Abstract
Protein synthesis in mitochondria is initiated by formylmethionyl-tRNAMet (fMet-tRNAMet), which requires the activity of the enzyme MTFMT to formylate the methionyl group. We investigated the molecular consequences of mutations in MTFMT in patients with Leigh syndrome or cardiomyopathy. All patients studied were compound heterozygotes. Levels of MTFMT in patient fibroblasts were almost undetectable by immunoblot analysis, and BN-PAGE analysis showed a combined oxidative phosphorylation (OXPHOS) assembly defect involving complexes I, IV and V. The synthesis of only a subset of mitochondrial polypeptides (ND5, ND4, ND1, COXII) was decreased, whereas all others were translated at normal or even increased rates. Expression of the wild-type cDNA rescued the biochemical phenotype when MTFMT was expressed near control levels, but overexpression produced a dominant-negative phenotype, completely abrogating assembly of the OXPHOS complexes, suggesting that MTFMT activity must be tightly regulated. fMet-tRNAMet was almost undetectable in control cells and absent in patient cells by high-resolution northern blot analysis, but accumulated in cells overexpressing MTFMT. Newly synthesized COXI was under-represented in complex IV immunoprecipitates from patient fibroblasts, and two-dimensional BN-PAGE analysis of newly synthesized mitochondrial translation products showed an accumulation of free COXI. Quantitative mass spectrophotometry of an N-terminal COXI peptide showed that the ratio of formylated to unmodified N-termini in the assembled complex IV was ∼350:1 in controls and 4:1 in patient cells. These results show that mitochondrial protein synthesis can occur with inefficient formylation of methionyl-tRNAMet, but that assembly of complex IV is impaired if the COXI N-terminus is not formylated.
Introduction
Mammalian mitochondria maintain a translation machinery that is dedicated to the synthesis of the 13 structural subunits of the mitochondrial oxidative phosphorylation (OXPHOS) complexes encoded by the mitochondrial genome (mtDNA). Except for the 22 tRNAs and 2 rRNAs that are mtDNA-encoded, all other components of the translation machinery are nuclear-encoded and must be imported into the mitochondrion. Although there are major differences in the composition of bacterial and mitochondrial ribosomes, the core translation apparatus in mammalian mitochondria closely resembles that in bacteria, reflecting the evolutionary origins of mitochondria from α-proteobacteria.
Translation in both systems is initiated by formylmethionyl tRNAMet (fMet-tRNAMet). The formylation reaction is carried out by methionyl-tRNA transformylase (MTF) in bacteria (1), and by the homologue methionyl-tRNA formyltransferase (MTFMT) in mammalian mitochondria (2). Although bacteria possess two distinct tRNAMet species for translation initiation and elongation (1), a single tRNAMet functions in both roles in mammals (3). As mitochondrial tRNAMet has a dual role in translation, the ratio of fMet-tRNAMet to Met-tRNAMet has to be regulated to meet the needs of both the translation initiation and elongation. The ratio of the two aminoacylated tRNAMet species is thought to be determined by competition between MTFMT and the elongation factor EF-Tumt, which delivers aminoacyl-tRNAs to the acceptor site of the ribosome (4,5). EF-Tumt has no detectable affinity in vitro for fMet-tRNAMet, whereas the initiation factor IF2mt exhibits a 50-fold preference for fMet-tRNAMet over Met-tRNAMet in promoting initiator tRNA binding to mitochondrial ribosomes (6), suggesting that formylation of Met-tRNAMet is required for efficient initiation of mitochondrial translation.
In bacteria, there is a strict requirement for formylation of the initiator tRNAMet (1); however, yeast mitochondria can initiate translation without fMet-tRNAMet (7), albeit with the help of an accessory factor (8), raising questions about the essential role of formylation in eukaryotes.
Defects in the mitochondrial protein translation are among the most frequent causes of mitochondrial disease in humans, leading predominantly to early-onset, severe and usually fatal clinical phenotypes (9). Mutations in the MTFMT gene were first described in two families with Leigh syndrome and combined mitochondrial respiratory chain deficiency (10). Recently, 12 other cases with MTFMT mutations and mitochondrial respiratory chain complex deficiencies have been described (11–13).
Here we have investigated the molecular basis for pathogenesis in three new MTFMT cases presenting with Leigh Syndrome or cardiomyopathy. We show that although MTFMT is barely detectable in patient fibroblasts, the mitochondrial translation defect is restricted to a subset of mtDNA-encoded polypeptides. Despite this, there is a severe combined assembly defect in all of the OXPHOS complexes containing mtDNA-encoded subunits, except complex III. Quantitative mass spectrometry analyses showed nearly a 100-fold enrichment of formylated COXI in the assembled complex IV holoenzyme, demonstrating that an N-formyl methionine residue on this subunit is crucial for assembly of the COX holoenzyme complex.
Results
Identification of mutations in MTFMT subjects by exome sequencing
Whole-exome sequencing identified four different heterozygous mutations in MTFMT, the gene encoding mitochondrial methionyl-tRNA formyltransferase, in three patients with combined OXPHOS assembly defects in fibroblasts (Table 1). The mutations were confirmed by Sanger sequencing of genomic DNA, which identified an additional c.242T>C mutation in patient 2. The c.626C>T mutation was previously described in four patients with Leigh syndrome (10,11). The mutation predicts a p.S209L amino acid substitution, however it eliminates two exonic splicing enhancers located at the 3′ end of exon 4, which leads to skipping of exon 4 and to a premature stop codon at p.R181SfsX5. The c.994C>T mutation has previously been reported in two patients with Leigh syndrome (11,12). This nonsense mutation leads to a premature stop codon at p.R332X. In addition, two novel deletions, c.219_222del and c.247_253del, were identified in our patients. Both of the deletions are frameshift mutations leading to a premature stop codon in exon 2, predicting a stop at p.E74KfsX3 and p.V83QfsX12.
Table 1.
MTFMT mutations in the three patients
| Patient | Mutation | Amino acid substitution | Clinical phenotype |
|---|---|---|---|
| P1 | (1) c. 626C>T (2) c. 994C>T |
p.R181SfsX5 p.R332X |
Leigh syndrome |
| P2 | (1) c. 242T>C+c.247_253del (2) c. 994C>T |
p.L81P+p.V83QfsX12 p.R332X |
Hypertrophic cardiomyopathy |
| P3 | (1) c.219_222del (2) c. 626C>T |
p.E74KfsX3 p.R181SfsX5 |
Leigh syndrome |
The allelic distribution of the mutations and the expression levels of the alleles containing the different MTFMT mutations were investigated by sequencing the cDNAs from immortalized patient fibroblasts. As previously reported (10), the skipping of exon 4 caused by c.626C>T mutation is not complete, with the result that there is a small residual amount of the mRNA carrying the mutation (data not shown). Almost all mRNA in P1 and P2 contained c.994C>T (data not shown) indicating a compound heterozygous inheritance pattern of the MTFMT mutations in these cases. In P3, we were not able to amplify enough cDNA for Sanger sequencing, suggesting that only small amounts of the MTFMT mRNA remained, the result of nonsense-mediated RNA decay, and that the two frameshift mutations identified in this subject are present on separate alleles.
