Abstract
Hypoxia is a critical microenvironment in tumor pathogenesis. There is a close relationship between hypoxia, tumor metastasis and poor prognosis. Hypoxia has been shown to induce epithelial-mesenchymal transition and high levels of lactic acid production, through which cancer cells gain migratory capability. Here, we present a high-throughput microfluidic platform with a controlled oxygen environment to specifically monitor mesenchymal migration under hypoxic conditions. We found that, combined with a slightly alkaline microenvironment, such a platform can help to improve the efficiency of antimetastatic drugs. We also use this platform to study primary and rare cells from mice and demonstrate the correlation between on-chip results and in vivo outcome. This device may provide a new opportunity for biologists and clinicians to better perform assays that evaluate cancer cell behaviors related to metastasis.
Introduction
Cancer mortality is mainly caused by the spread of cancer cells within the host in a process called metastasis 1. Cell metastasis involves a multistep cascade process starting with local invasion, of the lymphatic or vascular system, survival in the systemic circulation, and then colonization of the metastatic site2. Increasing evidence has shown that activated mesenchymal migration is a key process of the metastatic cascade and cancer cells usually gain migratory capability through epithelial to mesenchymal transition (EMT)3. Therefore, insights into mesenchymal migration and blocking of this process should aid in the prevention of cancer metastasis, improve prognosis, and lead to more effective cancer treatments. Additionally, much evidence has suggested that cell migration is a social behavior related to cell numbers in culture4. Characterization of mesenchymal-mode migration and quantitation of cell migratory capability in relation to cell numbers may provide a powerful tool to more accurately study cell invasiveness and metastasis.
Hypoxia is a condition in which the body or a region of the body is deprived of adequate oxygen supply and is a critical microenvironment in tumor pathogenesis5. Tumor metastasis occurs in a series of distinct steps that include tumor cell invasion, intravasation, extravasation, and proliferation6. There is a close relationship between hypoxia and tumor metastasis and then poor prognosis. Several mechanisms have been proposed to explain how hypoxia might lead to a poor prognosis in the clinical settings, none of which are mutually exclusive. For example, hypoxia induces EMT via activation of Snail by hypoxia-inducible factor-1α (HIF-1α) in hepatocellular carcinoma; it also stimulates migration and increases the metastatic ability of breast cancer cells7. The low pH of hypoxic tumors as a result of high levels of lactic acid can promote tumor cell invasion by destruction of adjacent non-cancerous tissue8. These studies indicate that hypoxia may increase metastatic potential via the induction of EMT and activated mesenchymal migration. The validation of this unconfirmed theory to explain metastasis requires a powerful platform to aid in analysis. The ideal assay to study tumor cell migration under hypoxic conditions would allow for precise control of the oxygen content, real-time monitoring, discrimination of the cell morphological mode, and require only a small number of cells. To meet these challenges, a high throughput mesenchymal-mode migration assay (M-Chip, containing 3120 microchambers, Figure 1A) has been recently developed in our laboratory for antimetastatic drug screening 9. Combining microfluidic and imaging techniques along with statistical evaluation, we studied how varying oxygen partial pressure (pO2) from 21% (ambient) to 1% (hypoxia) influences mesenchymal-mode migration at different cell densities10. Using the M-Chip, we demonstrated that the migration velocity and percentage of migrating cells was increased in a hypoxic microenvironment. The more numbers of cells were cultured in the microchamber, the higher percentage of cells migrated. We then found that this phenomenon was related to the acidic extracellular pH in the microchambers. Increasing the cell numbers will lead to lower PH values. The acidic extracellular pH promotes mesenchymal-mode migration. We also used the M-Chip to screen antimetastatic drugs and study the migratory capacity of primary cells. The M-Chip and its cell assay capability may provide a new avenue for biologists to better deliver cell metastatic assay and drug selection.
Fig. 1.
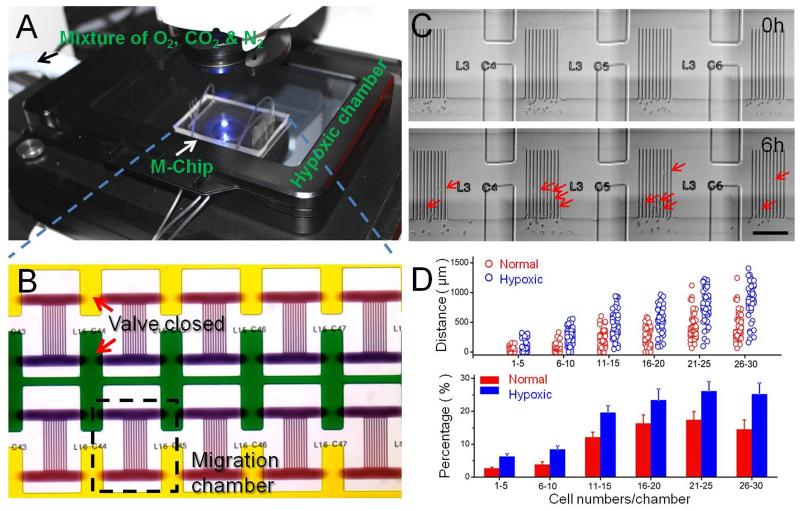
Hypoxia cell culture and imaging system, M-Chip design, and representative data. (A) This design permits incubation of the cells within controlled pO2 environments, followed by quantitative assays of mesenchymal-mode cell migration (migration velocity and percentage) from quantized cell populations. (B) Bright-field micrograph of several micro-chambers shows the chemotaxis gradient. (C) Optical micrographs of a SUM-159 tumor cell migrating across the channels taken at 6 h intervals. Scale bar: 200μm. (D) Migratory potential of cells derived from SUM-159 cell lines in a 1% O2 environment. The columns represent the average value of percentages, which follow a Gaussian distribution. Error bars represent the standard deviation of replicates (n=40 chambers). All results were measured at 6th hour of the on-chip culture.
