Abstract
Three common diseases, isolated cleft lip and cleft palate (CLP), hypothyroidism and thyroid cancer all map to the FOXE1 locus, but causative variants have yet to be identified. In patients with CLP, the frequency of coding mutations in FOXE1 fails to account for the risk attributable to this locus, suggesting that the common risk alleles reside in nearby regulatory elements. Using a combination of zebrafish and mouse transgenesis, we screened 15 conserved non-coding sequences for enhancer activity, identifying three that regulate expression in a tissue specific pattern consistent with endogenous foxe1 expression. These three, located −82.4, −67.7 and +22.6 kb from the FOXE1 start codon, are all active in the oral epithelium or branchial arches. The −67.7 and +22.6 kb elements are also active in the developing heart, and the −67.7 kb element uniquely directs expression in the developing thyroid. Within the −67.7 kb element is the SNP rs7850258 that is associated with all three diseases. Quantitative reporter assays in oral epithelial and thyroid cell lines show that the rs7850258 allele (G) associated with CLP and hypothyroidism has significantly greater enhancer activity than the allele associated with thyroid cancer (A). Moreover, consistent with predicted transcription factor binding differences, the −67.7 kb element containing rs7850258 allele G is significantly more responsive to both MYC and ARNT than allele A. By demonstrating that this common non-coding variant alters FOXE1 expression, we have identified at least in part the functional basis for the genetic risk of these seemingly disparate disorders.
Introduction
Isolated cleft lip with or without cleft palate (CL/P) and cleft palate (CP) are common structural birth defects with a strong genetic component that remains incompletely understood. While periconceptional and gestational environmental influences contribute (smoking, alcohol and nutrition), twin, family and population-based studies have clearly established a genetic basis for CL/P etiology. For instance, there is a 40-fold increased prevalence among first-degree relatives compared with within the general population (1). Moreover, genome-wide association studies (GWAS) have identified at least 18 loci in which certain alleles of specific single nucleotide polymorphisms (SNPs) are significantly associated with isolated CL/P (2–5). However, coding mutations have been identified in <10% of patients. This follows the trend of GWAS of other diseases, where a meta-analysis of over 150 GWAS studies revealed only 12% of associated SNPs were in a haplotype block containing coding exons (6). Mutations landing in non-coding DNA are presumed, in many or most cases, to disrupt cis-regulatory elements (CREs) (i.e. promoters and enhancers). However, our understanding of how sequence variation alters the function of regulatory elements remains in its infancy. Consequently, very few mutations that directly elevate risk for CL/P have been identified, hindering progress in the investigation of disease mechanisms (7–8).
We have recently identified a strong peak of association for both CL/P and CP at 9q22, encompassing the gene FOXE1 (9). This association has been replicated in a variety of Caucasian populations (10,11) and Hispanics from Honduras (12). Importantly, FOXE1 is likely to be the relevant gene, because homozygous FOXE1 mutations cause Bamforth–Lazarus syndrome which is characterized by cleft palate, choanal atresia, bifid epiglottis, thyroid agenesis or disgenesis, hypothyroidism and spikey hair (13–15). Inactivation of FOXE1 in mice causes a similar phenotype (16). We have found FOXE1 coding mutations in only 1% of subjects, despite exhaustive mutation screens (9). This is incongruent with estimates that FOXE1 has a 25–38% attributable risk, suggesting that causal variants occur in nearby cis-regulatory elements.
Consistent with thyroid defects in patients with Bamforth–Lazarus syndrome, a variety of common thyroid diseases also map to FOXE1, but causal variants for these disorders at this locus remain largely unknown. Thyroid disorders mapping to 9q22 include congenital hypothyroidism due to thyroid dysgenesis (17–19), hypothyroidism (20,21), goiters, non-medullary thyroid cancer (22,23), papillary thyroid carcinoma (24–27), radiation induced papillary thyroid carcinoma as a result of the Chernobyl accident (28,29), and thyroid cancer (30–32). Also, a number of biomarkers of thyroid metabolism are also associated at this locus (27,30,33–37). These observations are consistent with evidence that FOXE1 is involved in thyroid homeostasis, negatively regulating thyroglobulin and thyroperoxidase expression (38). Interestingly, a SNP associated with papillary thyroid cancer, rs1867277, is located in the FOXE1 promoter and alters binding of the USF1/USF2 transcription factors (26). However, rs1867277 is unlikely to explain all of the risk for thyroid cancer at this locus, because other SNPs show more significant association, and because the haplotype block structure in the region suggests more than one risk allele is present for these diseases. Although FOXE1 coding mutations may be present in a subset of patients with thyroid conditions mentioned earlier, it is probable that additional causative variants in non-coding DNA remain to be identified.
We hypothesized that causal mutations for orofacial clefting and thyroid diseases occur within cis-regulatory elements at the human FOXE1 locus. To that end, we screened evolutionarily conserved sequences of human DNA at the FOXE1 locus (Fig. 1) for craniofacial and thyroid enhancers using both transgenic zebrafish and mouse strategies that have been shown to effectively identify cis-regulatory elements (39,40). We identified three such elements, including one containing a SNP that is associated with CL/P, hyperthyroidism and thyroid cancer. In vitro quantitative reporter assays in embryonic oral epithelial and thyroid cells demonstrated that different alleles of this SNP altered the level of enhancer activity of the element.
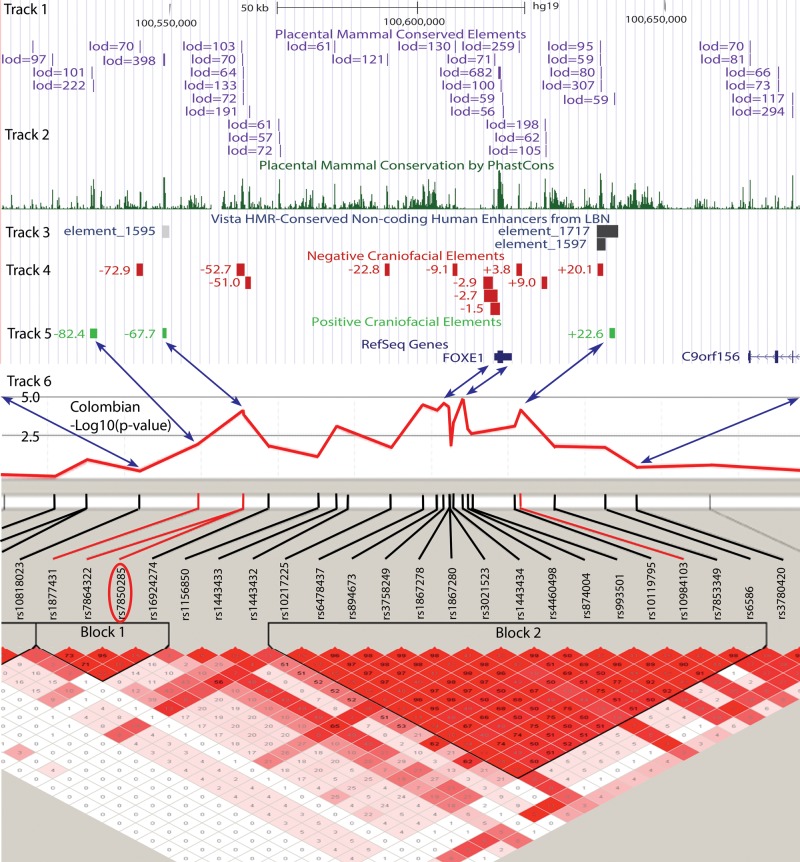
Figure 1.
Genomic overview of the FOXE1 locus. Shown is a view from the UCSC Genome Browser of a 152 kb region bracketed by the SNPs rs10818023 and rs3780420. Track 1 shows genomic regions with a mammalian PhastCons score of >400. Track 2 is a histogram plot of mammalian conservation. Track 3 is from the Vista Enhancer indicating tissue specific enhancers (http://enhancer.lbl.gov/). Tracks 4 and 5 show the location of the tested conserved elements relative to the FOXE1 start codon and depicted by green and red boxes, representing negative and positive enhancer activity, respectively. Track 6 shows association with CL/P in the Colombian population. The linkage disequilibrium (LD) plot shows the D′ correlation between any given SNP pair, red indicating a maximum D′ of 1, pink shades ranging through values D′ < 1 with white indicating no correlation. Blue arrows show the correspondence between the physical genomic and recombinant genetic distances. Circled in red is SNP rs7850258 that in addition to being the most significantly associated SNP with CLP in block 1 is also the most significantly associated SNP in genome-wide association studies of hypothyroidism or thyroid cancer.