Moderate decreases in mitochondrial translation in MTFMT fibroblasts
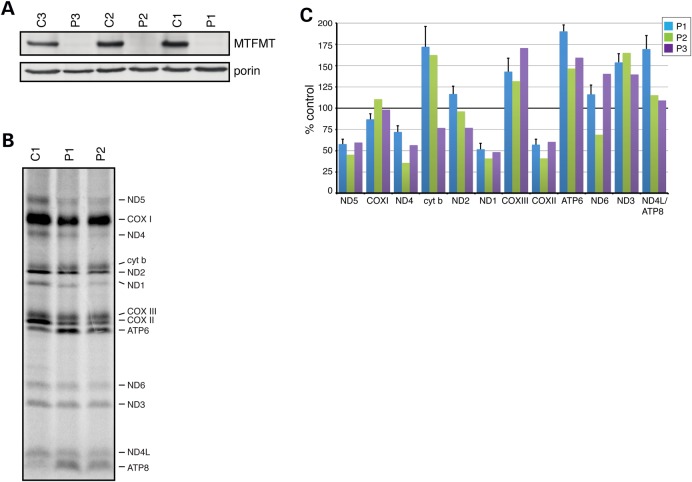
MTFMT protein was barely detectable by immunoblot analysis of mitochondria isolated from immortalized fibroblasts in all three patients (Fig. 1A), indicating that the missense mutations in MTFMT destabilize the protein. Despite the very low steady-state levels of MTFMT, pulse translation experiments showed that mitochondrial protein synthesis was relatively moderately reduced (globally by about 35%) in patient fibroblasts compared with control (Fig. 1B). However, there were obvious differences in the synthesis of different polypeptides: several (cytb, ND3, ATP6, ATP8/ND4L) polypeptides were synthesized at control rates or even higher, whereas those most affected (ND1, ND4, ND5 and COXII) were synthesized at about 50% of control levels (Fig. 1C).
Figure 1.
Immunoblot analysis and mitochondrial translation assay in fibroblasts from three patients with mutations in MTFMT (A). Immunoblot with an anti-MTFMT antibody showing virtually undetectable levels of MTFMT in patient fibroblasts. Porin was used as a loading control. (B) Mitochondrial translation experiment in fibroblasts from patients P1 and P2 and a control C1. The seven subunits of complex I (ND), one subunit of complex III (cyt b), three subunits of complex IV (COX) and two subunits of complex V (ATP) are indicated at the right of the figure. (C) Quantitation of the protein synthesis abnormalities in the three patients. Rates of synthesis of individual polypeptides were calculated for each patient and expressed relative to controls at 100%. Data for P1 (N = 4) are shown ± SEM relative to controls (N = 3). Data from P2 and P3 are the means of two separate experiments.
Combined mitochondrial OXPHOS assembly defects in MTFMT subjects
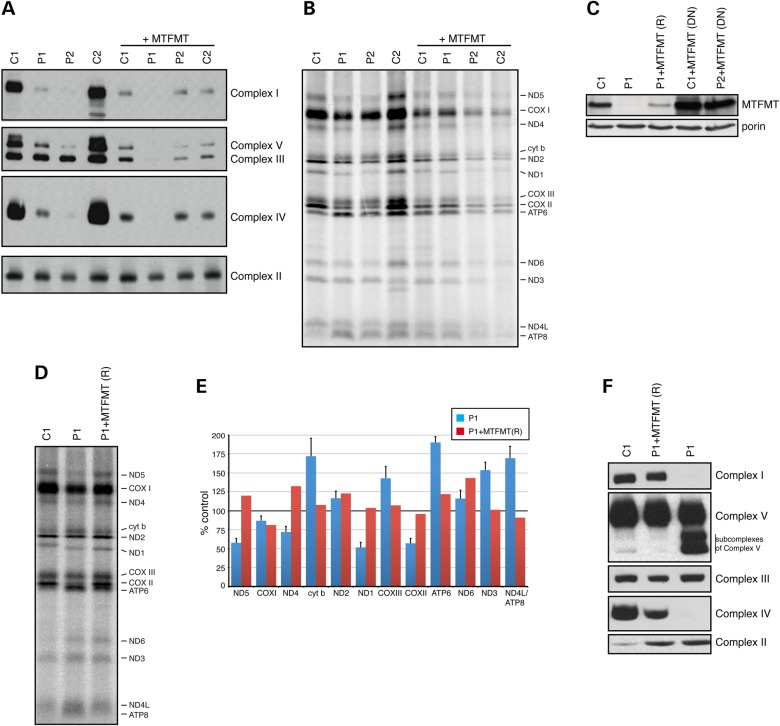
To study the consequences of the MTFMT mutations, we further investigated the assembly of OXPHOS enzyme complexes by BN-PAGE in patient fibroblasts (Fig. 2A). The assembly of complexes I, IV and V was markedly decreased in fibroblasts and subcomplexes of complex V accumulated, but complex III appeared to assemble normally. As expected, complex II, which has no mtDNA-encoded subunits, was unaffected in patient fibroblasts.
Figure 2.
BN-PAGE analysis of patient and control fibroblasts and rescue of the biochemical phenotype. (A) BN-PAGE analysis showing severe combined deficiency in the assembly of complexes I, IV and V, and the dominant negative effect of overexpressing MTFMT in two controls (C1, C2) and patients P1 and P2. (B) Mitochondrial translation experiment using the same controls and patients as in (A) showing the dominant negative effect of MTFMT overexpression. (C) Immunoblot analysis of MTFMT levels in control, patient P1, rescued patient clonal cell line (P1+MTFMT (R)) and control and patient cells with a dominant negative phenotype (DN). Porin was used as a loading control. (D) Mitochondrial translation experiment showing the rescue of the translation phenotype in a clone expressing near control levels of wild-type MTFMT. (E) Quantitation of results in (C). (F) BN-PAGE analysis of a rescued patient clonal cell line (P1+MTFMT (R)). (A, F) Antibodies against an individual subunit of each of the OXPHOS complexes were used.