Results and Discussion
We used multilayer soft lithography to design a polydimethylsiloxane (PDMS) microfluidic device featuring 3120 microchambers, each with a volume of 2.4 nL. The details of the fabrication process can be found in our previous work9. The PDMS microfluidic device has been widely used for cell-culture applications and provides high gas permeability for the efficient exchange of oxygen and carbon dioxide5a, 11. Each microchamber has an upper compartment that serves as a chemoattractant reservoir; a second reservoir, connected to upper compartment by 10 parallel migration microchannels, serves as a cell culture reservoir (Figure 1B). The dimension of these channels was optimized to 10×10×400 μm (W×H×L), mimic the capillaries and pores of tissue or vasculature that allow tumor cell migration. The chemoattractant gradient established in these microchannels can be maintained for as long as 4h under our experimental conditions. Carefully replacing the medium at 3 h intervals maintains a chemoattractant gradient during the 6 h experiments 9. The only migratory manner for the tumor cells to pass through these microchannels was mesenchymal-mode migration. Before loading cells, the channels of the M-Chip were gently flushed with 70% (vol/vol) ethanol for sterilization, followed by deionized water and PBS. A 50 μg/ml Basement Membrane Extracts (BME) solution was loaded into the chip and incubated overnight at 4° C for physisorption. BME coated chambers and microchannels can be used to mimic in vivo-like microenvironment. Cells are loaded from an upstream inlet into the M-Chip and distributed randomly among the microchambers. Controlling the cell loading density ensures that most of the microchambers contain 1–30 cells.
After cell loading, the microchip is placed into the microscope equipped with the cell culture chamber, in which the O2 environment can be controlled. An O2 sensor (0.1% accuracy) measured the level and equilibration rate of the pO2 (Figure S1). We chose to work with the highly metastatic SUM-159 cell line. For this cell line, we collected cell migration data at 21% and 1% pO2 and assayed ~1000 microchambers under each condition. After imaging and statistical evaluation, scatter plots of cell migration data were compared under the different O2 conditions. Figure 1C and S2 shows the optical images of SUM-159 cells migrating across channels after 6 h in a hypoxic environment. The migration distance of each cell was tracked using the center of the nucleus (or the cell center if the nucleus could not be seen clearly) as the tracking point. The total distance migrated and percentage of migrated cells in each chamber was calculated and compared to the data collected in a normal O2 environment. At all cell densities, the migration distance of SUM-159 cells, as well as the percentage of migrated cells, in 1% pO2 was greater than that in 21% pO2 (Figure 1D). For example, at lower cell density (1–5 cells/chamber) the percentage of migrating SUM-159 cells in 1% pO2 was greater than 2-fold of the percentage migrating in 21% pO2. In chambers with a higher cell density (>26–30 cells/chamber), the percentage of migrating cells was about 1.5 times higher than that found under normal conditions (Figure 1D). The migration velocity for individual cells migrating in the channel was calculated using the distance migrated in 6h. The average migration velocity was calculated either for a whole chamber or for multiple chambers loaded with the same number of cells. We found the velocity of the single cell migration has followed the normal distribution and has no significant difference with varying cell density. Either in the single cell chamber or in the high cell density chamber, the velocity of cell migration is close. The velocity is mainly determined by the cell types in our system. The average migration velocity of SUM-159 cells (in the channel) at 1% pO2 was 19.3 μm/h (S.D. =5.24 μm/h) and the average velocity of SUM-159 cells at 21% pO2 was approximately 15.4 μm/h (S.D. =5.77 μm/h).
