Abstract
Endogenous circadian clocks are poorly understood within early-diverging animal lineages. We have characterized circadian behavioral patterns and identified potential components of the circadian clock in the starlet sea anemone, Nematostella vectensis: a model cnidarian which lacks algal symbionts. Using automatic video tracking we showed that Nematostella exhibits rhythmic circadian locomotor activity, which is persistent in constant dark, shifted or disrupted by external dark/light cues and maintained the same rate at two different temperatures. This activity was inhibited by a casein kinase 1δ/ε inhibitor, suggesting a role for CK1 homologue(s) in Nematostella clock. Using high-throughput sequencing we profiled Nematostella transcriptomes over 48 hours under a light-dark cycle. We identified 180 Nematostella diurnally-oscillated transcripts and compared them with previously established databases of adult and larvae of the symbiotic coral Acropora millepora, revealing both shared homologues and unique rhythmic genes. Taken together, this study further establishes Nematostella as a non-symbiotic model organism to study circadian rhythms and increases our understanding about the fundamental elements of circadian regulation and their evolution within the Metazoa
The phrase “timing is everything” is often accurate. Since the beginning of life on this planet, organisms have evolved under periodic cycles of light and temperature, caused by the Earth’s rotation and revolution. In response to these cyclic changes, endogenous clocks have evolved in many organisms, allowing them to anticipate daily and seasonal environmental rhythms and to adjust their biochemical, physiological, and behavioral processes accordingly1,2. The most widely studied endogenous biological clock is the circadian clock, an endogenous self-sustained system that drives daily physiological and behavioral rhythms. Broadly, circadian clocks are built from three components: 1) environmental sensors in the clock input pathway through which entraining signals from the environment (e.g., light and temperature) are perceived, 2) transcriptional-translational feedback loops in the core oscillator, which maintain the clock pacing and transmit rhythmic signals to downstream components3 and 3) clock-controlled genes (CCGs), which respond to core oscillator pacing signals and coordinate circadian responses within cells4. In addition,post-translational mechanisms, such as phosphorylation of PERIOD proteins in bilaterian animals by casein kinase 1 family members, are also involved in the clock regulation5. Circadian clocks have been characterized in cyanobacteria, fungi, plants, and animals; however, there is little conservation in clock pathway architecture among these different taxonomic groups6, indicating that circadian rhythmicity is a key adaptive element that evolved independently in metazoans and in several non-metazoan groups7. Within the bilaterian animals, a great deal has been learned about circadian signaling through studies conducted in well-characterized model organisms. Through such studies, investigators have identified both components that are shared among bilaterian animals and those that are restricted to specific lineages. However, findings in these earlier studies also indicate that every model system has its own set of adaptations, specializations, and caveats6,8. Thus, to further expand our understanding of the evolutionary history of circadian behavior and rhythmic gene expression, study of these processes in species that diverged at informative points in evolution are required.
Cnidarians are ecologically important marine and aquatic organisms that arose about 740 million years ago9 and possess a worldwide distribution. They are the simplest extant animals to possess a true tissue-grade of organization (Eumetazoa) and are particularly informative in making inferences about the gene content of the common metazoan ancestor10. An understanding of rhythmic regulation of behavior in cnidarians would provide insight both into the evolution of animal circadian clocks and into the physiology of this key animal group.
The starlet sea anemone, Nematostella vectensis, has emerged as a powerful cnidarian model with a sequenced genome and a growing suite of available molecular resources and tools11,12. Nematostella is widely distributed in brackish environments and unsurpassed for the ease with which its entire life cycle is maintained in the laboratory13,14. As proof of its utility, Nematostella has already provided a first glance into the evolution of the metazoan circadian clock15,16. Several recent studies have indicated that Nematostella and reef-building corals share homologues of some core clock genes with bilaterians15,17,18,19. In addition, microarray studies of the coral Acropora millepora have identified groups of genes including antioxidants, metabolic enzymes, and chaperones that exhibit daily oscillations in expression and may be regulated by circadian mechanisms20. However, many questions remain regarding the mechanism of circadian regulation as well as physiological and behavioral significance of the circadian clock in cnidarians. While Acropora and Nematostella are both members of the class Anthozoa, they exhibit substantial physiological differences. In particular, Acropora and other reef-building corals typically host algal symbionts, which are likely to possess their own circadian clocks and which introduce strong diurnal metabolic signals associated with photosynthesis21. Because Nematostella lacks algal symbionts, it provides a simpler cnidarian model of circadian regulation.
Here we have characterized the Nematostella circadian locomotor activity using a video tracking system under light dark cycles (LD) and under free- running conditions of constant darkness (DD) and constant light (LL). In addition, we have demonstrated that selective inhibition of casein kinase signaling disrupts the circadian locomotor activity under DD free-running conditions. Finally, to characterize the molecular rhythmic actors of Nematostella, RNA-seq and whole transcriptome analysis were conducted during day and night.
Results
Nematostella locomotor activity is rhythmic and is controlled by endogenous circadian clock
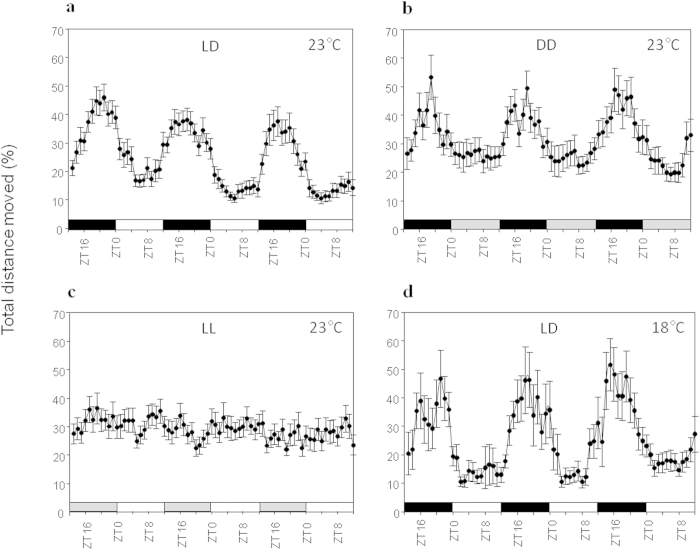
The behavioral rhythms of Nematostella were studied by monitoring the locomotor activity of individuals using a tracking system, which was equipped with an infrared (IR) camera and time-controlled white LED illumination that can be set to different intensities (Fig. 1). In parallel with the locomotor activity tracking, some of the experiments were recorded with a video camera in order to enable visualization of the different movement patterns. Three major movement types were recorded (Supplementary Video S1): head movement from side to side, body banding and constant peristaltic movement along the body axis. Behavioral rhythms were initially characterized over 3 days under 12 h light: 12 h dark (LD) conditions at 23 °C. Automated infrared tracking showed that Nematostella exhibits greater locomotor activity during the subjective night. Under LD conditions, during the night (ZT12–ZT24), the tested animals (n = 35) moved a total of 188.6 cm on average (standard error (SE) = 34.3), compared to only 84.5 cm on average (SE = 4.1) during light hours (ZT0–ZT12; Fig 2A). The averaged Nematostella locomotor activity peaked between four and nine hours after dark onset (ZT16–ZT21). Within this time period, the animals moved on average 101.9 cm (SE = 20.8) compared to only 16.3 (SE = 3.9) in the equivalent time period during light hours (ZT4–ZT9). Fourier analysis of the locomotor activity average ratios during the three days in LD conditions resulted in a single significant periodogram peak at 23.99 h (n = 35), indicating circadian frequency (Fig. 2B in red). To normalize the differences in the absolute distance covered between Nematostella individuals that may originate from differences in size or metabolic rate, we have calculated the relative locomotor activity as a percentage of the maximum locomotor activity recorded for each animal (in LD the average relative locomotor activity ranged between a minimum of 11.2% during light hours and a maximum of 46% during dark hours, Fig. 3A). Similar to the LD results, a single significant frequency peak at 23.98 h was identified in constant dark free-running conditions (DD; n = 20; Fig. 2B in blue) with average relative locomotor activity ranging from 19.9% to 53.4% and following the same oscillation pattern as in LD (Fig 3B). These results support the existence of endogenous clock oscillator; however, in contrast with previous observation [16], our results didn’t show any significant circadian oscillation frequency during the constant light free-run (LL; n = 30; Fig. 2B in green). Under LL conditions, the average relative locomotor activity ranged from 22.2% to 32.1% with no significant dominant frequency (Fig. 3C).
