Abstract
We have previously shown that relative potency factors and DNA adduct measurements are inadequate for predicting carcinogenicity of certain polycyclic aromatic hydrocarbons (PAHs) and PAH mixtures, particularly those that function through alternate pathways or exhibit greater promotional activity compared to benzo[a]pyrene (BaP). Therefore, we developed a pathway-based approach for classification of tumor outcome after dermal exposure to PAH/mixtures. FVB/N mice were exposed to dibenzo[def,p]chrysene (DBC), BaP, or environmental PAH mixtures (Mix 1-3) following a 2-stage initiation/promotion skin tumor protocol. Resulting tumor incidence could be categorized by carcinogenic potency as DBC >> BaP = Mix2 = Mix3 > Mix1 = Control, based on statistical significance. Gene expression profiles measured in skin of mice collected 12 h post-initiation were compared with tumor outcome for identification of short-term bioactivity profiles. A Bayesian integration model was utilized to identify biological pathways predictive of PAH carcinogenic potential during initiation. Integration of probability matrices from four enriched pathways (P < .05) for DNA damage, apoptosis, response to chemical stimulus, and interferon gamma signaling resulted in the highest classification accuracy with leave-one-out cross validation. This pathway-driven approach was successfully utilized to distinguish early regulatory events during initiation prognostic for tumor outcome and provides proof-of-concept for using short-term initiation studies to classify carcinogenic potential of environmental PAH mixtures. These data further provide a ‘source-to-outcome’ model that could be used to predict PAH interactions during tumorigenesis and provide an example of how mode-of-action-based risk assessment could be employed for environmental PAH mixtures.
Keywords: polycyclic aromatic hydrocarbons, toxicogenomics, modeling, skin cancer, mixtures
Polycyclic aromatic hydrocarbons (PAHs) are a class of over 1500 chemicals formed as incomplete combustion products and released into the environment from both natural (e.g. forest fires) or anthropogenic (e.g. burning of fossil fuels, tobacco, charbroiled meats) sources. Several PAHs, particularly those with more than 4 rings such as benzo[a]pyrene (BaP), dibenzo[def,p]chrysene (DBC), have been designated as Class 1 known or Class 2A probable human carcinogens by the International Agency for Research on Cancer (IARC, 2010). While much of the research on PAH carcinogenicity focuses on individual PAHs and BaP, in particular, most human exposures to PAHs result from chemical mixtures through dietary, inhalation, or dermal routes of exposure. Primary sources of environmental exposure to these PAHs include wood smoke, creosote, and burning of fossil fuels and tobacco (IARC, 2010). Recently, diesel engine exhaust was added to the list of Class 1 known human carcinogens and certain other PAH-containing mixtures, including air pollution, have been designated as probable or possible Class2A/B carcinogens in humans (IARC, 2010, 2014).
One of the most difficult challenges for risk assessment is the evaluation of health hazards from exposure to environmental chemical mixtures. Currently, significant data gaps exist for understanding carcinogenicity of PAH mixtures and complex environmental mixtures containing PAHs. Further, little is known about the mechanisms of tumorigenesis for PAH mixtures. Current assessment of cancer risk for PAHs involves testing compounds in the 2-year rodent bioassay, which is not practical for screening large numbers of compounds or mixtures due to expense and time. Therefore, alternative approaches are typically utilized for evaluating the carcinogenic potential of PAHs and PAH-containing mixtures. Currently, the primary method for assessing cancer risk of complex mixtures is the relative potency factor (RPF) approach in which complex mixtures are evaluated based on a subset of individual component PAHs compared with BaP as a surrogate or reference (US EPA, 2010). However, we and others have found this approach inadequate for predicting carcinogenicity of mixtures and certain individual PAHs, particularly those that function through alternate pathways or exhibit greater promotional capacity compared to BaP (Courter et al., 2008; Siddens et al., 2012).
Significant challenges have also been identified in utilizing such reference-based approaches for estimating risk from exposure to PAHs in air pollution or waste sites. Complex environmental mixtures subjected to weathering and aging processes can contain many different PAHs, including alkyl-, N-, S-, and O-substituted forms, along with other unknown chemicals; however, only a limited number of unsubstituted PAHs have been characterized for use in RPF calculations. Mixture toxicity for risk assessment is calculated based on select individual components and assumes additivity through a common mechanism of action for PAHs compared to BaP as a standard. Therefore, the RPF approach does not take into consideration mechanistic information about the different pathways, cells, and tissues affected by PAHs during initiation and promotion. This approach is also insufficient for predicting carcinogenicity of complex real-world environmental mixtures of unknown composition.
In this study, we propose an innovative model for determining carcinogenic risk of PAH mixtures using mechanistic approaches. We hypothesize that a chemical bioactivity profile measured after short-term exposure to individual and mixture PAHs from global transcriptional profiling can be used to discriminate future carcinogenic potential based on important mechanistic differences among exposures. The bioactivity profile acts as a unique fingerprint for genes and pathways activated by chemicals and mixtures postexposure and can be used for predicting long-term consequences such as cancer outcome. An important aspect of the bioactivity profile is that the gene signatures are linked to chemical mechanism of action and can also provide insight into alternate mechanisms of PAH carcinogenesis and related mechanisms for complex mixtures. Based on preliminary data, we demonstrate that long-term cancer outcome for PAHs and mixtures can be predicted from high-content genomic evaluation of bioactivity after short-term exposure.
MATERIALS AND METHODS
Chemicals
BaP and DBC were handled in accordance with National Cancer Institute Guidelines. All pure PAHs and mixtures were prepared under UV depleted light as described in Siddens et al. (2012). The PAHs and environmental PAH mixtures utilized for initiation of skin carcinogenesis in animal models are summarized in Table 1. PAH mixture 1 (Mix 1) consisted of 5 mg/ml diesel particulate exhaust (DPE) in vehicle (toluene containing 5% DMSO). PAH mixture 2 (Mix 2) consisted of 5 mg/ml each DPE and coal tar extract (CTE) in vehicle. PAH mixture 3 (Mix 3) consisted of 5 mg/ml DPE, 5 mg/ml CTE, and 10 mg/ml cigarette smoke condensate (CSC).
TABLE 1.
