Abstract
Objective:
The aim of this study was to evaluate the antibiotic resistance pattern of Psedomonas aeruginosa and its prevalence in patients with urinary tract infections (UTI) for effective treatment in a developing country like Pakistan.
Methods:
This is an observational study conducted for a period of ten months which ended on December 2013 at the Dr. Essa Laboratory and Diagnostic Centre in Karachi. A total of 4668 urine samples of UTI patients were collected and standard microbiological techniques were performed to identify the organisms in urine cultures. Antibiotic susceptibility testing was performed by Kirby-Bauer technique for twenty five commonly used antimicrobials and then analyzed on SPSS version 17.
Results:
P. aeruginosa was isolated in 254 cultures (5.4%). The most resistant drugs included Ceclor(100%) and Cefizox (100%) followed by Amoxil/Ampicillin (99.6%), Ceflixime (99.6%), Doxycycline (99.6%), Cefuroxime (99.2%), Cephradine (99.2%), Cotrimoxazole (99.2%), Nalidixic acid (98.8%), Pipemidic acid (98.6%) and Augmentin (97.6%).
Conclusion:
Emerging resistant strains of Pseudomonas aeruginosa are potentially linked to injudicious use of drugs leading to ineffective empirical therapy and in turn, appearance of even more resistant strains of the bacterium. Therefore, we recommend culture and sensitivity testing to determine the presence of P.aeruginosa prior to specific antimicrobial therapy.
KEY WORDS: Pseudomonas aeruginosa, Urinary tract infections, Antibiotic resistance pattern
INTRODUCTION
Urinary Tract Infection (UTI) is one of the leading causes of infection worldwide. Although E-coli is predominantly associated with the etiology of UTI,1 other organisms such as Klebsiella pneumonia, Proteus mirabilis, Staphylococcus aureus and most importantly P.aeruginosa are on the rise.2
Multi-drug resistant (MDR) bacteria have been defined as ‘resistant to one agent in three or more antibiotic categories’.3 Unfortunately, P.aeruginosa has proven to be one of them in our study, thus making its treatment ineffective. It is a gram-negative rod that can survive in myriad of environments such as aquatic and terrestrial.4 It contributes strongly to nosocomial infections and affects immunocompromised patients.4 The inherent antimicrobial resistance mechanisms of P. aeruginosa which include lower outer membrane permeability, increased expression of efflux pumps of different specificity and presence of Amp C beta-lactamase wreak havoc by granting the bacteria resistance mechanisms to commonly used antibiotics namely Penicillin G, first and second generation of cephalosporins and quinolones.5
We are confronted with limited therapeutic options due to the acquisition of resistance mechanisms by these pathogens chiefly because of the indiscrete use of antibiotics, failure to complete therapeutic regimens and variation in the doses administered along with over the counter availability.6 This may explain the fact that about 150 million cases of UTI are witnessed annually, making it the third most diagnosed infection.7,8 This collectively results in higher rates of morbidity and mortality, and accounts for the immense cost incurred on our healthcare system. Therefore, there is an increased need for newer antibiotics to treat UTI efficaciously.
Our study focuses on the prevalence and antibiotic resistance pattern of P.aeruginosa. We have assessed and compared our findings with the national and international antimicrobial patterns and have investigated drugs for the effective treatment of P.aeruginosa. We also briefly draw attention to the relation of antimicrobial patterns with the intrinsic and acquired antimicrobial mechanisms of P.aeruginosa.
METHODS
Research subject and design
The study includes both nosocomial and community- acquired isolates of P.aeruginosa. A total of 4668 UTI cases were reviewed in this study from March 2013 to December 2013. Midstream specimen of urine, preferably of first morning void was collected of known UTI patients. The samples were then taken to the lab within two hours and standard microbiological procedures were performed to detect the presence of P.aeruginosa species.