Figure 1. Nematostella locomotor activity tracking.
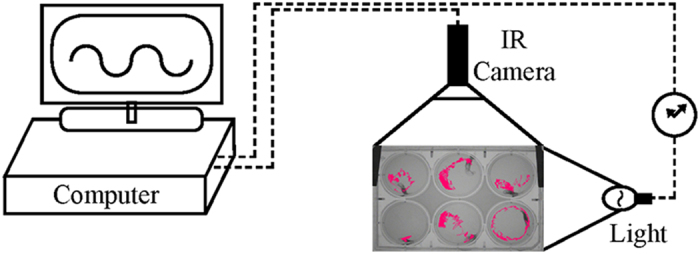
Adult Nematostella were separated in wells of six-well plates and were constantly monitored by an infrared camera inside the Noldus DanioVision XT tracking device. White light was automatically turned on and off as required during the 72–108 hours of the experiments. The red dots within the wells’ arenas illustrate the movement paths of the six anemones. Significant differences in the total distance moved were recorded between individuals; therefore, we used the ratios from the maximal value recorded for each animal in each of the experiments.
Figure 2. Nematostella locomotor activity shows circadian oscillations with nocturnal maxima.
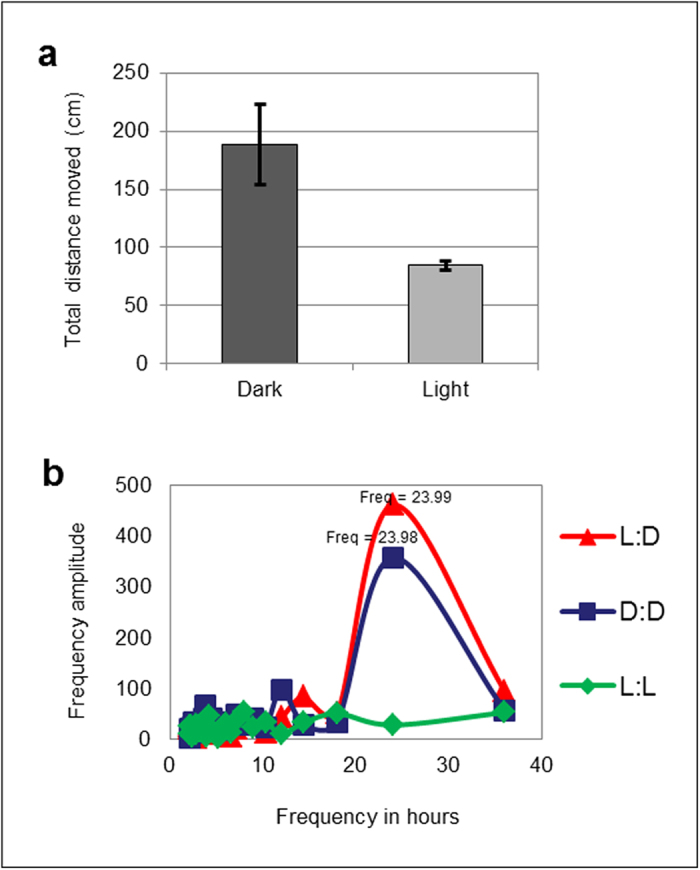
(A). The average total distance moved in dark hours (ZT12–ZT24) was 188.6 cm (n = 35; SE = 34.3) compared to 84.5 cm (SE = 4.1) during light hours (ZT0–ZT12). (B). In the LD and DD experiments, the major oscillation frequency peak identified through Fourier analysis is almost exactly 24 h. In the LL experiment no significant oscillation frequency was identified.
Figure 3. Nematostella locomotor activity has an endogenous rhythm.
(A). Nematostella locomotor activity under a 12 : 12 h light : dark cycle (LD) at 23 °C. (B). Nematostella locomotor activity in constant dark (DD) at 23 °C. (C). Nematostella movement in constant light (LL) at 23 °C. (D). Nematostella locomotor activity in LD at 18 °C. White bars indicate light hours, black bars indicate dark hours, and gray bars indicate illumination conditions different from an LD cycle (dark instead of light; light instead of dark).
As a first approach to study whether Nematostella oscillator exhibits temperature compensation, we inquired if the rhythms observed in LD conditions were also maintained at a lower temperature, we monitored the locomotor activity during three days under LD conditions at 18 °C (5 °C below the temperature of all other experiments). We observed a similar locomotor activity oscillation pattern as in the LD and DD experiments with locomotor activity ratios averages of 10.4% to 55.7% (Fig. 3D). The tested approach may have some limitations associated with potential masking effects of light. Nevertheless, the obtained results suggest potential temperature compensation of Nematostella circadian system.
The Nematostella locomotor activity cycle can be shifted by dark pulse or disrupted by light pulse
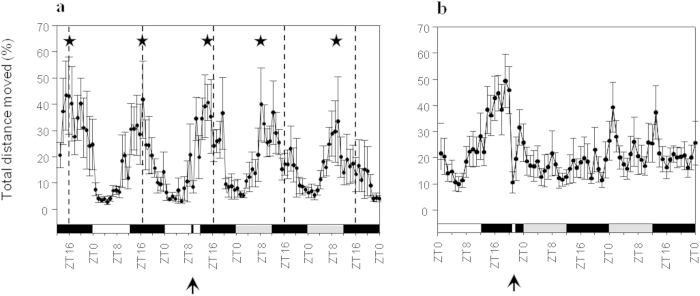
We tested the effect of 1 h dark and 1 h light pulses on the locomotor activity oscillation. The pulses were performed during normal LD conditions, while the effect was tested under DD free-run in order to prevent entrainment by a light cue. When a 1 h dark pulse was applied between ZT9 and ZT10 (2 h before the entrained dark onset), the oscillation phase was advanced and changes were observed in the cycle length. The observed locomotor activity cycle length (based on peak locomotor activity) was advanced by 2 h on the next day (ZT14), by 8 h on the second day after the pulse (ZT8) and by 6 h on the third day after the pulse (ZT10) (n = 11; Fig. 4A). In contrast, a 1 h light pulse, between ZT21 and ZT22 (2 h before the entrained light onset) caused a complete disruption of the locomotor activity cycle during the following dark free-run for the rest of the experiment (Fig. 4B). Due to the nature of the experimental system, prolonged behavioral monitoring was not possible, so it is not possible to determine whether the observed disruption is transient or permanent.
Figure 4. Activity phase shift and cycle disruption following dark or light pulses.
(A). Locomotor activity oscillation phase is shifted following one hour of darkness (dark pulse) between ZT9 and ZT10 (of the second experiment day). (B). Locomotor activity oscillation is completely disrupted following 1 h of light pulse between ZT21 and ZT22 (of the first experiment day). White bars indicate light hours, black bars indicate dark hours, and gray bars indicate illumination conditions different from an LD cycle (dark instead of light; light instead of dark), stars indicate the locomotor activity peak, arrows indicate light/dark pulse.
Rhythmic locomotor activity is inhibited by a pan-CK1δ/ε inhibitor, but not by a CK1δ-selective inhibitor in Nematostella
The casein kinase I (CK1) family consists of serine/threonine protein kinases, some of which are key regulators of circadian timing in bilaterian animals, fungi and green algae22. CK1-like genes have previously been identified in both Acropora and Nematostella and were suggested as components of circadian gene network in these organisms23. Reciprocal BLASTx searches of human and Drosophila CK1 sequences against predicted proteins in the Nematostella JGI genomic database revealed six CK1 family members in Nematostella. Three of the Nematostella CK1 sequences grouped into a clade with Drosophila Doubletime as well as human CK1δ and CK1ε (NvCK1_12115, NvCK1_12051, NvCK1_88486) Two others (NvCK1_159193 and NvCK1_161273) grouped with Drosophila CK1 and human CK1α genes, and the final Nematostella gene (NvCK1_192152) grouped with human CK1γ1 and CK1γ3 (Supplementary Fig. S1).