Results
Endogenous foxe1 expression during zebrafish development
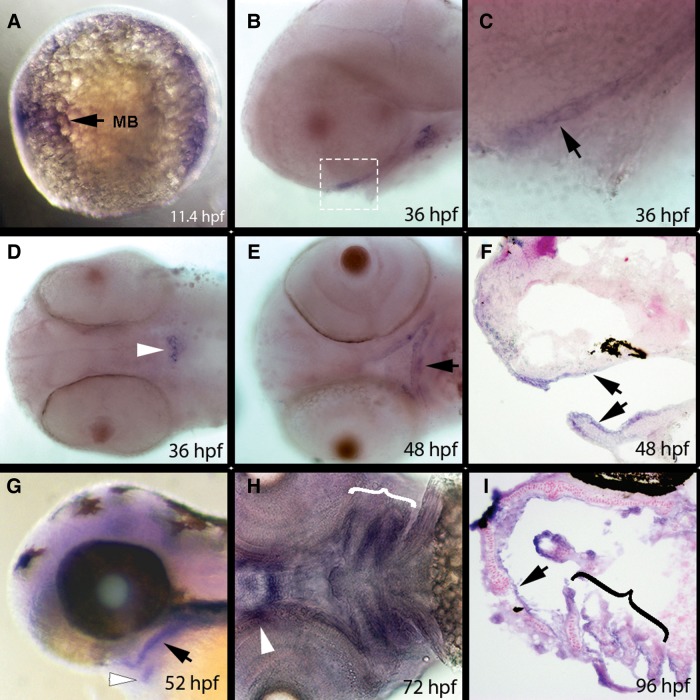
We reasoned that oral and thyroid enhancers near human FOXE1 would be more likely to be functional in zebrafish if the fish ortholog, foxe1, were expressed in oral tissues and thyroid gland. Indeed, RNA in situ hybridization revealed foxe1 expression in the central nervous system at 11.4 h post fertilization (hpf) (Fig. 2A), in oral epithelium at 36 hpf (Fig. 2B, C, E and F, data not shown) and thyroid gland (Fig. 2D and G) at 36 hpf and later stages. We detected foxe1 expression in pharyngeal arch epithelium at 72 and 96 hpf (Fig. 2H and I). Furthermore, we observed foxe1 expression in the developing heart at 48 hpf, the pectoral fin starting at 72 hpf and the gill rakes (data not shown). These observations confirm and extend a previous expression analysis (41).
Figure 2.
Endogenous foxe1 expression during zebrafish development. In situ hybridization of foxe1 on embryos fixed at the indicated stages. All embryos are presented with anterior to the left. (A) Lateral view of an 11.4 hpf embryo. Expression is evident in the central nervous system at approximately the level of the midbrain (mb). (B and C) Lateral views of a 36 hpf embryo. Expression is evident in oral epithelium (arrow in C). White box in B indicated region shown in C. (D) Ventral view of a 36 hpf embryo. Expression is visible in the developing thyroid (arrowhead). (E) Ventral view of a 48 hpf embryo. Expression is evident in oral epithelium (arrow). (F) Sagittal section of a 48 hpf embryo. Expression is seen in oral epithelium (arrowheads). (G) Lateral view of a 52 hpf embryo. Expression is seen in oral epithelium (oe, black arrow), and in thyroid (white arrowhead). (H) Ventral view of a72 hpf embryo. Expression is seen in epithelium of the pharyngeal arches (bracket) and in oral epithelium around mouth (arrowhead). (I) Transverse section of a 96 hpf embryo. Expression is seen in the oral epithelium (black arrowhead) and pharyngeal arches (bracket). MB, midbrain. hpf, hours post fertilization.
Enhancer screen results
We amplified from human DNA, 15 human conserved non-coding elements (hsCNEs) in a 152 kb region spanning from 101 kb upstream and 51 kb downstream of FOXE1 and tested them for enhancer activity in transient transgenic zebrafish (Fig. 1; Supplementary Table 1). The hsCNEs are named by their position relative to the FOXE1 start codon, negative being upstream and positive downstream. Our zebrafish enhancer screen was based on an eGFP reporter, pT2cfosGW eGFP (39). This reporter construct is designed to clone the human test DNA upstream of the mouse minimal cfos promotor that is unable to independently initiate mRNA transcription. To confirm this, we injected >200 single cell embryos with the reporter lacking any human sequences and did not detect any GFP expression. In contrast, of 15 hsCNEs, three had enhancer activities capable of directing transcription in a pattern consistent with endogenous foxe1 expression (hsCNE+22.6, hsCNE-67.7 and hsCNE-82.4; Supplementary Material, Table S1), as summarized below. These positive hsCNEs acted independently and without the FOXE1 promoter.
FOXE1 hsCNE+22.6 is active in the developing pharyngeal arches and heart
In embryos injected with FOXE1 hsCNE+22.6 kb element (hg19 chr9: 100 638 752–100 639 947), we detected a reproducible pattern of eGFP expression in a mosaic pattern within pharyngeal muscles, a subset of cranial muscles (see Fig. 3 legend), heart and pharyngeal arch epithelium (n > 800 embryos scored) (Fig. 3). We established five independent, stable transgenic lines and all exhibited non-mosaic GFP expression in the same structures that expressed GFP mosaically in transient transgenic G0 embryos. In stable transgenic embryos, GFP was evident in the developing mouth by 24 hpf (data not shown) and by 72 hpf the muscles of the second and branchial arch were labeled along with the branchial arch cartilages including the gill rakes (Fig 3A–C). Moreover, this element also directed GFP expression in the developing heart and pectoral fins (Fig. 3A–C). This pattern was confirmed by in situ hybridization with an eGFP probe. While aspects of this pattern matched that of foxe1 expression, for instance in epithelium of ceratobranchial arches (e.g. compare Fig. 3D to Fig. 1H), however expression of foxe1 mRNA was not detected in pharyngeal muscles. If human FOXE1 is also absent from pharyngeal muscles, we infer that additional regulatory elements must normally prevent the enhancer at +22.6 kb from driving FOXE1 expression in this tissue. Alternatively, through divergence in tissue specific expression of upstream regulatory factors, this enhancer may be active in pharyngeal muscles only in fish, although ENCODE data show this region has chromatin marks of enhancer activity in human skeletal muscle myoblasts.
Figure 3.
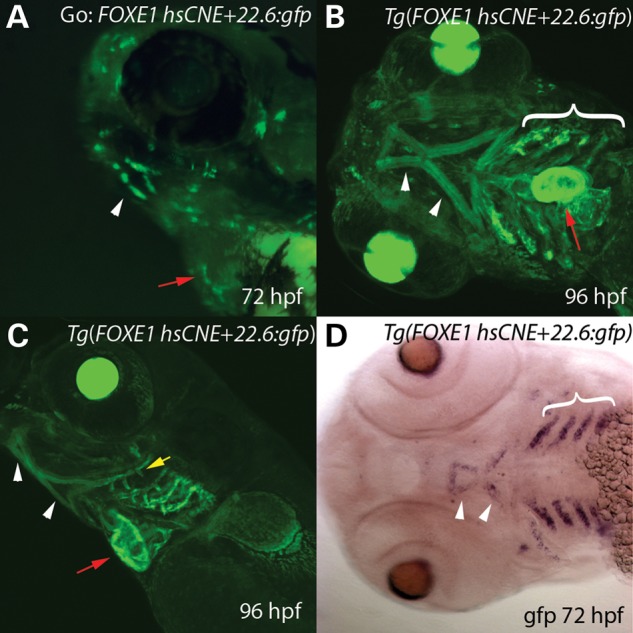
FOXE1 hsCNE+22.6 kb element drives GFP expression in pharyngeal muscles, the heart and pharyngeal epithelium. (A–C) Compressed Z-stacks from confocal imaging of live (A) transient (G0), and (B,C) stable (F1) transgenic Tg(foxe1+22.6:gfp) animals. (A) Oblique lateral view of a G0 72 hpf embryo mosaic for Tg(foxe1+22.6:gfp). GFP is visible in cells in the region of the developing mouth (white arrow) and in the heart (red arrow). (B) Ventral and (C) lateral views of a 96 hpf larvae showing eGFP expression in the pharyngeal arches (bracket), heart (red arrow) and the ventral pharyngeal muscles (arrowheads) including the hyohoideus muscle, interhyoideus, intermandibularis anterior and intermandibularis posterior muscles. Also labeled are the gill rakes (yellow arrow). (D) Ventral view of 72 hpf Tg(foxe1+22.6:gfp) embryo processed to reveal gfp expression by RNA in situ hybridization, which is evident in the pharyngeal arches (bracket) and ventral pharyngeal muscles (arrowheads). The heart, which was positive for gfp expression, was removed in the specimen shown here.
