Abstract
A physiologic signature can be defined as a consistent and robust collection of physiologic measurements characterizing a disease process and its temporal evolution. If a library of physiologic signatures of impending cardiopulmonary instability were available to clinicians caring for inpatients, many episodes of clinical decompensation and their downstream effects could potentially be averted. The development and resolution of cardiopulmonary instability are processes that take time to become clinically apparent, and the treatments provided take time to have an impact. The characterization of dynamic changes in hemodynamic and metabolic variables is implicit in the concept of physiologic signatures. Changes in vital signs such as blood pressure and heart rate, as well as measures of flow such as cardiac output are some of the standard variables used by clinicians to determine cardiopulmonary instability. When these primary variables are collected with high enough frequency to derive new variables, this data hierarchy can be used to development physiologic signatures. The construction of new variables from primary variables, and therefore the creation of physiologic signatures requires no new information; additional knowledge is extracted from data that already exists. It is possible to create physiologic signatures for each stage in the process of clinical decompensation and recovery to improve patient outcomes.
Keywords: data hierarchy, fused parameter, physiologic signature, cardiopulmonary instability, machine learning
Introduction
Cardiopulmonary instability can occur in any disease process when the body’s metabolic needs are not being met with adequate supply. Cardiopulmonary equilibrium is achieved in the presence of adequate oxygenation, preload, contractility, and vasomotor tone. While the body may be able to compensate for a significant change in any one of these components from baseline, any change may still lead to significant morbidity and mortality. Each component can contribute to cardiopulmonary instability. In the setting of trauma, there is a loss of adequate preload due to hemorrhage. Hemorrhage accounts for 50% of deaths within the first 24 hours of hospitalization for a traumatic injury.1 Vasomotor tone is the most prominent derangement in sepsis, though these patients can also experience hypovolemia with reduced preload, and decreased contractility due to myocardial suppression. Inflammatory and apoptotic mediators contribute significantly to the pathophysiology of all three components in sepsis.2 For patients with global tissue hypoxia, as evidenced by elevated lactate levels or hypotension, mortality can range from 36% to 46.5%.3–6 In addition to global circulatory function, organ and microcirculatory function should also be addressed.7 Early identification and management of threats to physiologic equilibrium, preferably before instability is clinically apparent, may prevent untoward patient outcomes.
In recent years, advances in hemodynamic monitoring have ushered the concept of physiologic signatures, specific physiologic profiles describing a disease process through time. Such profiles are constructed using an expanded set of physiologic variables and can be used to identify and manage critical illness in a timely manner. In this chapter we will summarize many of the physiologic variables available in current clinical practice, the successes and challenges of protocolized care that use many of these physiologic variables (goal-oriented therapy), as well as ways to address some of those challenges by building physiologic signatures. The substrate for these signatures is created through the use of a data hierarchy (table 1), or the idea that new variables can be created from existing clinical variables collected at different frequencies. The goal of signature creation would be to identify a patient’s location on the spectrum of critical illness and continuously assess the response to therapy.
Table 1.
Levels of data hierarchy
| Data hierarchy | Examples | Notes |
|---|---|---|
| Primary variables | HR, MAP, CVP, ScvO2, SV, SpO2 |
|
| Secondary (derived) variables | HRV measuresa, PPV, SVV |
|
| Advanced waveform analyses | Morphologic changesb, harmonic analysesb |
|
HR=heart rate; MAP=mean arterial pressure; CVP=central venous pressure; ScvO2=central venous oxygen saturation; SV=stroke volume; SpO2= arterial oxygen saturation measured by pulse oximetry; HRV=heart rate variability; PPV=pulse pressure variability; SVV=stroke volume variation; ABP=arterial blood pressure
The term HRV measures represents dozens of independent variables
Represent potentially hundreds of variables
Diagnosis and management of critical illness through contemporary monitoring is good, but can be improved
Cardiopulmonary parameters used in clinical practice
There are many simple variables currently available to assess cardiopulmonary function and the balance between global oxygen supply and demand, but they may be nonspecific or late markers of cardiopulmonary compromise. Blood pressure is a primary determinant of organ blood flow,8 and also provides some insight into cardiac afterload. Combined with other traditional vital signs such as heart rate- a measure of sympathetic response to physiologic stress7- clinicians can get an overall sense of cardiopulmonary health via noninvasive variables. Tachycardia is the most sensitive of all vital signs for detecting hemodynamic anomalies, but it is nonspecific and may not be present until 15% of blood volume is lost.9
In addition to traditional vital signs, many more reliable variables that were once only measureable through invasive devices have become available in less invasive ones. These parameters, many of them highlighted in table 2, can serve to complement simple vital signs. Cardiac output is a measure of global blood flow, and therefore systemic oxygen delivery. Cardiac output changes to match metabolic demands.7 Since cardiac output normally varies with changing end organ requirements, cardiac output measurements must be interpreted in context. One must know the changes in tissue oxygen extraction to get a more complete picture of the balance between metabolic need and demand. The central venous oxygen saturation (ScvO2) is a minimally invasive metric used to quantify this balance. An increase in ScvO2 after volume expansion reflects volume responsiveness.10
Table 2.
Abbreviated list of available primary physiologic parameters
| Physiologic parameter | Interpretation | Device used to obtain measurement | Level of invasiveness |
|---|---|---|---|
| HR | Sympathetic tone | ECG monitor | Noninvasive |
| SBP DBP MAP PP |
Sympathetic tone, cardiac contractility, vasomotor tone, & volume status | Sphygmomanometry, finger plethysmography, or Arterial catheter | Noninvasive or Minimally invasivea |
| SPAP DPAP MPAP PAPP |
Pulmonary vascular tone, right heart contractility, and volume status | Pulmonary artery catheter | Highly invasive |
| Pra or CVP Ppaob |
Preload (static measures) | Central venous catheter, or pulmonary artery catheter | Minimally invasive or Highly invasivec |
| SV CO |
Cardiac contractility | Finger plethysmographd, Arterial catheterd, thoracic bioimpedence, pulmonary artery catheter, etc. | Noninvasive, Minimally invasive or Highly invasivec |
| SpO2 | Arterial oxygenation | Pulse oximetry | Noninvasive |
| SvO2 | Total body oxygen extractione | Pulmonary artery catheter | Highly invasive |
| Arterial pH paO2 SaO2 pCO2 Hemoglobin |
Arterial oxygenation and carrying capacity | Arterial blood gas analysisf | Minimally invasive |
| Central venous pH pcvO2 ScvO2 pcvCO2 |
Approximate total body oxygen extraction & delivery, and metabolic clearancee,g | Central venous catheter | Minimally invasive |
| Central venous pH pvO2 SvO2 pvCO2 |
Total body oxygen extraction & delivery, and metabolic clearancee | Pulmonary artery catheter | Highly invasive |
| petCO2 VCO2 |
Cardiac contractilityh | Capnograph | Noninvasive |
| Global DO2 Global VO2 |
Global oxygen delivery Global oxygen consumption |
Pulmonary artery catheter | Highly invasive |
HR=Heart rate; ECG=electrocardiogram; SBP=systolic blood pressure; DBP=diastolic blood pressure; MAP=mean arterial pressure; PP=pulse pressure; SPAP=systolic pulmonary artery pressure; DPAP=diastolic pulmonary artery pressure; MPAP=mean pulmonary artery pressure; PAPP=pulmonary artery pulse pressure; Pra=right atrial pressure; CVP=central venous pressure; Ppao=pulmonary artery occlusion pressure; SV=stroke volume; CO=cardiac output; SpO2=arterial oxygen saturation measured by pulse oximetry; SvO2=mixed venous oxygen saturation; paO2=partial pressure of oxygen; SaO2=arterial oxygen saturation; pCO2=partial pressure of carbon dioxide; pcvO2=partial pressure of central venous oxygen; ScvO2=central venous oxygen saturation; pcvCO2=partial pressure of central venous carbon dioxide; pvO2=partial pressure of mixed venous oxygen; SvO2=mixed venous oxygen saturation; pvCO2=partial pressure of mixed venous carbon dioxide; petCO2=end-tidal partial pressure of carbon dioxide; VCO2=volume of exhaled carbon dioxide
Noninvasive if obtained from sphygmomanometry, and minimally invasive if from arterial catheter
Ppao is obtained from a pulmonary artery catheter only
Noninvasive if measured by thoracic bioimpedence or finger cuff, minimally invasive if obtained from central venous catheter or arterial catheter; Highly invasive if from pulmonary artery catheter or esophageal doppler (latter not mentioned in table)
Any measures of SV or CO obtained from the finger plethysmograph or arterial catheter are from approximate calculations from arterial pressure waveform analysis. These measurements can only be made by attaching an additional monitoring device to the arterial catheter.
Oxygen delivery (and CO2 production/clearance) can be calculated as the difference in arterial and mixed/central venous values (ΔpvCO2)
From arterial vessel puncture or an arterial catheter.