Antimicrobial resistance testing
Antimicrobial resistance testing was performed and analyzed by Kirby Bauer technique on Mueller-Hinton agar plates using twenty five antibiotics according to their respective break points. NCCLS (1987) and WHO guidelines were followed to determine the zone diameters consistent with the Zone Size Interpretative Chart. Data was analyzed using SPSS version 17.
RESULTS
E-coli was the predominant organism (42.9%). Out of the total 4668 samples, P.aeruginosa grew in 254 cultures (5.4%). Other organisms detected were Klebsiella spp (18.4%), Staphylococcus spp (18.1%), Enterococci (4.8%), Enterobacter (3.5%), Salmonella spp (0.77%) and Proteus spp (0.64%). Candida was found in 5.4% of the cases.
Amongst the 254 P.aeruginosa infected UTI patients, 64.71% were females and 35.29% were males. Highest number of cases were seen in the age group 61-80 followed by the age group 21-40 while patients ranging from 81 to 100 years were least affected (Fig.1).
Fig.1.
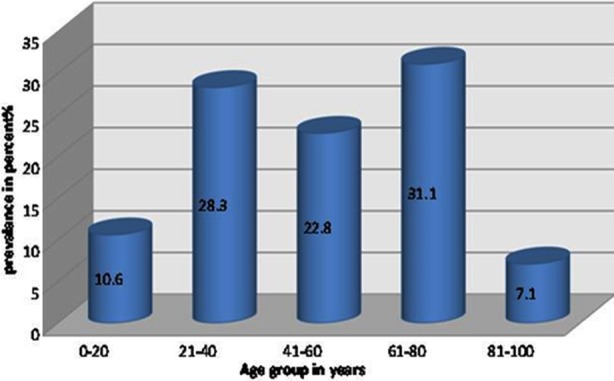
Prevalence of Pseudomonas aeruginosa in different age groups.
Interestingly, certain cases showed the presence of other organisms along with P.aeruginosa. Enterococcus spp was found in 11 cases (4.3%), Staphylococcus spp in 6 (2.4%), E-coli in 3 (1.2%) and Candida in 11 (4.3%). Table-I displays the overall patterns of antimicrobial resistance of P.aeruginosa. Analysis of the disk-diffusion tests in vitro revealed that P.aeruginosa was sensitive to Imipenem, Piperacillin/Tazobactam and Amikacin. Resistance was observed to Ceclor, Cefizox, Amoxil/Ampicillin, Ceflxime, Doxycycline, Cefuroxime, Cephradine, Cotrimoxazole, Nalidixic acid, Pipemidic acid and Augmentin.
Table-I.
Resistivity pattern of P.aeruginosa towards various antibiotics. (n = 254).
| Resistance | Drugs | ||||
|---|---|---|---|---|---|
| (%) | Number(percentage) | ||||
| 100 | Ceclor | ||||
| 100 | Cefizox | ||||
| 99.6 | Amoxil/Ampicillin | ||||
| 99.6 | Ceflxime | ||||
| 99.6 | Doxycycline | ||||
| 99.2 | Cefuroxime | ||||
| 99.2 | Cephradine | ||||
| 99.2 | Cotrimoxazole | ||||
| 98.8 | Nalidixic acid | ||||
| 98.6 | Pipemidic acid | ||||
| 97.6 | Augmentin | ||||
| 63.9 | Cefepime | ||||
| 63.9 | Fosfomycin | ||||
| 61.7 | Ceftriaxone | ||||
| 58.4 | Tobramycin | ||||
| 56.1 | Ceftazidime | ||||
| 50.4 | Enoxacin | ||||
| 50.2 | Sparfloxacin | ||||
| 50 | Ciprofloxacin | ||||
| 49 | Ofloxacin | ||||
| 45.5 | Cefotaxime | ||||
| 35.3 | Gentamicin | ||||
| 25.3 | Amikacin | ||||
| 19.6 | Piperacillin/Tazobactam | ||||
| 10.4 | Imipenem |
DISCUSSION
Majority of the P.aeruginosa related UTI cases occurred in females as compared to males. Chances of contracting UTI increase by 1% per decade for women.8 In our study, patients in the age group of 61-80 were most affected. This could be explained by an increase in the incidence of prostatic hypertrophy in men after the age of 55. Women, on the other hand, are prone to conditions such as vaginitis, atrophy and prolapse of womb or vagina which set up the path for UTI.8,9
The age group 21-40 was second most affected possibly due to the increase in sexual activity, higher number of pregnancies and use of certain types of contraception that can cause UTI for example diaphragm or spermicide.8 It is reported that persistent forms of UTI affect less than 2% of the school-going girls.8