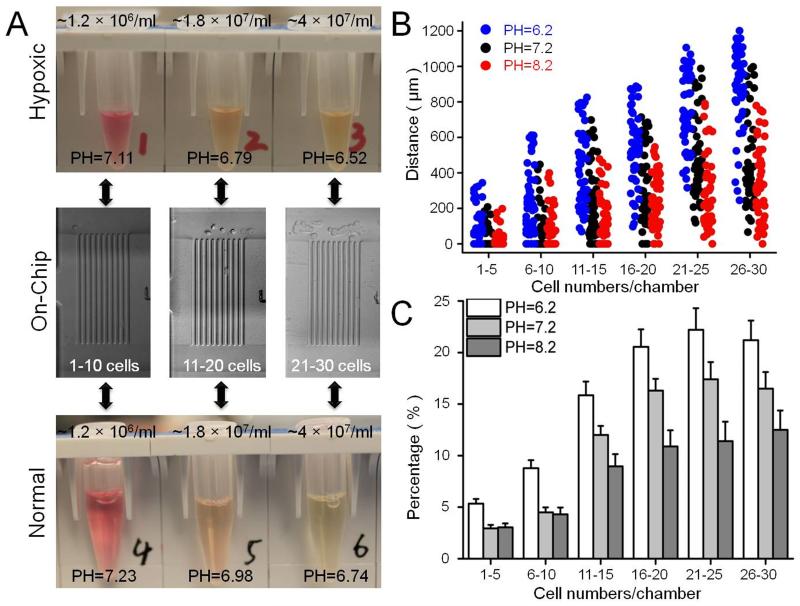
An important consequence of hypoxic conditions is the switch to glycolytic metabolism and increased lactic acid production12, a condition that cannot be controlled in the microchamber. To circumvent this issue, we first measured the pH of cell culture medium under conditions of increasing cell density after 8h in both 21% and 1% pO2. The selection of cell densities corresponded to the cell numbers in a microchamber. As shown in Figure 2A, increasing cell density resulted in decreased pH under both O2 environments. Compared to normal conditions at any cell density, the pH of the culture medium in hypoxic conditions was lower. Similar conclusions can be extended to the M-Chip. In order to prove that the acidic extracellular pH influences mesenchymal-mode cell migration, we cultured the cells and loaded them into the chip with medium at different pHs (pH= 6.2, 7.2, and 8.2). At all cell densities, the migration distance of SUM-159 cells under slightly acidic conditions was greater than that in a neutral or slightly alkaline environment. Migration distance under neutral conditions was greater than in a slightly alkaline environment (Figure 2B). We also studied cell migration velocity under these three medium conditions. The average velocity of SUM-159 cells in acidic medium was approximately 21.4 μm/h (SD=6.37 μm/h), faster than in neutral medium (14.6 μm/h, S.D. =4.64 μm/h) and in alkaline medium (13.2 μm/h, S.D. =4.41 μm/h). The average migration velocity at different cell densities did not show an appreciable difference. We also studied the percentage of migrating cells under these conditions. At a lower cell density (1–10 cells/chamber) the percentage of migrating cells in acidic medium was about 2-fold greater than that in neutral or slightly alkaline medium. At higher cell density, the percentage of migrating cells in acidic medium was only slightly higher than that in neutral medium, but still about 2-fold greater than that in slightly alkaline medium. Similar results were also observed by another breast cancer cell line SUM-149, which was shown in Figure S3. These results indicate that an acidic extracellular pH promotes mesenchymal-mode migration that depends both on a greater percentage of migrating cells and a higher velocity per cell. These results also indicate that one of the reasons hypoxia increases the migratory capacity of breast cancer cells is an acidic environment, even in the absence of lactic acid.
Fig. 2.
(A) Measuring the PH value in the micro-chambers at different cell numbers at 21% and 1% O2 environments, in the 1% O2 environments, the PH value of the medium is lower than 21% O2 environments. (B, C) Comparison of the migration distances and percentages of migrating SUM-159 cells at different pH in a 21% O2 environments. Data in the bar graph were analyzed in the same condition as Figure 1D.
We next examined the effect of an acidic extracellular pH on cell polarizability and morphology, using circularity of cell morphology to quantitate. To measure the effects of acidic pH over longer times, we seeded SUM-159 cells on fibronectin-coated polystyrene dishes and allowed them to adhere for 24 hours before changing the media to pH 6.2, 7.2, or 8.2. Six hours after the pH change, many cells at pH 6.2 had developed an elongated morphology and longer protrusions, while the cells cultured in medium at pH 8.2 exhibited cobblestone-like morphology. The morphology was quantified by calculating cell circularity, which ranges from 0–1. Circularity close to 1 indicates that the cell has a rounded morphology, while circularity close to 0 indicates an elongated or dendritic morphology 13. Consistent with observations from images, the mean cell circularity was significantly lower at pH 6.2 than at pH 7.2 or 8.2 (Figure S4, S5). We also found the mean cell circularity under conditions of hypoxia was similar to that found at pH 6.2. Such morphology is more conducive to mesenchymal-mode migration, distinguished by elongated cell morphology with cell polarity (Figure S6). In our device, increasing the cell numbers will rise up lactic acid production and lead to lower PH values in the microchamber. The mean cell circularity at acidic microenvironment is lower than the one at normal condition. The cancer cells in higher cell density will prefer to exhibit elongated morphology and longer protrusions. Such morphology favors mesenchymal-mode migration.