Central venous values only take into account the metabolic activity of the upper body, but the differences between them and their mixed venous counterparts is marginal
petCO2 and VO2 used as a surrogate for CO
Some surrogate markers that are commonly used to provide hemodynamic information may be inadequate. For instance, central venous pressure (CVP) is often used as a surrogate for cardiovascular preload. However, CVP is the back-pressure to, and not a synonym for venous return.8 Independent of the volume of blood returning to the right heart, factors intrinsic to cardiac performance and structure can influence CVP. It is therefore not surprising that CVP is a poor marker for circulating blood volume and volume responsiveness.11–13
Other variables are used in clinical practice to quantify global metabolic demands. One multicenter study showed that lactate clearance of 10% of the initial value was as effective as ScvO2 in the protocolized resuscitation of patients during early septic shock.14 Central venous-to-arterial partial pressure difference of carbon dioxide (ΔpCO2) is another useful measure for determining hemodynamic status. According to the Fick principle, changes in ΔpCO2 are inversely related to changes in CO, if we assume constant total body CO2 production. The addition of ΔpCO2 to ScvO2 may predict outcome in patients with septic shock better than ScvO2 alone.15, 16
Early detection of cardiopulmonary instability in contemporary practice through static variables
Early detection of critical illness through the use of isolated hemodynamic and laboratory-based measures can be a first step toward improving outcome in patients at risk for, or those experiencing cardiopulmonary decompensation.
One observational study demonstrated that patients who developed shock later than 48 hours after hospital admission had a 15.6% higher ICU mortality compared with those who developed shock within 48 hours of admission.17
Zhen et al18 showed that septic patients identified from the emergency department had lower in-patient mortality, less mechanical ventilation in the first 24 hours following onset of shock, and a shorter time to achieve a target ScvO2 than those identified later.
In the very early stages of cardiopulmonary instability, prior to the development of hypotension or respiratory failure, clinicians may be uncertain about the presence of abnormal physiology. Generally, the triggers to initiate therapeutic interventions are quite crude, often using some combination of static vital signs and laboratory tests indicative of global tissue hypoperfusion such as lactate. These methods lack sensitivity for detecting cardiopulmonary instability at its very early stages. However, these methods have proven moderately successful in clinical practice.
Observational studies have shown that trauma patients with elevated base deficit, and strong ion difference- both measures of abnormal tissue perfusion- were most correlated with mortality.19, 20 Interestingly, neither lactate20 nor lactate clearance21 have been shown to be predictive of outcomes in trauma patients.
Measures of exhaled CO2 such as end-tidal CO2 (petCO2) and volume of exhaled CO2 (VCO2) can be used as surrogate measures of CO because of their dependence on pulmonary capillary blood flow. Young et al22 showed that petCO2 and VCO2 were both associated with volume responsiveness in patients with shock if they had no baseline lung disease. Furthermore, Dunham et al23 showed that low petCO2 was correlated with low CO in trauma, and was therefore associated with higher injury severity scores, hypotension, major blood loss, and death.
Early goal-oriented therapies: the current gold-standard for dynamic cardiopulmonary assessment and optimization
Goal-oriented therapeutic strategies are systematic approaches to the identification and management of cardiopulmonary decompensation that ensure the use of dynamic physiologic assessment and reassessment. While it is very important to identify impending or obvious cardiopulmonary compromise early with instantaneously measured variables, it is crucial to follow the effects of therapy with repeated measures of these variables. Early identification of impending critical illness will not itself have an effect on outcome unless it is tied to therapeutic interventions which affect outcome.7 In applying these strategies, one must know whether they are providing the desired effect(s), or if the management strategy should be changed. Goal-oriented therapies encompass a group of proven, widely applied management strategies that use physiologic data to not only identify pathology early, but to track disease and its response to therapy over time.
Goal-oriented therapies are often multi-step strategies that combine the information gained from physiologic variables obtained by noninvasive and minimally invasive means (table 2) to optimize cardiopulmonary performance. It has become important in many areas of medical practice.3, 24–32 They have proven useful for a number of reasons: (1) Most studies assessing the use of goal-oriented therapies highlight the need to correct cardiopulmonary collapse early to minimize end organ injury and ischemia;3, 24–29, 31, 32 (2) Goal-oriented approaches provide clear targets for resuscitation, with the intention of avoiding many of the complications, morbidity, and mortality of excessive resuscitation; and (3) Goal-oriented approaches often prioritize the most important physiologic problem to correct at any given moment, with the aim of focusing clinician resources.
Goal-oriented approaches to therapy, when applied as early as possible in the disease course, positively affect outcome:
Recent meta-analyses demonstrated that goal-oriented approaches implemented preoperatively or perioperatively decreased the likelihood of complications in both cardiac24, 26 and noncardiac surgeries33–35 using a range of physiologic variables.
Other meta-analyses showed that perioperative hemodynamic optimization through use of parameter targets decreased postoperative GI and renal dysfunction.25, 36 The benefits to renal function were seen among the highest-risk surgical patients. Moreover, when patients were stratified by the therapeutic strategy design- fluids and inotropes vs. fluids alone- the benefit of goal-oriented therapy on postoperative renal function was only statistically significant with the fluids and inotropes strategy.
Dalfino et al37 showed in another meta-analysis that early goal-oriented therapy based on flow parameters such as cardiac output decreased the risk of postoperative infections, including pneumonia, urinary tract infections, and surgical site infections.
A few studies have shown that goal-oriented therapies decrease perioperative wound-healing and length of stay.27, 38, 39
Rivers et al demonstrated a 16% ARR in mortality (RRR 34%) when early goal-directed therapy (EGDT) was applied to patients with severe sepsis and septic shock.3 Their approach targeted the problems of hypovolemia, vasomotor tone, oxygen carrying capacity, and cardiac dysfunction that can be present in the septic population.
EGDT and its dynamic use of physiologic variables is a key component of many so-called “sepsis bundles” that have revolutionized care of patients with severe sepsis/septic shock. Sepsis bundles involve the protocolization of every aspect of sepsis management, including not just hemodynamic optimization but also (early) antibiotic administration. One meta-analysis showed that early implementation of sepsis bundles can decrease morbidity and mortality in patients with severe sepsis and septic shock.40 Early hemodynamic optimization is the most important feature in bundles to improve patient outcome.41 Early hemodynamic optimization received some of the highest recommendations in the most recent edition of the Surviving Sepsis Campaign Guidelines.4
After a patient is identified as being at risk for, or is experiencing cardiopulmonary decompensation, therapeutic interventions used to correct the problem can lead to further problems if done in excess.
One study showed an association between positive fluid balance and mortality in postoperative noncardiac surgery patients.42
Among patients enrolled in the Vasopressin in Septic Shock Trial (VASST), those with the highest fluid balance after volume resuscitation had the highest adjusted mortality, particularly among those who had impairment in abdominal visceral perfusion secondary to profound volume overload.43
A few studies have shown that a restrictive goal-oriented fluid management strategy decreased post-operative complications when compared to a more liberal approach.44, 45
Hayes et al46 demonstrated that goal-oriented care in critically ill patients using supranormal targets of oxygen delivery caused an absolute risk increase in mortality of 19% in the treatment group.
The aim of any goal-oriented approach to therapy should be to resuscitate only to what is physiologically necessary. The end-points of resuscitation can be defined in a number of ways. One approach is to use prespecified “hard” targets for physiologic variables without regard for individual metabolic demand. One example of this would be a fluid resuscitation strategy that only targets a prespecified CVP range. While this approach may streamline the process and allow for broad, easy implementation of a goal-oriented resuscitation strategy, it may lead to either over- or under-resuscitation depending on a patient’s other underlying disease(s). A better, more common approach in goal-oriented care is to: (1) correct oxygen debt, as determined by the reversal of lactic acidosis and/or base deficit; and (2) match metabolic supply with demand using cardiac output and/or ScvO2, and clinical markers of end organ perfusion such as urine output and mental status assessment. This practice is highlighted in a number of goal-oriented approaches with proven utility in clinical practice.3, 30, 47
In the event that multiple physiologic issues contribute to clinical decline, goal-oriented approaches streamline the management strategy for ease of execution. For instance, early goal-directed therapy in the management of septic patients is designed to first reverse decreased organ perfusion and global tissue hypoxia by addressing hypovolemia. Once volume status is optimized (as determined by appropriate increase in CVP to a prespecified target range), vasomotor tone is increased using vasopressors to a target mean arterial pressure. If there are still signs of oxygen supply-demand mismatch as evidenced by a ScvO2 less than 70%, only then is the patient transfused or given inotropes to increase oxygen delivery to tissues.3
While an ordered approach of therapy allows for easy execution of resuscitation and efficient mobilization of resources, it may not represent an optimal strategy. Many of the goal-oriented approaches we have discussed collect continuous beat-to-beat data. When data is collected at this frequency, clinicians can exploit interactions between physiologic variables that could be used to identify impending cardiopulmonary instability. These variable interactions could not have been utilized if abnormalities in physiologic variables are addressed sequentially. If high-frequency data is used in new and innovative ways, taking advantage of inter-variable (and thus inter-organ) interaction, clinicians may discover new derived variables from these interactions that could be incorporated into physiologic signatures of critical illness.