PAH treatments
| Treatment | Components | |||
|---|---|---|---|---|
| Control | 200 µl toluene | |||
| BaP | 200 µl toluene | 400 nM BaP | ||
| DBC | 200 µl toluene | 4 nM DBC | ||
| Mix 1 | 200 µl toluene | 1 mg DPE | ||
| Mix 2 | 200 µl toluene | 1 mg DPE | 1 mg CTE | |
| Mix 3 | 200 µl toluene | 1 mg DPE | 1 mg CTE | 2 mg CSC |
BaP, Benzo[α]pyrene (100 µg) (Midwest Research Institute, Kansas City, Missouri); DBC, Dibenzo[def,p]chrysene (1.2 µg) (Midwest Research Institute); DPE, Diesel particulate extract (SRM 1650b, National Institute of Standards and Technology, Gaithersburg, Maryland); CTE, Coal tar extract (SRM 1597a, National Institute of Standards and Technology); CSC, Cigarette smoke condensate (provided by Hollie Swanson, University of Kentucky).
Animal studies and tumor analysis
FVB/N mice were exposed to PAHs or PAH mixtures following a 2-stage tumor-promotion protocol in skin. All procedures were conducted according to National Institutes of Health guidelines and were approved by the Oregon State University Institutional Animal Care and Use Committee. Six-week-old, female FVB/N inbred mice obtained from the NCI-Fredrick’s Animal Production Program (Frederick,Maryland) were fed AIN93-G pellets (Research Diets, Inc., New Brunswick, New Jersey) throughout the experiment. At 7.5 weeks of age, mice (groups of 36) were initiated with PAH treatments (summarized in Table 1) by application to shaved skin in 200 µl toluene vehicle. Animals for microarray analysis (N = 4 or 5 per treatment) were killed 12 h after treatment and skin was collected for RNA isolation. Two weeks post-initiation, a 25-week promotion regimen was begun with remaining animals, treating animals twice weekly with 6.5 nmol 12-O-tetradecanoylphorbol-13-acetate in 200 μl acetone. Mice were observed and tumor incidence recorded weekly throughout the 25-week promotion interval. Following promotion, all animals were euthanized and necropsied. Tumors were removed, fixed in formalin, and prepared for histopathology of hematoxylin and eosin-stained sections to determine stage of progression. Tumor incidence was measured as the percent incidence for each treatment based on tumor type. Statistical significance among the treatment groups was calculated by ANOVA with Newman–Keuls multiple testing correction.
Microarrays and gene expression analysis
Individual mouse dermal samples were analyzed by Agilent microarray after initiation with PAHs (N = 4 biological replicates, Table 1) or toluene control (N = 5 biological replicates). RNA was isolated from flash frozen skin samples in Trizol Reagent (Life Technologies, Carlsbad, California) followed by clean-up with Qiagen RNeasy mini prep kit (Valencia, California) according to manufacturer protocols. RNA quality and quantity were assessed by Agilent Bioanalyzer (Santa Clara, California) and Nanodrop spectrophotometry (Thermo Fisher Scientific, Waltham, Massachusetts) analysis, respectively. Samples with A260/280 ratios of 1.9–2.2 and RNA integrity values 6.5 or greater were selected for microarray analysis. For microarrays, RNA was labeled with Agilent’s 2 color Quickamp kit for hybridization to the Agilent 8 X 60K mouse array. Raw intensity data were quantile normalized by RMA summarization (Bolstad et al., 2003) and subject to pairwise analysis of variance (Kerr et al., 2000) with Tukey’s post hoc test and 5% false discovery rate calculation (Benjamini, 1995).
Bioinformatics
Unsupervised hierarchical clustering of microarray data was performed using Euclidean distance metric and centroid linkage clustering to group gene expression patterns by similarity. The clustering algorithms, heat map visualizations, and centroid calculations were performed with Multi-Experiment Viewer (Saeed et al., 2003) software based on Log2 expression ratio values. Functional enrichment analysis was performed in MetaCore (GeneGO, Thomson Reuters) based on mappings of the significant (P < .05) genes in each treatment group onto built-in functional network processes and Gene Ontology biological process categories. Analyses were performed for each database independently. Metacore’s knowledgebase, which is derived through manual annotation and curation from the literature, was used for the biological network processes. Statistical significance for enrichment was calculated using a hypergeometric distribution, where the P-value represents the probability of a particular mapping arising by chance for experimental data compared with the background, which included all genes on the Agilent platform (Nikolsky et al., 2009). All processes included more than 15 genes. Gene Ontology biological processes were further filtered to include only the top 10 most significant (P < 5E−7) processes for each treatment group that were categorized greater than level 2 in the gene ontology tree to reduce redundancy. To identify major transcriptional regulators of gene expression by PAHs, the Statistical Interactome tool was used in MetaCore to measure the interconnectedness of genes in the experimental dataset relative to all known interactions in the background dataset. Statistical significance of overconnected interactions was also calculated using a hypergeometric distribution. Networks were constructed in MetaCore for experimental data using an algorithm that identifies the shortest path to directly connect nodes in the dataset to transcription factors. Network visualizations were created in Cytoscape (Shannon et al., 2003) utilizing the spring-embedded layout. PAH treatments were classified based on tumor outcome with Visual Integration for Bayesian Evaluation (VIBE) v2.0 (Beagley et al., 2010) in which Bayesian integration of significantly enriched (P < .05) pathways was performed using k-nearest neighbors statistical learning algorithm (Atiya, 2005) with leave-one-out cross validation. VIBE performs Bayesian integration of the experimental datasets (i.e. pathways) and provides a classification accuracy based on the integrated probability model (Webb-Robertson et al., 2009).