While hsCNE+22.6 was able to activate lacZ expression in human embryonic oral epithelial cell lines, no difference was observed between the two rs10984103 alleles (Supplementary Material, Fig. S1).
FOXE1 hsCNE-67.7 drives expression in the developing oral epithelium, hyoid arch, branchial arches, thyroid and heart
In transient transgenic embryos injected with the FOXE1 -67.7 element (hg19 chr9: 100 548 460–100 549 280), we reproducibly detected GFP expression in the jaw, hyoid and pharyngeal arches, thyroid and heart (data not shown). The same pattern was observed in five independent transgenic lines and in these, expression in the oral epithelium was more evident than in transient transgenics (Fig. 4A and B). Expression in oral epithelium and thyroid precursors was detectable by 24 hpf, while heart expression became evident at 48 hpf (data not shown). Staining with the thyroid specific T4 antibody demonstrated coincident signal with eGFP within the thyroid in the hsCNE-67.7 transgenic fish (data not shown).
Figure 4.
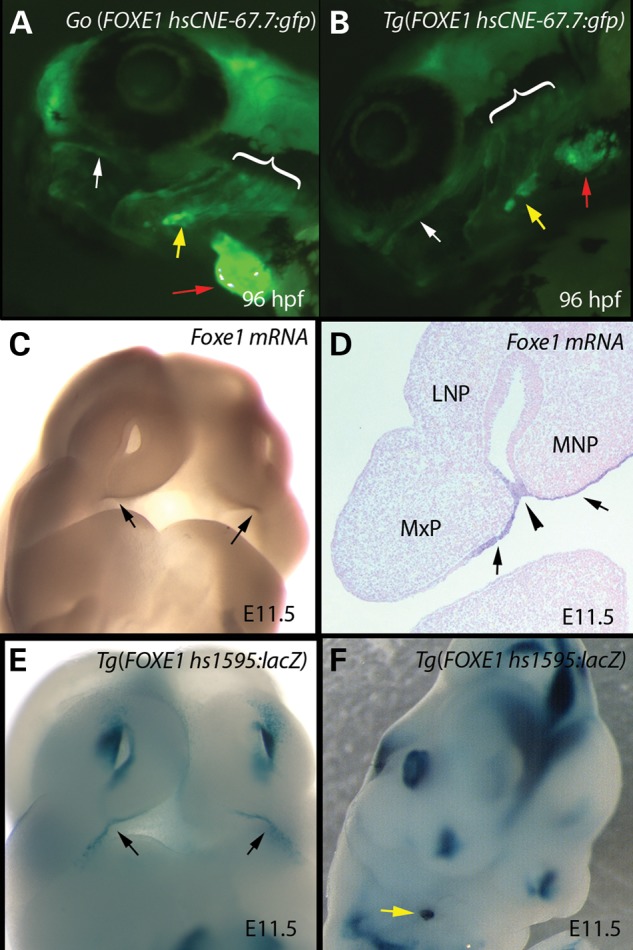
FOXE1 hsCNE-67.7 kb element drives GFP expression in the oral epithelium, pharyngeal arches, thyroid and heart tissues. (A and B) Compressed Z-stacks from confocal imaging of live stable transgenic Tg(foxe1-67.7:gfp) animals from two independent lines. (A and B) Oblique lateral views of 96 hpf larvae showing GFP expression in the oral epithelium (white arrows); pharyngeal arches (brackets), thyroid (yellow arrows) and heart (red arrows). (C) Oblique frontal view of E11.5 mouse embryo showing endogenous Foxe1 expression in the oral epithelium at the juncture of the medial nasal and maxillary processes (arrows). (D) Frontal section of E11.5 mouse embryo showing endogenous Foxe1 expression in the oral epithelium of the medial nasal and maxillary processes (arrows), including the point of fusion (arrowhead) between them that results in the formation of the upper lip. (E and F) Mouse embryos transiently transgenic Tg(hs1595:lacZ) stained for B-galactosidase. hs1595 corresponds to hg19 chr9: 100 548 557–100 549 640) overlapping the Tg(foxe1-67.7:gfp) tested in zebrafish. (E) Oblique frontal view of E11.5 mouse embryo showing β-galactosidase staining in the oral epithelium at the juncture of the medial nasal and maxillary processes (arrows). (F) Oblique view of E11.5 mouse embryo showing β-galactosidase staining in the developing thyroid (yellow arrow).
We next tested an overlapping region, (hg19 chr9: 100 548 557–100 549 640) in transient transgenic mouse embryos using a β-galactosidase reporter with an Hsp68 minimal promotor. This region, Vista Enhancer hs1595, drove reporter expression in a pattern matching endogenous foxe1 expression in the oral epithelium and developing thyroid in three of four mouse embryos (Fig. 4C–F, Supplementary Material, Fig. S2). In summary, the FOXE1 hsCNE-67.7 element has enhancer activity in multiple oral tissues, in the heart and the thyroid gland during development in zebrafish, and an overlapping human DNA element has similar activity in mouse embryos.
FOXE1 hsCNE-82.4 directs expression in the oral epithelium in zebrafish embryos
For the two hsCNEs just described, the expression patterns in stable transgenic lines could be accurately predicted from summing the expression patterns in G0 embryos. Therefore, we screened additional elements solely in G0 embryos. G0 embryos injected with the FOXE1 hsCNE-82.4 element (hg19 chr9: 100 533 759–100 535 187) showed eGFP expression initially in the presumptive oral epithelium at 24 hpf until at least 96 hpf, with additional signal in the pharyngeal region (Fig. 5) (n > 370 embryos injected). We observed such expression in 7–19% of embryos in seven different injection batches, a rate comparable to those observed for the elements above (Table 1).
Figure 5.
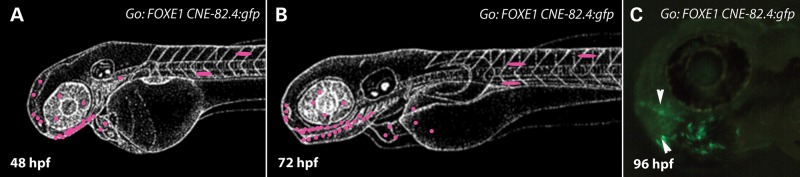
FOXE1 hsCNE-82.4 directs GFP expression in the oral epithelium. (A and B) Lateral view line drawings representing a composite of transient G0 embryos at 48 and 72 hpf, showing eGFP labeled cells highlighted in pink. GFP expression is primarily in the oral epithelium. (C) Lateral view of a representative G0 larvae at 96 hpf showing expression in the oral epithelium (white arrowheads). In all over 370 embryos were screened from seven different injection batches.
Table 1.
Scoring of expression in Go embryos
| FOXE1 hsCNE | Total live embryos | Expressers (%) | Expression in oral or pharyngeal regionsa |
Number transgenic lines | |||
|---|---|---|---|---|---|---|---|
| 24 hpf (%) | 48 hpf (%) | 72 hpf (%) | 96 hpf (%) | ||||
| +22.6 | 822 | 41 | 29 | 18 | 13 | 11 | 5 |
| −67.7 | 262 | 30 | ND | ND | ND | ND | 5 |
| −82.4 | 453 | 20 | 19 | 17 | 13 | 7 | 0 |
aPercent of live embryos.