Towards an earlier diagnosis of cardiopulmonary collapse: Derived variables from high-frequency continuous data
Until now, we have only discussed the use of primary variables, the first level in the physiologic data hierarchy (table 1). If these variables are collected at high frequency, then derived variables can be constructed to aid in earlier, more accurate diagnosis of cardiopulmonary collapse (table 3).
Table 3.
Derived (secondary) variables and their source data
| Derived variables | Type | Source/primary data | Calculation |
|---|---|---|---|
| PPV | Arterial pressure variation | Pulse pressurea | (PPmax − PPmin) / [(PPmax − PPmin) × 0.5] |
| SVV | Arterial pressure variation | SV | (SVmax − SVmin) / SVmean |
| SPV | Arterial pressure variation | SBP | SBPmax − SBPmin SBPexp − SBPminb |
| ΔPOP | Plethysmographic variation | Pulse oximeter waveformc | (POPmax − POPmin) / [(POPmax − POPmin) × 0.5] |
| PVI | Plethysmographic variation | Pulse oximeter waveformc | [(PImax − PImin) / PImax]×100 |
| HRV | Variability analysis | ECG R-R interval | Variable [See table 4 for further details] |
| RRV | Variability analysis | Respiratory rate | Variable |
| BPV | Variability analysis | SBP, DBP, MAP | Variable |
| Temperature variability | Variability analysis | Continuous temperature | Variable |
| Glucose variability | Variability analysis | Continuous blood glucose | Variable |
PPV=pulse pressure variation; PPmax=maximum pulse pressure over a single respiratory cycle; PPmi n=minimum pulse pressure over a single respiratory cycle; SVV=stroke volume variation; SV=stroke volume; SVmax=maximum stroke volume over a given time interval; SVmin=minimum stroke volume over a given time interval; SVmean=mean stroke volume over a given time interval; PPV=systolic pressure variation; SBP=systolic blood pressure; SBPmax=maximum systolic blood pressure over a single respiratory cycle; SBPmin=minimum systolic blood pressure over a single respiratory cycle; SBPexp=systolic blood pressure during an expiratory hold; ΔPOP=Pulse oximeter plethysmographic waveform amplitude; POPmax=maximum pulse oximeter plethysmographic amplitude; POPmin=minimum pulse oximeter plethysmographic amplitude; PVI=pleth variability index; PImax=maximum pleth variability index value over one respiratory cycle; PImin=minimum pleth variability index value over one respiratory cycle; HRV=heart rate variability; ECG=electrocardiogram; RRV=respiratory rate variability; BPV=blood pressure variability; DBP=diastolic blood pressure; MAP=mean arterial pressure
Blood pressure parameters are measured from an arterial catheter only
This difference is also referred to as the Δdown in the literature
Plethysmograph variation is not dependent on the raw pulse oximetry value- i.e. arterial oxygen saturation. It is calculated from the relative changes in the pulse oximeter pleth waveform
Variability analysis
Organ cross-talk constantly takes place between multiple organs along anatomical, neural, and endocrine channels.48 This interaction of organs forms a highly structured and tightly regulated system which on the surface seems chaotic in view of the various physical and time scales involved, but serves to couple organs for more efficient function of the entire organism.49, 50
When the body encounters a disease process that acts as a systemic stressor, communication between organs, and consequently variability in organ system read-out (as measured using a set of primary and secondary physiological signals) decreases considerably or ceases altogether.49, 51 Saturation phenomena in autonomic response is the most commonly proposed explanation for decreased variability in critical illness,52–55 but there could be additional explanations from other aspects of organ interaction manifested at different time scales. Heart rate variability (HRV) is a well-known domain comprising many secondary variables that describe various aspects of the beat-to-beat interval time series. Because beat-to-beat intervals can be acquired from any monitor that provides continuous heart rate measurements, the computation of all of these secondary variables is easily implemented at the bedside if one has the appropriate computer software.56
A decrease in HRV represents increased regularity in the beat-to-beat interval time series, as measured by one or more secondary variables of the HRV domain. The association between decreased HRV and poor outcome in cardiovascular disease have been known for decades,57 and stimulated work in other disease processes such as sepsis58 and trauma.53, 59
There is a growing amount of literature linking reduced heart rate variability and sepsis in adults. Chen et al60 showed in 81 emergency department (ED) patients with early sepsis that HRV can be a useful method to predict impending septic shock.
A study of 15 ED patients demonstrated that HRV decreased in all patients who decompensated.61
A pilot study of 17 bone marrow transplant patients demonstrated that a consistent drop in HRV occurred as sepsis developed in these patients- approximately 30 hours before conventional vital signs.62
Fathizadeh et al53 showed that pathological changes in autonomic function occurred prior to tachycardia among a cohort of trauma patients without severe injury.
Growing evidence from human data59 and animal models63 suggests that HRV is superior to traditional vital signs in detecting hemodynamic decompensation from trauma. Decreased HRV is associated with mortality among trauma patients in the pre-hospital setting64 and in the trauma ICU.59
While the evidence linking decreased HRV and other variability analyses to clinical decompensation is growing, there are still many challenges limiting their clinical use. Most bedside monitors do not hold in memory the continuous vitals data that are displayed at a degree of granularity necessary to compute secondary variables characterizing HRV in real-time. However, variability analyses can be performed in real-time with appropriate software.56 Even if one is able to perform variability analysis, HRV and respiratory rate variability (RRV) can be affected by medications such as sedatives or vasopressors. One study showed that HRV and RRV can still be reliably identified in mechanically ventilated patients on sedation.65 Finally, HRV can be characterized by dozens of derived variables (table 4) with varying degrees of correlation between these variables. There is no clear evidence demonstrating the superiority of any HRV-related variable, and there are no guidelines as to how to obtain an overall assessment of HRV. This extends to variability analyses of all other primary physiologic signals such as blood pressure, oximetry, respiratory rate, or temperature. The potential benefits of variability analysis needs to be clarified and confirmed in more rigorous study among different populations at risk for clinical deterioration.
Table 4.
HRV domain groups and an abbreviated list of derived variable examples
| Domain | Comments | Variable examples |
|---|---|---|
| Statistical | Describe statistical features of time-series data; assumes the state of subsequent data is determined independent of prior data | SDNN, RMSSD, NN50, pNN50, IQRNN |
| Frequency | Deconstructs R-R interval sequences into their spectral components to construct the power distribution of the time series | Total power, ULF, VLF, LF, HF, LF/HF |
| Geometric | Identifies and creates a “shape” from the histogram representation of some specified property in an R-R interval series (see indices column). | NN interval length distribution, Poincare plot, Differential index, TINN, HTI |
| Nonlinear methods | Describes properties that demonstrate fractality, and other characteristics that do not vary in time and space | SampEn, ApEn, Shannon entropy, DFA, Lyapunov exponents, Dispersion analysis |
HRV=heart rate variability; NN= The interval between two normal R-waves (i.e., from non-ectopic beats); RMSSD=Squared root of the mean squared differences of successive; NN50=number of interval differences of successive NN intervals >50ms; pNN50=proportion derived by diving NN50 by the total NN intervals; IQRNN=Interquartile range of NN; SDSD=Standard deviation of the first derivative of the time series; ULH= “ultralow” frequency (</=0.003 Hz); VLF= very low range (0.003–0.04 Hz); LF= low frequency (0.04–0.15 Hz); HF=high frequency (0.15–0.4 Hz); TINN= Triangular interpolation of NN interval histogram; HTI= HRV triangular index; SampEn= sample entropy; ApEn= approximate entropy; DFA=Detrended fluctuation analysis; FDDA= fractal dimension by dispersion analysis; FDCL= fractal dimension of the signal
Arterial pressure variation and plethysmograph variability
When physiologic variables are collected on a beat-to-beat, or continuous basis (> 100Hz), a larger number of additional variables can be computed from these primary signals. Together with the time series of primary physiologic variables, these secondary, or derived, variables could potentially identify impending cardiopulmonary collapse earlier than the variables from which they are derived. It can be speculated that such predictive secondary variables reflect deep physiologic interactions that are perturbed early in the process of cardiovascular instability.
Variables calculated from arterial pressure variation are dynamic and therefore estimate preload dependence, the key factor in predicting volume responsiveness. Preload dependence is superior to static measures of preload such as CVP because preload is not the only determinant of preload dependence.66 A fluid bolus will lead to an increase in cardiac output only if: (1) the patient has good baseline cardiac function; and (2) the patient’s cardiovascular status places him/her on the “steep”, or preload-dependent portion of the Frank-Starling curve. Volume responsiveness is best estimated with dynamic assessment of physiologic variables collected at high frequency.