RESULTS
Classification of PAH Treatments Based on Tumor Outcome
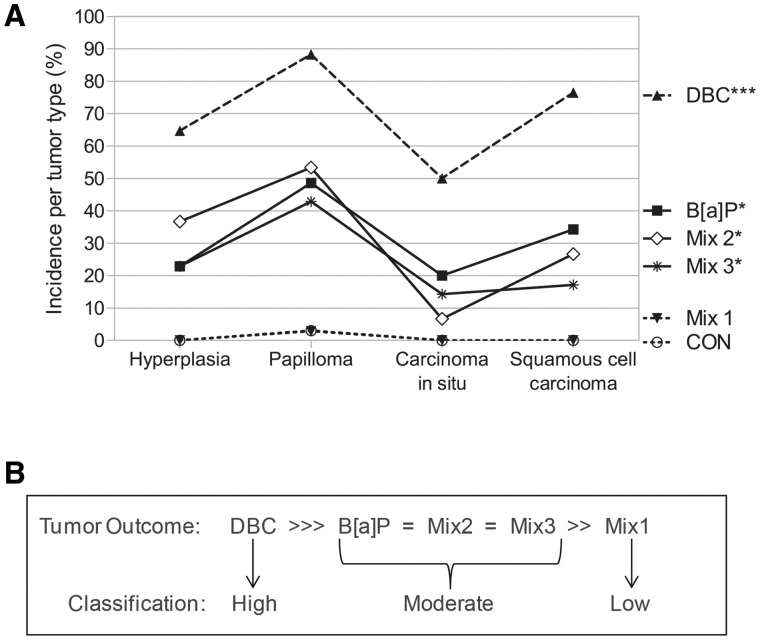
PAHs and environmental PAH mixtures were classified into low, moderate, or high categories based on their ability to induce tumorigenesis following a 2-stage initiation/promotion skin tumor protocol. Classification was based on statistical evaluation of tumor incidence calculated as the percent incidence per tumor type, which was determined by histology from the progression from hyperplasia to papilloma, carcinoma in situ or squamous cell carcinoma. Overall, exposure of FVB/N mice to BaP, DBC, or 1 of 3 environmental PAH mixtures resulted in treatment-specific tumor incidence profiles; although the relative amounts of each tumor type was similar across all PAH treatments (Fig. 1A). The percent incidence of papillomas was greatest for all PAHs and PAH mixtures, while carcinoma in situ was the least prevalent tumor type. In animals initiated with vehicle control or Mixture 1, only one papilloma was detected resulting in 3% tumor incidence for each group. Tumor incidence was highest after initiation with DBC (P < .001 compared with control), ranging from 50 to 90% depending on tumor type. Tumor incidence was similar for BaP, Mix 2, and Mix 3, all of which were significant from controls (P < .05) and were not significantly different from each other. Actual percent tumor incidence, number of animals per treatment group, and individual P-values for each tumor type are provided in Supplementary Data S1. The carcinogenic potential for each PAH treatment was ranked as DBC >> BaP = Mix2 = Mix3 > Mix1 = Control based on statistical evaluation of tumor incidence, which was consistent with that previously reported for time until tumor event and tumor multiplicity for these treatments in mouse skin (Siddens et al., 2012). Based on this ranking, PAH treatments were categorized as having low (Mix 1), moderate (BaP, Mix 2, Mix 3), or high (DBC) carcinogenic potential (Fig. 1B) for evaluation of mechanisms driving PAH-mediated carcinogenesis in skin.
FIG. 1.
Classification of PAH and PAH mixture carcinogenic potential based on tumor incidence. Exposure of female FVB/N mice to PAHs following a 2-stage initiation/promotion skin tumor protocol resulted in (A) tumor incidence profiles of DBC >>> BaP= Mix2= Mix3 >> Mix1 = Control, based on statistical significance (***P < .0001, *P < .05 by 1-way ANOVA with Newman–Keuls multiple testing correction). Tumor incidence was calculated as the percent incidence for each treatment based on tumor type. B, Based on this ranking, PAH treatments were categorized as having low, moderate, or high carcinogenic potential in mouse skin.
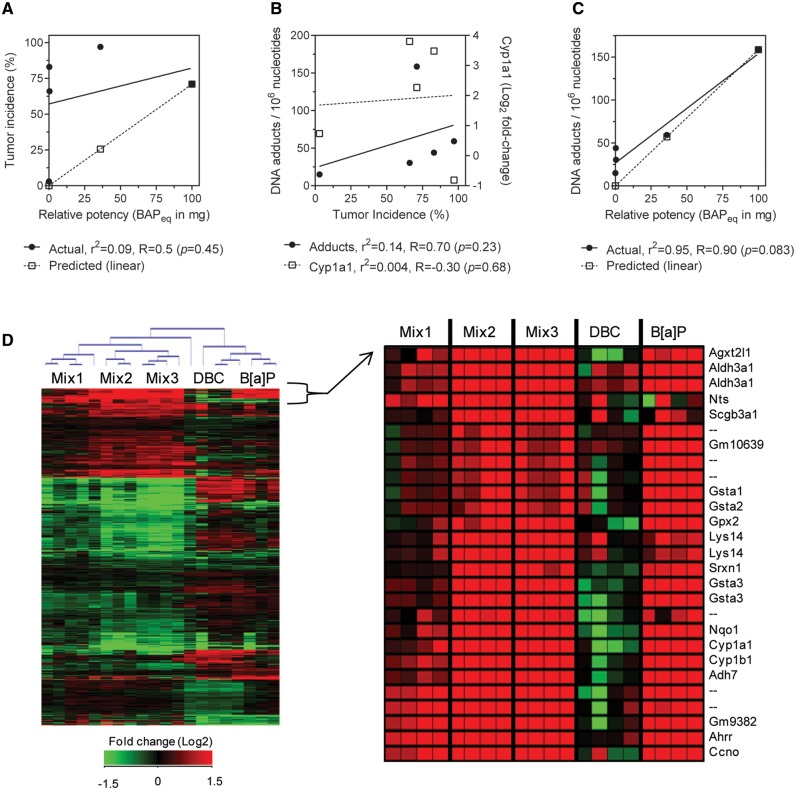
Overall tumor incidence did not correlate with relative potency calculated based on BaP equivalency (BaPeq) in Siddens et al. (2012) in which mixture RPFs are determined using reported RPFs (US EPA, 2010) for known components. Figure 2A shows correlation of actual tumor incidence (black circles, r2 = 0.09, R = 0.5, P = .45) compared with predicted tumor incidence from RPFs by Spearman rank. RPF calculations underestimated carcinogenicity of DBC and the coal-tar containing mixtures (Mix 2 and 3). Induction of Cyp1a1 gene expression measured by microarray at 12 h postinitiation also correlated very poorly with tumor incidence by treatment group (r2 = 0.004, R = −0.30, P = .68). Further, DNA adduct formation measured previously in skin postinitiation (Siddens et al., 2012) did not significantly correlate with tumor incidence (r2 = 0.14, R = 0.70, P = .68) as shown in Figure 2B. DNA adducts were more accurately predicted by RPFs than tumor incidence (Fig. 2C), particularly for DBC treatment. Actual adduct formation correlated with calculated BaPeq (Spearman 0.90, P = .083; linear regression r2 = 0.95, P = .005).
FIG. 2.