To investigate a potential role for CK1 activity in circadian function in Nematostella, we characterized the effects of two specific pharmacological inhibitors of vertebrate CK1 activity on circadian behavioral rhythms in Nematostella. One of these inhibitors (PF-4800567) specifically targets CK1δ. The second (PF-670462) inhibits both CK1δ and CK1ε and has been shown to disrupt behavioral rhythms in distantly related organisms, such as the green alga Ostreococcus tauri22. The two inhibitors were tested at concentrations that have been shown to specifically inhibit circadian function in zebrafish24.
Twelve hours prior to the initiation of the locomotor tracking, Nematostella individuals were incubated in 1 μM of the pan-CK1δ/ε inhibitor or CK1δ-selective inhibitor. Over the next two days (48 h), locomotor activity tracking was performed under DD free-running conditions followed by inhibitor-free recovery of 1.5 days (36 h) under LD conditions. CK1δ/ε inhibitor-treated Nematostella lost their locomotor activity oscillation (n = 12, Fig. 5A), while CK1δ inhibitor-treated Nematostella maintained their original oscillation. (n = 12, Fig. 5B). The locomotor activity oscillation of CK1δ/ε inhibitor-treated Nematostella was successfully recovered after replacing the water with inhibitor-free water and changing the light conditions back to LD (Fig. 5A). This suggests that one or more CK1 family members may be involved in the regulation of circadian behavior in Nematostella.
Figure 5. Inhibition of Nematostella locomotor activity oscillation by a CK1δ/ε inhibitor.
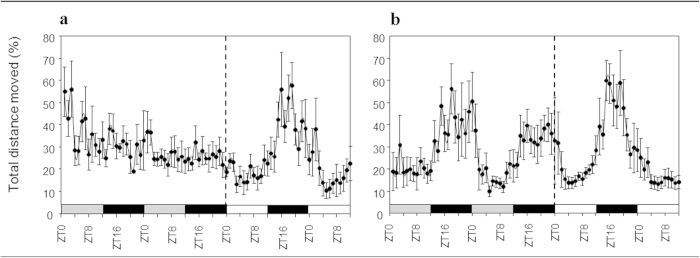
A. Nematostella locomotor activity was inhibited during DD free-run in water containing 1 μM pan-CK1δ/ε inhibitor. Dashed line indicates initiation of recovery following the replacement of the water medium and shifting back to LD illumination regime. B. Nematostella locomotor activity in water containing 1 μM CK1δ inhibitor in the same conditions as in A. No changes in locomotor activity were observed.
Expression of many Nematostella genes exhibit diel rhythmicity
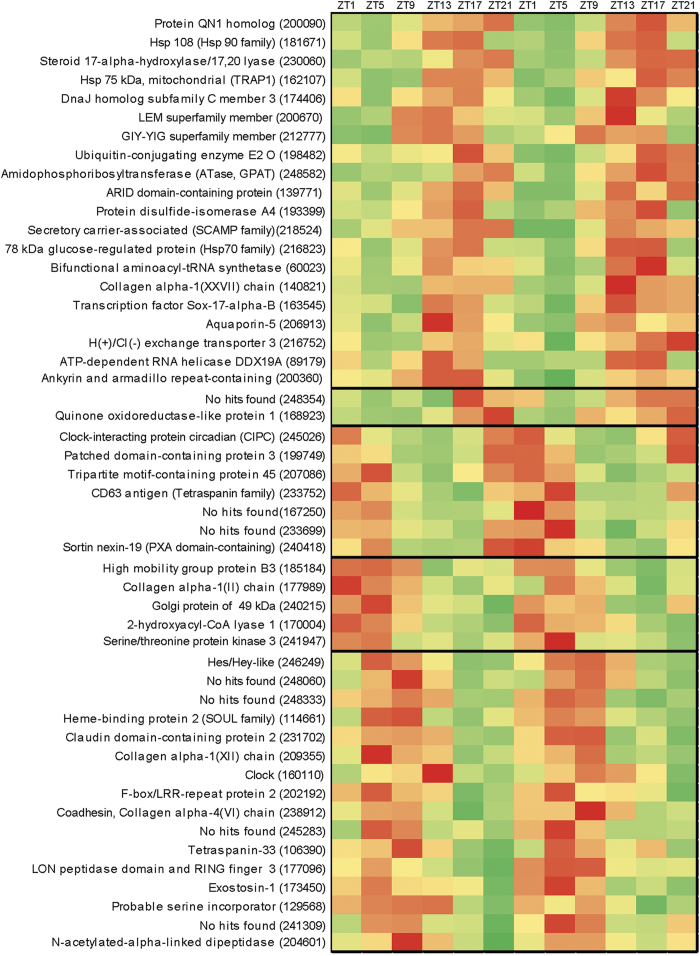
To better understand the molecular forces that regulate the circadian locomotor activity rhythm in Nematostella, we conducted transcriptional profiling using the Illumina HiSeq platform with samples collected every four hours over two days under LD conditions identical to those in the behavioral assay (BioProject accession number: PRJNA246707). Using Fourier analysis, the possible diel rhythmicity (i.e., 24-h periodicity) of all the genes was quantified, and the 180 transcripts exhibiting a g-factor >0.5 were further analyzed. Through K-means clustering, these transcripts were divided into 5 groups, each with a characteristic peak expression time. The 50 transcripts exhibiting the strongest diel rhythm are shown in Fig. 6, and expression data for all 180 genes are listed in Supplementary Table S1; we subsequently refer to these as diel cycle genes (DCGs). Because these genes were identified based on their oscillations under LD conditions they were characterized as “diel control genes” (DCGs) rather than as “clock-controlled genes” (CCGs), which have been specifically demonstrated to maintain a cycle under constant conditions.
Figure 6. Heat map showing major expression of the 50 transcripts with strongest diel rhythmicity (highest g-factor) in Nematostella.
Provisional annotation was based on the top hit to the Swissprot database. Some gene names were edited based on phylogenetic analysis of the Nematostella genes, and some family or domain names were added parenthetically. See supplementary Table S1 for more complete annotation. Color scale ranges from red to green (highest to lowest relative expression). The x-axis indicates time of sampling, where Zeitgeber time (ZT) is the number of hours since the light cue was turned on (lights were turned on at 7 am and off at 7 pm; 8 am is ZT1). Heavy black lines within the heat map indicate genes with similar expression patterns, as identified through K-means clustering.
We annotated 143 of the DCGs through BLASTp-based searches of the SwissProt database. In addition, we identified putative homologues for 59% (22/37) of the unannotated genes through BLAST searches of the Acropora millepora genomic database. These may represent taxonomically restricted genes. GO terms were associated with 135 of the DCGs; however, none of these GO terms were statistically enriched in comparison with the Nematostella transcriptome.
In the present study, NvClock, NvCry1a, NvCry1b and NvCry2 exhibited diel periodicity with similar timing of peak expression to that reported by Reitzel et al.15 (Table 1). Specifically, NvClock expression peaked late in the day (ZT9-13), Cry1a and Cry1b peaked during mid-day (ZT4-11 and ZT5-9, respectively), and Cry2 peaked during early morning or late night (ZT0-4 in15 and ZT21 in the present study). Both Clock and cryptochromes play central roles in regulating circadian cycles in bilaterians. Some cryptochromes are light sensitive and act to directly coupling the circadian clock with exogenous light cues25.
Table 1. Comparison of peak expression times of selected core clock genes between Nematostella and Acropora millepora. All experiments were conducted using a 12 : 12 h light : dark cycle. Peak expression indicated as Zeitgeber time (ZT), which in this case indicates the number of hours after the lights were turned on. Data from qPCR15,19, Illumina (this study), and microarray20.
| Nematostella vectensis | Acropora millepora | |||||
|---|---|---|---|---|---|---|
| Gene | Accession numbers | Reitzel et al. 2010 | This study | Accession numbers | Levy et al. 2011 | Brady et al. 2011 |
| Clock | JGI: 160110 XP_001639742 | ZT11 | ZT9-13 | None/Unknown | NA | ZT14 |
| NvCry1a | JGI: 168581 XP_001631029 | ZT4-ZT11 | ZT5 | CRYb (DY585180; SeqIndex10300; probe A031-G12) | ZT5 | NA |
| NvCry1b | JGI: 16062 XP_001632849 | ZT7 | ZT5-9 | CRY2 (EF202590; SeqIndex 10301) | NA | ZT2 |
| NvCry2 | JGI: 194898 XP_001623146 | ZT0-4 | ZT21 | CRY1 (EF202589; SeqIndex 10302, probes C018-C3, D027-B12) | ZT5 | ZT6 |
Comparative transcriptomic analysis reveals diel cycle genes shared between Nematostella and corals
Nematostella and the scleractinian coral Acropora millepora are both anthozoan cnidarians, but they differ profoundly in terms of habitat and symbiont composition. Individual Nematostella polyps lack algal symbionts and colonize salt marsh environments, while A. millepora forms calcified colonies on tropical reefs through an obligate symbiosis for dinoflagellates. Genes with circadian expression patterns in both taxa are likely to serve fundamental roles in circadian physiology of cnidarians.