Measures of arterial pressure variation- namely pulse pressure variation (PPV), systolic pressure variation (SPV), and stroke volume variation (SVV)- utilize the normal changes that occur in the arterial waveform during mechanical ventilation to assess hypovolemia. Early in a positive pressure breath, the increase in intrathoracic pressure will cause compression of pulmonary veins. This leads to an increase in left atrial pressure, and left ventricular (LV) preload. The concomitant decrease in left sided afterload causes an increase in left ventricular stroke volume (SV) and blood pressure. Meanwhile, the increased intrathoracic pressure decreases venous return, and thus right sided preload and SV. Towards the end of inspiration or beginning of expiration, the lower SV from the right ventricle (RV) will reach the left heart, causing a drop in LVSV and blood pressure.67 This normal heart-lung interaction will be exaggerated in someone who is hypovolemic.
In one meta-analysis, arterial pressure variation was shown to be sensitive and specific in predicting volume responsiveness, outperforming static measures of preload such as CVP.68 PPV seemed to perform slightly better than SVV (AUC 0.94 vs. 0.84, respectively).
Michard et al69 showed that PPV of 13% or more identified volume responders with a sensitivity of 94% and specificity of 98%. While their study was conducted on patients with rather large tidal volumes, these findings were validated in an ARDS population getting lower tidal volumes and high positive end-expiratory pressure (PEEP).70
Zhang et al71 showed in one meta-analysis that SVV had an diagnostic odds ratio of 18.4 to predict volume responsiveness in the OR and ICU, with good sensitivity and specificity. The literature supports measuring SVV only in patients on a control mode ventilation with tidal volumes of 8cc/kg or more.71 However, many studies in this paper measured SVV in patients receiving smaller tidal volumes and published AUCs of 0.8 or greater for predicting volume responsiveness. The authors did not report the positive end-expiratory pressure (PEEP) for the patients in these studies.
A number of studies have validated the use of arterial pressure variation as a marker of volume responsiveness in goal-oriented therapeutic protocols which decreased many complications, including organ failure.33–35, 72
Multiple studies have looked at SPV as a marker of volume of responsiveness, with conflicting results.69, 73–76 However, Tevernier et al77 showed that when systolic blood pressure (SBP) is measured during a clinician-initiated expiratory pause on the ventilator, the difference between that SBP measurement and the minimum SBP in the next respiratory cycle- termed the Δdown- can predict volume responsiveness.
Prior to the development of new noninvasive devices that can produce arterial tracings78–80 investigators have studied whether plethysmograph variation can be a noninvasive surrogate for the arterial pressure variation taken from an arterial catheter.
Canneson et al81 showed that changes in pulse oximetry plethysmograph tracings correlated highly with PPV.
Forget et al82 demonstrated lower lactate levels among patients in whom plethysmograph variation was used as a marker of volume responsiveness, compared with patients who had no dynamic measure of preload dependence.
Finally, one meta-analysis involving 326 critically-ill and perioperative patients showed that the pleth variability index (PVI)- one measure of plethysmograph variation- was able to identify preload responsive patients (diagnostic odds ratio 16.0; AUROC 0.87).83 This was particularly true among adults and those on mechanical ventilation.
The usefulness of pulse pressure variation as a clinically relevant and actionable secondary variable is appealing from elementary physiological considerations. Many more variables derived from primary signals may also be useful predictors of instability that relate to deeper, more complex, but no less important physiological disruptions.
Advanced waveform analysis: untapped secondary variables
PPV, SVV, and SPV are all detected via very simple analyses of arterial waveforms, but there is a wealth of information that potentially could be gathered from more sophisticated analyses of digitized waveform data. Physiologic waveforms contain information collected at a much higher frequency than is available from intervals- such as the R-R interval in HRV. There is potential to uncover more about impending cardiopulmonary decompensation than what could be gathered from beat-to-beat-dependent variables. It is thus at the highest level in the data hierarchy constructed to create physiologic signatures of critical illness (table 1). Moreover, it is possible to extract important information not only from arterial waveforms (as is already available clinically through arterial pressure variation) but also from CVP and other waveforms that are currently underutilized. The information collected from these secondary signals could play a key role in the construction of physiologic signatures.
One straightforward, yet potentially productive approach is to track changes in the morphology of waveforms. All physiologic waveforms have a characteristic shape for one “normal” cycle. Certain disease processes cause changes to the shape of these waveforms which, while not pathognomonic, can identify the presence of pathology.67, 84 For instance, cardiac tamponade is one well-known cause of the pulsus paradoxus pattern in the arterial waveform which morphologically is very distinct from the normal shape and structure of the arterial wave. Similarly, in the presence of an atrial arrhythmia or tricuspid regurgitation, the CVP waveform also undergoes changes from its normal morphology.67 In contemporary practice, the CVP waveform is used simply to extract the CVP value at end expiration.85 With the appropriate technology85 other secondary variables can be extracted from CVP which could be used to predict cardiopulmonary decompensation by expressing the underlying physiology.
Roy et al86 showed that it is possible to measure the difference in size of the wave components in one cycle of CVP through time. While they found no difference between fluid responders and nonresponders, they did not look at how these wave components change in size with time, potentially providing additional key information.
A more complicated, but more comprehensive approach for morphologic analysis and secondary predictor extraction from waveforms would be to use harmonic analysis. This approach breaks a waveform down into a superposition of sinusoidal waves of decreasing wavelengths, which in turn undergo signal processing to extract useful information. Fourier transform (FT) is the most commonly used signal processing approach performed on physiologic waveforms.87 FT yields a power spectrum, a summary of the contribution of sine waves of varying frequencies to the original waveform. The goal in advanced waveform analysis is to detect changes in the underlying physiology over time. FT requires additional modifications to be applied on dynamically changing waveforms, and therefore it would be more useful to use a signal processing approach that inherently takes this into account.
Wavelet analysis incorporates the temporal evolution of a signal, potentially allowing one to better identify physiologic decompensation. DeMelis et al88 have shown that it is possible to implement these transforms on arterial pressure waveform data. Wavelet transform offers the added advantage of analyzing time windows of different lengths.88
The challenge of data granularity
Any tool used to create physiologic signatures of critical illness should retrieve the most meaningful variables at a specific point in disease progression, and present them to clinicians to inform decision-making. For instance, if PPV and SVV are elevated (indicating volume responsiveness) in a patient who is at risk for pneumonia, this may indicate the early stages of sepsis (simple hypovolemia). However, if a clinician encounters the same patient with a high PPV and SVV, abnormal variables extracted from the CVP waveform, low CO, and some of the HRV variables are decreased indicating some aberrancy in autonomic tone, then this patient may be much further along the disease process. The physiologic signature of the former state may even be able to predict when the latter state would occur.
There are a number of potential variables at different data hierarchy levels that could be incorporated into a physiologic signature at any given point in the evolution of a disease process (table 1). Tables 2 and 3 review some variables that are already used in contemporary practice. If we add to that the dozens of available variability variables (table 4), and the hundreds of variables that could be extracted from each waveform, it would be impossible for a busy bedside clinician to manually synthesize all of this potentially useful data to find those that are abnormal, and then make decisions about where patients are in the critical illness spectrum.89
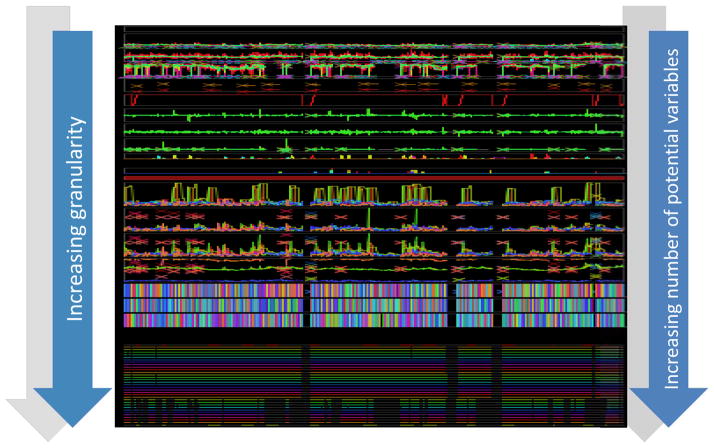
The challenge of data granularity- the density of information that can be used in a prediction model- is illustrated in figure 1. The top of the figure represents raw data (as outlined in table 2) collected at different frequencies- some intermittent, and some waveform data. As more secondary variables are extrapolated from the primary signals (table 2 and variables from advanced waveform analyses), the data granularity increases. When critical mass is reached (represented by the very bottom of figure 1), there are hundreds, if not thousands of secondary variables. How does one process all of this data to retrieve meaningful information to construct physiologic signatures of critical illness? We will summarize a number of existing approaches used to create physiologic signatures for early diagnosis and therapeutic management of critical illness.