Expression of MTFMT rescues the biochemical phenotype in subject cells, but overexpression produces a strong dominant negative phenotype
To confirm the pathogenicity of the MTFMT mutations, we transduced patient fibroblasts with a wild-type MTFMT cDNA expressed from a retroviral vector (pBABE) and established stable lines from P1 and P2. Unexpectedly, expression of the wild-type MTFMT did not rescue the assembly defect of the OXPHOS complexes, but rather produced a strong dominant negative phenotype, exacerbating the combined OXPHOS complex assembly and mitochondrial translation defect, even in control fibroblasts (Fig. 2A and B). However, the steady-state levels of MTFMT were much greater than control levels in the transduced cells (Fig. 2C). In order to decrease the level of MTFMT expression, we employed a different retroviral vector (pLXSH), plated the cells at low density and picked clones to identify those with MTFMT expression levels near control. The abnormal pattern of protein synthesis (Fig. 2D) and the OXPHOS assembly defect (Fig. 2F) were completely rescued in clones expressing near control levels of MTFMT (Fig. 2C). These results confirm the pathogenicity of the MTFMT mutations and demonstrate that MTFMT activity is crucial for normal mitochondrial protein synthesis.
Formylation of mitochondrial methionyl-tRNAMet
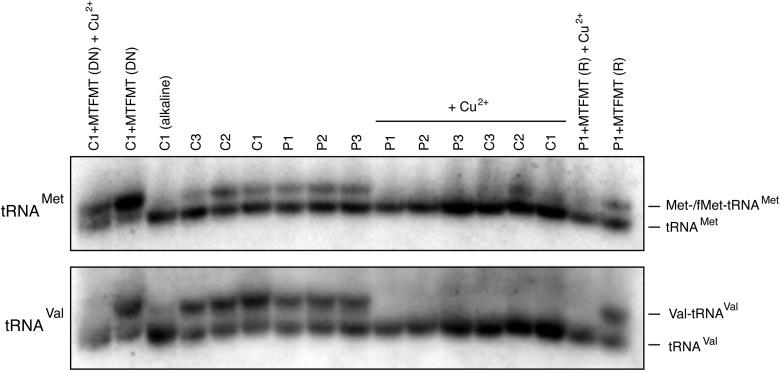
To investigate the levels of fMet-tRNAMet in subject fibroblasts, we ran total RNA isolated from patient and control fibroblasts under acidic conditions on a high-resolution acid-urea PAGE gel, followed by northern blotting (Fig. 3). This analysis showed no major differences in the levels of uncharged and aminoacylated Met-tRNAMet between patients and controls. To assess the levels of fMet-tRNAMet in the patient and control cell lines, we treated the RNA samples with copper which should specifically deacylate the Met-tRNAMet but not fMet-tRNAMet (10). In the patient cell lines, we were unable to detect fMet-tRNAMet ; however, we could not unequivocally resolve fMet-tRNAMet in most control cell lines (except C2) by this analysis, or in the rescued patient line, suggesting that the steady-state level of fMet-tRNAMet is kept very low even in control fibroblasts. Overexpression of MTFMT clearly drove the accumulation of fMet-tRNAMet at the expense of unacylated tRNAMet in cells with a dominant negative phenotype.
Figure 3.
High-resolution northern blot of tRNAMet species in control and patient fibroblasts. Treatment with alkali deacylates the tRNA and copper (Cu2+) treatment selectively deacylates the non-formylated species. Analysis of RNA from cells expressing a dominant negative phenotype (DN) resulting from MTFMT overexpression is shown in first two lanes (left) and from a patient clonal cell line in which the biochemical phenotype was rescued (R) in the last two lanes (right). The position of charged and uncharged tRNAs are shown. Hybridization with mt-tRNAVal was used as a loading control.
Silencing peptide deformylase increases the rate of mitochondrial protein synthesis
Initiation of translation with fMet-tRNAMet should produce polypeptides with an N-terminal formyl-methionine. In most bacterial proteins, the N-terminal formyl group is removed by the peptide deformylase (PDF) (14). The removal of N-terminal formyl group by PDF is a co-translational event and it has an important role in protein maturation. A methionine amino peptidase then cleaves the N-terminal methionine, and further processing with other peptidases occurs corresponding to the N-end rule (15). A human homologue of the bacterial PDF enzyme has been identified in mitochondria; however, the slow kinetic deformylation properties make the functional role of human PDF unclear (14,16,17).
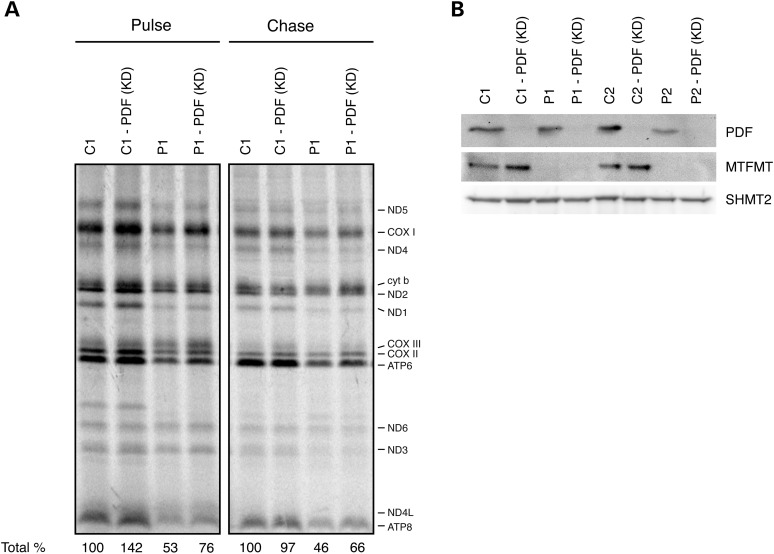
To test whether modulating PDF activity could suppress the biochemical phenotypes produced by the lack of MTFMT activity in patient fibroblasts, PDF was transiently silenced by siRNA to levels that rendered the protein undetectable by immunoblot analysis (Fig. 4B), and the effect on mitochondrial translation and the stability of newly synthesized mitochondrial polypeptides were investigated (Fig. 4A). Silencing PDF resulted in a ∼50% increase in overall mitochondrial translation in both control and patient fibroblasts; however, this was not associated with any amelioration of the OXPHOS assembly defect in MTFMT patient fibroblasts (data not shown). To investigate the stability of the newly synthesized polypeptides, we chased the label for 17 h (Fig. 4A). Rather unexpectedly, newly synthesized mitochondrial proteins in the MTFMT patient did not turnover as rapidly as did those in the control, whereas the turnover of newly synthesized mitochondrial translation products in PDF-silenced P1 fibroblasts was reduced compared with control.
Figure 4.
Pulse-chase mitochondrial translation experiment in control and patient fibroblasts in which the PDF was silenced using siRNA. (A) Suppression of PDF (KD) globally increased the rate of protein synthesis in the pulse by ∼50% in both control and patient cell lines. The migration of individual OXPHOS subunits is indicated on the right of the figure. (B) Immunoblot showing siRNA-mediated silencing of PDF in fibroblasts from controls and MTFMT patients. Serine hydroxymethyltransferase 2 (SHMT2) was used as a loading control.