In our study, percentage of P.aeruginosa was 5.4% which was close to the percentage reported in a European study as 6.9%.10 In Kashmir, P.aeruginosa accounted for 7.6% of the urine samples11 while in Nepal, P.aeruginosa encompassed 1.20% of the total cultures.12 Study from India showed prevalence of P.aeruginosa to be 9.3%7 while 9% of the bacterial dominance was found in Palestine.13 Retrospective studies in Pakistan show prevalence of the bacteria to be 9.2% and 3.27%.16,17 According to our study, P.aeruginosa in UTI patients can be best treated with Imipenem with minimum resistance (10.4%), followed by Piperacillin/Tazobactam (19.6%) and then Amikacin (25.3%). A similar study by Naeem et al from Pakistan showed 99-100% effectiveness of Amikacin and improved therapeutic outcomes with Imipenem and Piperacillin/Tazobactam against P.aeruginosa. 14 However a study from India reported resistance of P.aeruginosa to Amikacin which was in contradiction to our study and that of Naeem et al.14,15 Among other aminoglycosides, Gentamicin’s level of resistance was 35.3% against P.aeruginosa while two studies from Pakistan showed this antibiotic’s efficacy to be 69.2% and 83%.16,17 In a European study, nosocomial isolates of P.aeruginosa showed 72% resistivity towards Gentamicin, 69.2% to Tobramycin and 40% to Amikacin.10
This could be associated with the growing resistance of gram-negative bacteria including P.aeruginosa to Gentamicin. Amikacin has been used sparingly only in severe forms of the disease owing to high treatment costs and the intravenous nature of administration. Therefore, drug resistance has been slow to emerge.
P.aeruginosa resists antimicrobial drugs either through acquiring beta-lactamases especially extended-spectrum enzymes, carbapenemases or aminoglycoside-modifying enzymes.18 Resistance is conferred by the transfer of plasmids which carry genes to produce antimicrobial enzymes. Plasmids are passed on to the bacterial progeny by replication and to other members by conjugation, generating a bacterium populace that becomes harder to treat with the passage of time.19,20
Antibiotic resistance correlates well with the frequency of drug use and in a state like Pakistan where drugs are easily available over-the-counter, bacterial resistance to antibiotics grows rapidly, putting our health care system in a conundrum. Drug resistance facilitates growth and increases prevalence of persistent pathogens which become difficult to exterminate. Studies previously have shown the emergence of resistant genes in bacteria within 5 years of antibiotic intervention.21
Imipenem (carbapenem) shows remarkable activity in our study and this could be attributed to its infrequent use in the treatment of UTI patients, however, Carbapenem-hydrolyzing strains of P.aeruginosa have been isolated in clinical settings of some developed countries such as Italy, China, Japan and Singapore.5 Carbapenem-hydrolyzing strains of P.aeruginosa resist carbapenems (Imipenem and Meropenem).5 The only monobactam possibly effective against carbapenem-hydrolyzing strains of P.aeruginosa is Aztreonam.5 Frequent use of Imipenem changes the outer membrane permeability or in seldom cases when given in collaboration with Piperacillin, modifies the target sites of P.aeruginosa which then becomes resistant to carbapenems.5
Growing resistance of Ciprofloxacin and Cefotaxime was found in P.aeruginosa in previous studies which was quite similar to the resistivity pattern we found of Cefotaxime (45.5%) and Ciprofloxacin (50%).15