Figure 1.
Data granularity
An illustration of how data granularity can dramatically increase with secondary variable derivation. The top of this diagram shows approximately 1.5 patient-years of data. As derived variables are extracted from primary parameters- intermittent, continuous, and waveform data- there is a remarkable increase in data granularity. The bottom of the figure represents >7000 variables- more than the amount of pixels needed to display all of them on a screen.
Creating physiologic signatures of critical illness using machine learning
Machine learning refers to a rich discipline in computer science dedicated to the design and implementation of automated computer-based methods and algorithms to identify patterns in typically large datasets. Standard statistical analysis such as logistic regression can be construed as a subset of those methods. The application of a machine learning approach can efficiently deal with densely granular data. While there are many different approaches that could be employed (table 5), there is a common process that can be applied to the identification of physiologic signatures in critical illness as depicted in figure 2:
Table 5.
Some commonly used machine learning techniques
| Technique | Overview | Comments |
|---|---|---|
| Regression analysis | Determines the probable expectation of a dependent variable based on training data from an independent variable(s) in a subject sample. Dependent variables can be dichotomous or continuous, depending on the type of regression. | Every type of regression has assumptions- for instance, about linear/nonlinear relationships between dependent and independent variables. Care must be taken to know these assumptions before application |
| Decision tree learning | Uses decision trees to classify data. Algorithm determines the most informative attribute given a set of observations, and splits the dataset according to this attribute (“divide-and-conquer” algorithm). Process repeated recursively. | Overfitting is common; prevented by “pruning” algorithms |
| Support Vector Machine (SVM) | Based on linear optimization; subjects are classified in a way that maximizes the “distance” between the observations and a separation hyperplane (hyperplane margin). | |
| k-Nearest Neighbor (kNN) | Given an unlabeled (test) observation, kNN looks for the k most similar observations in the training cohort. k and the definition of “similarity” are defined by the user. The most represented class of labeled observations from the training cohort is the output | Relatively simple to construct |
| k-means clustering | Iterative process used to partition data into k clusters. Clusters initiated by picking k centroids, or cluster focal points. Iteration involves assigning new data points to the “closest” centroid (closeness is user-defined), then reweighting each cluster mean to the geometric center of the new cluster. | Simple to understand and execute. Sensitive to initiation and therefore may change with every execution. Algorithm may fail if clusters aren’t distinct when the process is complete. Optimal k often tested by trial and error |
| Artificial neural networks (ANN) | Models simulate brain organization. “Neurons” (nodes) receive weighted inputs, and output a transfer function. Groups of these building blocks form a network. Training data adjust input weights and build/destroy connections. | Exhibit complex/nonlinear behavior based on the connection network. Can be used for supervised (involving experts) or unsupervised (automated) learning |
| Ensemble learning algorithms | Learns sets of classifiers and merges their outputs. Classifiers are trained independently on specific sets of training observations. In boosting, each subsequent training set emphasizes importance of training samples that have been problematic for the models that are already part of the ensemble. Some ensembles (e.g. Random Forest) utilize a bagging (bootstrap aggregation) approach. Separate decision trees are learned from independent samples of the training data. Multiple random samples- of subjects or attributes- yield the ensemble of models. | Robust; can handle small number of samples |
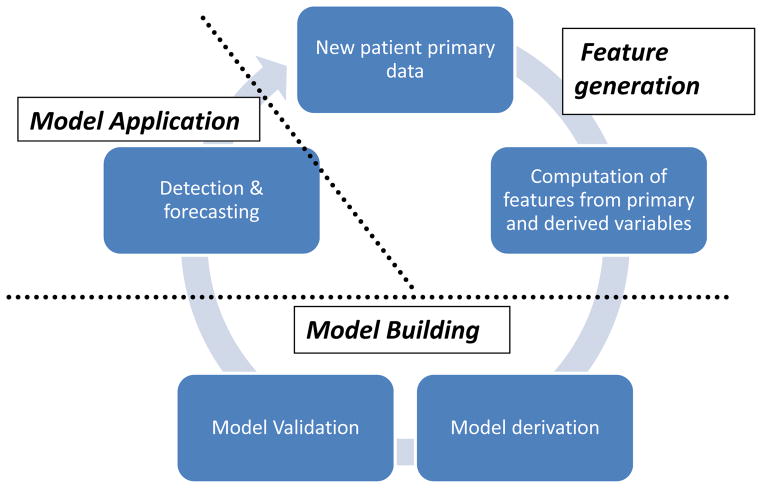
Figure 2. Generation of a machine learning-based model.
An overview of the application of the machine learning approach as it relates to physiologic signature generation. New patient data is featurized to create input from physiologic variables. The model is derived and internally validated on a cohort of training data. It is then applied to a test case for the detection and prediction of clinical instability. New data may be added to the training set to refine model performance.
Any machine learning approach has to generate models that are generalizeable, therefore learning must proceed on a patient cohort of a sufficient size as to include several instances of signatures representative of disease evolution. The learned signatures are disease process-specific.
All primary, secondary variables, and extracted variables from waveforms are obtained or computed from the primary patient data.
All variables in the data hierarchy (table 1) will be used as input to machine learning algorithms. Different machine learning approaches have different tools for classifying and predicting. The algorithms generate models that offer a prediction given primary data. The nature of the prediction depends on the learning task. A typical learning task relevant to the identification of clinically applicable physiologic signatures is to estimate a normal trajectory of predictor variables and classify a new case as abnormal if one or more predictors deviate from that trajectory.90
Since it is not necessary, and would likely be inefficient for the model to use all data for accurate classification, the algorithm will choose the most informative features at a particular point in time in the disease process.
A set of algorithms are applied to maximize external validity of the predictions.
In a typical application, the model is applied to data from a test case, continuously updated as data accrues, and generates an instantaneous predictive forecast of what is expected to happen over a specified time horizon.
More patients and their data could then be added to the derivation/validation cohorts for model refinement.
Static forms of machine learning algorithms have already been applied in many fields, including weather forecasting91 and infectious disease biosurveillance.92 It has also been applied in medicine.93–96 Machine learning is applicable to a broad scope of acute care settings, including ED triage,97 and the ICU.98, 99 It is also applicable to specific diseases like sepsis100–103 and traumatic hemorrhage.95, 100, 104–106 Applications of machine learning to learn physiological signatures represents a step forward from static classifications and predictions, in that it is dynamic in nature; predictions are updated, and models can potentially continue to learn adaptively as data is accrued.
The machine learning approach has a few advantages: first, machine learning can utilize all available physiologic data- primary data collected intermittently or continuously, and variables derived from beat-to-beat data (PPV, SVV, HRV, etc.) and advanced waveform features. Second, once a subject is classified, a model can demonstrate other aspects of the disease process, such as the amount of fluid/blood that was lost. Glass et al107 was able to identify not only bleeding, but the amount of blood lost under experimental conditions of controlled hemorrhage in an animal model using machine learning. The same findings were confirmed in simulated “hemorrhage” in patients using lower body negative pressure to reduce central blood volume.108 This would be critical to an approach used to identify the physiologic signatures at all points in the evolution of impending critical illness. Third, many algorithms deal effectively with correlated or missing data. Finally, model building through machine learning is an iterative process. New patient data is incorporated into older versions of the model to refine prediction. Model-building from machine learning represents best clinical practice; clinicians identify and treat disease more effectively when they encounter more patients with the disease.
Machine learning algorithms are utilized by a number of existing models in critical illness prediction. Many models build fused parameters that function as an effective simplification of multiparameter physiologic signatures. Fused parameters incorporate multiple data inputs and integrate them to form a single value used in decision-making. We will highlight three such parameters from the literature.
The Visensia Stability Index
The Visensia Stability Index (VSI) is a fused parameter built by the Visensia software (OBS Medical, Oxford, UK) designed as an integrated monitoring tool. Visensia automatically processes and integrates simple noninvasively measured vital sign data in real-time. It converts multiparameter monitoring into a single parameter for interpretation by health care professionals (figure 3).
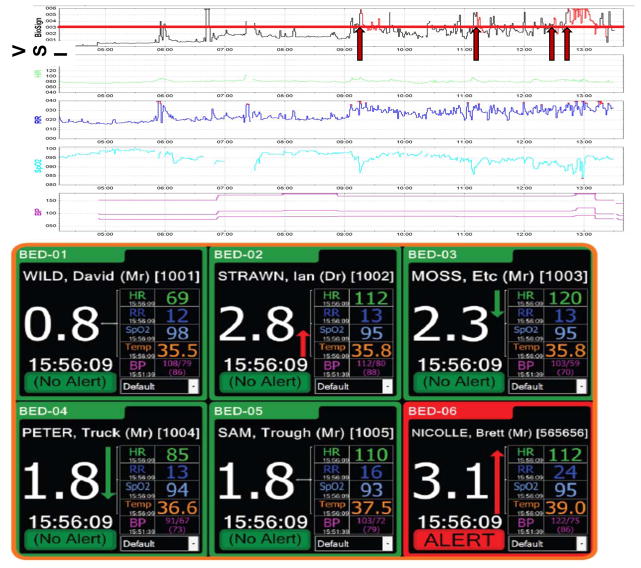
Figure 3. The Visensia Stability Index.