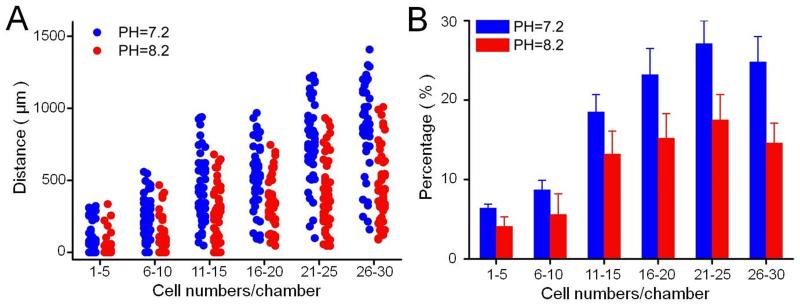
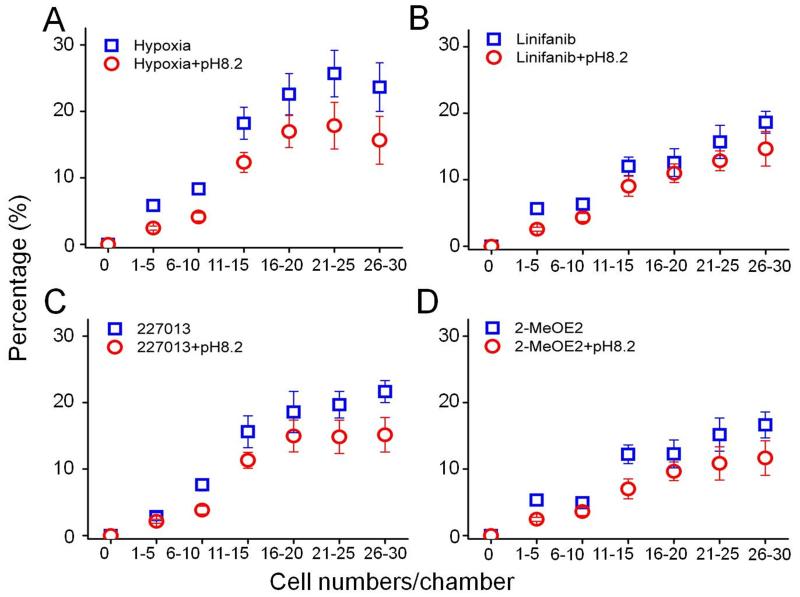
Hypoxia increases lactic acid production and results in an acidic extracellular pH; therefore, neutralization of the acidic microenvironment may inhibit mesenchymal-mode migration. We first cultured the cells in slightly alkaline medium with 1% pO2 and then introduced the cells into the M-Chip, keeping the medium slightly alkaline. After 6 h, the images were collected and data was analyzed. At all cell densities, the migration distance of SUM-159 cells at neutral pH was greater than in a slightly alkaline environment (Figure 3A). The average velocity of SUM-159 cells at pH 8.2 was approximately 17.4 μm/h (SD=3.37 μm/h), which is slightly slower that at pH 7.4 (SD=19.3 μm/h). Similar to the normal O2 microenvironment, the percentage of migrating cells was lower in slightly alkaline medium than at pH 7.4 at any of the cell densities tested. These results indicated that neutralization of an acid microenvironment reduced both migratory velocity and percentage of migrating cells. Combined with the appropriate antimetastatic drug, an alkaline microenvironment may aid in the inhibition of mesenchymal-mode migration. Therefore, we tested three small-molecule compounds that inhibit specific chemokines and growth factors related to hypoxia and breast-cancer metastasis. The migratory velocity and percentage of migrating cells treated with these inhibitors was plotted in Table 1 and Figure 4. HIF-1α is a pivotal transcriptional factor in the cellular response to hypoxia14. In our platform, the HIF-1α inhibitor 2-MeOE2 reduced both velocity and percentage of migrating cells; in an alkaline medium, the velocity and percentage of migrating cells were further reduced. We also treated SUM-159 cells with linifanib (colony stimulating factor 1 receptor, CSF-1R inhibitor) or the drug 227013 (C-C chemokine receptor type 4, CCR4 inhibitor), both of which inhibited cell migration and the percentage of migration-competent cells in the M-Chip. Similar to 2-MeOE2, in an alkaline medium, the velocity and percentage of migrating cells was further reduced. Our screening results are consistent with the findings of other research groups showing that inhibition of HIF-1α reduces the migratory potential of tumor cells. However, our device allows more accurate and higher-throughput screening of migration-inhibiting drugs under hypoxic conditions and in controlled medium.
Fig. 3.
Comparison of migration distance and percent migrating cells of SUM-159 cells at different pH in a 1% O2 environment. Data in the bar graph were analyzed in the same condition as Figure 1D.
Table 1.
Migration velocity of SUM-159 cells treated with the inhibitors. Velocity unit is μm/h.
| Inhibitors | Normoxia | Hypoxia | Linifanib | 2-MeOE2 | 227013 |
|---|---|---|---|---|---|
|
Velocity
(pH=7.4) |
15.4 | 19.3 | 15.4 | 15.1 | 17.1 |
|
Velocity
(pH=8.2) |
14.6 | 13.6 | 10.8 | 11.4 | 12.1 |
Fig. 4.
Screening small molecule inhibitors of mesenchymal-mode migration at 1% pO2: the percentage of migrated cells inhibited by the small-molecule compounds. The points represent the average value of the percent migrated cells, which follows a Gaussian distribution. Error bars represent the standard deviation of replicates (n=40 chambers).