Differential enhancer activity for rs7850258 alleles in FOXE1 hsCNE-67.7
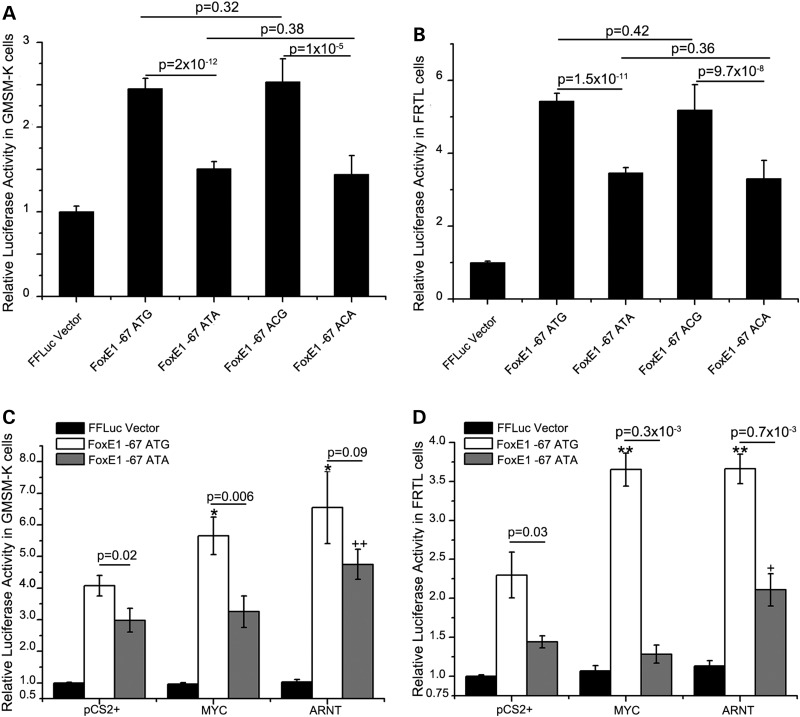
Interestingly, rs7850258, for which one allele (G) is associated with hypothyroidism and CLP, and another (A), with thyroid cancer, is present within the FOXE1 hsCNE-67 element. We engineered variants of this element, containing one or the other alleles of SNP rs7850258, into a vector containing a minimal promoter and the gene encoding firefly luciferase. Since Foxe1 is expressed in the oral epithelium during facial development (9), an embryonic oral epithelial cell line that endogenously expresses FOXE1, GMSM-K (human fetal oral epithelial cells), was used in these assays. For assessment in the context of thyroid function, the rat FRTL epithelial thyroid cell line that expresses Foxe1 under hormonal control was used (42). In both cell types, the enhancer activity of the hsCNE-67 was confirmed with significant increases (1.5–5.5-fold) above the empty vector (Fig. 6 A and B). Moreover, in both cell types, the rs7850258 G allele had statistically significant greater enhancer activity, ranging from 1.2- to 1.7-fold more than the rs7850258 A allele (Fig. 6A and B). rs7864322 is another SNP within the hsCNE-67.7 enhancer that is also associated with CL/P (9), raising the possibility that it has functional effects. Yet testing all allelic combinations with rs7850258 in both GMSM-K and FRTL cells revealed no difference between the two rs7864322 alleles (C/T) whether they were on either the rs7850258 A or G alleles (Fig. 6A and B). These results suggest that rs7850258 is the functional variant for both oral epithelial and thyroid diseases. Transient transgenic zebrafish assays, scored by two individuals blinded to the rs7850258 genotype, did not reveal any statistically significant differences in GFP expression patterns, supporting conclusion that variation at rs7850258 alters the level of expression rather than the tissue specificity of expression (data not shown).
Figure 6.
rs7850258 alleles differentially affect FOXE1 hsCNE-67 enhancer activity in human embryonic oral epithelial and rat thyroid cell lines. Different alleles of rs7864322 (C/T) and rs7850258 (A/G) in the FOXE1 hsCNE-67.7 element were cloned pTol2-cFos-FLuc (firefly luciferase vector with a c-fos minimal promoter). The same genotype for the other SNP in hsCNE-67, rs1006125 (A), was maintained in all constructs. The clones are named ATG, ATA, ACG and ACA designating the sequential genomic order of rs1006125, rs7864322 and rs7850258 within the constructs. The ATA and ATG constructs along with Renilla luciferase (pTol2-cFos-RLuc) were separately transfected into (A and C) GMSM-K human oral epithelia cells and (B and D) FRTL rat thyroid cells. Luciferase measurements were normalized across transfections using Renilla luciferase readings and then the firefly luciferase readings were normalized to the empty Firefly luciferase vector. (A and B) In each cell line, the rs7850258 G allele had increased enhancer activity compared with the A allele. (C and D) rs7850258 alleles differentially respond to MYC and ARNT. Since there was no differrence between the ATG versus ACG or ATA versus ATA, only the ATG and ATA clones described earlier were tested for differential response to cotransfected human MYC and ARNT. In both cell lines, the rs7850258 G allele had increased enhancer activity compared with the A allele in the presence of either MYC or ARNT. */+ P-value <0.05 and **/++ P-value <0.01. The data in each column represent the mean ± SEM of three measurements each of four biological replicates.
Using JASPAR (43), it was predicted that there was a gain of E-box transcription factor binding site to the rs7850528 G allele that could be bound by the basic-helix-loop-helix transcription factors MYC and ARNT (Supplementary Material, Table S2). Co-transfection of either human MYC or ARNT along with the reporter construct of hsCNE-67.7 showed differential allelic activation with the G allele being more strongly activated by both transcription factors in both human embryonic oral epithelial and rat thyroid cell lines (Fig. 6C and D).
Discussion
Zebrafish and mouse orthologs of FOXE1 are expressed in the oral epithelium and in the thyroid gland consistent with role for FOXE1 in diseases of both tissues
We have confirmed in zebrafish that foxe1 is expressed in the developing oral and pharyngeal epithelium, thyroid gland and pectoral fins. Furthermore, foxe1 is expressed in the heart, and in the developing midbrain to hindbrain. While foxe1 expression in central nervous system has not been explicitly mentioned previously, it is evident in an earlier study in zebrafish [see Fig. 1 of (41)]. Similarly, in mice Foxe1 expression in the developing central nervous system and heart has not been previously reported, but a review of the literature shows expression in the midbrain and hindbrain on embryonic Day 10.5 (44). Furthermore, in the public domain via EuroExpress, Foxe1 mRNA expression is evident in the midbrain, cerebellum and heart outflow tracts in E14.5 mouse embryos (Supplementary Material, Fig. S3) (45). This is consistent with RT-PCR data showing in zebrafish adults very strong expression in the heart and brain (41).
Identification of human tissue specific enhancers at the FOXE1 locus
Using an strategy based primarily on sequence conservation, we tested 15 regions of human DNA totaling 14.9 kb, encompassing 9.8% of the human FOXE1 locus as defined by the CL/P critical region (9). Of these, three showed positive tissue specific enhancer activity in patterns consistent with endogenous foxe1 expression. All three enhancers, hsCNE+22.6, hsCNE-67.7 and hsCNE-82.4 shared the same capacity to independently regulate expression in the oral and pharyngeal epithelium, and pharyngeal arch structures. Two of these, hsCNE+22.6 and hsCNE-67.7 are also heart and pectoral fin enhancers. Unique is the ability of hsCNE-67.7 to direct expression in the developing thyroid. The oral epithelial and thyroid activity of the -67.7 element was also confirmed in transient transgenic mice. In contrast, the enhancer activity of hsCNE-82.4 is primarily confined to the oral epithelium.
In several previous studies, human DNA, which is not detectably conserved to zebrafish, has been shown to nonetheless possess enhancer activity in zebrafish. Examples include enhancers for RET, a gene for Hirschsprung disease (46), for PHOX2B, which is associated with neuroblastoma and Crohn's disease (47–49), and one for NOS1AP that contains a functional SNP associated with cardiac QT interval (50). Similarly, the FOXE1 enhancers that we have identified here lack detectable conservation to zebrafish, the longest stretch of 100% identity being 26 bp and for any identity >85% being 47 bp of which none map to the zebrafish foxe1 locus (Supplementary Material, Table S1). While the interpretation of CNE-82.4 as a facial enhancer is based entirely on transient transgenic zebrafish embryos, our experience and published work has shown expression patterns in transient transgenic zebrafish embryos accurately reflect that found in founder lines (51,52). Nonetheless, there is no guarantee that every element conserved in mammals will have enhancer activity in zebrafish, and therefore negative results, as we observed for 12 of 15 tested elements, cannot be strongly interpreted. Alternative explanations for the negative results could be those elements act as silencers or insulators, which would not have been detected using the reporter construct in this study. Additional craniofacial enhancers for FOXE1 are likely to exist; criteria other than conservation, for instance chromatin marks in appropriate cell lines, may be of use to identify them.
In retrospect to see if we could improve upon our 20% success rate when choosing human candidate elements based on conservation, we compared our results to available ENCODE data. Only the hsCNE+22.6 enhancer had chromatin marks and an abundance of ChIP-Seq peaks, clearly indicating regulatory activity. Beyond that, we could not identify a matching pattern for either positive or negative elements with chromatin marks for regulatory function in any cell type. This underscores the need to have such data available on the relevant cell type. This is especially true for studying embryonic structural birth defects for which there is a paucity of data resources.
Relevance to human diseases
The expression of FOXE1 in the developing heart suggests it may be associated with cardiac defects. To date, variations at the FOXE1 locus have not been associated with cardiac anomalies. Yet tangential data exist in that among infants with congenital hypothyroidism there is a 3–5-fold over representation of heart defects (53,54), suggesting they may have shared developmental processes.