Correlation of traditional endpoints with tumor incidence in mouse skin after exposure to PAHs and PAH mixtures. A, Comparison of actual tumor incidence measured in skin to predicted tumor incidence calculated from BaP equivalency (BaPeq). Actual tumor incidence did not significantly correlate with calculated RPFs (Spearman R = 0.50, P = .45; linear regression r2 = 0.09, P = .62). B, Correlation of DNA adduct formation (circles) and expression of Cyp1a1 transcripts (squares) with tumor incidence by Spearman rank. Linear regression was not significant from zero (P > .43). C, Comparison of actual DNA adducts measured in skin by 32P-postlabeling (Siddens et al., 2012) to predicted adducts calculated from BaPeq. Actual adduct formation correlated with calculated RPFs with Spearman R = 0.90 (P = .08) and linear regression r2 = 0.95 (P = .005). D, Global gene expression in mouse skin 12 h postinitiation. Unsupervised clustering of 922 genes differentially expressed (P < .05, 5% FDR) across all treatments. Enlarged heatmap shows gene cluster of highly differentially expressed genes in BaP, Mix2, and Mix3 groups. Values are log2-fold change for all treatments compared with control; red, green, and black represent upregulated, downregulated, and unchanged genes, respectively.
PAHs and PAH Mixtures Have Unique Gene Signatures Postinitiation
Global gene expression was evaluated in mouse skin by microarray 12 h postinitiation with BaP, DBC, and 3 environmental PAH mixtures in order to identify gene signatures during initiation associated with PAH-induced skin carcinogenesis. Overall, 922 genes were differentially expressed (P < .05) in skin after treatment with any PAH or PAH mixture compared with vehicle control; including 137, 246, 97, 428, and 521 genes for BaP, DBC, Mix 1, Mix 2, and Mix 3, respectively (Supplementary Data S2). Comparison of significant genes among treatments is visualized as a 5-way Venn diagram in Supplementary Data S2. Raw and normalized Agilent data files are available online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39455. Microarray results were confirmed using RT-qPCR on a subset of 6 genes with decreased, increased, and no significant change in expression levels relative to control (Larkin et al., 2013). Unsupervised bidirectional hierarchical clustering of all differentially expressed genes resulted in distinct clustering of biological replicates based on treatment group with clear separation between the individual PAH exposures (BaP and DBC) and the environmental mixtures (Fig. 2D). Gene signatures did not cluster based on tumor outcome suggesting they were indicative of treatment-specific responses in skin that were not necessarily contributing to tumorigenesis. This is further supported by the fact that the total number of genes differentially regulated by each treatment group did not correlate with overall tumor incidence (Spearman R = 0.3, P = .68) and linear regression of these endpoints was not significant from 0 (r2 = 0.22, P = .43). In particular, the environmental PAH mixtures containing CTE (Mix 2 and Mix 3) altered the largest number of transcripts in skin postinitiation; although, did not result in the highest incidence of skin tumors. Instead, DBC treatment resulted in the highest tumor incidence while only causing moderate gene expression in skin postinitiation. Based on the strong similarity in both the gene expression patterns and overall tumor incidence by Mix 2 and Mix3, it is apparent that their response was either driven by the CTE alone or by the cumulative effect of diesel exhaust and CTE present in the mixtures with minimal impact from the addition of CSC to Mix 3.
Even though the overall transcriptional response was unrelated to tumor outcome, there were clusters of genes with gene expression patterns similar to the tumor profiles for these PAHs suggesting that a subset of the transcriptional data may be predictive of tumor outcome. The enlarged heatmap in Figure 2D shows 1 example cluster of genes that are highly differentially expressed for BaP, Mix 2, and Mix 3 with a distinct pattern of response from DBC indicating that this particular gene cluster may be relevant for initiation of PAH-induced skin cancer. Genes in this cluster included several phase I and II metabolizing enzymes known to be involved in metabolism of PAHs, including Gsta1, Gsta2, Gsta3, Gpx2, Cyp1a1, Cyp1b1, and Nqo1. Therefore, in order to identify the subset of gene changes during initiation that may be predictive of tumor outcome, we used the full gene expression dataset to systematically model gene changes driving carcinogenesis.
Pathway-Based Classification of Tumor Outcome
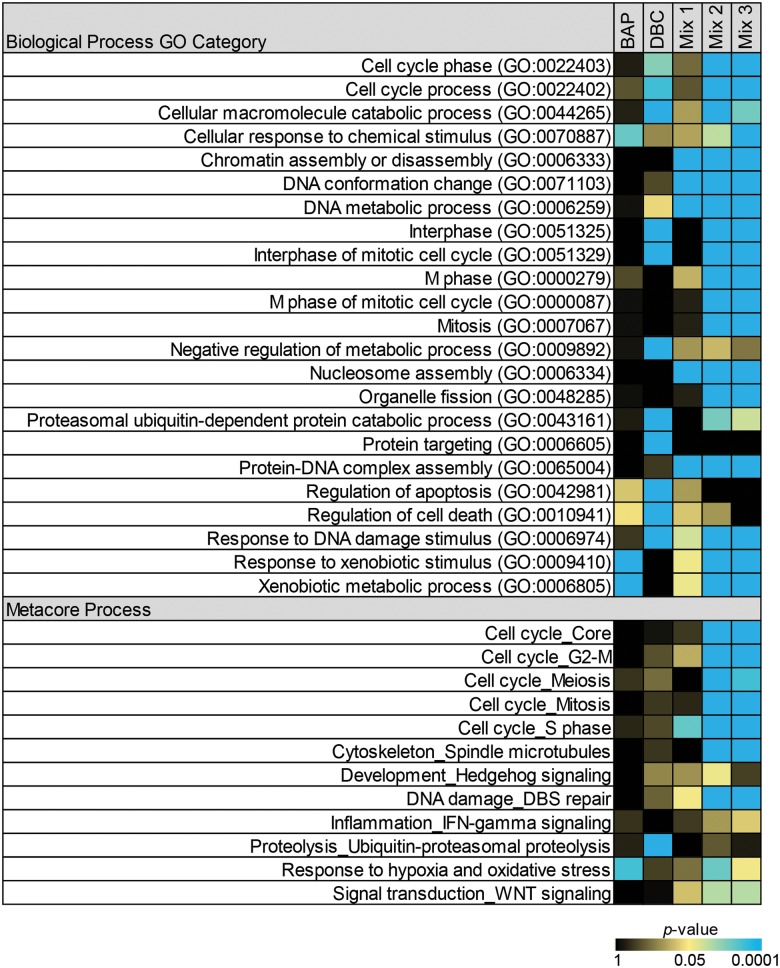
We hypothesized that PAH-induced gene regulation from biological pathways most closely associated with induction of carcinogenesis could be predictive of tumor outcome after exposure. Further, we hypothesized that the mechanism-based gene signatures associated with these pathways could be used to classify potential carcinogens based on their carcinogenic potential. The biological processes that met significance criteria (as described in the Methods section) for Gene Ontology and Metacore processes are shown in Figure 3 as a heatmap in which the most significant functions for each treatment are colored blue and the least significant are colored black. Actual enrichment P values are provided in Supplementary Data S3. Overall, the functions enriched in skin after initiation with Mix 2 and 3 are very similar to each other and mostly unique from the functions enriched for the individual PAH treatments of BaP and DBC. The most significant processes for mixtures 2 and 3 include those associated with cell cycle, mitosis, and response to xenobiotic or DNA damage stimulus. Fewer biological processes are significant postinitiation with BaP and include those associated with xenobiotic metabolism and response to chemical stimulus and oxidative stress. There is little overlap in the processes significant between BaP and DBC and those enriched postinitiation with DBC include cell cycle, apoptosis, interphase of mitosis, and ubiquitin-dependent catabolic processes. While significant enrichment of these functions postinitiation by PAHs provides a basis for understanding their individual mechanisms of action, they do not necessarily indicate which pathways are linked to PAH carcinogenic response. In fact, Mix 1, which did not induce skin tumors, significantly altered several pathways in common with Mix 2/3 associated with DNA–protein complex assembly or nucleosome assembly, suggesting that these processes are not associated with carcinogenic outcome (Fig. 3).