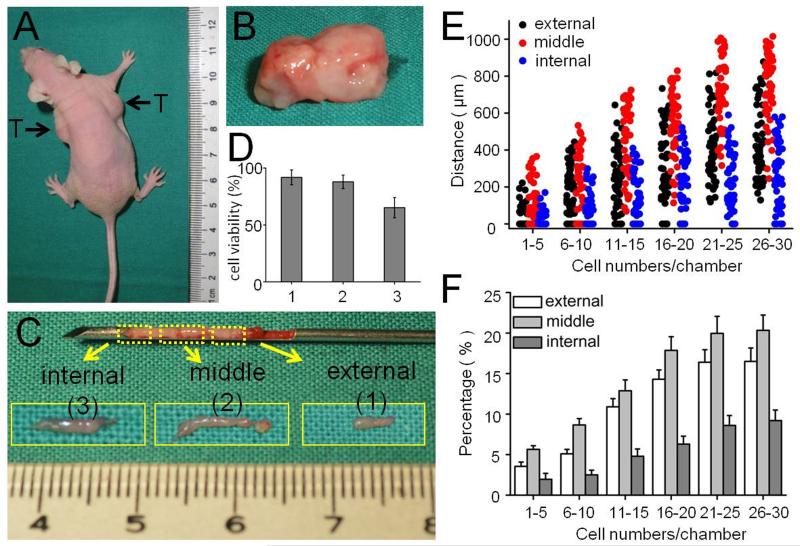
Another important advantage of the M-Chip is the need for only a small sample size and the ability to study different cell types in one experiment. (The device can use <10000 cells; in such cases, we use only one or two of twenty groups of channels by applying cells directly from the outlet access holes.) This allows detection and analysis of the metastatic potential of primary or rare cells. It has been reported that, in many cases, the tumor interior and periphery respond differently to chemotherapy15. The needle biopsy method utilizes a small needle to aspirate cells from the tumor and is more cost effective and less invasive than open biopsy, requiring only sedation and local anesthetic. However, needle biopsy may not give enough tissue for a standard diagnosis. Using needle biopsy methods we can collect cancer cells from different layers of a solid tumor, from the periphery to the interior (Figure 5A-C). In vivo, directional movement of cancer cells develops at distances typically 100–150 μm beyond the diffusion capacity of chemokines and oxygen from blood vessels. In Figure S7 we show that the tumor periphery contains many tiny blood vessels, however, the tumor interior has fewer blood vessels suggesting that cells located inside the tumor may tolerate a low oxygen environment. We loaded cells derived from different positions within the tumor into the M-Chip and observed the cell migration. We first studied cell viability and showed that cells collected from peripheral (external) or middle (midway between the periphery and the center) positions grew well. However, a small portion of cells obtained from the internal portion of the tumor died in the M-Chip (Figure 5D). At all cell densities tested, the migration distance of SUM-159 cells from the middle portion of the tumor was greater than cells from the periphery or the interior part of the tumor (Figure 5E). The average velocity of SUM-159 cells from the middle was approximately 18.4 μm/h (SD=4.17 μm/h), slightly, but not significantly, faster than from the periphery, (17.9 μm/h, SD=3.91 μm/h). However, the average velocity of cells from the interior of the tumor was slower (15.4 μm/h, SD=2.88 μm/h). We also studied the percentage of migrating cells collected from the three positions. At all cell densities tested, the percentage of migrating cells from the middle position was only slightly higher than that from peripheral tumor, but about 2-fold greater than that from the central tumor interior (Figure 5F). This could be due to the fact that the cell viability was lower in the interior than at the peripheral or middle positions. We next examined the cell morphology and migration mode of the cells obtained from the different positions within the tumor. The cells derived from peripheral and middle tumor generated many protrusions in the M-Chip microchambers; most cells exhibited mesenchymal-mode migration (Figure S8-S10) and spread very well in the M-Chip. On the other hand, cells from the tumor interior generated fewer protrusions and did not spread well in the M-Chip, morphology not conducive to mesenchymal-mode migration. These results may explain why the cells from the tumor interior travelled less distance than other cells.
Fig. 5.
Comparison of migratory capacity of tumor cells dissociated from the peripheral (external), middle (between the periphery and the center of the tumor) and internal portions of the tumor. (A, B) Images of nude mice with tumors and excised tumor. (C) Images of carcinoma specimens collected by puncture needle. (D) The viability of cancer cells collected from external, middle and internal positions of the tumor. (E, F) Comparison of the migration distances and percentages of migrating cells collected from different depths of tumor. Data in the bar graph were analyzed in the same condition as Figure 1D.