At the FOXE1 locus, based on association peaks and LD blocks in different populations, there is evidence for three CL/P risk haplotypes relative to the start codon: −89 to −59 kb; −19.2 to +0.4 kb; and +7.0 to +81 kb (9). We postulate that rs7850258 within hsCNE-67.7 explains entirely or in part the risk for CL/P associated with the most upstream risk haplotype. The facial enhancer, hsCNE+22.6, containing rs10984103, the SNP most strongly associated with CL/P in the Filipino population, likely has a role in the most downstream risk haplotype. However, there was no statistical difference between the two rs10984103 alleles.
The discovery of the hsCNE-67.7 thyroid enhancer properties is noteworthy given the number of thyroid diseases ascribed to the FOXE1 locus. It contains rs7850258, that has the most significant association in GWAS studies of hypothyroidism [P = 2.5 × 10−19 (20); P = 3.96 × 10−9 (21)] and papillary thyroid cancer (P = 1.7 × 10−9) (21,29). We show here that rs7850258 alters enhancer function in both oral epithelial and thyroid cell lines. Of interest is that the G allele is associated with increased risk for CLP and hypothyroidism, while the A allele is associated with thyroid cancer. Relative to the pathogenesis of CLP, that the G allele has more enhancer activity is consistent the gain of function mutation characterized in one patient with Bamforth–Lazarus syndrome (55). Additionally, over expression of Foxe1 in mice also leads to cleft palate (56). One possible mechanism proposed for the association with hypothyroidism is that ectopic FOXE1 induces apoptosis in the developing thyroid, leading to agenesis or dysgenesis (55).
The decreased enhancer activity of the A (cancer associated) allele of rs7850258 also implies that FOXE1 is a tumor suppressor. Evidence for this exists in that the RET/PTC3 proto-oncogene caused by chromosomal rearrangements leading to thyroid papillary carcinoma is known to decrease FOXE1 expression (57). Furthermore, somatic loss of the 9q22 region containing FOXE1 is frequent in squamous cell carcinoma (58). Moreover, hypermethylation of the FOXE1 promoter and/or decreased FOXE1 expression have been observed in a number of cancers including squamous cell carcinoma, (59), pancreatic cancer (60), adenoid cystic carcinoma of salivary gland (61), breast cancer (62), colorectal cancer (63) and anaplastic carcinomas (64). Additionally, another risk allele at rs965513 for papillary thyroid carcinoma is associated with decreased FOXE1 expression in thyroid tissue (65). In total, these reports are also consistent with FOXE1 being pro-differentiation in the developing thyroid and differentiation maintenance in the adult thyroid (44,65).
rs7850258 is predicted to disrupt ARNT and MYC transcription factor binding and consistent with this our data shows a significant response to either factor when the binding site is present in contrast to no response when absent. rs7850258 affects an E-box binding site, so it is possible that other E-box binding transcription factors may be involved as well. That being said there is evidence for both ARNT and MYC being involved in orofacial clefting and thyroid diseases.
ARNT and ARNT2 are basic helix-loop-helix transcription factors that are expressed in the palatal shelf epithelium (66–68). Furthermore, ARNT2 is expressed in the developing thyroid (66). ARNT and ARNT2 regulate gene expression through dimerization with the aryl hydrocarbon nuclear receptor (AHR). AHR responds to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a well-known teratogen that causes cleft palate in mice (69) and is associated with clefting in humans through agricultural chemical exposures (70). TCDD is also found in tobacco smoke that through maternal gestational exposure attributes to 11% of orofacial clefts (71). Moreover, a genetic variation at the ARNT locus is associated with CL/P in humans (72). While there is convincing data in animal models demonstrating TCDD exposure interferes with fetal and newborn thyroid function, epidemiological studies with equivocal results have not shown a consistent correlation in humans (73). There have not been any reports of association between ARNT, ARNT2 or AHR with hypothyroidism or thyroid cancer in any human studies.
c-MYC is also a b-HLH protein that is expressed in both the epithelia and mesenchyme of the facial processes (74,75). Conditional inactivation in cranial neural crest cells results in craniofacial anomalies (76) and deletion of a facial enhancer at the Myc locus results in cleft lip in conjunction with decreased facial Myc expression. Increased MYC expression is associated with undifferentiated thyroid cancer and the loss of thyroid differentiation markers, PAX8 and TTF1 (77). So in theory, the rs7850528 cancer associated allele A would be less responsive to MYC, resulting in relative decrease of FOXE1 expression compared with the non-risk cancer allele G. This model is consistent with FOXE1 being a tumor suppressor.
A final caveat in this discussion is that until there is association data based on comprehensive sequencing across the region in individuals with CLP and thyroid diseases, it is not possible to state whether rs7850258 is the main contributing factor or if there are additional functional variants on a shared haplotype that in total comprise the genetic risk for these disorders at this locus.
In summary, using zebrafish and mouse transgenesis, we have identified craniofacial and thyroid enhancers at the FOXE1, including one that contains functional risk alleles for both CLP and thyroid diseases. These results reveal the pathogenesis of these disorders associated with the FOXE1 locus. We anticipate that there are additional genetic and biological mechanisms at this locus yet to be discovered that contribute to risk for these disorders. Given that thyroid diseases and orofacial clefting converge on the same SNP within a thyroid and facial enhancer, there may a shared risk for both diseases within families or a population.
Materials and Methods
Zebrafish husbandry
Parental fish were housed at room temperature overnight prior to breeding. Embryos were raised at 28.5°C. The ethical use of animals for research was approved by the University of Iowa The Institutional Animal Care and Use Committee.
In situ hybridization
A 870 bp Foxe1 probe was cloned using primers [forward—ATGCCTGTGGTTAAAGTGGAGAGT (chr1: 25 668 976–25 668 999; Zv9/danRer7) and reverse—GCACTCAGCATTATGGGCCA (chr1: 25 668 130–25 668 149; Zv9/danRer7)] to amplify from a zebrafish cDNA library. The probe was sequenced and by BlastN search of the Zebrafish genome, 100% identity over the entire 870 bp probe was observed for only foxe1 (ref|XM_690973.3|) at chr1: 25 668 130–25 668 999 in Zv9/danRer7. The next level of BlastN identity was 79% with Foxe3 for only 328 base pairs. Based on these results and those published by Nakada et al. (41), there is only one zebrafish foxe1 gene and that this gene has been given the gene ID number ENSDARG00000079266 in Ensembl. In situ hybridization was performed as previously described (78). Given the stringency conditions used, it is highly unlikely that hybridization of the full length probe would occur with anything other than the endogenous foxe1 mRNA.
Selection of potential regulatory elements
To be conservative relative to boundaries established by our CL/P association data, we have targeted a 152 kb interval between hg19 Chr9: 100 514 919–100 666 931 (rs10818023 to rs3780420) (Fig. 1). We chose 15 conserved non-coding human sequences (hsCNE) using publicly available sequence data (UCSC) and the Phastcons algorithm (79) with a mammalian Phastcons score of >400 as inclusion criteria (Supplementary Material, Table S1). Fourteen of our chosen elements met these criteria. One additional element, hsCNE-2.9, was added since it contained a region overlapping with the rat Foxe1 promotor (80). For all 15 regions there was no identity within the Zebrafish genome for more than 30 base pairs.
Zebrafish enhancer screen
Potential regulatory elements were PCR amplified from human BAC clones RP11-10012 or RP11-151120, cloned into the Gateway® (Invitrogen) pENTR/D-TOPO vector and transferred to the pT2cfosGW eGFP reporter construct that is constructed on the Gateway® system (39). The pT2cfosGW eGFP reporter is designed to clone the human test DNA upstream of the mouse minimal cfos promotor that is unable to independently initiate mRNA transcription. Also, built into the pT2cfosGW vector are Tol2 recombination sites bracketing the entire construct that facilitate integration of the expression construct into the zebrafish genome thus reducing cell mosaicism within the injected embryos. All PCR products and plasmid constructs were sequenced and the sequence results were aligned onto the human genome using the UCSC tool BLAT (81) to ensure fidelity of these steps. Tol2 mRNA was transcribed from the plasmid pCS-TP (82).
Constructs were injected along with the tol2 mRNA into 100–200 zebrafish embryos at the 1 cell stage. The developing embryos (generation zero; Go) were screened at 24, 48, 72 and 96 hpf for eGFP expression patterns consistent with endogenous foxe1 expression. A consistent pattern of expression in a minimum of 10% of injected fish was the criteria for tissue specific enhancer activity (83). The use of recombinant DNA was approved by University of Iowa Institutional Biosafety Committee.