Assembly of the complex IV holoenzyme
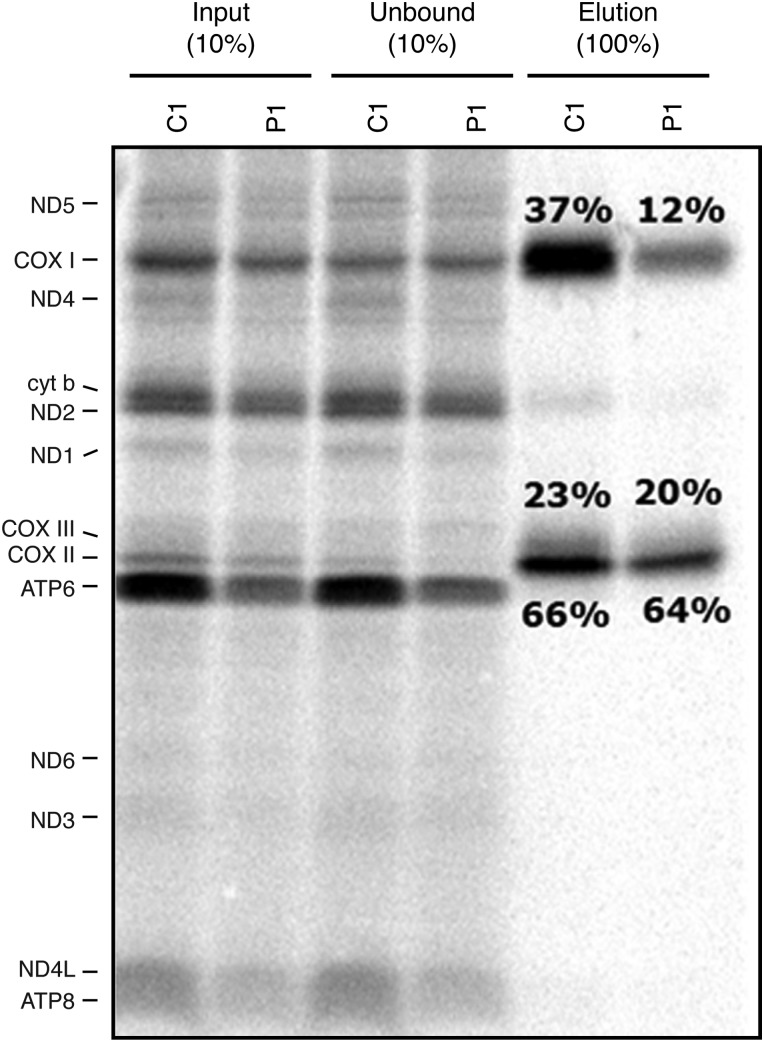
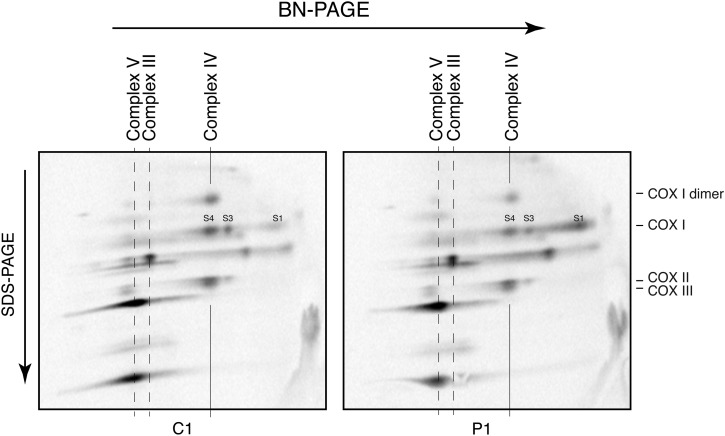
Because the fibroblasts with MTFMT mutations showed polypeptide-specific decreases in translation of mtDNA-encoded proteins, but markedly decreased levels of fully assembled OXPHOS complexes, we decided to study the assembly of newly synthesized polypeptides comprising the catalytic core of complex IV (COXI, II, III) in some detail. We first immunoprecipitated the complex IV using an anti-COXII antibody (which quantitatively immunoprecipitates the holoenzyme complex (18)) after a 1 h pulse with [35S]-methionine/cysteine followed by an 18 h chase and measured the relative incorporation of the three core mtDNA-encoded subunits into the holoenzyme complex. This analysis showed that only one-third of newly synthesized COXI was assembled into COXII containing complexes relative to the control (Fig. 5). To further evaluate the complex IV assembly pathway, we performed the same pulse-chase experiment and analysed the samples by two-dimensional BN-PAGE to follow the assembly of newly synthesized subunits into the holoenzyme complex. In P1 fibroblasts, the free COXI subunit (S1) accumulated compared with control and there was reduced incorporation into the mature complex (S4), indicating that the early assembly of complex IV was impaired (Fig. 6). We conclude that the low levels of complex IV in MTFMT patients are caused by a failure to efficiently incorporate newly synthesized COXI into the holoenzyme complex.
Figure 5.
Analysis of the incorporation of the core catalytic subunits into complex IV. Mitochondrial translation products were pulse-labelled for 1 h, chased for 18 h and then the holoenzyme was precipitated with an anti-COXII antibody. Individual fractions from the immunoprecipitation experiment were analysed on an SDS–PAGE gel and the relative incorporation of the three COX subunits was determined and is indicated as percent of the total synthesized product (elution/input).
Figure 6.
Two-dimensional BN-PAGE analysis of assembly of complex IV. Mitochondrial translation products were pulse-labelled for 1 h, chased for 18 h and analysed on a two-dimensional BN-PAGE gel. Free COXI (S1) accumulated in patient fibroblasts relative to the fully assembled complex (S4). The migration of complexes V, III and IV in the one-dimensional BN-PAGE is indicated on the top of the figure. The position of individual complex IV subunits (COXI, COXII, COXIII and COXI dimer) is indicated on the right. S1, S3 and S4 are subassemblies of complex IV.
Quantitative mass spectrometry of N-formylated COXI in complex IV holoenzyme
To test whether the inefficient assembly of COXI into the enzyme complex was because of a lack of an N-formyl methionine, we quantitatively determined the ratio of N-terminal formylated to unmodified COXI using heavy isotope-labelled COXI peptides with or without an N-terminal methionine as internal standards. As described above, we immunoprecipitated complex IV using an anti-COXII antibody, excised the band corresponding to COXI from an sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gel, which was subjected to mass spectrometry analysis as detailed in Materials and Methods. This analysis showed that the ratio of formylated to unmodified COXI was ∼350:1 in the complex immunoprecipitated from control cells, but only 4:1 in cells from P1 (Table 2). We conclude that the N-formyl group plays an important role in signalling assembly competent COXI.
Table 2.