Tumor cell migration is conventionally understood as the migration and invasion of individual cells or cell clusters that detach from the primary tumor. However, cancer therapeutics targeting adhesion receptors or proteases to inhibit cell migration and metastasis have yet to demonstrate effectiveness in clinical trials. Invasion mechanisms of cancer cells are still unclear; factors such as hypoxia, angiogenesis, and cell reprogramming may be involved, allowing cells to maintain migratory properties via morphological and functional de-differentiation. For example, the mode of invasion may transition from mesenchymal movement to amoeboid movement16. Therefore, the implementation of an effective assay to quantify and evaluate the migratory capacities of cancer cells may significantly aid in the development of novel cancer therapeutics. Compared to traditional techniques, our method provides more desirable features including real-time monitoring, effective discrimination of migration modes, fine controlled gradients, and the minimal requirement of sample cells. In this study, we verified the utility of the M-Chip for studying cancer metastasis by performing real-time mesenchymal-mode migration assays using breast cancer cell lines. By calculating and analyzing the percentage and velocity of migrating cells in each microchamber, we quantified the migratory capacity of breast cancer cells over a wide range of cell densities that may correspond to different stages of malignancy. The small sample size used with the M-Chip allows analysis of the metastatic potential of primary or rare cells. By using inhibitors of receptor signaling related to hypoxia and breast cancer metastasis, we found that a slightly alkaline microenvironment can help to improve the efficiency of antimetastatic drugs, which may provide a more effective method to reduce cell invasion and metastasis. For example: increasing fruits and vegetables in an alkaline diet blocks lactic acid releasing from cancer cells. Using the M-Chip, we can adjust the microenvironment easily, such as oxygen content, PH value and controlled gradient. This device provides a high-throughput platform for biologists and clinicians to better perform assays that evaluate cancer cell behaviors related to metastasis.
Experimental
Materials and reagents
Silicon wafers (4-inch) were purchased from Corning Inc. (Corning, NY). SPR-220, SU-8 2015 photoresist, and SU-8 developer were purchased from MicroChem Corp. (Newton, MA). MF-CD26 was obtained from Rohm and Haas Electronic Materials (Marlborough, MA). Polydimethylsiloxane (PDMS RTV615) was obtained from Momentive Performance Materials (Waterford, NY). Ham’s F12 medium and fetal bovine serum (FBS) was obtained from Invitrogen (Grand Island, NY). Basement Membrane Extract (BME) was obtained from Trevigen (Gaithersburg, MD). Phosphate-buffered saline (PBS; 0.1 M, pH 7.4) was obtained from Lonza Inc. (Allendale, NJ). All M-Chip devices were designed as computer graphics using AutoCAD software and then printed out as 10-μm–resolution chrome masks by Photo Science Inc. (Torrance, CA). The SUM-159 cell line was obtained from Asterand (Detroit, MI). Linifanib and 2-MeOE2 were obtained from Selleckchem (Houston, TX). Drug 227013 was obtained from EMD Millipore (Darmstadt, Germany).
Photolithography
Microstructure patterns were first designed with AutoCAD software and fabricated using standard photolithography and molding processes, as shown in Figure S1. To create the master for the flow layer, the silicon wafer was first spin-coated with a layer of SPR-220 photoresist and then baked at 95°C for 5 min. The photomask and silicon wafer were aligned and exposed to ultraviolet light for 30 s. The silicon wafer was developed by immersing in SPR developer solution (MF-CD26) for 3–5 min to dissolve the exposed areas. The wafer was thoroughly rinsed with Millipore water and dried with nitrogen gas. The silicon masters were baked at 115°C for 15 min and treated with an anti-adhesive agent, trimethylchlorosilane (TMCS), via vapor reaction for 1 h. The master for the control layer was prepared by spin coating SU-8 2015 negative photoresist onto a silicon wafer and crosslinking with UV for 180 s. Subsequently, the designed pattern was developed using SU-8 developer (Microchem Corp, Newton, MA) and cleaned with isopropyl alcohol and nitrogen gas. The silicon masters were baked at 150°C for 30 min and treated with the anti-adhesive agent TMCS via vapor reaction for 4 h. The SPR 220 mold was ~10 μm in height after rounded by a thermal treatment. The SU-8 mold was ~30 μm in height.
Fabrication of Migration Chip
The M-Chip is composed of two layers. The flow layer was fabricated by spin-casting the pre-polymer of GE RTV 615 PDMS part A and part B (20:1) onto a SPR 220 mold at 1500 rpm for 30 s and then curing at 80°C for 45 min. The control layer was fabricated by casting the pre-polymer of GE RTV 615 PDMS part A and part B (10:1) onto a SU-8 mold and curing at 80°C for 45 min. The control layer was carefully aligned and placed onto the flow layer, followed by a thermal bonding step at 80° C for 60 min. This PDMS slab was cut and peeled off the mold and bonded onto a glass slide (as shown in Figures S1).The channels of the M-Chip were gently flushed with 70% (vol/vol) ethanol for sterilization followed by deionized water and, finally, with PBS. A 50 μg/ml BME solution was loaded into the chip and incubated overnight at 4°C for physisorption. During the BME incubation period, all valves were closed and the areas under the valves remained uncoated, reducing the chance of cell loading and adhesion in these areas. Prior to each experiment, BME solution was replaced by cell culture medium and the device was warmed to 37°C in the incubator.
Cell Culture and Treatment
SUM-159 cells were cultured at 37°C in an atmosphere of 5% CO2 in Ham’s F12 medium with 10% FBS, insulin (5 μg/ml), and hydrocortisone (1 μg/ml). Cells were harvested by treatment with 0.25% trypsin-EDTA. After centrifugation, cells were resuspended in FBS-free medium at a final concentration of 1×106 cells/ml. Before screening small-molecule inhibitors, the SUM-159 cells were treated with inhibitors for 12 h.