We first compared the set of 180 Nematostella DCGs with a set of CCGs that were identified from a previous microarray-based study of Acropora millepora20. Of note, the A. millepora genes exhibited daily oscillations both under LD conditions and under DD free-run. We mapped the differentially expressed microarray probes to 99 unique A. millepora transcripts, 9 of which are putative homologs of Nematostella DCGs (Table 2). Among the shared genes (Nematostella DCGs and A. millepora CCGs) were two cryptochromes. Nematostella and A. millepora each contain two Type I cryptochromes and one Type II cryptochrome. In each species, the Type II cryptochrome and one of the Type I cryptochromes exhibited diel oscillations (i.e., were identified as DCGs in Nematostella and as CCGs in A. millepora, Fig. 7a). The Acropora Type I and Type II cryptochromes have a similar oscillation pattern, which generally overlaps with a Nematostella Type I cryptochrome (NvCry1a), peaking at 12 pm (ZT18) but not with Nematostella Type II cryptochrome (NvCry2), which peaks at 4 pm (ZT10; Fig. 7a).
Table 2. Homologous gene pairs exhibiting light-entrained diel expression cycles in both Nematostella (present study) and Acropora millepora20.
| Gene number | g-factor | Annotation | Peak | Acropora SeqIndex | Acropora peak (# probes) |
|---|---|---|---|---|---|
| 246249 | 0.9231 | Hes/Hey-like | 12 pm | 18661 | 8 am (1/1) |
| 185184 | 0.8102 | High mobility group protein B3 | 12 pm | 7362 | 4 am (1/1) |
| 114661 | 0.8014 | Heme-binding protein 2 (SOUL family) | 4 pm | 16238 | 12 pm (30/32) |
| 181671 | 0.7892 | Heat shock protein 108 (Hsp90 family) | 12 am | 2404 | 4 pm (5/5) |
| 167250 | 0.7128 | No hits | 8 am | 18512 | 8 pm (2/3) |
| 193399 | 0.692 | Protein disulfide-isomerase A4 | 12 am | 2399 | 4 pm (1/1) |
| 216823 | 0.6703 | 78 kDa glucose-regulated protein (Hsp70 family) | 12 am | 12749 | 4 pm (2/2) |
| 194898* | 0.5128 | Cryptochrome 2 | 4 am | 10302 (Cry1) | 12 pm (2/2) |
| 168581* | 0.5076 | Cryptochrome 1a | 12 pm | 10301 (Cryb) | 12 pm (1/1) |
Annotations are based on BLASTp results of the Swissprot database as well as phylogenetic analyses of cryptochromes17 and basic helix-loop-helix genes i.e., Hes/Hey-like;26. Columns labeled “peak” indicate times of maximum expression. In many cases, multiple microarray probes corresponded to a single SeqIndex. The fraction of probes showing the described circadian pattern is indicated parenthetically.
Figure 7. Temporal expression patterns of selected genes exhibiting a rhythmic periodicity in Nematostella.
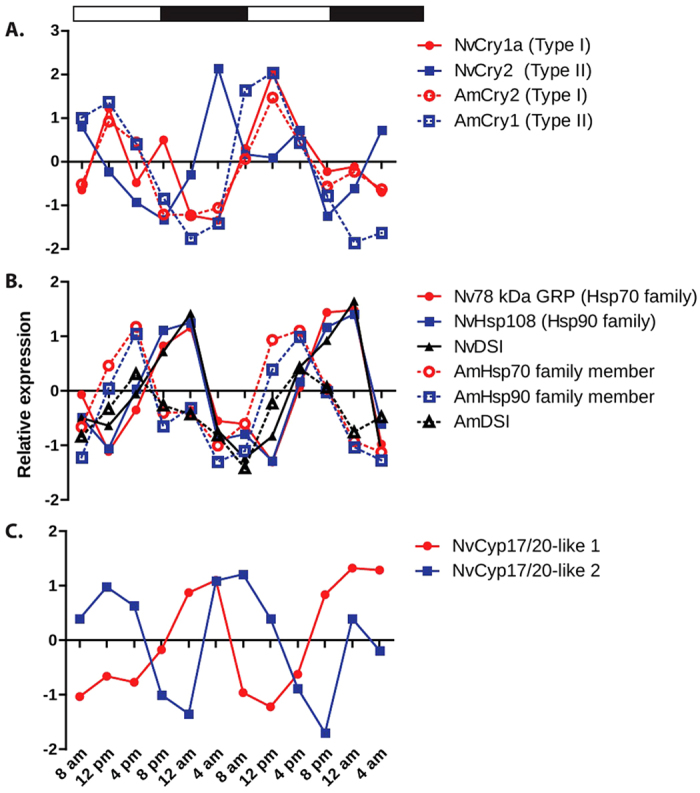
Where available, expression patterns of corresponding genes from Acropora millepora are shown using the same colors with dotted lines and open symbols. Expression values were standardized to facilitate visualization of genes with different expression levels on the same axes. (A) Type I cryptochromes (NvCry1a and AmCry2, red symbols) exhibit peak expression around noon in both species. Type II cryptochrome expression (NvCry2, AmCry1, blue symbols) peaks during subjective night in Nematostella (4 am) and during the day (noon) in A. millepora. Accession numbers in Table 2. (B) Heat shock proteins and disulfide isomerase oscillate strongly both in A. millepora and Nematostella, but the timing differs between the two taxa. For brevity, the heat shock proteins are labeled as Hsp70 and Hsp90 because they are members of these large families. See text for additional discussion. Accession numbers in Table 2. (C) Two cytochrome P450 genes that are most closely related to the CYP17/20 family exhibit diel oscillations that are out of phase with one another (JGI Accession numbers: Cyp17/20-like 1: 230060, Cyp17/20-like 2: 164939).
Additional genes that exhibited diel oscillations in both species were two heat shock proteins (members of the Hsp70 and Hsp90 families) and protein disulfide isomerase, all of which act as chaperones to maintain correct protein folding. In A. millepora, expression of these three genes peaked at 4 pm (ZT10), which was hypothesized to correspond to diel patterns of stress20. In contrast, in Nematostella, all three genes exhibited peak expression during subjective night (12 am, ZT17, Fig. 7b).
Four additional diurnally oscillated genes in Nematostella and A. millepora
Four additional genes exhibited diel oscillations in both Nematostella and A. millepora: Hes/Hey-like, a heme-binding protein in the SOUL family, a high mobility group B protein (HMGB), and a transcript with no similarity to genes of known function. The unannotated gene did exhibit significant similarity (40–50% amino acid identity, e-values around 1 × 10−40) to predicted proteins of unknown function from diverse metazoans. Hes/Hey-like exhibited a strong diel expression pattern in both species, with peak expression at noon for Nematostella and 8 am for A. millepora. Shoguchi et al.26 showed that Nematostella Hes/Hey-like falls within a clade of basic-helix-loop-helix transcription factors that contain an orange domain (bHLH-O). The SOUL family member exhibited similar expression in both species (daytime maxima), but the HGMB and unannotated gene did not.