Top panel: The time series of a modified VSI (without temperature) utilized in one academic center is shown for one step-down unit patient, alongside the vital sign components of the VSI. A VSI of 3.2 (red line) was the cutoff value selected based on a training dataset of a similar patient population. If the VSI was consistently above this value, the alert would be triggered (red arrows) and would stay activated as long as the VSI stayed above 3.2 (red portions of VSI tracing). A medical emergency team (MET) was called to see this patient at 13:29; the VSI alert was triggered 4 times beforehand based often on subtle changes in one or more vital signs. (The first alert was over 4 hours prior to the MET activation.) Bottom panel: A monitor showing the VSI and its component vitals from multiple patients simultaneously. The monitor shows not only the current VSI value for a given patient, but the trend based on prior values. (Note that the cutoff value in this patient population was 3.0.) (Reprinted with permission from OBS Medical, Ltd.)
Development of the VSI is data-driven and specific to the patient population. Unlike many of the existing goal-directed strategies for early identification of critical illness discussed in previous sections, Visensia does not depend on an artificial cut-off of vital signs for abnormality. Instead, it employs k-means clustering (table 5) which is trained on a robust cohort of similar patients before the VSI is used for decision-making purposes. It identifies normality and then quantifies departures from this, and an alert is triggered if one vital sign is ±3 standard deviations (SD) from normal, or if 2 or more vital signs are outside of normal range by a smaller amount.109 The appropriate cutoff value for the VSI is determined from the training dataset based on these training-specific norms. An alert is triggered when the VSI value goes over this cutoff for 80% of the time in a time window of a set length. The algorithm’s major strength is that it triggers an alert before patients show signs of obvious abnormality such as hypotension.
To date, there are two major publications that discuss the VSI, and its role in instability identification:
Watkinson et al110 randomized 402 high-risk medical and surgical ward patients to usual monitoring versus VSI monitoring. Their primary outcome was the proportion of patients experiencing major adverse events, including the activation of a rapid response service, transfer to an ICU, and mortality. They found no difference in these outcomes between the two groups. Note that the study design did not institute a protocol to address an emergency if one arose, so no intervention was tied to the monitoring. Moreover, many patients in the control arm had multiparameter monitoring- e.g. an ECG monitor and pulse oximeter. There was likely “contamination” of the control group if clinicians recognized instability that the VSI also would have recognized.
Hravnak et al111 studied the difference in the incidence of cardiopulmonary instability before and after implementation of the Visensia system in a step-down-unit population. They found that the number of alerts decreased by 58%, and the amount of time patients spent with unstable vital signs decreased by 60%. The number of instability episodes- defined by the local medical emergency team (MET) protocol and not the VSI- decreased by 70%. Lastly, in those patients with both VSI alerts and unstable vitals (again defined by the local MET) the VSI alert preceded the unstable vital sign by an average of 9 minutes.
Survival probability
Bayard et al devised a search and display, or stochastic program that outputs a survival probability. The survival probability (SP) is another fused parameter derived from a probabilistic model that integrates a number of physiologic predictors representative of global (CO, SpO2) and regional perfusion (transcutaneous oxygen and CO2 tensions).
Similar to the VSI, the SP is derived from a training set of patients with similar clinical and physiologic states, defined by their primary diagnoses, comorbidities, and hemodynamics, among other factors. The machine learning approach of choice was k nearest neighbors (table 5). The algorithm looks at 40 or more similar nearest neighbor states, and predicts survival of a test case based upon these training set examples.
When the SP system was applied to a cohort of 396 severely ill trauma patients, the SP was 25% lower among nonsurvivors compared with survivors.112 It accurately classified survivors and nonsurvivors 91.4% of the time.
Unlike the VSI, there is a decision-support component built into the system that creates the SP. One can quantify the relative efficiency of a therapy used in the nearest neighbors case to inform decision-making.113
Compensatory reserve index
The compensatory reserve index was devised to identify acute volume loss. It is a fused parameter calculated from waveform analysis, SV, SpO2, petCO2, along with vital sign data. The CRI is calculated by comparing the patient’s arterial waveform features to that of a similar patient in the training set. The model estimates the CRI for a given patient based on the CRI value of those in the training set with similar input features.
Convertino et al114 assessed if the CRI was able to identify persons with low stressor tolerance (fainters) and high stressor tolerance (nonfainters) among 101 participants exposed to lower body negative pressure (LBNP). (LBNP was used to simulate a decrease in central blood volume, and thus intravascular blood volume.) CRI was able to identify low-tolerance patients with hemodynamic decompensation when SV was not decreased. From these results, the authors inferred that the CRI is an estimate of cardiovascular reserve.
Creating physiologic signatures of critical illness from heuristic models: the Rothman Index
Fused parameters can be calculated from rule-based approaches applied on a broad scale that serve similar goals as machine learning algorithms- i.e., these rule-based algorithms can select the most useful data from the wealth of information that is clinically available to come up with the best “guess” of what may be happening to a patient at any given moment. The Rothman index (RI) is a heuristic model that uses not only physiologic data (vital signs), but also standardized nursing assessments of organ systems, laboratory data, and cardiac rhythm information from hemodynamic monitors to construct a fused parameter. Unlike fused parameters that use machine learning the goal is not to forecast what could happen, the goal of the RI is simply to describe a patient’s current condition.115
According to Rothman et al, the RI takes 43 continuously streaming clinical variables from a range of sources in the electronic medical records of patients and applies risk functions, or mathematical equations to their behavior with respect to some outcome. The creators of the RI defined “excess risk” as a percent increase in 1-year all-cause mortality associated with a given value of a variable when compared with the minimum possible mortality of that variable. These mortality risks were determined from a derivation cohort. The goal was not to predict mortality but to use an easily determined outcome that closely correlated with discharge condition. The model was constructed by summing the excess risk input from the 26 variables that independently affected 1-year all cause mortality; the excess risk values for each variable were subtracted from 100 to calculate the RI. The granularity of certain data sources such as laboratory values may be low. The RI controlled for that by applying smoothing functions that would scale the relative importance of data based on its age; newer values for certain variables weighted more with respect to their excess risk input compared with older values of other variables. Scores were calculated on a derivation cohort and validated in five separate cohorts.
The RI can identify clinical instability based on a number of studies:
Among a cohort of medical and surgical patients, Bradley et al116 showed that patients with highest-risk RIs (RI<70) and moderate-risk RIs (RI 70–79) had 2.65 higher odds, and 2.40 higher odds of readmission, respectively, compared with those with the lowest-risk RIs.
Another study showed that the RI, when compared with the Modified Early Warning Score (MEWS; an established early warning system designed to predict impending cardiopulmonary arrest), correlated better with 24-hour mortality (ROC 0.82 and 0.93, respectively).117
Tepas et al118 showed that initial RI values correlated with postoperative complication rates in a cohort of surgical patients. Moreover, as complications ensued in any given patient, the RI decreased, suggesting progressive physiologic dysfunction.
The RI’s strength is that it gives a longitudinal view of patient condition with the goal of early detection of pathologic trends.
Creating physiologic signatures of critical illness from heart rate characteristics: the HeRO score
Some fused parameters may utilize a portion of the available variables that could be used to create physiologic signatures. The HeRO score is a fused parameter used to detect neonatal sepsis, a rather prevalent disease process in the neonatal ICU with a significant mortality reaching 20% in preterm infants.119 The score is not derived from a large number of components, but it is a powerful physiologic signature for identifying impending neonatal critical illness.
The HeRO score incorporates two common HRV variables- standard deviation of R-R intervals from two nonectopic heartbeats (SDNN) and sample entropy (SampEn). (SampEn is a robust measure of the “irregularity”, or randomness in a time series.120, 121) The HeRO score also incorporates information about heart rate accelerations and decelerations. Sample asymmetry refers to the relative frequency of heart rate accelerations and decelerations. Sudden transient decelerations in heart rate are pathologic phenomena of unexplained etiology unique to the neonatal population, causing an increase in sample asymmetry.120
The HeRO score is a composite metric derived from SDNN, SampEn, and sample asymmetry. It measures the patient’s probability of developing sepsis in 24 hours, normalized to the average probability of a similar patient population. It is displayed as the fold-increase in probability of developing sepsis, with ≤1 being normal or low risk and 5 representing a 5-fold increased risk of developing sepsis over the next 24 hours.120
In the years since the creators first noted the association between transient heart rate decelerations and the clinical diagnosis of sepsis122, the HeRO score has proven itself to be a highly useful early detection tool in neonates:
Griffin et al123 showed that reduced variability and transient decelerations- the latter being specific to the neonatal population- can identify culture-positive and culture-negative sepsis up to 5 days prior to the clinical suspicion of sepsis determined by traditional vital sign measurements.