FIG. 3.
Pathways significantly enriched (P < .05) in skin postinitiation by PAH and PAH mixtures. Functional enrichment analysis was performed in MetaCore (GeneGO, Thomson Reuters) based on mappings of the significant (P < .05) genes in each treatment group onto built-in functional network processes and Gene Ontology biological process categories. Statistical significance for enrichment was calculated using a hypergeometric distribution. All processes included more than 15 genes. Gene Ontology biological processes were further filtered to include only the top 10 most significant (P < 5E−7) processes for each treatment group that were categorized greater than level 2 in the Gene Ontology tree.
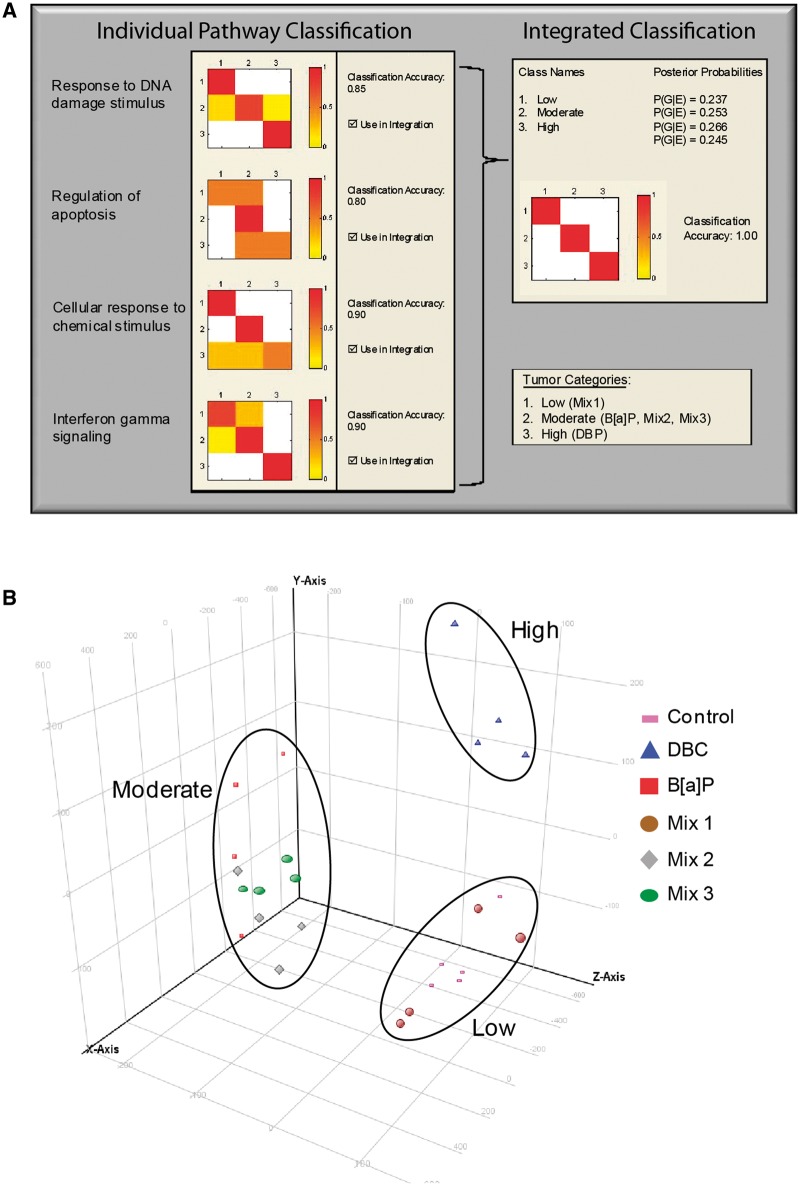
Therefore, in order to systematically filter the significant pathway list in Figure 3 to only those associated with skin carcinogenesis, the microarray transcripts from enriched Gene Ontology and MetaCore processes were evaluated for their ability to classify the PAH treatment groups based on tumor outcome utilizing a Bayesian integration framework. This approach evaluates the ability of the genes differentially expressed in each pathway to classify the PAH exposures based on tumor outcome (low, moderate, or high) utilizing the k-nearest neighbors statistical learning algorithm to build likelihood probability models for each pathway. A classification accuracy was calculated for each pathway based on the number of correctly classified samples compared with the total number of samples. In this case, each sample is an individual animal or biological replicate in the study. Since it is likely that multiple pathways are contributing to the carcinogenic potential of the different PAHs and environmental PAH mixtures, we integrated the posterior probabilities of each pathway utilizing a Bayesian approach to further identify the subset of pathways that result in the highest classification accuracy when integrated together. As shown in Figure 4A, 4 pathways have high individual classification accuracies, ranging 0.80–0.90, including (1) Response to DNA damage stimulus, (2) Regulation of apoptosis, (3) Cellular response to chemical stimulus, and (4) Interferon gamma signaling. When integrated together, the overall classification accuracy improves and the 4 pathways above predict tumor outcome with 100% classification accuracy indicating their importance for the carcinogenic potential of PAHs during initiation.