The Nematostella transcripts that exhibited diel oscillations in expression were also compared with an Illumina-based study of gene expression during the day and night in Acropora millepora larvae19. Of the 180 Nematostella genes with diel oscillations in expression, we identified putative homologues of 108 genes within the A. millepora data set. Six of these A. millepora transcripts exhibited ≥3-fold higher expression during the night, and eight exhibited ≥3-fold higher expression during the day (Table 3). Three of these genes (AmCry1, AmCry2, Hes/Hey-like) also exhibited circadian expression patterns in the A. millepora microarray. In larvae, both AmCry1 and AmCry2 were expressed at higher levels during the day, as they were in the microarray study of adult corals. Hes/Hey-like was also expressed most highly during the day in coral larvae. Comparison of the larval dataset with the Nematostella DCGs revealed additional shared genes that were not identified in the microarray study. For example, Clock expression in larvae was about four times higher during the day compared with the night. A putative homolog of the clock-interacting circadian pacemaker (CIPC) exhibited greatly elevated expression during the night in A. millepora larvae and also exhibited peak expression during the night in Nematostella. CIPC is a mammalian protein that regulates period length by forming complexes with CLOCK, leading to enhanced phosphorylation and degradation27,28. Although CIPC was initially described as absent from invertebrates, similar predicted protein sequences are present in urchins and molluscs (e.g., XP005109657 Aplysia californica and XP800566 Strongylocentrotus purpuratus). It is unknown whether the CIPC-like protein from Nematostella or other invertebrates forms complexes with CLOCK and/or performs a circadian function. Interestingly, in contrast to adult A. millepora, the larvae did not show significant (≥3-fold) transcription change between day and night in any chaperone homologues, this may be related to the fact that in the larvae was sampled only once during the daytime and once during the nighttime (ZT10 and ZT22, respectively).
Table 3. Homologous genes exhibiting daily variation in expression both in the present study of Nematostella and in a study of Acropora millepora larvae. See text for additional details. Peak refers to the time of maximal expression within the Nematostella study.
| Gene number | g-factor | Annotation | Peak | Acropora SeqIndex | Day/Night Counts |
|---|---|---|---|---|---|
| Homologues of genes upregulated during day in coral larvae | |||||
| 246249 | 0.9231 | Hes/Hey-like | 12 pm | 18661 | 407/90 |
| 160110 | 0.7001 | Clock | 8 pm | 10199 | 681/216 |
| 241935 | 0.5782 | Signal transducer and activator of transcription 5A | 8 am | 70856 | 9/3 |
| 243788 | 0.5448 | Pleckstrin homology domain-containing family G member 5 | 12 pm | 61779 | 50/14 |
| 194898* | 0.5128 | Cryptochrome (NvCry2, AmCry1) | 4 am | 10302 | 5724/353 |
| 168581* | 0.5076 | Cryptochrome (NvCry1a; AmCRYb) | 12 pm | 10301 | 1843/309 |
| Homologues of genes upregulated during night in coral larvae | |||||
| 245026 | 0.9255 | Clock-interacting circadian pacemaker (CIPC) | 4 am | 90172 | 7/181 |
| 167250 | 0.7128 | No hits | 8 am | 18512 | 98/1513 |
| 163545 | 0.636 | Transcription factor Sox17αB) | 8 pm | 3863 | 36/449 |
| 240625 | 0.6005 | NIPA-like protein | 12 pm | 10904 | 2/7 |
| 242499 | 0.5467 | Sortilin-related receptor | 4 pm | 13077 | 2/9 |
| 212997 | 0.5288 | Protein BZZ1 | 12 pm | 18943 | 1/3 |
| 192745 | 0.5265 | Putative adenosyl-homocysteinase 3 | 12 p m | 13339 | 2/14 |
| 98402 | 0.5059 | Conserved oligomeric Golgi complex subunit 5 (COG5) | 4 pm | 15726 | 4/18 |
Day/Night counts refers to the number of counts mapped to a given gene in Acropora millepora larvae sampled during the day and night, respectively.
Nematostella Cyp17- and Cyp21-like genes have a phase-shifted diel oscillation
Two transcripts belonging to the superfamily of cytochrome P450 mono-oxygenases (CYPs) exhibited diel periodicity in expression, but with a 4–8 hour phase-shift from one another (Fig. 7c). The difference in timing might indicate that the two enzymes catalyze different steps within a metabolic pathway, producing metabolites that cycle out of phase with one another. In a phylogenetic analysis of animal CYPs, these two Nematostella CYPs fell into a clade that included the vertebrate steroidogenic Cyp17 and Cyp21 genes29. Synthesis of vertebrate-type steroids requires side-chain cleavage of cholesterol by the vertebrate-specific CYP11; CYP17 and CYP21 then act catalyze downstream steps in the synthesis of sex steroids and corticosteroids30. Several mammalian CYPs, including Cyp17, exhibit circadian oscillations in expression, which result in daily cycles in cholesterol homeostasis and hormone concentrations31,32. While the substrate of the Nematostella Cyp17-like genes is unknown, mammalian CYP17 is able to metabolize a variety of substrates including the steroid precursor squalling33.
Profiling of Nematostella reveals genes not previously implicated in cnidarian transcriptional oscillations
Several Nematostella transcripts exhibited strong diel oscillations that had not previously been implicated in cnidarian circadian signaling. Among these, a transcript (NV_200090) similar to QN1 (Centrosomal protein quail neuroretina 1) exhibited strong cycling with peak expression at night (Supplementary Table S1, Fig 6). In vertebrates QN1 helps to regulate the cell cycle during retinal development and serves a motor protein during mitosis34. Of the 50 transcripts oscillating with the strongest diel periodicity (Supplementary Table S1, Fig. 6), four were collagen family members. Collagen transcripts undergo circadian cycles in mammalian cartilage35, but rhythms in collagen expression have not been previously identified in cnidarians. Also of note, many of these strongly oscillating genes (7 of 50), exhibited no significant similarity to annotated genes, or could only be weakly annotated as possessing a conserved domain (e.g, LEM superfamily member, GIY-YIG superfamily member, ARID domain-containing protein). Clearly a great deal remains to be learned regarding the function of these cyclic genes.
K-means clustering demonstrated that distinct groups of DCGs exhibit peak expression throughout the day and night. As previously mentioned, three chaperone proteins exhibited peak expression during subjective night (Fig. 6, top cluster; Supplementary Table S1, cluster 1). Beyond this grouping, genes with similar apparent functions did not necessarily cluster together. For example the genes identified as likely circadian regulators (Clock, CIPC, Cryptochromes, Hes/Hey-like) are distributed broadly among clusters. Because circadian regulation is characterized by feedback from intersecting transcriptional/translational loops, it makes sense that expression patterns of regulatory components will be offset. The four collagen-like DCGs were distributed among three expression clusters. While the reason for this offset is unknown, it’s possible that serial expression of different collagen forms helps to stabilize total collagen levels or that the different forms are necessary for specific components that are produced during on a daily cycle.
Discussion
Through the use of locomotor activity tracking, pharmacological manipulations and transcriptional profiling, we have demonstrated that Nematostella maintains a circadian behavioral cycle, revealed a likely role for CK1 in circadian regulation, and identified novel genes with a diel transcriptional cycle.
The automated locomotor activity tracking approach used in this study provides high spatial and temporal resolution. We found that the use of gray scale analysis with an average center point recorded every second was very informative in this study because Nematostella exhibited frequent peristaltic contractions and bending movements that often resulted in little or no net distance advancement. These movement types may be missed during still image analysis since single frames are unlikely to capture small repetitive changes.
Our locomotor activity recordings indicate, in accordance with a previous report16, that Nematostella is a nocturnal animal with daily oscillations in activity that are controlled by an endogenous clock. However, in contrast to previous observation16, we found that light completely inhibits Nematostella locomotor rhythmicity as no rhythmicity was identified under LL free-run conditions and rhythmicity was lost in response to a light pulse under DD free-run conditions. This difference in results may be due to differences in Nematostella populations used, the light and incubation conditions, or the method of recording. Our work also demonstrates that the clock exhibits phase advance in response to a dark pulse during the entrained light period. It also points to the ability of Nematostella to maintain a consistent behavioral oscillation period under LD conditions at two different temperatures (18 and 23 °C). This observation suggests that Nematostella behavioral rhythms exhibit temperature compensation within a 5 °C range. Temperature compensation is an important feature that corrects for the natural tendency of biochemical reaction rates to change with temperature and thus permits the clock mechanism to have the necessary flexibility to accurately maintain time under changing environmental conditions. The ability to maintain the clock periodicity by compensating temperature is especially important due the rapid climate change and global warming influencing aquatic and marine organisms.