Formylation state of the N-terminal peptide of COXI (MFADRWLSFSTNHK) in complex IV from control (C1) and patient (P1) fibroblasts
| MFADRWLSFSTNHK |
|||
|---|---|---|---|
| Unformylated (unmodified + oxidation M) | Formylated (no oxidation + oxidation M) | Ratio formylated/unmodified | |
| Average of ratios Endogenous/heavy label | Average of ratios endogenous/heavy label | ||
| C1 COX 1 | 0.4120 | 143.1782 | 347.5 |
| P1 COX1 | 6.1004 | 29.1766 | 4.8 |
Discussion
The results of this study with patient-derived fibroblasts harbouring MTFMT mutations show that mitochondrial protein synthesis can be initiated without efficient formylation of Met-tRNAMet, but that the lack of an N-terminal formyl-methionine in the newly synthesized polypeptides, at least in the case of COXI, impairs incorporation into the holoenzyme complex.
The loss of MTFMT activity had a variable effect on the synthesis of individual mtDNA-encoded polypeptides in our studies. Several complex I subunits (ND1, ND4, ND5) and one complex IV subunit (COXII) were most affected, whereas the synthesis of many others was either not significantly impaired (COXI) or even enhanced (cytb, COXIII, ATP6, ND3, ND4L) relative to control. This result contrasts sharply with the severe translation defect observed in the first reported MTFMT patients, in which the authors concluded that MTFMT activity is required for mitochondrial translation (10); however, our results are similar to those in the only other case we are aware of in which mitochondrial translation was investigated (12). It is not clear what might explain this difference as some of the pathogenic alleles are shared among the patients investigated.
Initiation has remained the least well-understood aspect of mammalian mitochondrial translation owing to the distinctive features of mammalian mitochondrial mRNAs, such as the absence of significant 5′-UTRs, and the fact that translation occurs with a single species of tRNAMet. Initiation of translation requires two factors, IF2mt and IF3mt, both of which have been cloned and sequenced in humans (19,20). The current model suggests that IF3mt interacts and helps dissociate the subunits of the 55S monosome. This leads to the formation of an IF3mt 28S complex, which then recruits IF2mt GTP, forming a complex onto which the mRNA feeds through a specialized entrance tunnel. IF1 in bacteria blocks the acceptor (A) site during translation initiation, and although an independent human IF1mt has not been identified, expression of IF2mt rescues a double deletion of IF1 and IF2 in Escherichia coli (21), suggesting that it serves a dual function in the mammalian mitochondria.
IF2mt positions the initiator tRNA over the initiator codon on the small ribosomal subunit and displays a strong preference (∼50-fold) for fMet-tRNAMet over Met-tRNAMet in vitro (6). On the other hand, EF-Tumt, which brings charged tRNAs to the A site for chain elongation strongly discriminates against fMet-tRNAMet. These observations have suggested that formylation has an important role in translation initiation and that the balance between the steady-state levels of formylated and unformylated tRNAMet is essential for normal translation. Indeed, the results we present here demonstrate that overexpression of MTFMT, which drives the formation of fMet-tRNAMet, produces a severe dominant negative phenotype. We were not able to observe detectable levels of fMet-tRNAMet in all control cell lines, or in any of the patients, suggesting that the levels of this species are kept very low relative to the entire tRNAMet pool and that the majority of the aminoacylated tRNAMet pool is unformylated. We did not observe any remarkable differences in the total pool or in the relative abundance of charged tRNA between controls and patients. These results contrast somewhat with a previous report that showed an increase in the total and aminoacylated tRNAMet pool in patient fibroblasts, but curiously an apparently complete lack of the charged unformylated form in controls (10).
Mitochondrial translation in yeast does not require FMT1, the yeast methionyl-tRNA formyltransferase homologue (7). Mitochondrial translation in ΔFMT1 strains is indistinguishable from that in wild-type cells, and the cells assemble the OXPHOS complexes normally and remain respiration competent, implying that the N-terminal formyl group is not essential for the assembly of the OXPHOS complexes in yeast (7,8). The first indication that mitochondrial translation initiation without formylation might be possible in mammalian cells came from a study in which bovine IF2mt was shown to rescue defective mitochondrial protein synthesis in yeast strains lacking both IFM1 (the yeast homologue of IF2mt) and FMT1 (22), suggesting that IF2mt might have an appreciable affinity for Met-tRNAMet in vivo. The ability of ΔFMT1 strains to carry out mitochondrial translation, however, requires an additional factor, Aep3p, a pentatricopeptide repeat containing protein (PPR) that physically interacts with IF2mt and promotes binding of non-formylated initiator tRNAMet (8). Although the exact molecular mechanism is unknown, all studied PPR proteins are RNA-binding proteins that mediate some aspect of RNA metabolism (23), so one idea is that Aep3p mediates the recruitment of the mRNA and initiator tRNA to the initiation complex. Aep3p is essential for the growth of yeast on non-fermentable carbon sources only when non-formylated initiator tRNAMet is available. Whether such a suppressor exists in mammals is unknown, as a convincing human Aep3p homologue has not been identified.
In most eukaryotes (but not yeast), mitochondria have an N-terminal methionine excision pathway, apparently homologous to that in bacteria where it is used to remove the formyl group and the initiating methionine by PDF and mitochondrial methionine amino peptidase, respectively (15). However, in mammals, only one of the mitochondrial polypeptides, COXIII, has been shown to lack formyl-methionine (24) and the PDF has very low catalytic activity on mitochondrial peptides, so the true physiological function of these enzymes in mammals remains unclear. This prompted us to investigate whether depleting the deformylase activity could suppress the loss of function MTFMT mutations. Although depletion of PDF in both patient and control cell lines caused a global increase in mitochondrial protein synthesis of the translation products, this did not influence subsequent assembly of the OXPHOS complexes. Elucidation of the role of PDF in mitochondrial biology will require further investigation.
The most striking observation in this study is the lack of concordance between the severity of the translation defect and the OXPHOS assembly defect in fibroblasts from MTFMT patients. This prompted us to hypothesize that an N-terminal formyl methionine plays a crucial role in post-translational incorporation of at least some of the newly synthesized peptides into a holoenzyme complex. We tested this by a quantitative mass spectrometry analysis of the amount of formylated versus unmodified COXI in intact complex IV using heavy isotope labelled standards. This analysis showed a nearly 100-fold difference in the presence of formylated COXI in the holoenzyme complex between controls and MTFMT patients, sharply contrasting with a semi-quantitative analysis of COXI formylation which concluded that formylated COXI was the predominant species in the enzyme complex from controls and patients (10). These results further indicate that most translation in MTFMT patient cells initiates without formyl methionine, consistent with a recent study showing that the activity of the mutant MTFMT proteins found in patients is at least an order of magnitude less than in controls (25).