In our study, Ofloxacin showed the greatest level of activity in quinolones against the resistant bacterium being 51% sensitive, followed by Ciprofloxacin which was 50% sensitive, Sparfloxacin (49.8%) and Enoxacin (49.6%). However, another study from Pakistan showed resistance pattern of Ciprofloxacin to be 25%.22 The same study showed the resistance of Nalidixic acid and Pipemidic acid to be 87% and 25% respectively.22 However in our study the resistance of these two drugs was found to be 98.8% and 98.6% respectively, validating the stifled therapeutic action of the quinolones due to frequent use.22 A study from Nigeria found the resistance levels of Ciprofloxacin, Sparfloxacin and Ofloxacin to be 77.1%, 44.8% and 19.8% respectively.23
Quinolones are preferred empiric therapy for UTI before the physicians receive culture results mainly because of the easy availability in oral forms and reasonable cost.24 However, emerging quinolone resistant strains of P.aeruginosa have surfaced owing to the changes in target enzymes of the bacteria and active efflux pumps generated to prevent the entry of the drugs.5
Fosfomycin was considered a sensitive drug at 66% against P.aeruginosa in another study from Pakistan while the level of resistance in our study was 63.9%.17 Among the cephalosporins, Cefotaxime was most effective against P.aeruginosa with only 45.5% resistivity level, followed by Ceftazidime (56.1%) and Cefepime (63.9%). Ceftriaxone’s resistance level was 80% in a study from India while in our study it was 61.7%.2
A study from Palestine, showed resistivity level of Ceftazidime to be 13.3% and that of Cefuroxime to be 53.3% while in our study it was 56.1% and 99.2%, respectively.13 Although susceptibility levels of Ceftazidime and Ceftriaxone found by Paryani et al which were 97% and 87% respectively, differs greatly with that found in our study, it does prove the relative efficacy of these drugs amongst the cephalosporins against P.aeruginosa. 17 The resistance rate of Cotrimoxazole is 99.2% in our study which was similar to that found by Manjunath GN et al. (100%).7
Production of beta-lactamase by P.aeruginosa cleaves the amide bond on the beta-lactam ring, rendering beta-lactams ineffective.5 Production of Amp C-beta lactamase (cephalosporinase) enables P.aeruginosa to resist third generation cephalosporins and surprisingly, the amount of cephalosporinase produced rises 100 to 1000 times in the presence of carbapenems like Imipenem.5
Thus, the only carbapenem (Imipenem) tested in our study showed the greatest level of efficacy although effectiveness of Meropenem has been noted in other studies.25 Aminoglycosides were the next best treatment in Pseudomonas aeruginosa associated UTI, followed by quinolones while cephalosporins did not fare well and similar comparisons have been reported by Paryani et al.17 However 4th generation cephalosporins were not tested in our study, which do show a good salutary response against some strains of P.aeruginosa as noted in different studies.5 Lastly, penicillins were highly ineffectual against P.aeruginosa except for Piperacillin/Tazobactam possibly because of the beta-lactamase inhibitor in addition to the extended-spectrum nature and scarce use of the drug.
While growing resistance of bacteria itself puts the future of our health care in uncertainty, what poses a greater threat is the emergence of multi-drug resistant (MDR) pathogens that would render most antibiotics ineffective by obtaining newer and complex resistance mechanisms. Some factors contributing to the rise of MDR bacteria are immunosuppression, extensive use of wide spectrum drugs and prolonged hospitalization.25
The antibiotic susceptibility trends investigated narrow down the most sensitive drugs from the most resistant ones and our study can be used to provide a more sound approach in improving the antibiotics therapy against the MDR P.aeruginosa in UTI patients.
