in protein sequence space (roughly 1 in 1011) that they may be discovered by entirely stochastic means, such as presumably operated when proteins were first used by living organisms. However, this frequency is still low enough to emphasize the magnitude of the problem faced by those attempting de novo protein design.
Methods
Library construction, in vitro selection and amplification
Library construction, and the preparation and purification of mRNA-displayed proteins have been described4,9,10. In vitro translation yielded 7 × 1013 independent random-sequence mRNA-displayed proteins; after purification and reverse transcription 6 × 1012 were recovered for input into the first selection step. Subsequent rounds were carried out at a 2–10-fold reduced scale. For rounds 1–9, we used a butyl-agarose pre-column (Sigma) and incubated the flow through with the ATP-affinity column. Rounds 14–16 included two ATP-agarose selection steps, and rounds 17 and 18 included three ATP-agarose selection steps. For reiterated selection steps the eluted material was purified away from ATP on a denaturing Ni-NTA column and reverse transcribed again before the subsequent selection step. For a more detailed description of the selection protocol and the full sequences of the selected proteins, see Supplementary Information.
Mutagenic PCR amplification
Mutagenic PCR11,12 was used in rounds 10–12, with an average mutagenic rate of 3.7% at the amino-acid level as determined by DNA sequencing. Serial transfer mutagenic PCR was carried out to an average mutagenic extent of 3.7% at the amino-acid level, with aliquots being combined to give a broad range of mutagenic extents.
MBP fusion protein expression
Selected clones were ligated into pIADL14 and modified versions of pIADL14 (pTAG2K), and protein expression was carried out as described13. The proteins were further purified on a Ni-NTA column under denaturing conditions.
Kd determinations
We made Kd measurements by equilibrium dialysis or spin-filtration14, with equivalent results, using purified MBP fusion proteins exchanged into selection binding buffer by gel filtration. We determined the Kd for ATP and ATP analogues by displacing bound 32P-ATP from the protein by increasing concentrations of competitor. The stoichiometry of the protein–ATP interaction was determined by a procedure adapted from ref. 15.
Deletion analysis
Deletion analyses were done with both DNA-tagged proteins5 and MBP fusion proteins. Primers designed to anneal to specific internal parts of clone 18–19 were synthesized. For the DNA-tagged proteins, 5′ primers encoded the TMV enhancer and T7 promoter sequences, with MD as the common N terminus, and 3′ primers encoded the peptide MGMSGS as the common C terminus. These primers were used to generate truncated mRNA-displayed proteins from clone 18–19 as described above. Subsequent purification on oligo(dT)cellulose, incubation with RNase A, and a second purification on oligo(dT)cellulose yielded purified DNA-tagged proteins5. The fraction of each DNA-tagged protein that was competent to bind ATP-agarose was determined under selection conditions as described above. MBP fusion proteins were generated in a similar manner by PCR of internal fragments of the protein 18–19 coding sequence, followed by cloning into the MBP fusion expression plasmid, expressing and purifying the MBP fusion deletion constructs, thrombin cleavage to release the deletion fragment from the MBP, and measuring solution Kd by equilibrium dialysis or spin-filtration.
Gel-filtration chromatography
Protein 18–19 was expressed as an MBP fusion as described above. The time taken for the protein to traverse a Sepharose 200 FPLC column in selection binding buffer was measured on an AKTA FPLC by analysis of column fractions on silver-stained SDS–PAGE gels. The molecular weight of the protein was calculated by comparison with known protein standards.
Supplementary Material
Acknowledgements
We thank members of the Szostak laboratory and especially D. Wilson, G. Cho, G. Short, J. Pollard, G. Zimmermann, R. Liu, J. Urbach and R. Larralde-Ridaura for their helpful advice. This work was supported in part by the NASA Astrobiology Institute and the NIH. J.W.S. is an investigator at the Howard Hughes Medical Institute.
Footnotes
Supplementary information is available on Nature's World-Wide Web site (http://www.nature.com) or as paper copy from the London editorial office of Nature.
The DNA sequences encoding the consensus protein sequences of families A, B, C, D, 18predom and clone 18–19 have been deposited in GenBank under accession codes AF306524 to AF306529, respectively.
References
- 1.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl Acad. Sci. USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 4.Cho G, Keefe AD, Liu RL, Wilson DS, Szostak JW. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol. 2000;297:309–319. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DS, Keefe AD, Szostak JW. In vitro selection of high affinity protein-binding peptides using mRNA display. Proc. Natl Acad. Sci. USA. doi: 10.1073/pnas.061028198. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedman LP, et al. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Considine DM, editor. Van Nostrand’s Scientific Encyclopedia. 7th edn. New York: Van Nostrand Reinhold; 1989. p. 3067. [Google Scholar]
- 9.Liu R, Barrick J, Szostak JW, Roberts RW. Optimized synthesis of RNA–protein fusions for in vitro protein selection. Methods Enzymol. 2000;318:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- 10.Keefe AD. In: Current Protocols in Molecular Biology. Ausubel FM, et al., editors. Unit 24.5. New York: Wiley; 2001. [Google Scholar]
- 11.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DS, Keefe AD. In: Current Protocols in Molecular Biology. Unit 8.3. Ausubel FM, et al., editors. New York: Wiley; 2000. [Google Scholar]
- 13.McCafferty DG, Lessard IAD, Walsh CT. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal d-Ala-d-Ala dipeptidase VanX. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 14.Jenison RD, Gill SC, Pardi A, Polisky B. High resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, von Hippel PH. Escherichia coli transcription termination factor Rho. J. Biol. Chem. 1993;268:13947–13955. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.