All of the mtDNA-encoded polypeptides are hydrophobic subunits of the OXPHOS complexes and it is thought that their synthesis occurs adjacent to the mitochondrial inner membrane where they are co-translationally inserted during the assembly of the holoenzyme complexes (26). COXI nucleates the assembly of complex IV by forming an early assembly intermediate in association with the nuclear-encoded subunits COXIV and COXVa. Our results show that newly synthesized free COXI accumulates in MTFMT patient fibroblasts, presumably because of a relative decrease in N-terminal methionine formylation. How the presence of a formyl group is important in this process remains unknown. Clearly the formyl group does not act as a signal to prevent proteolysis of unincorporated subunit. One possibility is that the lack of formylation might influence chaperoning of the COXI subunit from the exit tunnel of the ribosome and insertion into the inner membrane. The assembly and the amount of complex III was not at all affected in MTFMT patient fibroblasts, suggesting that N-terminal formylation of cyt b does not play a role in the assembly process of complex III. This could simply reflect the fact that cyt b is the single mtDNA-encoded subunit in the holoenzyme complex. Further investigations will be required to resolve this issue.
Materials and Methods
Cell culture
Immortalized patient and control fibroblasts were grown in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, at 5% CO2 and 37°C.
Exome sequencing analysis
Whole-exome sequencing was used to search for pathogenic mutations in our three patients. Genomic DNA extracted from patient fibroblasts was fragmented and exomes were captured with the TruSeq Exome Enrichment Kit (Illumina, San Diego, CA). Subsequently, each captured library was sequenced on a Illumina Hiseq 2000 sequencer at the high-throughput sequencing platform of the McGill University and Genome Quebec Innovation Centre, Montreal, Canada. The bioinformatic analysis of exome sequencing data was carried out as previously described (27,28). In brief, all raw 100 bp paired-end reads were aligned to the human reference genome (UCSC hg19) using BWA (version 0.5.9) (29) and realigned locally around small insertions or deletions (indels) by the Genome Analysis Toolkit (GATK, version 1.0.5) (30). Duplicate reads were then removed by using Picard tools (http://picard.sourceforge.net) and the depth of coverage was calculated with GATK. The average coverage of consensus coding sequences (CCDS) was 80X (5097), 70X (48 248) and 65X (55 191), and 90% of bases were covered by ≥10 reads. The single-nucleotide variants and indels were detected using Samtools (v. 0.1.17) (31) mpileup and bcftools and were annotated with in-house annotation pipeline that uses ANNOVAR and some custom scripts. Three in silico prediction tools were used for predicting the damaging effect of the identified mutations (SIFT PolyPhen-2 (32,33) and MutationTaster (34)). To distinguish rare variants from common polymorphisms and sequencing noises, we excluded the annotated variants with minor allele frequency of >5% in public databases (the 1000 Genomes or in the Exome Variant Server (EVS)) or seen in >20 individuals in our in-house exome database (contains ∼1000 previously sequenced samples). Finally, all coding (frameshift indels, nonsense, missense) and canonical splice site variants were annotated with known and predicted mitochondrial proteins by using the Mitocarta database and were considered for further investigation.
Sanger sequencing of the MTFMT gene (NM_139242.3) in cDNA and gDNA from the three patients was used to confirm the exome sequencing results. Total RNA was isolated from fibroblasts using the RNeasy Kit (Qiagen, Toronto, Ont, Canada) and the cDNA was amplified with specific primers using the one-step reverse transcriptase-polymerase chain reaction (RT-PCR) kit (Qiagen). Total genomic DNA was extracted from fibroblasts using the DNAeasy Kit (Qiagen) and primers specific for exons 2, 4 and 9 of MTFMT were used to amplify the gDNA.
Immunoblotting
Whole-cell protein extracts or purified mitochondrial fractions were prepared in 1.5% n-dodecyl maltoside in phosphate-buffered saline, then 20 μg of protein per sample was used for Tris–glycine SDS–PAGE. The proteins were transferred to a nitrocellulose membrane and used for detection of MTFMT (Abcam), PDF (Abcam), SHMT2 (Sigma) or porin (EMD Calbiochem).
Mitochondrial translation assay
Mitochondrial translation products in cultured fibroblasts were pulse-labelled for 60 min with 200 μCi/ml of [35S]-methionine/cysteine (PerkinElmer, Waltham, MA) in DMEM lacking methionine and cysteine, and containing 100 μg/ml of the cytoplasmic translation inhibitor emetine (35). To test the stability of newly synthesized mitochondrial proteins, we pulse labelled the mitochondrial translation products in the presence of a reversible cytosolic translation inhibitor (anisomycin) and chased the newly synthesized proteins for 17 h. Analysis of the mitochondrial translation products was then further carried out on a 15–20% polyacrylamide gradient gel as described in detail in (35) or by immunoprecipitation or BN-PAGE followed by two-dimensional SDS–PAGE.
Acid-urea PAGE and northern blotting
Total RNA was isolated from fibroblasts under acidic conditions using TRIzol (Invitrogen, Carlsbad, CA) according to the instructions of the manufacturer. Aminoacyl-tRNAs were deacylated in 0.1 M Tris–HCl (pH 9.0) at 37°C for 1 h. Alternatively, aminoacyl-tRNAs were treated with 5 mm CuSO4 in 0.1 M Tris–HCl (pH 8.0) at room temp for 15 min, to specifically hydrolyse the unformylated aminoacyl-tRNAs, but not the formylated aminoacyl-tRNAs. Then 16 µg of the treated/untreated RNA was mixed 1:1 with loading buffer (8 M urea, 0.1 M sodium acetate pH 5, 0.05% bromophenol blue, 0.05% xylene cyanol), boiled, placed on ice and loaded on a 12% polyacrylamide (19:1 acrylamide:bisacrylamide) gel containing 8 M urea in 0.1 sodium acetate buffer (pH 5.0). The RNA samples were electrophoresed at 800 V for 70 h at 4°C followed by transfer onto a Hybond N+ membrane (GE Healthcare). Prehybridization and hybridization were carried out in EXPRESSHyb solution (Clontech) according to the manufacturer's instructions. The oligonucleotides used for the generation of the 32P-labelled probes by the end-labelling technique had the sequences 5′-TGGTAGTACGGCAAGGGTATAACC-3′ for mt-tRNAMet and 5′-TGGTCAGAGCGGTCAAGTTAAGTT-3′ for mt-tRNAVal.
BN-PAGE and two-dimensional SDS–PAGE
BN-PAGE was used to separate the individual mitochondrial OXPHOS complexes. Mitoplasts, prepared from fibroblasts by treatment with 0.8 mg of digitonin per milligram of protein, were solubilized with 1% n-dodecyl maltoside. Solubilized samples (10–20 μg) were run in the first dimension on 6–15% polyacrylamide gradient gels as described in detail elsewhere (36). Incorporation of the newly synthesized mtDNA-encoded subunits into the holoenzyme complexes was assessed by two-dimensional BN-PAGE. For this analysis, 30 μg of [35S]methionine/cysteine-labelled mitochondrial translation products was run in the first dimension on 6–15% non-denaturing polyacrylamide gradient gels, followed by a separation on 10–16% Tricine–SDS gradient gels as described in detail in (37).