In the largest clinical trial of very low birth weight (VLBW) infants to date, Moorman et al124 randomized 3003 VLBW neonates to traditional monitoring versus HeRO monitoring. They found that simply providing health care providers with the HeRO monitor caused a statistically significant relative risk reduction (RRR) in mortality of 20.5% (ARR=2.1%) among VLBW infants. Similar findings were seen in extremely low birth weight (ELBW) infants (RRR=25%; ARR=4.4%).
The HeRO score stands out as a physiologic signature of disease. The overwhelming majority of clinical decompensation seen in neonates is due to sepsis,120 and the interventions are usually very specific (blood culture with or without antibiotic administration). So while there are no specific therapeutic interventions tied to early detection, it appears that the score’s success lies in its ability to draw attention to a particular patient for the initiation of those interventions. Also, clinicians using the HeRO monitor are encouraged to incorporate the HeRO score into routine clinical assessment. One study prepared a scorecard to help predict sepsis, incorporating information from the HeRO score and routinely used clinical signs. The HeRO score incrementally improved accurate prediction of sepsis in those with low risk determined from clinical evaluation.125
Because the HeRO score doesn’t utilize the scope of all potentially useful data, there is no need for machine learning approaches in its implementation. It is constructed from three variables that the designers proved would provide the most valuable information in the neonatal population.121 These variables exemplify the complex nonlinear behavior of physiology and organ-organ interaction that may be lost in the process of clinical decompensation, specifically in the development of neonatal sepsis.
Creating physiologic signatures of critical illness from multiorgan monitoring
Seely et al126 describe the Continuous Individualized Multiorgan Variability Analysis (CIMVA™) software as a tool designed to take multiorgan input- mainly cardiac (ECG) and respiratory (end-tidal capnography)- and perform variability analyses. The output from the system is the following: (1) a matrix of numerical results (the variability measures) organized in chronological order of analysis windows; and (2) a summary report. The creators expect that by producing these variability measures at every point in the evolution of disease, users would gain some insight into changing organ-organ dynamics, identifying new emergent properties of this complex system that would be informative.
There are preliminary studies that demonstrate the potential utility of a technology like CIMVA for creating physiologic signatures of critical illness:
In a study of 33 patients, Green et al127 assessed the trajectory of CIMVA multiorgan output during the development of shock, and the resolution of respiratory failure. HRV and RRV variables started to decrease about 12–18 hours before the onset of shock. Patients who were successfully extubated had higher HRV measurements before and after extubation. RRV showed an upward trend beginning 10 hours before extubation indicating the resolution of pathology.
Seely et al126 assessed whether there was a correlation between multi-organ monitoring and failed extubation after a spontaneous breathing trial (SBT). Patients who failed extubation after SBT had a greater loss of certain RRV measures compared to patients who passed extubation. (Some HRV measures showed a similar reduction that was not statistically significant.)
CIMVA could constitute a feasible128 and viable platform from which physiologic signatures can be generated.
Challenges to the generation of physiologic signatures in critical illness
The techniques described for physiologic signature creation in the critically ill are cutting-edge; most have not been executed in widespread clinical practice, and none of them is considered standard of care for cardiopulmonary monitoring in any disease process. Potential challenges must be overcome before this groundbreaking research can be applied on a large scale.
Current hardware and network configurations are not readily amenable to data intensive, third party applications, which may need to draw from physiological data streams and the electronic health record. Improving system interoperability is a necessary step towards the implementation of advanced physiologic monitoring tools on standard monitoring platforms.
Some clinicians and hospitals may be skeptical of using machine learning algorithms to drive identification and therapeutic management of critical illness. They may fear the application of a process that occurs, from their perspective, in a black-box. Some may even feel that they are being replaced by these algorithms. However, as available data grows in number, it will eventually become impossible for clinicians in a busy work environment to select the most important variables to use for medical decision-making at any given time.129 Hospitals and health-care professionals will need to be in-serviced on the benefits of integrating machine learning approaches into everyday practice.
Clinicians are not accustomed to seeing physiologic data “merged” to form new values, making it difficult to interpret the output of numerical fused parameters derived from machine learning techniques, the Rothman index or the HeRO score. Perhaps one way to deal with fused parameters is to provide a visual tool, an innovative way that can explain what component of the cardiopulmonary system is wrong. This could provide clinicians some structure on how to therapeutically correct the abnormalities detected. Some fused parameters already provide some graphical or diagrammatic platform,130 including the HeRO score (figure 4) and VSI. The key is to provide the means by which clinicians can apply/change management in response to what they see. It is possible that the application of certain fused parameters did not show the expected outcomes because this was not done.110
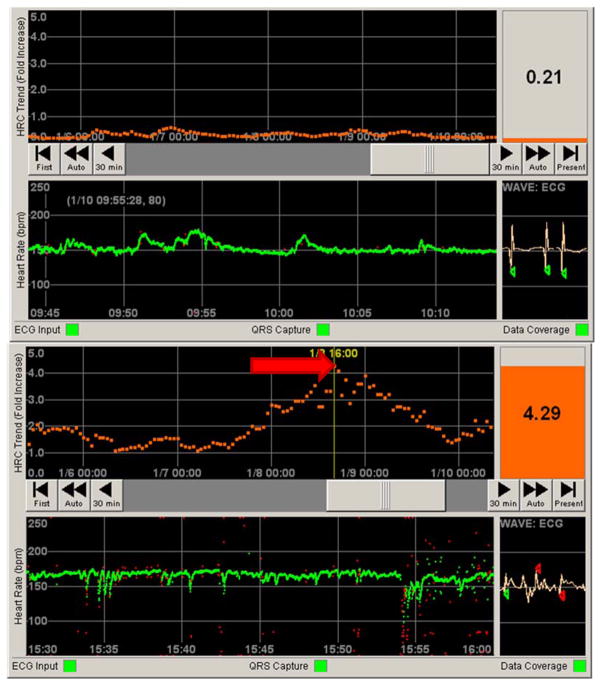
Figure 4. Contrasting normal and abnormal patient data on the HeRO (HRC) monitor.
Top panel: Still photo of patient in a NICU. The bottom half represents 30 minutes of heart rate data (green tracing, in beats per minute). Note the normal variation in heart rate. The top half represents 5 days of continuous HRC output, representing a fold increase in risk of sepsis over the next 24 hours (orange tracing). The HeRO score (HRC index) at the time was less than 1, indicating a low risk of developing sepsis. Bottom panel: By contrast, a still photo of a different patient shows a heart rate tracing with frequent decelerations. The corresponding HRC tracing has a spike (red arrow) corresponding to a HeRO score of 4.29. When this photo was taken, this patient was over four times more likely to develop sepsis in the next 24 hours when compared to similar patients in a control sample.
Summary
Physiologic data are the building blocks for the physiologic signatures of critical illness. It is the dynamic behavior of physiologic variables that make them ideal for this purpose. The numeric value of a physiologic variable and its relation to prior measurements can identify the presence of pathology, and inform clinicians about a patient’s current position in the evolution of a disease process. By looking at physiologic data in new ways, an increasing number of derived variables are being created from primarily-measured data like CO. Derived variables, many already available for bedside use, provide additional information previously unavailable to clinicians about the physiologic behavior of certain diseases. Waveforms contain physiologic data that is grossly underutilized, and may be more useful than the static, intermittent values that are extracted from them. There are many ways to construct physiologic signatures from available data to identify and manage impending critical illness in a timely manner. If all data is to be utilized, eventually the granularity, or density of data will reach a point where novel techniques of data synthesis will be essential. Machine learning approaches can be very helpful for that purpose. Fused parameters can be constructed to synthesize the data into a useable format, though machine learning may not be needed if the data isn’t highly granular. Future study is necessary to assess the use of physiologic signatures in improving patient outcome, particularly when signature use is tied to interventions. It is clear that timely identification and management of critical illness or impending decompensation is important. Clinicians should utilize all available data to their full potential to reach this goal.
Keypoints.
Physiologic monitoring of dynamic changes is more useful than static variables for the early detection of critical illness, and the guidance and appropriate cessation of therapeutic interventions.
Physiologic monitoring techniques that take advantage of complex organ-organ interaction- such as heart rate variability, arterial pressure variation, and secondary variables from hemodynamic waveforms- are valuable, but an underutilized resource for identifying critical illness.