Mouse enhancer assay
The methods for evaluating conserved non-coding sequences in transgenic mice have been previously described as part of the Vista Enhancer project (84). Briefly, target regions were amplified from human genomic DNA, cloned into an HSP68-LacZ reporter vector, and sequence validated. The linearized vector was injected into the pronucleus of FVB oocytes to generate transgenic embryos. LacZ activity was assessed at E11.5 for craniofacial staining. The results for Vista Enhancer hs1595 have not been previously reported in detail.
Luciferase reporter constructs, transfections and luciferase assays
To assess differential allelic activity for the rs10984103 CL/P risk SNPs within hsCNE+22.6, the alleles (A/C) two were introduced by PCR mediated mutagenesis (85) into the hsCNE+22.6 enhancer and subsequently subcloned into pTol2-cFos-FLuc (firefly luciferase vector with a c-fos minimal promoter) using LR reaction according to the manufacture's protocol (Life Technologies). In a similar fashion different combinations of the alleles for rs7850258 (A/G) and rs7864322 (C/T) were introduced into hsCNE-67.7, keeping the same allele (A) for the other SNP, rs1006125 that also resides in this enhancer. Sequencing was used to ensure clone integrity. Transient transfections were performed using X-tremeGENE HP (Roche, Germany) into GMSM-K or FRTL cells with every firefly luciferase vector. For each construct, three independent transfections were performed with Renilla luciferase (pTol2-cFos-RLuc) co-transfection, as an internal control for transfection efficiency. The Dual-Luciferase Reporter Assay System (Promega, Madison, USA) and a luminometer were used to measure luciferase activity in cell lysates. All quantified results are presented as mean ± SEM. Three luciferase measurements were made on each of three or four independent biological replicates. A two-tailed unpaired Student's t-test was used to determine statistical significance.
Predictions of differential allelic transcription factor binding for rs7850258 were performed using JASPAR (43). Ten base pairs surrounding rs7850258 GCCAGa/gTGTCT was evaluated and relative score of >0.85 was used as a threshold for significance.
The human ARNT expression vector is from GeneCopoeia (Rockville, MD, USA) plasmid #EX-C0312-M68 and the human MYC expression vector is from Addgene (Cambridge, MA, USA) plasmid #18773: MSCV MYC T58A puro (86).
Cell culture
GMSM-K a human fetal oral epithelial cell line (a kind gift from Dr Daniel Grenier) (87) was maintained in keratinocyte serum-free medium (Life Technologies) supplemented with EGF 1–53 and bovine pituitary extract (Life Technologies) in a 5% CO2 atmosphere at 37°C. GMSM-K cells constitutively express FOXE1. FRTL a rat thyroid cell line was purchased from ATCC (CRL-1468; 42) and cultured in Ham's F-12K (Kaighn's, Life Technologies) supplemented with 10 mU/ml TSH (Sigma, St Louis, USA), 0.01 mg/ml insulin (Sigma), 10 nm hydrocortisone (Sigma), 0.005 mg/ml transferrin (Sigma), 10 ng/ml somatostatin (Sigma), 10 ng/ml glycyl-l-histidyl-l-lysine acetate (Sigma), and 0.5% FBS. In order to induce the expression of Foxe1, the concentration of TSH was increased to 30 mU/ml.
Supplementary Material
Funding
This publication was supported by the National Institutes of Health (RO1 DE014667), the University of Iowa Department of Orthodontics and the National Center for Advancing Translational Sciences through Grant 2 UL1 TR000442-06 (A.C.L.). RO1 DE021071 and the National Science Foundation IOS-1147221 (R.A.C.). U01DE020060 (A.V.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplementary Material
Acknowledgements
We thank Dr Shannon Fisher for sharing the pT2cfosGW eGFP reporter construct. Also, Dr Koichi Kawakami of Division of Molecular and Developmental Biology, National Institute of Genetics and Department of Genetics, Graduate University of Advanced Studies, Mishima, Shizuoka, Japan generously provided the Tol2 plasmid pCS-TP. Dr Daniel Grenier at the Université Laval, Québec graciously provided the GMSM-K human fetal oral epithelial cell line. Youssef Kousa aided with Figure construction. We greatly appreciate Drs Rick Domann and Adam Dupuy for sharing the ARNT and MYC expression plasmids.
Conflict of Interest statement. None declared.
References
- 1.Mitchell L.E., Christensen K., et al. (1997) Evaluation of family history data for Danish twins with nonsyndromic cleft lip with or without cleft palate. Am. J. Med. Genet., 72, 120–121. [DOI] [PubMed] [Google Scholar]
- 2.Grant S.F., Wang K., Zhang H., Glaberson W., Annaiah K., Kim C.E., Bradfield J.P., Glessner J.T., Thomas K.A., Garris M., et al. (2009) A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr., 155, 909–913. [DOI] [PubMed] [Google Scholar]
- 3.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M., et al. (2010) Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet., 42, 24–26. [DOI] [PubMed] [Google Scholar]
- 4.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D., et al. (2010) A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet., 42, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E., et al. (2012) Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet., 44, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio T.A. (2010) Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med., 363, 166–176. [DOI] [PubMed] [Google Scholar]
- 7.Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. (2011) Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet., 12, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leslie E.J., Marazita M.L. (2013) Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet., 163, 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno L.M., Mansilla M.A., Bullard S.A., Cooper M.E., Busch T.D., Machida J., Johnson M.K., Brauer D., Krahn K., Daack-Hirsch S., et al. (2009) FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet., 18, 4879–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikopensius T., Kempa I., Ambrozaitytė L., Jagomägi T., Saag M., Matulevičienė A., Utkus A., Krjutškov K., Tammekivi V., Piekuse L., et al. (2011) Variation in FGF1, FOXE1, and TIMP2genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mol. Teratol., 91, 218–225. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig K.U., Böhmer A.C., Rubini M., Mossey P.A., Herms S., Nowak S., Reutter H., Alblas M.A., Lippke B., Barth S., et al. (2014) Strong association of variants around FOXE1 and orofacial clefting. J. Dent. Res., 93, 376–381. [DOI] [PubMed] [Google Scholar]
- 12.Lennon C.J., Birkeland A.C., Nuñez J.A.P., Su G.H., Lanzano P., Guzman E., Celis K., Eisig S.B., Hoffman D., Rendon M.T.G., et al. (2012) Association of candidate genes with nonsyndromic clefts in Honduran and Colombian populations. Laryngoscope, 122, 2082–2087. [DOI] [PubMed] [Google Scholar]
- 13.Castanet M., Park S.-M., Smith A., Bost M., Leger J., Lyonnet S., Pelet A., Czernichow P., Chatterjee K., Polak M. (2002) A novel loss-of-function mutation in TTF-2 is associated with congenital hypothyroidism, thyroid agenesis and cleft palate. Hum. Mol. Genet., 11, 2051–2059. [DOI] [PubMed] [Google Scholar]
- 14.Clifton-Bligh R.J., Wentworth J.M., Heinz P., Crisp M.S., John R., Lazarus J.H., Ludgate M., Chatterjee V.K. (1998) Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet., 19, 399–401. [DOI] [PubMed] [Google Scholar]
- 15.Baris I., Arisoy A.E., Smith A., Agostini M., Mitchell C.S., Park S.M., Halefoglu A.M., Zengin E., Chatterjee V.K., Battaloglu E. (2006) A novel missense mutation in human TTF-2 (FKHL15) gene associated with congenital hypothyroidism but not athyreosis. J. Clin. Endocrinol. Metab., 91, 4183–4187. [DOI] [PubMed] [Google Scholar]
- 16.De Felice M., Ovitt C., Biffali E., Rodriguez-Mallon A., Arra C., Anastassiadis K., Macchia P.E., Mattei M.G., Mariano A., Scholer H., et al. (1998) A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat. Genet., 19, 395–398. [DOI] [PubMed] [Google Scholar]
- 17.Hishinuma A., Ohyama Y., Kuribayashi T., Nagakubo N., Namatame T., Shibayama K., Arisaka O., Matsuura N., Ieiri T. (2001) Polymorphism of the polyalanine tract of thyroid transcription factor-2 gene in patients with thyroid dysgenesis. Eur. J. Endocrinol., 145, 385–389. [DOI] [PubMed] [Google Scholar]