Retroviral transduction
To confirm the pathogenicity of the MTFMT mutations, we used retroviral vectors to express the wild-type MTFMT cDNA in fibroblasts from the patients. The MTFMT cDNA was amplified by one-step RT-PCR (Qiaqen) with specific primers and was cloned into pBABE and pLXSH, both modified to be Gateway-compatible. Retroviral constructs were transiently transfected into the Phoenix packaging cell line using HBS/Ca3(PO4)2, and the fibroblasts were transduced 48 h later with virus-containing medium in the presence of 4 μg/ml polybrene as in Ref. (38).
RNAi-mediated knockdown of PDF
RNA interference was used for transient knockdown of PDF in control and MTFMT patient fibroblasts. Two stealth RNA interference duplex constructs against human PDF, designed using Block-iT RNAi Express (http://rnaidesigner.invitrogen.com/rnaiexpress), were used. The different RNAi constructs were transiently transfected into the fibroblasts with Lipofectamine RNAimax (Invitrogen) at a final concentration of 24 nM. The cells were re-transfected on day 3, harvested and analysed on day 6.
Immunoprecipitation of radiolabelled COX subunits
Fibroblasts from control or from one MTFMT patient (one 100 mm cell culture dish, 80–90% confluent) were incubated in culture medium containing 40 μg/ml chloramphenicol for 22 h, then labelled for 60 min with [35S]-methionine/cysteine as described and chased for 18 h. Cells were subsequently collected by trypsinization, washed twice in PBS and then extracted in 100 μl of extraction buffer (1% n-dodecyl maltoside, 50 mm HEPES pH 7.6, 150 mm NaCl and protease inhibitors), for 45 min on ice, with occasional vortexing. The extract was cleared by centrifugation at 20 000g, 4°C, for 40 min, and the supernatant was diluted to 0.5% n-dodecyl maltoside (DDM) by mixing with an equal volume of extraction buffer without DDM. Immunoprecipitation of radiolabelled COX subunits was then carried out using a polyclonal anti-COXII antibody (a kind gift from N. Kennaway), with Dynabeads protein A (Invitrogen) according to the manufacturer's instructions, except that the incubations were carried out overnight. The immunoprecipitate was eluted from the beads in a Laemmli sample buffer (BioRad) with β-mercaptoethanol for 15 min, at 45°C, and then fractions (input, unbound, eluate) were run on a 15–20% gradient polyacrylamide gel, which was subsequently dried and exposed to PhosphorImager cassette.
Quantitative mass spectrometry
Protein digestion
Gel pieces were washed with water for 5 min and destained twice with the destaining buffer (50 mm ammonium bicarbonate, acetonitrile) for 15 min. An extra wash of 5 min was performed after destaining with a buffer of ammonium bicarbonate (50 mm). Gel pieces were then dehydrated with acetonitrile. Proteins were reduced by adding the reduction buffer (10 mm dithiothreitol, 100 mm ammonium bicarbonate) for 30 min at 40°C and then alkylated by adding the alkylation buffer (55 mm iodoacetamide, 100 mm ammonium bicarbonate) for 20 min at 40°C. Gel pieces were dehydrated and washed at 40°C by adding ACN for 5 min before discarding all the reagents. Gel pieces were dried for 5 min at 40°C and then re-hydrated at 4°C for 40 min with enzyme solution. Lys-C digestion was performed with a 2.5 ng/µl solution of mass spectrometry grade lysyl endopeptidase from Wako in 25 mm ammonium bicarbonate buffer, incubated at 30°C for 18 h and stopped with 15 µl of 1% formic acid/2% acetonitrile. The supernatant was transferred into a 96-well plate and peptide extraction was performed with two 30-min extraction steps at room temperature using the extraction buffer (1% formic acid/50% ACN). All peptide extracts were pooled into the 96-well plate and then completely dried in vacuum centrifuge. The plate was sealed and stored at 220°C until LC-MS/MS analysis.
LC-MS/MS analysis
Heavy-labelled standards of all targeted peptides were bought from JPT Peptide Technologies. Prior to LC-MS/MS, protein digests were re-solubilized under agitation for 15 min in 15 µl of a solution containing 1% ACN, 1% formic acid and an average concentration of 3 fmol/μl of all heavy-labelled peptide standards. The LC column was a PicoFrit fused silica capillary column (15 cm × 75 µm i.d.; New Objective, Woburn, MA), self-packed with C-18 reversed-phase material (Jupiter 5 µm particles, 300 Å pore size; Phenomenex, Torrance, CA) using a high-pressure packing cell. This column was installed on the Easy-nLC II system (Proxeon Biosystems, Odense, Denmark) and coupled to the Q Exactive (ThermoFisher Scientific, Bremen, Germany) equipped with a Proxeon nanoelectrospray Flex ion source. The buffers used for chromatography were 0.2% formic acid (buffer A) and 100% acetonitrile/0.2% formic acid (buffer B). Peptides were loaded on-column at a flow rate of 600 nl/min and eluted with a 2-slope gradient at a flow rate of 250 nl/min. Solvent B first increased from 2 to 40% in 19 min and then from 40 to 80% B in 5 min. LC-MS/MS data were acquired using a targeted selected ion monitoring method using an inclusion list containing the endogenous peptide ions of interest and their heavy-labelled counterparts. The mass resolution for full MS scan was set to 70 000 (at m/z 200) and a lock mass was used to improve mass accuracy. The AGC target value was set to 1E6, the maximum ion fill time (IT) to 200 ms and the isolation window to m/z 2.0. The normalized collision energy value for each peptide had previously been optimized with the heavy-labelled standards. The capillary temperature was 250°C and the nanospray and S-lens voltages were set to 1.3–1.7 kV and 50 V, respectively.
Data analysis
LC-MS peptide quantification was performed by manual integration of SIM chromatograms using Qual Browser/Xcalibur.
Conflict of Interest statement. None declared.
Funding
This research was supported by a grant from the Canadian Institutes of Health Research (MT15460) to E.A.S. and by grants from the Research Council for Health of the Academy of Finland (135956 and 266498), the Sigrid Juselius Foundation, the Emil Aaltonen Foundation, a Marie Curie International Outgoing Fellowship of the European Union′s Seventh Framework Programme under the grant agreement number 273669 (BioMit) and University of Oulu.