FIG. 4.
Classification of PAHs and PAH mixture treatments based on tumor outcome. A, Bayesian integration of pathways using k-nearest neighbors statistical learning algorithm with leave-one-out cross validation improves classification accuracy of PAH treatments based on tumor outcome. The color scale for the heatmaps indicates accuracy for actual versus predicted classification of treatments into the low, moderate, and high tumor categories. Highest classification accuracy (100%) is indicated in dark shades and lowest (0%) in white. The panel on the left-hand side shows classification accuracy for each pathway individually and the panel on the right-hand side shows the classification accuracy for all four pathways integrated. B, PCA of the predictive gene set shows separation of treated animals based on tumor outcome.
A total of 172 genes are represented in the pathways from Figure 4A and were differentially expressed in skin postinitiation by PAHs. The list of genes from the predictive pathways is provided in Supplementary Data S4. Principal components analysis (PCA) on this gene set allows for visualization of how these particular genes, reduced from 55K on the Agilent array, may be driving tumor response after PAH initiation (Fig. 4B). Clustering of the samples by PCA clearly distinguishes the samples based on carcinogenic potential, such that the control and Mix 1 samples group together (low), the Mix 2, Mix 3, and BaP samples group together (moderate), and the DBC samples group together (high). In addition to predicting carcinogenesis of the PAHs tested in this study, our data suggest that this approach could also be used to predict carcinogenic potential of unknown PAHs or environmental PAH mixtures in skin based on short-term exposure assessment with additional evaluation and validation.
Distinct Transcriptional Regulators Driving PAH-Mediated Gene Expression in Predictive Pathways
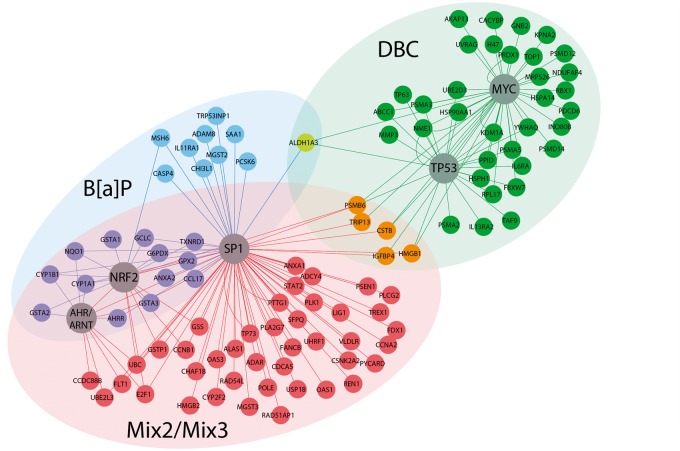
To understand how the pathways predictive of PAH carcinogenesis are regulated in skin during initiation, we performed transcription factor enrichment analysis on the significant genes differentially expressed (out of 172 genes) by each PAH treatment within the predictive pathways. Table 2 lists the transcription factors for each treatment that are significantly (P < .05) overconnected (i.e. transcription factors with a significant number of downstream target genes that are differentially expressed in the gene list compared with that calculated by chance). The most significant transcription factors regulating gene expression after treatment with BaP, Mix 2, and Mix 3 include Arnt, Nrf2, and Sp1. In contrast, DBC-treated genes were most significantly regulated by Myc and p53 resulting in a relatively higher tumor response. These results indicate that there are distinct mechanisms regulating gene expression postinitiation leading to moderate and high levels of skin tumors after PAH exposure. The gene regulatory networks associated with each treatment are shown in Figure 5. Through investigation of the subnetworks for BaP and the PAH mixtures 2 and 3, it is apparent that even though they regulate transcription through the same transcription factors, there are many differentially expressed genes that are unique to each treatment group. The genes that are regulated in common between BaP and the mixtures primarily include Phase I and Phase II enzymes important for the activation and metabolism of PAHs. Most of these genes were not significant after treatment with Mix 1 and none were significant postinitiation with DBC. Overall, however, the treatments associated with a moderate tumor response are more similar at the pathway level than at the gene level suggesting that gene regulation within pathways make better predictors of tumor outcome than a suite of individual gene biomarkers. Transcriptional regulation of genes associated with a high tumor outcome was mostly unique to DBC treatment (Fig. 5).
TABLE 2.
Transcription factor analysis
| Transcription factor | BaP | DBC | Mix 1 | Mix 2 | Mix 3 |
|---|---|---|---|---|---|
| ARNT | *** | ** | *** | **** | |
| NRF2 | **** | **** | **** | ||
| SP1 | **** | * | **** | **** | |
| P53 | **** | ||||
| C-MYC | **** |
****P < .00001, ***P < .0001, **P < .001, *P < .05.
FIG. 5.
Network analysis of pathways predictive of PAH carcinogenic potential during initiation in mouse skin. Gene networks for predictive pathways are visualized for DBC (green), BaP (blue), and Mix2/3 (red). Transcription factors significantly overconnected (P < .05) by hypergeometric distribution to downstream gene expression networks were identified for each PAH treatment (Table 1) and are highlighted (circles) in the network figure. In particular, DBC displays unique gene expression and regulation compared with BaP and the PAH mixtures.
DISCUSSION
Environmental mixtures containing PAH chemicals are of continued and emerging concern because of the existing significant data gaps for understanding their carcinogenic potential and their modes of action as carcinogens. Certain individual PAHs, including BaP and DBC used in our study, are known to produce tumors in mouse skin, lung, liver, and breast and were recently elevated to Class 1 known and Class 2A probable human carcinogens, respectively (IARC, 2010). However, most human PAH exposures result from chemical mixtures of multiple PAHs. Current risk assessment of PAHs primarily relies on the reference-based approach of applying RPFs compared with BaP equivalents for estimating carcinogenicity, which assumes a common mode of action for PAH-induced tumors. We have previously identified tumor profiles for several individual and mixture PAHs that did not correlate with calculated RPF values or with formation of DNA adducts in the 2-stage mouse skin tumor model (Siddens et al., 2012, Figs 2A and B). For the most part, calculated RPFs based on BaPeq underestimated potency in skin. In particular, DBC, which has a reported RPF of 30, was found to be over 100-fold more potent than BaP in our study. Also, the PAH mixtures containing CTE (Mix 2 and Mix 3) induced tumors with similar incidence, multiplicity, and latency to BaP despite calculated BaPeqs of 0.34 and 0.47, respectively, which suggested much lower potency. We also found that the addition of CSC in Mix 3 did not produce an elevated tumor response above Mix 2 as was predicted based on the relative BaPeq. These data support the idea that RPFs do not accurately reflect carcinogenicity of certain individual PAHs or PAH mixtures, which likely involve more complex interactions among PAHs than can be predicted based on BaPeq additivity resulting in either an under or over estimation of carcinogenic potential. We therefore decided to evaluate the mechanisms for initiation of skin tumors by BaP and DBC using gene expression profiling and determine if reference mixtures reflect similar or distinct mode of action compared to the individual PAHs.