The light level used in our behavioral experiments (200 lux) is low relative to light levels Nematostella could naturally experience. At the sediment water interface in Sippewissett Marsh, MA, a site with a natural Nematostella population, we frequently measure levels above 20,000 lux (Tarrant, unpublished data). It is difficult to know exactly how Nematostella perceives the light environment because the animals are able to burrow into the sediments, which would greatly attenuate their exposure to light. Temperature also produces strong daily cycles in tidepool environments, fluctuating by as much as 20 °C within a single day in Sippewissett Marsh. In addition, Nematostella experiences tidal cycles that affect temperature, salinity, oxygen content and prey availability. It is currently unknown which of these potential zeitgebers act to entrain the endogenous clock within natural environments or how these multiple entraining factors may interact.
In Nematostella we identified 6 members of the casein kinase I (CK1) family of serine/threonine kinases. Several members of this family have been shown to regulate circadian timing in model organisms through phosphorylation of target proteins, including PERIOD (PER) in bilaterians and FREQUENCY (FRQ) in Neurospora36. In bilaterian animals, the CK1 clade containing CK1δ and CK1ε (vertebrates) and Doubletime (Drosophila) plays a well-documented role in clock function24,37,38. CK1ε regulates the circadian negative feedback loop by periodically binding to and phosphorylating the PERIOD proteins, which form complexes with cryptochromes and regulate transcription by the CLOCK/BMAL1 heterodimer. CK1ε can also phosphorylate other circadian proteins including BMAL1 and cryptochromes39. In the golden hamster, mutation of CK1δ (tau mutant) is associated with a shortened behavioral cycle40. In Drosophila, mutations in doubletime (DBT) alter both behavioral rhythmicity and molecular oscillation through interaction with PER proteins38.
We have shown that incubation of Nematostella with a pharmacological inhibitor of CK1δ/ε (PF-670462) signaling disrupts the free-running behavioral rhythm. The same treatment with a CK1δ specific inhibitor resulted in no behavioral rhythm change. In bilaterian animals, CK1-mediated phosphorylation of clock components, especially of PERIOD proteins, helps to regulate circadian period. In studies conducted in mammalian systems, PF-670462 exposure resulted in phase shifts or changes in circadian period41,42. However, similar to our observations with Nematostella complete loss of circadian cycling has been observed in zebrafish following exposure to PF-67046243; the reasons for these differences among studies and model organisms are unknown. Nematostella contains multiple CK1 isoforms, none of which are orthologous to mammalian CK1δ or CK1ε, so it is not clear which form or forms the inhibitor directly targets. Thus, we can only hypothesize that a CK1 family member targeted by the CK1δ/ε inhibitor may be involved in circadian regulation in Nematostella, although a potential toxic effect of the CK1δ/ε inhibitor cannot be ruled out. The CK1δ inhibitor is more specific in its targeting of mammalian CK1 genes, and it appears none of the genes regulating circadian behavior in Nematostella are sufficiently similar to mammalian CK1δ to be affected by the inhibitor. Also, since homologues of period genes have not been identified in Nematostella or other cnidarians, it is difficult to predict the targets for CK1 activity although our behavioral data showed arrhythmicity in the presence of the inhibitor and full recovery in the absence of the inhibitor, as found in studies with other model organisms (e.g.22,42,43).
Through high-throughput sequencing, we identified a subset of genes that exhibited diel variation in transcript expression. These included transcripts such as Clock and cryptochromes that have been identified in previous studies15,44. Others, like CIPC and bHLH-O genes, have well-described roles in bilaterian circadian regulation. Genes in these groups (CIPC-like and Hes/Hey-like) exhibited daily oscillations both in the present study and in one or more studies of A. millepora; however, these genes have not been explicitly discussed as potential regulatory components of the cnidarian clock. bHLH-O proteins generally serve as transcriptional repressors in bilaterians to regulate diverse processes including neurogenesis, vasculogenesis and segmentation45. In Drosophila, the bHLH-O protein CWO (clockwork orange, mammalian homologues DEC1 and DEC2) competitively binds E-box regulatory elements to modulate CLOCK activity46. Similarly, in mammalian systems, HES1 modules CLOCK activity by binding E-box like clock-related elements (EL-boxes)47. Thus, we hypothesize that in Nematostella HES/HEY-like competitively binds to E-boxes and other regulatory elements to modulate signaling by CLOCK and CYCLE. Because CIPC regulates phosphorylation and degradation of mammalian CLOCK27,28, we further hypothesize that the Nematostella CIPC-like protein also forms complexes with CLOCK and affects its phosphorylation status.
A heme binding gene in the SOUL family and a HMGB gene also exhibited diel cycles both in Nematostella and Acropora; members of both of these gene families exhibit circadian cycles in other organism, but they are not known to act as core circadian regulators. Heme-binding genes in the SOUL family were originally identified in a screen for genes that were specifically expressed in the chicken retina and pineal gland, two tissues strongly entrained to circadian rhythms48. In vertebrates, heme plays an important role in circadian regulation through signaling by the Rev-erb nuclear receptors49. However, Rev-erb homologs are not found in cnidarians, and the role of heme, if any, in cnidarian circadian regulation is unknown. High mobility group B (HMGB) proteins act as DNA chaperones to facilitate complex formation between DNA and proteins including repair enzymes and transcription factors50. Circadian expression of some HMGB proteins has been observed in both plants51 and animals52, and they have been proposed to play a role in temperature compensation53.
Transcripts corresponding to chaperone proteins in the Hsp90, Hsp70 and disulfide isomerase families also show consistent daily oscillations in expression in both adult corals and Nematostella. Peak expression of these transcripts in late afternoon in the coral A. millepora has previously been attributed to defense against oxidative stress related to photosynthesis by symbionts in the corals20. Because daily transcriptional patterns in corals reflect the emergent physiology of the host and symbiont (i.e., the ‘holobiont’), interpreting patterns in Nematostella can be less complicated. Our observations in Nematostella suggest that cycles in chaperone expression may be more fundamentally rooted in circadian regulation. Indeed, studies in mammalian models suggest that some Hsp90 isoforms regulate BMAL1 cellular protein levels54, and heat shock proteins have been implicated in both the entrainment and output of the central oscillator55,56.
In conclusion, this work integrates behavioral studies with transcriptional profiling to investigate the circadian clock of Nematostella, a cnidarians species which arose about 700 million years ago11. Features shared between the circadian clocks of Nematostella and bilaterian animals were most likely present in the earliest metazoans. Our findings show that Nematostella meets all major conditions for the function of a true endogenous clock, and can serve as a valuable model organism to study the evolution of animal circadian clock and to understand its function in the cnidarian lineage.
Materials and methods
Nematostella culture
Laboratory-bred Nematostella were maintained in plastic containers with one-third strength artificial sea water (33% ASW, Reef crystals) at 18 °C under a 12 : 12 h (7 am–7 pm/7 pm–7 am) LD cycle. Animals were fed five times per week with freshly-hatched brine shrimp, and water was renewed weekly. Animals were gradually acclimated to 23 °C and starved for two days prior to behavioral experiments and transcriptional profiling.
Behavioral assays
Locomotor activity of individual Nematostella were monitored using two Noldus DanioVision XT tracking devices, each equipped with an IR camera and white LED illumination that can be set to different intensities and LD cycles (Fig. 1). The data collection and analysis were carried out by EthoVision XT8 video tracking software (Noldus information technology, Wageningen, Netherlands). Animals were isolated in wells of six-well plates, each of which was manually defined as a tracking ‘arena’ in the EthoVision software. Center-point detection with gray scaling (detection range of 25–77, contour erosion of 1 pixel, high pixel smoothing) was used to monitor movements, which were calculated according to the change in position of the average center pixel each second (Fig. 1).
Illumination was provided within the DanioVision tracking device by the integral white LED light with an intensity of 200 (+/−10) lux (25% of its maximum intensity) and did not significantly affect the experimental temperature (23 °C). When needed (as for the 18 °C experiment), a chiller pump was used to keep the water temperature fixed during the duration of the experiment. The illumination cycles were the same as used for culturing (12 : 12 h LD). Since this is the first application of this tracking system to measurement of sea anemone movements, we tested the system background noise using measurements of six immobilized (paralyzed with MgCl2) Nematostella individuals for 1 h. The recorded movement in this test was less than 1 cm, and was considered as insignificant background noise (compared with the average movement of the non-paralyzed animals). Parameters were optimized to ensure that organisms were detected throughout the entire observation period.