After centrifugation, cells were resuspended in FBS-free medium at a final concentration of 1×106 cells/ml. First, with the cell valve closed and the medium valve open, the cells were loaded in the device from the main inlets by the application of pressure via syringes. The flow is directed from the inlets to the outlets through the main chambers. Cells are too large to easily flow through the transverse micro-channels. The cell loading pressure was 3 psi and the pressure of control valves was 15-20 psi. Approximately 100 μl or less of the cell suspension was gently introduced into the chip to seed the cells in close proximity to the channels. Even with fewer than 10,000 cells, the cell migration assay can be performed in the M-Chip. We typically use only part of the device for assays of small samples. In such cases, we use one or two of the twenty groups of channels by applying cells from the access holes individually. The cell loading pressure was 0.5 psi and the pressure of the control valve was 15–20 psi.
During the small-molecule inhibitor screening, SUM-159 cells were treated with the inhibitors for 12 h. The treated cells were harvested by applying 0.25% trypsin-EDTA and centrifugation. The cells were then re-suspended in FBS-free medium at a final concentration of 1×106 cells/ml.
Whole Chip Imaging
Imaging was performed using a high-resolution camera connected to an inverted optical microscope (X81, Olympus, Tokyo, Japan) equipped with a cell culture chamber and objectives of 10×, 20×, and 40× magnification. During the experiment, the temperature of the cell culture chamber was maintained at 37°C. Time-lapse images of the cell migration process were captured, and the cell migration paths were plotted. Velocity of migration of individual cells was quantified by measuring the linear migration distance over time. Total migrated and unmigrated cells were counted and expressed as two fractions. It takes 10 min to take images of the entire chip (10×, bright field, 0.1s/frame, 2058 frames). To get higher resolution images and more detail regarding cell migration, the 20× or 40× objectives should be used for time-lapsed images, which require 30min to 1h for the entire chip.
Data collection and analysis
In our typical 6-h on-chip migration experiments, images of the entire device were taken every 2 h using a fluorescence microscope with stitching function. The images for the entire device at 0 h and 6 h were overlapped and cell movement was tracked by reading cells’ initial and final positions of cell nuclei. We were able to see the cell nuclei in microscopic images. In very rare circumstances, if the images were not sufficiently clear, we measured the distance by averaging the head-to-head and tail-to-tail distances in place of the inter-nuclear distance. Migration distance can thus be quantitated. The migration velocity (distance/time) of individual cells was calculated using the distance migrated at 6 h. The average migration velocity is the average value for a whole chamber or for multiple chambers with the same number of loaded cells. The percentage of migrating cells in each individual chamber was measured to evaluate the migratory capability. We used average migration velocity, percentage of cells migrating, and total migration distance to evaluate the migratory capacity of different cancer cells. We used the total migration distance to mimic the capacity of neoplastic cells to enter lymphatic and blood vessels for dissemination into the circulation. The total migration distance depends on both the migration velocity and percentage of migrating cells. Increases in velocity and percentage of migrating cells increase the total migration distance.
Tumor cell isolation from mouse tumor xenograft
All animal protocols and studies were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute. Orthotopic models from SUM-159 cell lines were established for these studies. Tumor models were established in Athymic nude mice (Charles River Laboratories, Wilmington, MA) by one-time injection of tumor cells/Matrigel 1:1 mixture into the mammary fat pad region. A volume of 100 μl of the cell suspension (1×107 cells/mL) was injected. When the volume of tumor reached approximately 1000 mm3, tumor-bearing mice were sacrificed and tumor was collected. The tumor tissue was digested with collagenase type 3 (2 mg/ml, Worthington Biochemical, Lakewood, NJ) for 1 h at 37 °C. The harvested cell suspension was filtered through a 40–μm nylon cell strainer (BD Biosciences), centrifuged at 200 × g for 5 min, and then re-suspended in PBS. After centrifugation, cells were suspended in DMEM supplemented with no FBS. Obtained tumor cells were loaded into microfluidic chip following the same procedure described above.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge support from the Cancer Prevention and Research Institute of Texas (CPRIT-R1007) and NIH-CA180083.