- 18.Carré A., Castanet M., Sura-Trueba S., Szinnai G., Vliet G., Trochet D., Amiel J., Léger J., Czernichow P., Scotet V., et al. (2007) Polymorphic length of FOXE1 alanine stretch: evidence for genetic susceptibility to thyroid dysgenesis. Hum. Genet., 122, 467–476. [DOI] [PubMed] [Google Scholar]
- 19.Szczepanek E., Ruchala M., Szaflarski W., Budny B., Kilinska L., Jaroniec M., Niedziela M., Zabel M., Sowinski J. (2011) FOXE1 polyalanine tract length polymorphism in patients with thyroid hemiagenesis and subjects with normal thyroid. Horm. Res. Paediatr., 75, 329–334. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson N., Tung J.Y., Kiefer A.K., Hinds D.A., Francke U., Mountain J.L., Do C.B. (2012) Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One, 7, e34442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny J.C., Crawford D.C., Ritchie M.D., Bielinski S.J., Basford M.A., Bradford Y., Chai H.S., Bastarache L., Zuvich R., Peissig P., et al. (2011) Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet., 89, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomaz R.A., Sousa I., Silva J.G., Santos C., Teixeira M.R., Leite V., Cavaco B.M. (2012) FOXE1 polymorphisms are associated with familial and sporadic nonmedullary thyroid cancer susceptibility. Clin. Endocrinol. (Oxf)., 77, 926–933. [DOI] [PubMed] [Google Scholar]
- 23.Bonora E., Rizzato C., Diquigiovanni C., Oudot-Mellakh T., Campa D., Vargiolu M., Guedj M., McKay J.D., Romeo G., Canzian F., et al. (2014) The FOXE1 locus is a major genetic determinant for familial nonmedullary thyroid carcinoma. Int. J. Cancer, 134, 2098–2107. [DOI] [PubMed] [Google Scholar]
- 24.Bullock M., Duncan E.L., O'Neill C., Tacon L., Sywak M., Sidhu S., Delbridge L., Learoyd D., Robinson B.G., Ludgate M., et al. (2012) Association of FOXE1 polyalanine repeat region with papillary thyroid cancer. J. Clin. Endocrinol. Metab., 97, E1814–E1819. [DOI] [PubMed] [Google Scholar]
- 25.Matsuse M., Takahashi M., Mitsutake N., Nishihara E., Hirokawa M., Kawaguchi T., Rogounovitch T., Saenko V., Bychkov A., Suzuki K., et al. (2011) The FOXE1 and NKX2–1 loci are associated with susceptibility to papillary thyroid carcinoma in the Japanese population. J. Med. Genet., 48, 645–648. [DOI] [PubMed] [Google Scholar]
- 26.Landa I., Ruiz-Llorente S., Montero-Conde C., Inglada-Pérez L., Schiavi F., Leskelä S., Pita G., Milne R., Maravall J., Ramos I., et al. (2009) The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet., 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan M., Chen G., Pan C.-M., Gu Z.-H., Zhao S.-X., Liu W., Wang H.-N., Ye X.-P., Xie H.-J., Yu S.-S., et al. (2014) Genome-wide association study identifies a novel susceptibility gene for serum TSH levels in Chinese populations. Hum. Mol. Genet., 23, 5505–5517. [DOI] [PubMed] [Google Scholar]
- 28.Damiola F., Byrnes G., Moissonnier M., Pertesi M., Deltour I., Fillon A., Le Calvez-Kelm F., Tenet V., McKay-Chopin S., McKay J.D., et al. (2014) Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int. J. Cancer, 134, 1659–1668. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi M., Saenko V.A., Rogounovitch T.I., Kawaguchi T., Drozd V.M., Takigawa-Imamura H., Akulevich N.M., Ratanajaraya C., Mitsutake N., Takamura N., et al. (2010) The FOXE1 locus is a major genetic determinant for radiation-related thyroid carcinoma in Chernobyl. Hum. Mol. Genet., 19, 2516–2523. [DOI] [PubMed] [Google Scholar]
- 30.Gudmundsson J., Sulem P., Gudbjartsson D.F., Jonasson J. G., Sigurdsson A., Bergthorsson J.T., He H., Blondal T., Geller F., Jakobsdottir M., et al. (2009) Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet., 41, , 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallel R., Belguith-Maalej S., Akdi A., Mnif M., Charfeddine I., Galofré P., Ghorbel A., Abid M., Marcos R., Ayadi H., et al. (2010) Genetic investigation of FOXE1 polyalanine tract in thyroid diseases: new insight on the role of FOXE1 in thyroid carcinoma. Cancer Biomark., 8, 43–51. [DOI] [PubMed] [Google Scholar]
- 32.Kohler A., Chen B., Gemignani F., Elisei R., Romei C., Figlioli G., Cipollini M., Cristaudo A., Bambi F., Hoffmann P., et al. (2013) Genome-wide association study on differentiated thyroid cancer. J. Clin. Endocrinol. Metab., 98, E1674–E1681. [DOI] [PubMed] [Google Scholar]
- 33.Medici M., van der Deure W.M., Verbiest M., Vermeulen S.H., Hansen P.S., Kiemeney L.A., Hermus A.R.M.M., Breteler M.M., Hofman A., Hegedüs L., et al. (2011) A large-scale association analysis of 68 thyroid hormone pathway genes with serum TSH and FT4 levels. Eur. J. Endocrinol., 164, 781–788. [DOI] [PubMed] [Google Scholar]
- 34.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W., et al. (2009) Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet, 5, e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One, 7, e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alul F.Y., Shchelochkov O.A., Berberich S.L., Murray J.C., Ryckman K.K. (2013) Genetic associations with neonatal thyroid-stimulating hormone levels. Pediatr. Res., 73, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porcu E., Medici M., Pistis G., Volpato C.B., Wilson S.G., Cappola A.R., Bos S.D., Deelen J., den Heijer M., Freathy R.M., et al. (2013) A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet, 9, e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santisteban P., Acebrón A., Polycarpou-Schwarz M., Di Lauro R. (1992) Insulin and insulin-like growth factor I regulate a thyroid-specific nuclear protein that binds to the thyroglobulin promoter. Mol. Endocrinol., 6, 1310–1317. [DOI] [PubMed] [Google Scholar]
- 39.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., Urasaki A., Kawakami K., McCallion A.S. (2006) Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc., 1, 1297–1305. [DOI] [PubMed] [Google Scholar]
- 40.Attanasio C., Nord A.S., Zhu Y., Blow M.J., Li Z., Liberton D.K., Morrison H., Plajzer-Frick I., Holt A., Hosseini R., et al. (2013) Fine tuning of craniofacial morphology by distant-acting enhancers. Science, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakada C., Iida A., Tabata Y., Watanabe S. (2009) Forkhead transcription factor foxe1 regulates chondrogenesis in zebrafish. J. Exp. Zool. B Mol. Dev. Evol., 312B, 827–840. [DOI] [PubMed] [Google Scholar]
- 42.Ambesi-Impiombato F.S., Parks L.A., Coon H.G. (1980) Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc. Natl Acad. Sci. USA, 77, 3455–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.-Y., Chou A., Ienasescu H., et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res., 42, D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zannini M., Avantaggiato V., Biffali E., Arnone M.I., Sato K., Pischetola M., Taylor B.A., Phillips S.J., Simeone A., Di Lauro R. (1997) TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO J., 16, 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I., et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol., 9, e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., McCallion A.S. (2006) Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science, 312, 276–279. [DOI] [PubMed] [Google Scholar]
- 47.McGaughey D., Stine Z., Huynh J., Vinton R., McCallion A. (2009) Asymmetrical distribution of non-conserved regulatory sequences at PHOX2B is reflected at the ENCODE loci and illuminates a possible genome-wide trend. BMC Genomics, 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. (2007) Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet., 39, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trochet D., Bourdeaut F., Janoueix-Lerosey I., Deville A., de Pontual L., Schleiermacher G., Coze C., Philip N., Frébourg T., Munnich A., et al. (2004) Germline mutations of the paired–like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet., 74, 761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapoor A., Sekar R.B., Hansen N.F., Fox-Talbot K., Morley M., Pihur V., Chatterjee S., Brandimarto J., Moravec C.S., Pulit S.L., et al. (2014) An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am. J. Hum. Genet., 94, 854–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadzhiev Y., Lele Z., Schindler S., Wilson S., Ahlberg P., Strahle U., Muller F. (2007) Hedgehog signaling patterns the outgrowth of unpaired skeletal appendages in zebrafish. BMC Dev. Biol., 7, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Royo J.L., Hidalgo C., Roncero Y., Seda M.A., Akalin A., Lenhard B., Casares F., Gómez-Skarmeta J.L. (2011) Dissecting the transcriptional regulatory properties of human chromosome 16 highly conserved non-coding regions. PLoS One, 6, e24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivieri A., Stazi M.A., Mastroiacovo P., Fazzini C., Medda E., Spagnolo A., De Angelis S., Grandolfo M.E., Taruscio D., Cordeddu V., et al. (2002) A population-based study on the frequency of additional congenital malformations in infants with congenital hypothyroidism: data from the Italian Registry for Congenital Hypothyroidism (1991–1998). J. Clin. Endocrinol. Metab., 87, 557–562. [DOI] [PubMed] [Google Scholar]