CONCLUSION
Ability of P. aeruginosa to resist antibiotics by their inherent or acquired antimicrobial mechanisms can be linked to the common practice of physicians to prescribe antimicrobials that are easily available over the counter. This trend is in conformity to global trends especially where there is no check on the purchase of drugs, confirming the rise of resistant strains of P.aeruginosa. Therefore we recommend careful empirical prescription, culture and sensitivity testing and selection of drugs accordingly.
Footnotes
Competing interests: The authors declare no competing interests.
Authors’ contributions
Dr. Farhan Essa Abdullah suggested the concept of this research. He provided the guidance and gave the technical support required for processing the samples meanwhile Dania Aijaz Shah and Shehnaz Wasim were involved in compilation and analysis of the results. Shehnaz Wasim tallied the data while Dania Aijaz Shah entered it on SPSS. Dania Aijaz Shah and Shehnaz Wasim wrote the manuscript after which Dr. Farhan Essa Abdullah reviewed it. The Final changes were made by Dania Aijaz Shah and Shehnaz Wasim. All authors have read and approved the manuscript.
REFERENCES
- 1.Tanvir R, Hafeez R, Hasnain S. Prevalence of multiple drug resistant Escherichia coli in patients of urinary tract infection registering at a diagnostic laboratory in Lahore, Pakistan. Pak J Zool. 2012;44(3):707–712. [Google Scholar]
- 2.Wasnik DD. Prevalence and antibacterial susceptibility pattern of urinary tract infection Causing Human Pathogenic Bacteria. Asian J Biomed Pharm Sci. 2012;2(15):1–3. doi:10.15272/ajbps.v2i15.130. [Google Scholar]
- 3.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drugresistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. doi:10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 4.Eguchi H, Miyamoto T, Kuwahara T, Mitamura S, Mitamura Y. Infectious conjunctivitis caused by Pseudomonas aeruginosa isolated from a bathroom. BMC Res Notes. 2013;6:245. doi: 10.1186/1756-0500-6-245. doi:10.1186/1756-0500-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strateva T, Yordanov D. Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J Med Microbiol. 2009;58(Pt 9):1133–1148. doi: 10.1099/jmm.0.009142-0. doi:10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 6.Bashir MF, Qazi JI, Ahmad N, Riaz S. Diversity of Urinary Tract Pathogens and Drug Resistant Isolates of Escherichia Coli in different age and gender Groups of Pakistan. Trop J Phar Res. 2008;7(3):1025–1031. [Google Scholar]
- 7.Manjunath GN, Prakash R, Annam V, Shetty K. Changing trends in the spectrum of antimicrobial drug resistance pattern of uropathogens isolated from hospitals and community patients with urinary tract infections in Tumkur and Bangalore. Int J Biol Med Res. 2011;2(2):504–507. [Google Scholar]
- 8.Najar MS, Saldanha CL, Banday KA. Approach to urinary tract infections. Indian J Nephrol. 2009;19(4):129–139. doi: 10.4103/0971-4065.59333. doi:10.4103/0971-4065.59333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urine Infection in Older people. http://www.patient.co.uk/health/urine-infection-in-older-people .