Locomotor activity data analysis
The total distance moved was summed in hourly bins and expressed as a percentage of the maximum hourly distance measured for each individual. The average and standard errors were calculated for all tested animals based on the normalized values of each hour. The oscillation frequencies were evaluated based on the average values of each experiment using Fourier analysis, as previously described57.
Casein Kinase inhibition
Nematostella individuals were monitored in 6-well plates containing one-third strength ASW with one of two casein kinase inhibitors; the pan-CK1δ/ε inhibitor PF-670462 or the CK1δ-selective inhibitor PF-4800567 (Pfizer Global Research and Development, TOCRIS Bioscience) dissolved in DMSO. In order to determine the effective concentrations, we performed an initial toxicity assay based on the range tested by Smadja Storz et al.24. We tested the viability of the animals based on response to mechanical touch 1, 3 and 10 days after adding the inhibitors to the water in the six-well plates to final concentrations of 0.1, 1 and 10 μM (n = 6). All Nematostella individuals survived up to 10 days after incubation in 0.1 and 1 μM of both inhibitors, but all died 10 days after incubation in 10 μM concentration of either inhibitor. Based on these results, all inhibition experiments were conducted in final concentrations of 1 μM. All controls were treated with identical concentrations of DMSO (0.05%).
RNA-seq
We used RNA-seq technology to identify diel cycle genes (DCGs) in Nematostella following an experimental design previously used in circadian studies of the coral Acropora millepora20. Anemones were acclimated and maintained during the experiment inside the Noldus DanioVision XT tracking device under identical light (LD) and temperature conditions as in the behavioral assay. Five anemones were sampled every 4 h over two consecutive days, starting at 8 am. Total RNA was extracted from pools of five individuals using the Qiagen RNeasy Mini Kit. The Illumina TruSeq protocol was used to prepare libraries from the RNA samples. We performed one biological replicate by constructing and sequencing two Illumina libraries from different samples of five animals collected at same time point (the second time point, 12pm). The libraries were multiplexed on 2 lanes of an Illumina HiSeq2000. On average, ~15 million 50 base-pair paired-end reads were obtained for each library. The data was deposited as an SRA BioProject (accession number: PRJNA246707). Reads were aligned to the Nematostella genome11 using TopHat58. Only reads that uniquely aligned to protein coding regions with up to two mismatches were retained. The Nematostella gene information was downloaded from Joint Genome Institute database ( http://genome.jgi-psf.org/Nemve1/Nemve1.info.html). A custom Perl script was used to parse the output from TopHat (Sequence Alignment/Map (SAM) format) and to convert it into raw number of reads aligned to each position in each Nematostella gene. The dataset was de-duplicated to remove multiple reads with identical start positions in the genome, as these might represent PCR artifacts59. Library quality was assessed in comparison with a benchmark library described by Levin and colleagues59. All library quality parameters met the benchmark standards, including mapping of reads to unique genome start sites and evenness in expressed gene coverage.
We tested the effect of biological variation by comparing two libraries derived from different anemones collected at the same time and light condition. The differences between the samples are close to the expected technical noise (96% of the genes are within the expected 99%-region of Poisson noise), as described recently for miRNA-seq multiplexing60.
The logarithmically-transformed gene expression values were normalized using a modification of the TMM method61, in which the mRNA profiles were scaled such that the log-fold changes of all the mRNAs are distributed around zero (after trimming the higher and lower quartiles of the log-fold changes). The scaling factor was thus set so that the trimmed mean of log-folds vanish. The mean was weighted using the inverse standard deviation, as estimated from Poisson distribution of counts60.
Fourier analysis for expression pattern
The time-dependent signal was converted into a frequency-dependent signal using the Fast Fourier Transform (FFT). We used in-house scripts that were previously found to be accurate in detecting circadian genes, as attested by ~90% true positive rate in independent validation experiments (20,62). The extent to which the original signal contains a 24-hr rhythm was quantified by the ratio (‘g-factor’) of the power (squared amplitude) of the frequency which corresponds to a 24-h period, to the sum of powers of all frequencies. The higher the g-factor, the higher is the confidence that the transcript exhibits a diel rhythm. Changing the definition of the g-factor by adding the powers of higher harmonics of the 24-h period to the numerator, gave similar results compared to the use of the definition above. The genes with the highest g-factor (g-factor greater than 0.5 was used as a cutoff) were sorted into five clusters with similar temporal expression patterns using a K-means clustering, implemented in Matlab as described by Levy et al.20.
Annotation of DCGs
Functional annotation of Nematostella transcripts, including predicted homologs within the Swissprot database and from the transcriptome of the coral Acropora millepora were downloaded from the Joint Genome Institute database. Annotations were manually curated for genes exhibiting strong diel periodicity in their expression patterns (50 genes with highest g-factor) and those identified through our comparative analysis (see below). Manual curation was based on BLASTp searches of the Swissprot and NR databases and, in a few cases, published phylogenetic analyses (cryptochromes, Hes/Hey-like).
Comparative transcriptomics
We compared the set of DCGs identified in our study with genes exhibiting circadian expression patterns or strong day/night differences in two published studies of the coral Acropora millepora. Brady et al.19 used Illumina-based transcriptional profiling to compare gene expression between coral larvae collected during day and night (12 : 12 h LD cycle, samples collected 10 hours after lights on (ZT10) and 10 hours after lights off (ZT22)). They reported the number of counts and fold change, but did not provide any further statistical analysis. From the 47,666 transcripts that they identified, we selected the 10,294 genes that exhibited a three-fold difference in expression between the day and night and identified potential homologs of the putative DCGs from Nematostella. Levy et al.20 used an experimental design similar to the present study: Acropora millepora colonies were sampled every 4 hours over 2 days under LD and DD conditions. They conducted expression profiling using a cDNA microarray. We selected 200 genes exhibiting the strongest circadian expression patterns (g-factor > 0.6468), identified the associated probe sequences in the NCBI Gene Expression Omnibus (GEO) database (Platform GPL6941), and annotated them using BLAST searches of a Acropora millepora larval transcriptome database hosted on SymBioSys (http: sequoia.ucmerced.edu/SymBioSys). We then identified potential homologs among the putative DCGs from Nematostella.
Additional Information
How to cite this article: Oren, M. et al. Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci. Rep. 5, 11418; doi: 10.1038/srep11418 (2015).
Supplementary Material
Acknowledgments
This work was supported by the Israel-US Binational Science Foundation to OL and AMT (Award 2011187). Additional support was provided by the WHOI Early Career Scientist Award to AMT. We would like to thank Mor Samuelson for his devoted caring of the animals and the help in the experiments, Dr. Adam Reitzel for helpful early discussions, Dr. Michal Sorek for her help in the Fourier analysis, Dr. Eldad Hoch, Dr. Modi Rupin and Dr. Tali Lerer for the technical and academic support.
Footnotes
Author Contributions M.O., L.A., O.L. conceived and designed the experiments. M.O., N.S.B., I.E. performed the experiments. A.M.T., S.A., M.O. analyzed the data. O.L., L.A., A.M.T. Contributed reagents, materials and research tools. M.O., A.M.T., S.A., L.A., O.L. wrote the paper.