Pathway-Based Classification of Carcinogenicity
In this study, we propose a method for predicting potency of PAH chemicals and environmental PAH mixtures based on a bioactivity profile derived from global transcriptional analysis short-term postexposure. Using our initial dataset in mouse skin as proof-of-concept, we provide evidence that a subset of genes and pathways are capable of classifying PAHs and mixtures by carcinogenic potency. This approach does not require a priori knowledge of individual components in mixtures nor does it assume a common mechanism of action for all PAHs and mixtures. Instead, we propose that chemical-specific signaling after exposure provides a unique signature or bioactivity profile for each PAH/mixture that is reflective of its mode of action and can be used to discriminate carcinogenic potency. Our current data suggest that gene expressions within four pathways related to DNA damage, apoptosis, response to chemical stimulus, and interferon gamma signaling were most important for describing variance in our skin model system associated with carcinogenesis of PAHs. When all four pathways were integrated together using a Bayesian framework, samples were classified correctly by potency nearly 100% of the time. Therefore, we provide evidence that short-term bioactivity profiles for a subset of pathways can be used to predict carcinogenic potential of unknown samples and mixtures.
The use of high-throughput data in toxicogenomics for identifying gene signatures and biomarkers associated with toxicity and disease phenotype is increasingly common; however, the application of systems approaches to risk assessment is still in the early stages of evaluation (Lesko et al., 2013). We believe that utilization of these approaches for complex environmental mixtures is an excellent case study for risk assessment due to the significant lack of knowledge regarding mixture toxicity and constituency. Similar genomic-based models have successfully been applied to individual chemicals after short-term exposure to identify modes of action for distinguishing hepatocarcinogens from noncarcinogens from in vivo rat and in vitro human models (Gusenleitner et al., 2014; Song et al., 2012). In particular, Gusenleitner et al. (2014) noted the tissue-specific responses observed when modeling carcinogenicity of a broad range of chemicals from short-term genomic responses. While our study only utilizes data from skin, it also more directly focuses on modeling responses to PAHs and PAH-containing mixtures. We believe that the results of this more focused dataset could be extended to other tissues and exposure routes. Transcriptional signatures have been used successfully to evaluate responses to complex and binary mixtures in multiple tissues and in a summary of comparative gene expression analyses induced by various complex PAH-containing mixtures in vitro and in vivo, several consensus pathways were identified associated with oxidative stress response, metabolism, and immune response that overlap with our predicted dataset (Huang, 2013; Sen et al., 2007). For each functional group, different genes were altered by the extracts supporting our finding that regulation within these pathways could be used to discriminate toxicity among complex PAH mixtures. Other studies that have modeled nonadditive effects of polycyclic aromatic compounds in mixtures on hepatotoxicity utilizing differential gene expression report the strong correlation of gene response with other toxicity endpoints in vivo, including histopathology, gross physiology (e.g. liver weight) and hepatic lipid composition (Kopec et al., 2010, 2011). These studies show the benefits of using gene expression to evaluate quantitative differences in mixture toxicity compared to individual components.
Use of Bioactivity Profiles for Understanding Toxicity Mechanisms
The bioactivity profiles identified through our classification approach reflect processes contributing toward PAH chemical mode of action. Network and transcription factor analysis of the predictive gene clusters further resulted in identification of the upstream transcriptional regulators associated with skin cancer. Overall, we observed distinct gene expression profiles linked to tumor outcome for PAHs and PAH mixtures. DBC treatment, which had the greatest tumor response, uniquely altered genes associated with cell cycle and DNA damage pathways mediated by p53 and c-Myc; while BaP and PAH mixtures containing coal tar were less carcinogenic and altered genes associated with metabolic and stress response pathways mediated by Arnt, Nrf2, and Sp1. The latter response is more typical of metabolic changes and induction in Phase I and II enzymes associated with exposure to PAHs, such as BaP, as shown in purple in the integrative network in Figure 5. The magnitude of gene expression for these enzymes was used, in part, to distinguish and classify PAHs and PAH mixtures based on carcinogenic potential, including the noncarcinogenic Mix 1 containing only the diesel exhaust particulate SRM. However, gene expression for other unique pathways was prognostic for DBC, which appears to function through alternate modes of action. The highly distinct mechanisms regulated by different PAHs short term after exposure suggests activation of unique stress–response pathways instead of a common mechanism of action for all PAHs.
These data help to support a whole mixture approach to risk assessment over a component-based approach, which requires chemical characterization of complex mixtures and assumes common mechanisms of actions for all PAHs. Whole mixture and comparative potency approaches have been proposed by the EPA and others (Jarvis et al., 2014; US EPA, 2010) as more appropriate for complex mixtures when chemical characterization is not possible. These approaches are also better suited for evaluating complex chemical interactions within mixtures because they do not rely on predicting the effects of interactions (e.g. additive vs inhibitory) based on knowledge of the individual components. As we observed in our study in the example of Mix 3, an additive response cannot be assumed. The addition of CSC to Mix 3 did not result in elevated tumor response as expected by RPF calculations. Others have reported similar lack of additive response with PAH mixtures on tumor outcome and suggested antagonistic effects on metabolizing enzymes as the cause (Courter et al., 2008). Instead, whole mixture assessment using mixture assessment factors (as discussed by Backhaus and Faust, 2010; Jarvis et al., 2014) compares the effects of whole mixtures based on a molecular biological endpoint, such as activation of DNA damage signaling. We propose that instead of focusing on a single endpoint, the whole mixture approach to risk assessment could be based on bioactivity profiles of predicted gene sets. Integration across several biological processes using a Bayesian approach improves overall classification accuracy. This approach could potentially be used to determine the quantitative relationships between modes of action so that better potency factors could be calculated for the purpose of evaluating risk among mixtures from various sources. The EPA Framework for use of genomics data provides that toxicogenomics data may be useful in a weight-of-evidence approach for assessing risk (Dix et al., 2006). As such, this pathway-driven approach was successfully utilized to distinguish early regulatory events during initiation linked to tumor outcome and shows the potential of using short-term initiation studies for prediction of carcinogenesis by environmental PAH mixtures. These data provide a ‘source-to-outcome’ model that could be used to predict PAH interactions during tumorigenesis and provide mode-of-action based risk assessment of environmental PAH mixtures.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences grants P42 ES016465, ARRA Supplement to Promote Diversity in Health Research P42 ES016465-S1 and P01 CA90890.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Hollie Swanson, from the University of Kentucky, for providing CSC and the Laboratory Animal Resources Center at Oregon State University for help with the animal studies. We also acknowledge Dr Chris Bradfield and Bradley Stewart at the University of Wisconsin EDGE3 Core Facility for processing of microarrays. We further thank Dr Derek Janszen for statistical analysis of tumor incidence by type. Pacific Northwest National Laboratory is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO1830.