References
- 1.RajBhandary U.L. (1994) Initiator transfer RNAs. J. Bacteriol., 176, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi N., Kawakami M., Omori A., Ueda T., Spremulli L.L., Watanabe K. (1998) Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J. Biol. Chem., 273, 15085–15090. [DOI] [PubMed] [Google Scholar]
- 3.Spremulli L.L., Coursey A., Navratil T., Hunter S.E. (2004) Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol., 77, 211–261. [DOI] [PubMed] [Google Scholar]
- 4.Schwartzbach C.J., Spremulli L.L. (1989) Bovine mitochondrial protein synthesis elongation factors. Identification and initial characterization of an elongation factor Tu-elongation factor Ts complex. J. Biol. Chem., 264, 19125–19131. [PubMed] [Google Scholar]
- 5.Liao H.X., Spremulli L.L. (1990) Identification and initial characterization of translational initiation factor 2 from bovine mitochondria. J. Biol. Chem., 265, 13618–13622. [PubMed] [Google Scholar]
- 6.Spencer A.C., Spremulli L.L. (2004) Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res., 32, 5464–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Holmes W.B., Appling D.R., RajBhandary U.L. (2000) Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J. Bacteriol., 182, 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C., Tibbetts A.S., Kramer G., Appling D.R. (2009) Yeast AEP3p is an accessory factor in initiation of mitochondrial translation. J. Biol. Chem., 284, 34116–34125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smits P., Smeitink J., van den Heuvel L. (2010) Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol., 2010, 737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tucker E.J., Hershman S.G., Kohrer C., Belcher-Timme C.A., Patel J., Goldberger O.A., Christodoulou J., Silberstein J.M., McKenzie M., Ryan M.T., et al. (2011) Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab., 14, 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haack T.B., Haberberger B., Frisch E.M., Wieland T., Iuso A., Gorza M., Strecker V., Graf E., Mayr J.A., Herberg U., et al. (2012) Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J. Med. Genet., 49, 277–283. [DOI] [PubMed] [Google Scholar]
- 12.Neeve V.C., Pyle A., Boczonadi V., Gomez-Duran A., Griffin H., Santibanez-Koref M., Gaiser U., Bauer P., Tzschach A., Chinnery P.F., et al. (2013) Clinical and functional characterisation of the combined respiratory chain defect in two sisters due to autosomal recessive mutations in MTFMT. Mitochondrion, 13, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haack T.B., Gorza M., Danhauser K., Mayr J.A., Haberberger B., Wieland T., Kremer L., Strecker V., Graf E., Memari Y., et al. (2014) Phenotypic spectrum of eleven patients and five novel MTFMT mutations identified by exome sequencing and candidate gene screening. Mol. Genet. Metab., 111, 342–352. [DOI] [PubMed] [Google Scholar]
- 14.Giglione C., Serero A., Pierre M., Boisson B., Meinnel T. (2000) Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J., 19, 5916–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giglione C., Boularot A., Meinnel T. (2004) Protein N-terminal methionine excision. Cell. Mol. Life Sci., 61, 1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M.D., She Y., Soskis M.J., Borella C.P., Gardner J.R., Hayes P.A., Dy B.M., Heaney M.L., Philips M.R., Bornmann W.G., et al. (2004) Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. J. Clin. Invest., 114, 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serero A., Giglione C., Sardini A., Martinez-Sanz J., Meinnel T. (2003) An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem., 278, 52953–52963. [DOI] [PubMed] [Google Scholar]
- 18.Mariottini P., Chomyn A., Doolittle R.F., Attardi G. (1986) Antibodies against the COOH-terminal undecapeptide of subunit II, but not those against the NH2-terminal decapeptide, immunoprecipitate the whole human cytochrome c oxidase complex. J. Biol. Chem., 261, 3355–3362. [PubMed] [Google Scholar]
- 19.Ma L., Spremulli L.L. (1995) Cloning and sequence analysis of the human mitochondrial translational initiation factor 2 cDNA. J. Biol. Chem., 270, 1859–1865. [DOI] [PubMed] [Google Scholar]
- 20.Koc E.C., Spremulli L.L. (2002) Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem., 277, 35541–35549. [DOI] [PubMed] [Google Scholar]
- 21.Gaur R., Grasso D., Datta P.P., Krishna P.D., Das G., Spencer A., Agrawal R.K., Spremulli L., Varshney U. (2008) A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol. Cell, 29, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibbetts A.S., Oesterlin L., Chan S.Y., Kramer G., Hardesty B., Appling D.R. (2003) Mammalian mitochondrial initiation factor 2 supports yeast mitochondrial translation without formylated initiator tRNA. J. Biol. Chem., 278, 31774–31780. [DOI] [PubMed] [Google Scholar]
- 23.Delannoy E., Stanley W.A., Bond C.S., Small I.D. (2007) Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans., 35, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 24.Walker J.E., Carroll J., Altman M.C., Fearnley I.M. (2009) Mass spectrometric characterization of the thirteen subunits of bovine respiratory complexes that are encoded in mitochondrial DNA (Chapter 6). Methods Enzymol., 456, 111–131. [DOI] [PubMed] [Google Scholar]
- 25.Sinha A., Kohrer C., Weber M.H., Masuda I., Mootha V.K., Hou Y.M., RajBhandary U.L. (2014) Biochemical characterization of pathogenic mutations in human mitochondrial methionyl-tRNA formyltransferase. J. Biol. Chem., 289, 32729–32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E.I., Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell, 123, 277–289. [DOI] [PubMed] [Google Scholar]
- 27.Fahiminiya S., Majewski J., Mort J., Moffatt P., Glorieux F.H., Rauch F. (2013) Mutations in WNT1 are a cause of osteogenesis imperfecta. J. Med. Genet., 50, 345–348. [DOI] [PubMed] [Google Scholar]
- 28.Fahiminiya S., Al-Jallad H., Majewski J., Palomo T., Moffatt P., Roschger P., Klaushofer K., Glorieux F.H., Rauch F. (2015) A polyadenylation site variant causes transcript-specific BMP1 deficiency and frequent fractures in children. Hum. Mol. Genet., 24, 516–524. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D. (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods, 7, 575–576. [DOI] [PubMed] [Google Scholar]
- 35.Sasarman F., Shoubridge E.A. (2012) Radioactive labeling of mitochondrial translation products in cultured cells. Methods Mol. Biol., 837, 207–217. [DOI] [PubMed] [Google Scholar]
- 36.Leary S.C., Sasarman F. (2009) Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol. Biol., 554, 143–162. [DOI] [PubMed] [Google Scholar]
- 37.Antonicka H., Leary S.C., Guercin G.H., Agar J.N., Horvath R., Kennaway N.G., Harding C.O., Jaksch M., Shoubridge E.A. (2003) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet., 12, 2693–2702. [DOI] [PubMed] [Google Scholar]
- 38.Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmuller H., Chevrette M., Kaufman B.A., Horvath R., et al. (2009) Mutation in TACO1, encoding a translational activator of COXI, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet., 41, 833–837. [DOI] [PubMed] [Google Scholar]