References
- Pittendrigh C. S. Temporal organization: Reflections of a Darwinian clock watcher. Annu Rev Physiol 55, 16–54 (1993). [DOI] [PubMed] [Google Scholar]
- Wijnen H. & Young M. W. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40, 409–448 (2006). [DOI] [PubMed] [Google Scholar]
- Hardin P. E., Hall J. C. & Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 (1990). [DOI] [PubMed] [Google Scholar]
- Dunlap J. C. Molecular bases for circadian clocks. Cell 96, 271–290 (1999). [DOI] [PubMed] [Google Scholar]
- Price J. L. Translational Regulation of the Drosophila Post-Translational Circadian Mechanism. PLoS genetics 10, e1004628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D. et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6, 544–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W. & Kay S. A. Time zones: a comparative genetics of circadian clocks. Nature Reviews Genetics 2, 702–715 (2001). [DOI] [PubMed] [Google Scholar]
- Zhu H. et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS biology 6, e4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. et al. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Molecular phylogenetics and evolution 62, 329–345 (2012). [DOI] [PubMed] [Google Scholar]
- Ball E. E., Hayward D. C., Saint R. & Miller D. J. A simple plan--cnidarians and the origins of developmental mechanisms. Nat Rev Genet 5, 567–577 (2004). [DOI] [PubMed] [Google Scholar]
- Putnam N. et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- Tarrant A. M. et al. Current directions and future perspectives from the third Nematostella research conference. Zoology Manuscript in press. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand C. & Uhlinger K. The culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biological Bulletin, Marine Biological Laboratory, Woods Hole 182, 169–176 (1992). [DOI] [PubMed] [Google Scholar]
- Stefanik D. J., Friedman L. E. & Finnerty J. R. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis. Nature Protocols 8, 916–923, 10.1038/nprot.2013.044 (2013). [DOI] [PubMed] [Google Scholar]
- Reitzel A., Behrendt L. & Tarrant A. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS ONE 5, e12805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks W. D., Byrum C. A. & Meyer-Bernstein E. L. Characterization of circadian behavior in the starlet sea anemone, Nematostella vectensis. PLoS ONE 7, e46843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel A. M., Tarrant A. M. & Levy O. Circadian clocks in the cnidaria: environmental entrainment, molecular regulation, and organismal output. Integrative and Comparative Biology 53, 118–130, 10.1093/icb/ict024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K. D., Szmant A. M. & Pyott S. J. Circadian clock gene expression in the coral Favia fragum over diel and lunar reproductive cycles. PLoS ONE 6, e19755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A. K., Snyder K. A. & Vize P. D. Circadian cycles of gene expression in the coral, Acropora millepora. PLoS ONE 6, e25072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O. et al. Complex diel cycles of gene expression in the coral-algal symbiosis. Science 331, 175 (2011). [DOI] [PubMed] [Google Scholar]
- Sorek M., Díaz-Almeyda E. M., Medina M. & Levy O. Circadian clocks in symbiotic corals: The duet between< i> Symbiodinium</i> algae and their coral host. Marine genomics (2014). [DOI] [PubMed]
- van Ooijen G. et al. Functional analysis of casein kinase 1 in a minimal circadian system. PLoS ONE 8, e70021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize P. D. Transcriptome analysis of the circadian regulatory network in the coral Acropora millepora. The Biological Bulletin 216, 131–137 (2009). [DOI] [PubMed] [Google Scholar]
- Storz S. S. et al. Casein kinase 1δ activity: A key element in the zebrafish circadian timing system. PLoS ONE 8, e54189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I. et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62, 335–364 (2011). [DOI] [PubMed] [Google Scholar]
- Shoguchi E., Tanaka M., Shinzato C., Kawashima T. & Satoh N. A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene 515, 426–431 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao W.-N. et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nature Cell Biology 9, 268–275 (2007). [DOI] [PubMed] [Google Scholar]
- Yoshitane H. et al. Roles of CLOCK phosphorylation in suppression of E-Box-dependent transcription. Mol Cell Biol 29, 3675–3686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Goldstone J. V. & Stegeman J. J. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome 450s. Phil Trans R Soc B 368, 20120474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov G. V. et al. Independent elaboration of steroid hormone signaling pathways in metazoans. Proceedings of the National Academy of Science USA 106, 11913–11918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamajuku D. et al. Regular feeding plays an important role in cholesterol homeostasis through the liver circadian clock. Circulation Research 105, 545–548 (2009). [DOI] [PubMed] [Google Scholar]
- Kosir R. et al. Circadian expression of steroidogenic cytochromes P450 in the mouse adrenal gland--involvement of cAMP-responsive element modulator in epigenetic regulation of Cyp17a1. FEBS J 279, 1584–1593 (2012). [DOI] [PubMed] [Google Scholar]
- Liu Y., Yao Z.-X. & Papadopoulos V. Cytochrome P450 17a hydroxylase/17,20 lyase (CYP17) function in cholesterol biosynthesis: identification of squalene monooxygenase (epoxidase) activity associated with CYP17 in Leydig cells. Molecular Endocrinology 19, 1918–1931 (2005). [DOI] [PubMed] [Google Scholar]
- Leon A., Omri B., Gely A., Klein C. & Crisanti P. QN1/KIAA1009: a new essential protein for chromosome segregation and mitotic spindle assembly. Oncogene 25, 1887–1895 (2006). [DOI] [PubMed] [Google Scholar]
- Honda K. K. et al. Different circadian expression of major matrix-related genes in various types of cartilage: modulation by light-dark conditions. J Biochem 154, 373–381 (2013). [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K. & Edery I. The Drosophila CLOCK Protein Undergoes Daily Rhythms in Abundance, Phosphorylation, and Interactions with the PER–TIM Complex. Neuron 21, 857–867 (1998). [DOI] [PubMed] [Google Scholar]
- Vielhaber E., Eide E., Rivers A., Gao Z. H. & Virshup D. M. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol 20, 4888–4899 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. L. et al. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95 (1998). [DOI] [PubMed] [Google Scholar]
- Eide E. J., Vielhaber E. L., Hinz W. A. & Virshup D. M. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Ie. J Biol Chem 277, 17248–17254 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey P. L. et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 2888, 483–492 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K. M. et al. Selective Inhibition of Casein Kinase 1ϵ Minimally Alters Circadian Clock Period. Journal of Pharmacology and Experimental Therapeutics 330, 430–439 (2009). [DOI] [PubMed] [Google Scholar]
- Meng Q.-J. et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proceedings of the National Academy of Sciences 107, 15240–15245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz S. S. et al. Casein kinase 1δ activity: a key element in the zebrafish circadian timing system. PloS one 8, e54189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres R. et al. Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone Nematostella vectensis. EvoDevo 5, 26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L. & Turner D. L. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20, 8342–8357 (2001). [DOI] [PubMed] [Google Scholar]
- Kadener S., Stoleru D., McDonald M., Nawathean P. & Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21, 1675–1686 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueshima T. et al. Identification of a new clock-related element EL-box involved in circadian regulation by BMAL1/CLOCK and HES1. Gene 510, 118–125 (2012). [DOI] [PubMed] [Google Scholar]
- Zylka M. J. & Reppert S. M. Discovery of a putative heme-binding protein family (SOUL/HBP) by two-tissue suppression subtractive hybridization and database searches. Molecular Brain Research 74, 175–181 (1999). [DOI] [PubMed] [Google Scholar]
- Yin L. et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007). [DOI] [PubMed] [Google Scholar]
- Štros M., Launholt D. & Grasser K. D. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Science 64, 2590–2606 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. D. & Zheng C. C. Abundance of mRNAs encoding HMG1/HMG2 class high-mobility-group DNA-binding proteins are differentially regulated in cotyledons of Pharbitis nil. Plant Mol Biol 37, 235–241 (1998). [DOI] [PubMed] [Google Scholar]
- Hoppe G., Rayborn M. E. & Sears J. E. Diurnal rhythm of the chromatin protein Hmgb1 in rat photoreceptors is under circadian regulation. J Comp Neurol 501, 219–230 (2007). [DOI] [PubMed] [Google Scholar]
- Podrabsky J. E. & Somero G. N. Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J Exp Biol 2237–54 (2004). [DOI] [PubMed] [Google Scholar]
- Schneider R., Linka R. M. & Reinke H. HSP90 affects the stability of BMAL1 and circadian gene expression. J Biol Rhythms 29, 87–96 (2014). [DOI] [PubMed] [Google Scholar]
- Buhr E. D., Yoo S. H. & Takahashi J. S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330, 379–385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H. C., Kay S. A. & Weber F. HSP90, a capacitor of behavioral variation. J Biol Rhythms 24, 183–192 (2009). [DOI] [PubMed] [Google Scholar]
- Sorek M. & Levy O. The effect of temperature compensation on the circadian rhythmicity of photosynthesis in Symbiodinium, coral-symbiotic alga. Sci Rep 2, 536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7, 709–715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S. et al. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res 21, 1506–1511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D. & Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovin A. et al. Systematic identification of rhythmic genes reveals camk1gb as a new element in the circadian clockwork. PLoS genetics 8, e1003116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.