- 10.Bouza E, San Juan R, Muñoz P, Voss A, Kluytmans J. Co-operative Group of the European Study Group on Nosocomial Infections: A European perspective on nosocomial urinary tract infections I. Report on the microbiology workload, etiology and antimicrobial susceptibility (ESGNI-003 study). European Study Group on Nosocomial Infections. Clin Microbiol Infect. 2001;7(10):523–531. doi: 10.1046/j.1198-743x.2001.00326.x. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad S. Pattern of urinary tract infection in Kashmir and antimicrobial susceptibility. Bangladesh Med Res Counc Bull. 2012;38(3):79–83. doi: 10.3329/bmrcb.v38i3.14330. [DOI] [PubMed] [Google Scholar]
- 12.Khatri B, Basnyat S, Karki A, Poudel A, Shrestha B. Etiology and antimicrobial susceptibility pattern of bacterial pathogens from urinary tract infection. Nepal Med Coll J. 2012;14(2):129–132. [PubMed] [Google Scholar]
- 13.Zakaria EA. Increasing Ciprofloxacin resistance among prevalent urinary tract bacterial isolates in Gaza Strip, Palestine. J Biomed Biotechnol. 2005;3:238–241. doi: 10.1155/JBB.2005.238. doi:10.1155/JBB.2005.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naeem M, Khan MA, Qazi SM. Antibiotic Susceptibility Pattern of Bacterial Pathogens Causing Urinary Tract Infection in a Tertiary Care Hospital. Ann Pak Inst Med Sci. 2010;6(4):214–218. [Google Scholar]
- 15.Hasan AS, Nair D, Kaur J, Baweja G, Deb M, Aggarwal P. Resistance patterns of urinary isolate in a tertiary Indian hospital. J Ayub Med Coll Abbottabad. 2007;19(1):39–41. [PubMed] [Google Scholar]
- 16.Gul N, Mujahid TY, Ahmad S. Isolation, Identification and Antibiotic Resistance Profile of Indigenous Bacterial Isolates from Urinary Tract Infection Patients. Pak J Biol Sci. 2004;7(12):2051–2054. [Google Scholar]
- 17.Paryani JP, Memon SR, Rajpar ZH, Shah SA. Pattern and Sensitivity of Microorganisms Causing Urinary Tract Infection at Teaching Hospital. JLUMHS. 2012;11(2):97–100. [Google Scholar]
- 18.Frank DW. Research Topic on Pseudomonas aeruginosa, Biology, Genetics, and Host–Pathogen Interactions. Front Microbiol. 2012;3:20. doi: 10.3389/fmicb.2012.00020. doi:10.3389/fmicb.2012.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neu HC. Emerging trends in antimicrobial resistance in surgical infections. A review. Eur J Surg Suppl. 1994;573:7–18. [PubMed] [Google Scholar]
- 20.Tomasz A. Multiple-antibiotic resistant pathogenic bacteria- a report on the Rockefeller university workshop. N Engl J Med. 1994;330(17):1247–1251. doi: 10.1056/NEJM199404283301725. doi:10.1056/NEJM199404283301725. [DOI] [PubMed] [Google Scholar]
- 21.Chakrabarty AN, Dastidar SG, Ganguli M, Chattopadhyay D. ‘DNA’ as contaminants in antibiotics and its capacity to transform bacteria to drug resistance. Indian J Exp Biol. 1990;28(1):58–62. [PubMed] [Google Scholar]
- 22.Khalil A. Imran. Prevalence and Antibiogram of Uncomplicated Lower Urinary Tract Infections in Human Population of Gilgit, Northern Areas of Pakistan. Pak J Zool. 2008;40(4):295–301. [Google Scholar]
- 23.Chikwendu CI, Amadi ES, Obi RK. Prevalence and antimicrobial resistance in Pseudomonas aeruginosa and Klebsiella pneumonia isolates from non-clinical urine samples. New York Sci J. 2010;3(11):194–200. [Google Scholar]
- 24.Drago L, De Vecchi E, Mombelli B, Nicola L, Valli M, Gismondo MR. Activity of levofloxacin and ciprofloxacin against urinary pathogens. J Antimicrob Chemother. 2001;48(1):37–45. doi: 10.1093/jac/48.1.37. [DOI] [PubMed] [Google Scholar]
- 25.Prakash D, Saxena RS. Distribution and Antimicrobial Susceptibility Pattern of Bacterial Pathogens Causing Urinary Tract Infection in Urban Community of Meerut City, India. ISRN Microbiol. 2013:749629. doi: 10.1155/2013/749629. doi:10.1155/2013/749629. [DOI] [PMC free article] [PubMed] [Google Scholar]