- 54.Devos H., Rodd C., Gagné N., Laframboise R., Van Vliet G. (1999) A search for the possible molecular mechanisms of thyroid dysgenesis: sex ratios and associated malformations. J. Clin. Endocrinol. Metab., 84, 2502–2506. [DOI] [PubMed] [Google Scholar]
- 55.Carre A., Hamza R.T., Kariyawasam D., Guillot L., Teissier R., Tron E., Castanet M., Dupuy C., El Kholy M., Polak M. (2014) A novel FOXE1 mutation (R73S) in Bamforth-Lazarus syndrome causing increased thyroidal gene expression. Thyroid, 24, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng T., Shi J.-Y., Wu M., Wang Y., Li L., Liu Y., Zheng Q., Huang L., Shi B. (2012) Overexpression of mouse TTF-2 gene causes cleft palate. J. Cell. Mol. Med., 16, 2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puxeddu E., Knauf J.A., Sartor M.A., Mitsutake N., Smith E.P., Medvedovic M., Tomlinson C.R., Moretti S., Fagin J.A. (2005) RET/PTC-induced gene expression in thyroid PCCL3 cells reveals early activation of genes involved in regulation of the immune response. Endocr. Relat. Cancer, 12, 319–334. [DOI] [PubMed] [Google Scholar]
- 58.Holmberg E., Rozell B.L., Toftgard R. (1996) Differential allele loss on chromosome 9q22.3 in human non-melanoma skin cancer. Br. J. Cancer, 74, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venza I., Visalli M., Tripodo B., De Grazia G., Loddo S., Teti D., Venza M. (2010) FOXE1 is a target for aberrant methylation in cutaneous squamous cell carcinoma. Br. J. Dermatol., 162, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 60.Sato N., Fukushima N., Maitra A., Matsubayashi H., Yeo C.J., Cameron J.L., Hruban R.H., Goggins M. (2003) Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res., 63, 3735–3742. [PubMed] [Google Scholar]
- 61.Bell A., Bell D., Weber R.S., El-Naggar A.K. (2011) CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer, 117, 2898–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weisenberger D.J., Trinh B.N., Campan M., Sharma S., Long T.I., Ananthnarayan S., Liang G., Esteva F.J., Hortobagyi G.N., McCormick F., et al. (2008) DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res., 36, 4689–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papadia C., Louwagie J., Del Rio P., Grooteclaes M., Coruzzi A., Montana C., Novelli M., Bordi C., de’ Angelis G.L., Bassett P., et al. (2014) FOXE1 and SYNE1 genes hypermethylation panel as promising biomarker in colitis-associated colorectal neoplasia. Inflamm. Bowel Dis., 20, 271–277. [DOI] [PubMed] [Google Scholar]
- 64.Nonaka D., Tang Y., Chiriboga L., Rivera M., Ghossein R. (2007) Diagnostic utility of thyroid transcription factors Pax8 and TTF-2 (FoxE1) in thyroid epithelial neoplasms. Mod. Pathol., 21, 192–200. [DOI] [PubMed] [Google Scholar]
- 65.He H., Li W., Liyanarachchi S., Jendrzejewski J., Srinivas M., Davuluri R.V., Nagy R., de la Chapelle A. (2015) Genetic predisposition to papillary thyroid carcinoma: involvement of FOXE1, TSHR, and a novel lincRNA gene, PTCSC2. J. Clin. Endocrinol. Metab., 100, E164–E172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aitola M.H., Pelto-Huikko M.T. (2003) Expression of Arnt and Arnt2 mRNA in developing murine tissues. J. Histochem. Cytochem., 51, 41–54. [DOI] [PubMed] [Google Scholar]
- 67.Abbott B.D., Probst M.R. (1995) Developmental expression of two members of a new class of transcription factors: II. Expression of Aryl Hydrocarbon Receptor Nuclear Translocator in the C57BL/6N Mouse Embryo. Dev. Dyn., 204, 144–155. [DOI] [PubMed] [Google Scholar]
- 68.Jain S., Maltepe E., Lu M.M., Simon C., Bradfield C.A. (1998) Expression of ARNT, ARNT2, HIF1α, HIF2α and Ah receptor mRNAs in the developing mouse. Mech. Dev., 73, 117–123. [DOI] [PubMed] [Google Scholar]
- 69.Pratt R.M., Dencker L., Diewert V.M. (1984) 2,3,7,8-Tetrachlorodibenzo-p-dioxin- induced cleft palate in the mouse: evidence for alterations in palatal shelf fusion. Teratog. Carcinog. Mutagen., 4, 427–436. [DOI] [PubMed] [Google Scholar]
- 70.García A.M., Fletcher T., Benavides F.G., Orts E. (1999) Parental agricultural work and selected congenital malformations. Am. J. Epidemiol., 149, 64–74. [DOI] [PubMed] [Google Scholar]
- 71.Wyszynski D.F., Duffy D.L., Beaty T.H. (1997) Maternal cigarette smoking and oral clefts: a meta-analysis. Cleft Palate. Craniofac. J., 34, 206–210. [DOI] [PubMed] [Google Scholar]
- 72.Kayano S., Suzuki Y., Kanno K., Aoki Y., Kure S., Yamada A., Matsubara Y. (2004) Significant association between nonsyndromic oral clefts and arylhydrocarbon receptor nuclear translocator (ARNT). Am. J. Med. Genet., 130A, 40–44. [DOI] [PubMed] [Google Scholar]
- 73.Giacomini S., Hou L., Bertazzi P., Baccarelli A. (2006) Dioxin effects on neonatal and infant thyroid function: routes of perinatal exposure, mechanisms of action and evidence from epidemiology studies. Int. Arch. Occup. Environ. Health, 79, 396–404. [DOI] [PubMed] [Google Scholar]
- 74.Yokoyama S., Ito Y., Ueno-Kudoh H., Shimizu H., Uchibe K., Albini S., Mitsuoka K., Miyaki S., Kiso M., Nagai A., et al. (2009) A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev. Cell, 17, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kitase Y., Shuler C.F. (2013) Microtubule disassembly prevents palatal fusion and alters regulation of the E-cadherin/catenin complex. Int. J. Dev. Biol., 57, 55–60. [DOI] [PubMed] [Google Scholar]
- 76.Wei K., Chen J., Akrami K., Galbraith G.C., Lopez I.A., Chen F. (2007) Neural crest cell deficiency of c-myc causes skull and hearing defects. Genesis, 45, 382–390. [DOI] [PubMed] [Google Scholar]
- 77.Zhu X., Zhao L., Park J.W., Willingham M.C., Cheng S.-Y. (2014) Synergistic signaling of KRAS and thyroid hormone receptor β mutants promotes undifferentiated thyroid cancer through MYC up-regulation. Neoplasia, 16, 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Otterloo E., Li W., Garnett A., Cattell M., Medeiros D.M., Cornell R.A. (2012) Novel Tfap2-mediated control of soxE expression facilitated the evolutionary emergence of the neural crest. Development, 139, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res., 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eichberger T., Regl G., Ikram M.S., Neill G.W., Philpott M.P., Aberger F., Frischauf A.-M. (2004) FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J. Invest. Dermatol., 122, 1180–1187. [DOI] [PubMed] [Google Scholar]
- 81.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suster M., Kikuta H., Urasaki A., Asakawa K., Kawakami K. (2009) Transgenesis in zebrafish with the Tol2 transposon system. In Cartwright E.J. (ed.), Transgenesis Techniques. Humana Press, New York, NY, Vol. 561, pp. 41–63. [DOI] [PubMed] [Google Scholar]
- 83.McGaughey D.M., Vinton R.M., Huynh J., Al-Saif A., Beer M.A., McCallion A.S. (2008) Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res., 18, 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature, 444, 499–502. [DOI] [PubMed] [Google Scholar]
- 85.Heckman K.L., Pease L.R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc., 2, 924–932. [DOI] [PubMed] [Google Scholar]
- 86.Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W. (2005) Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature, 436, 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilchrist E.P., Moyer M.P., Shillitoe E.J., Clare N., Murrah V.A. (2000) Establishment of a human polyclonal oral epithelial cell line. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod., 90, 340–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.