Reference
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.(a) Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]; (b) Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature reviews. Cancer. 2009;9(4):239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nature reviews. Cancer. 2011;11(8):573–87. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]; (e) Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature reviews. Cancer. 2003;3(6):453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.(a) Ferrer-Vaquer A, Viotti M, Hadjantonakis AK. Transitions between epithelial and mesenchymal states and the morphogenesis of the early mouse embryo. Cell Adhes Migr. 2010;4(3):447–457. doi: 10.4161/cam.4.3.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]; (d) Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–51. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Vedel S, Tay S, Johnston DM, Bruus H, Quake SR. Migration of cells in a social context. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):129–34. doi: 10.1073/pnas.1204291110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rosen P, Misfeldt DS. Cell density determines epithelial migration in culture. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(8):4760–3. doi: 10.1073/pnas.77.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Wei W, Shi Q, Remacle F, Qin L, Shackelford DB, Shin YS, Mischel PS, Levine RD, Heath JR. Hypoxia induces a phase transition within a kinase signaling network in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):E1352–60. doi: 10.1073/pnas.1303060110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) De Bock K, Mazzone M, Carmeliet P. Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nature reviews. Clinical oncology. 2011;8(7):393–404. doi: 10.1038/nrclinonc.2011.83. [DOI] [PubMed] [Google Scholar]; (c) Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15(1):R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(24):5928–35. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) He Q, Shi J. MSN anti-cancer nanomedicines: chemotherapy enhancement, overcoming of drug resistance, and metastasis inhibition. Advanced materials. 2014;26(3):391–411. doi: 10.1002/adma.201303123. [DOI] [PubMed] [Google Scholar]; (f) Barreto JA, O’Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Nanomaterials: applications in cancer imaging and therapy. Advanced materials. 2011;23(12):H18–40. doi: 10.1002/adma.201100140. [DOI] [PubMed] [Google Scholar]
- 6.Munoz-Najar UM, Neurath KM, Vumbaca F, Claffey KP. Hypoxia stimulates breast carcinoma cell invasion through MT1-MMP and MMP-2 activation. Oncogene. 2006;25(16):2379–92. doi: 10.1038/sj.onc.1209273. [DOI] [PubMed] [Google Scholar]
- 7.(a) Lundgren K, Nordenskjold B, Landberg G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. British journal of cancer. 2009;101(10):1769–81. doi: 10.1038/sj.bjc.6605369. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6392–7. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wilson GK, Brimacombe CL, Rowe IA, Reynolds GM, Fletcher NF, Stamataki Z, Bhogal RH, Simoes ML, Ashcroft M, Afford SC, Mitry RR, Dhawan A, Mee CJ, Hubscher SG, Balfe P, McKeating JA. A dual role for hypoxia inducible factor-1alpha in the hepatitis C virus lifecycle and hepatoma migration. Journal of hepatology. 2012;56(4):803–9. doi: 10.1016/j.jhep.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Dhup S, Dadhich RK, Porporato PE, Sonveaux P. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr Pharm Des. 2012;18(10):1319–30. doi: 10.2174/138161212799504902. [DOI] [PubMed] [Google Scholar]; (b) McCarty MF, Whitaker J. Manipulating tumor acidification as a cancer treatment strategy. Alternative medicine review: a journal of clinical therapeutic. 2010;15(3):264–72. [PubMed] [Google Scholar]; (c) Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfor K, Rofstad EK, Mueller-Klieser W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer research. 2000;60(4):916–21. [PubMed] [Google Scholar]
- 9.Zhang Y, Zhang W, Qin L. Mesenchymal-mode migration assay and antimetastatic drug screening with high-throughput microfluidic channel networks. Angewandte Chemie International Edition. 2014;53(9):2344–8. doi: 10.1002/anie.201309885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong B, Ren K, Shu Y, Chen Y, Shen B, Wu H. Recent Developments in Microfluidics for Cell Studies. Advanced materials. 2014;26(31):5525–32. doi: 10.1002/adma.201305348. [DOI] [PubMed] [Google Scholar]
- 11.(a) Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS, Guo D, Hood L, Mischel PS, Heath JR. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(2):419–24. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang W, Kai K, Choi DS, Iwamoto T, Nguyen YH, Wong H, Landis MD, Ueno NT, Chang J, Qin L. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):18707–12. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2480–7. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kamei K, Guo S, Yu ZT, Takahashi H, Gschweng E, Suh C, Wang X, Tang J, McLaughlin J, Witte ON, Lee KB, Tseng HR. An integrated microfluidic culture device for quantitative analysis of human embryonic stem cells. Lab on a chip. 2009;9(4):555–63. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]
- 12.Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, Dusetti NJ, Loncle C, Calvo E, Turrini O, Iovanna JL, Tomasini R, Vasseur S. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(10):3919–24. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Fardin MA, Rossier OM, Rangamani P, Avigan PD, Gauthier NC, Vonnegut W, Mathur A, Hone J, Iyengar R, Sheetz MP. Cell spreading as a hydrodynamic process. Soft Matter. 2010;6:4788–4799. doi: 10.1039/c0sm00252. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiong Y, Rangamani P, Fardin MA, Lipshtat A, Dubin-Thaler B, Rossier O, Sheetz MP, Iyengar R. Mechanisms controlling cell size and shape during isotropic cell spreading. Biophysical journal. 2010;98(10):2136–46. doi: 10.1016/j.bpj.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]; (b) Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Science’s STKE: signal transduction knowledge environment. 2007;2007(407):cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]; (c) Blagosklonny MV. Hypoxia-inducible factor: Achilles’ heel of antiangiogenic cancer therapy (review) International journal of oncology. 2001;19(2):257–62. doi: 10.3892/ijo.19.2.257. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, Crane C, He C, Gondek D, Agharkar P, Savellano MD, Hoopes PJ, Pogue BW. Disparity between prostate tumor interior versus peripheral vasculature in response to verteporfin-mediated vascular-targeting therapy. International journal of cancer. Journal international du cancer. 2008;123(3):695–701. doi: 10.1002/ijc.23538. [DOI] [PubMed] [Google Scholar]
- 16.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. The Journal of cell biology. 2003;160(2):267–77. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.