REFERENCES
- Atiya A. F. (2005). Estimating the posterior probabilities using the k-nearest neighbor rule. Neural Computat., 17, 731–740. [DOI] [PubMed] [Google Scholar]
- Backhaus T., Faust M. (2010). Hazard and risk assessment of chemical mixtures under REACH: state of the art, gaps and options for improvement. Swedish Chemicals Agency Bromma, Sweden, Report PM/3. [Google Scholar]
- Beagley N., Stratton K. G., Webb-Robertson B. J. (2010). VIBE 2.0: visual integration for bayesian evaluation. Bioinformatics, 26, 280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Courter L. A., Luch A., Musafia-Jeknic T., Arlt V. M., Fischer K., Bildfell R., Pereira C., Phillips D. H., Poirier M. C., Baird W. M. (2008). The influence of diesel exhaust on polycyclic aromatic hydrocarbon-induced DNA damage, gene expression, and tumor initiation in Sencar mice in vivo. Cancer Lett., 265, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix D. J., Gallagher K., Benson W. H., Groskinsky B. L., McClintock J. T., Dearfield K. L., Farland W. H. (2006). A framework for the use of genomics data at the EPA. Nat. Biotechnol., 24, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Gusenleitner D., Auerbach S. S., Melia T., Gomez H. F., Sherr D. H., Monti S. (2014). Genomic models of short-term exposure accurately predict long-term chemical carcinogenicity and identify putative mechanisms of action. PLoS One, 9, e102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C. (2013). The role of in vitro gene expression profiling in particulate matter health research. J. Toxicol. Environ. HealthB, 16, 381–394. [DOI] [PubMed] [Google Scholar]
- IARC (2010). Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. Monogr. Eval. Carcinog. Risks Hum., 92, 1–853. [PMC free article] [PubMed] [Google Scholar]
- IARC (2014). Diesel exhaust and gasoline engine exhausts and some nitroarenes. IARC Monogr. Eval. Carcinog. Risks Hum., 105, 1–714. [PMC free article] [PubMed] [Google Scholar]
- Jarvis I. W., Dreij K., Mattsson A., Jernstrom B., Stenius U. (2014). Interactions between polycyclic aromatic hydrocarbons in complex mixtures and implications for cancer risk assessment. Toxicology, 321, 27–39. [DOI] [PubMed] [Google Scholar]
- Kerr M. K., Martin M., Churchill G. A. (2000). Analysis of variance for gene expression microarray data. J. Comput. Biol., 7, 819–837. [DOI] [PubMed] [Google Scholar]
- Kopec A. K., Burgoon L. D., Ibrahim-Aibo D., Burg A. R., Lee A. W., Tashiro C., Potter D., Sharratt B., Harkema J. R., Rowlands J. C., et al. (2010). Automated dose-response analysis and comparative toxicogenomic evaluation of the hepatic effects elicited by TCDD, TCDF, and PCB126 in C57BL/6 mice. Toxicol. Sci., 118, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec A. K., D’Souza M. L., Mets B. D., Burgoon L. D., Reese S. E., Archer K. J., Potter D., Tashiro C., Sharratt B., Harkema J. R., et al. (2011). Non-additive hepatic gene expression elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 2,2',4,4',5,5'-hexachlorobiphenyl (PCB153) co-treatment in C57BL/6 mice. Toxicol. Appl. Pharmacol., 256, 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A., Siddens L. K., Krueger S. K., Tilton S. C., Waters K. M., Williams D. E., Baird W. M. (2013). Application of a fuzzy neural network model in predicting polycyclic aromatic hydrocarbon-mediated perturbations of the Cyp1b1 transcriptional regulatory network in mouse skin. Toxicol. Appl. Pharmacol., 267, 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesko L. J., Zheng S., Schmidt S. (2013). Systems approaches in risk assessment. Clin. Pharmacol. Therap., 93, 413–424. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y., Kirillov E., Zuev R., Rakhmatulin E., Nikolskaya T. (2009). Functional analysis of OMICs data and small molecule compounds in an integrated “knowledge-based” platform. Methods Mol. Biol., 563, 177–196. [DOI] [PubMed] [Google Scholar]
- Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques, 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Sen B., Mahadevan B., DeMarini D. M. (2007). Transcriptional responses to complex mixtures: a review. Mutat. Res., 636, 144–177. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddens L. K., Larkin A., Krueger S. K., Bradfield C. A., Waters K. M., Tilton S. C., Pereira C. B., Lohr C. V., Arlt V. M., Phillips D. H., et al. (2012). Polycyclic aromatic hydrocarbons as skin carcinogens: comparison of benzo[a]pyrene, dibenzo[def,p]chrysene and three environmental mixtures in the FVB/N mouse. Toxicol. Appl. Pharmacol., 264, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. K., Song M., Choi H. S., Kim Y. J., Park Y. K., Ryu J. C. (2012). Identification of molecular signatures predicting the carcinogenicity of polycyclic aromatic hydrocarbons (PAHs). Toxicol. Lett., 212, 18–28. [DOI] [PubMed] [Google Scholar]
- US EPA (2010). Development of a relative potency factor (RPF) approach for polycyclic aromatic hydrocarbon (PAH) mixtures. U.S. Environmental Protection Agency, Washington, DC, EPA/635/R-08/012A. [Google Scholar]
- Webb-Robertson B. J., McCue L. A., Beagley N., McDermott J. E., Wunschel D. S., Varnum S. M., Hu J. Z., Isern N. G., Buchko G. W., McAteer K., et al. (2009). A Bayesian integration model of high-throughput proteomics and metabolomics data for improved early detection of microbial infections. Pac. Symp. Biocomput., 2009, 451–463. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.