Using new tools to analyze available physiologic variables, it is possible to construct the physiologic signatures at every point in a disease process to identify and treat critical illness as early as possible.
Tools that integrate large amounts of physiologic data are complex to develop; their use requires collaboration with information technology experts.
The integration of physiologic predictors and applications in critical illness is an area of research still under intense investigation.
Acknowledgments
The work was supported by: NIH grants NR013912, HL07820 and HL67181
Footnotes
Conflicts of interest: none declared
References
- 1.Kauvar DS, Lefering R, Wade CE. Impact of hemorrhage on trauma outcome: An overview of epidemiology, clinical presentations, and therapeutic considerations. J Trauma. 2006;60:S3–S11. doi: 10.1097/01.ta.0000199961.02677.19. [DOI] [PubMed] [Google Scholar]
- 2.Otero RM, Nguyen HB, Huang DT, et al. Early goal directed therapy in severe sepsis and septic shock revisited: Concept, controversies, and contemporary findings. Chest. 2006;130:1579–1595. doi: 10.1378/chest.130.5.1579. [DOI] [PubMed] [Google Scholar]
- 3.Rivers EP, Nguyen B, Havstad S, et al. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 4.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro NI, Howell MD, Talmor D, et al. Serum lactate as a predictor of mortality in emergency department patients with infection. Ann Emerg Med. 2005;45:524–528. doi: 10.1016/j.annemergmed.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Trzeciak S, Chansky ME, Dellinger RP, et al. Operationalizing the use of serum lactate measurement for identifying high risk of death in a clinical practice algorithm for suspected severe sepsis. Acad Emerg Med. 2006;(Suppl 1):S150-b-1. [Google Scholar]
- 7.Pinsky MR. Hemodynamic evaluation and monitoring in the ICU. Chest. 2007;132:2020–2029. doi: 10.1378/chest.07-0073. [DOI] [PubMed] [Google Scholar]
- 8.Pinsky MR, Payen D. Functional hemodynamic monitoring. Crit Care. 2005;9:566–572. doi: 10.1186/cc3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.American College of Surgeons Trauma Committee. Advanced trauma life support for doctors. 8. Chicago, IL: American College of Surgeons; 2008. [Google Scholar]
- 10.Giraud R, Siegenthaler N, Gayet-Ageron A, et al. ScvO2 as a marker to define fluid responsiveness. J Trauma. 2011;70:802–807. doi: 10.1097/TA.0b013e3181e7d649. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Anel R, Bunnell E, et al. Pulmonary artery occlusion pressure and central venous pressure fail to predict ventricular filling volume, cardiac performance, or the response to volume infusion in normal subjects. Crit Care Med. 2004;32:691–699. doi: 10.1097/01.ccm.0000114996.68110.c9. [DOI] [PubMed] [Google Scholar]
- 12*.Marik PE, Baram M, Vahid B. Does central venous pressure predict volume responsiveness? A systematic review of the literature and the tale of seven mares. Chest. 2008;134:172–178. doi: 10.1378/chest.07-2331. [DOI] [PubMed] [Google Scholar]
- 13.Oohashi S, Endoh H. Does central venous pressure or pulmonary capillary wedge pressure reflect the status of circulating blood volume in patient after extended transthoracic esophagectomy? J Anesth. 2005;19:21–25. doi: 10.1007/s00540-004-0282-0. [DOI] [PubMed] [Google Scholar]
- 14.Jones AE, Shapiro NI, Trzeciak S, et al. Lactate clearance vs. central venous oxygen saturation as goals of early sepsis therapy. JAMA. 2010;303:739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du W, Liu D, Wang X, et al. Combining central venous-to-arterial partial pressure of carbon dioxide difference and central venous oxygen saturation to guide resuscitation in septic shock. J Crit Care. 2013;28:1110, e1–e5. doi: 10.1016/j.jcrc.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 16.Vallée F, Vallet B, Mathe O, et al. Central venous-to-arterial carbon dioxide difference: an additional target for goal-directed therapy in septic shock? Intensive Care Med. 2008;34:2218–2225. doi: 10.1007/s00134-008-1199-0. [DOI] [PubMed] [Google Scholar]
- 17.Sakr Y, Vincent J, Schuerholz T, et al. Early- versus late-onset shock in European intensive care units. Shock. 2007;28:636–643. [PubMed] [Google Scholar]
- 18.Zhen W, Schorr C, Hunter K, et al. Contrasting treatment and outcome of septic shock: presentation on hospital floors versus emergency department. Clin Med J. 2010;123:3550–3553. [PubMed] [Google Scholar]
- 19.Thom O, Taylor DM, Wolfe RE, et al. Pilot study of the prevalence, outcomes and detection of occult hypoperfusion in trauma patients. Emerj Med J. 2010;27:470–472. doi: 10.1136/emj.2009.073254. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan LJ, Kellum JA. Comparison of acid-base models for prediction of hospital mortality after trauma. Shock. 2008;29:662–666. doi: 10.1097/shk.0b013e3181618946. [DOI] [PubMed] [Google Scholar]
- 21.Jansen TC, van Bommel J, Mulder PG, et al. Prognostic value of blood lactate levels: Does the clinical diagnosis at admission matter? J Trauma. 2009;66:377–385. doi: 10.1097/TA.0b013e3181648e2f. [DOI] [PubMed] [Google Scholar]
- 22.Young A, Marik PE, Sibole S, et al. Changes in end-tidal carbon dioxide and volumetric carbon dioxide as predictors of volume responsiveness in hemodynamically unstable patients. J Cardiothorac Vasc Anesth. 2013;27:681–684. doi: 10.1053/j.jvca.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Dunham CM, Chirchella TJ, Gruber BS, et al. In emergently ventilated trauma patients, low end-tidal CO2 and low cardiac output are associated with hemodynamic instability, hemorrhage, abnormal pupils, and death. BMJ Anesthesiology. 2013;13:20. doi: 10.1186/1471-2253-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Aya HD, Cecconi M, Hamilton M, et al. Goal-directed therapy in cardiac surgery: a systematic review and meta-analysis. Br J Anesthesia. 2013;110:510–517. doi: 10.1093/bja/aet020. [DOI] [PubMed] [Google Scholar]
- 25*.Giglio MT, Marucci M, Testini M, et al. Goal-directed haemodynamic therapy and gastrointestinal complications in major surgery: a meta-analysis of randomized controlled trials. Br J Anaethesia. 2009;103:637–646. doi: 10.1093/bja/aep279. [DOI] [PubMed] [Google Scholar]
- 26*.Giglio M, Dalfino L, Puntillo F, et al. Haemodynamic goal-directed therapy in cardiac and vascular surgery. A systematic review and meta-analysis. Interact Cardiovasc Thorac Surg. 2013;15:878–887. doi: 10.1093/icvts/ivs323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghneim MH, Regner JL, Jupiter DC, et al. Goal directed fluid resuscitation decreases time for lactate clearance and facilitates early fascial closure in damage control surgery. Am J Surg. 2013;206:995–1000. doi: 10.1016/j.amjsurg.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Guiterrez MC, Moore PG, Liu H. Goal-directed therapy in intraoperative fluid and hemodynamic management. J Biomed Res. 2013;27:357–365. doi: 10.7555/JBR.27.20120128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas S, Eichhorn V, Hasbach T, et al. Goal-directed fluid therapy using stroke volume variation does not result in pulmonary fluid overload in thoracic surgery requiring one-lung ventilation. Crit Care Res Prac. 2012;2012:687018. doi: 10.1155/2012/687018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S, Huang C, Lin H, et al. A modified goal-directed protocol improves clinical outcomes in intensive care unit patients with septic shock: a randomized controlled trial. Shock. 2006;26:551–557. doi: 10.1097/01.shk.0000232271.09440.8f. [DOI] [PubMed] [Google Scholar]
- 31.Forget P, Lois F, de Kock M. Goal-directed fluid management based on the pulse oximeter-derived pleth variability index reduces lactate levels and improves fluid management. Anesth Analg. 2010;111:910–914. doi: 10.1213/ANE.0b013e3181eb624f. [DOI] [PubMed] [Google Scholar]
- 32.Hata JS, Scotts C, Shelsky C, et al. Reduces mortality with noninvasive hemodynamic monitoring of shock. J Crit Care. 2011;26:224.e1–e8. doi: 10.1016/j.jcrc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Benes J, Chytra I, Altmann P, et al. Intraoperative fluid optimization using stroke volume variation in high-risk surgical patients: results of prospective randomized study. Crit Care. 2010;14:R118. doi: 10.1186/cc9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzwedel C, Puig J, Carstens A, et al. Perioperative goal-directed hemodynamic therapy based on radial arterial pulse pressure variation and continuous cardiac index trending reduces postoperative complications after major abdominal surgery: a multi-center, prospective, randomized study. Crit Care. 2013;17:R191. doi: 10.1186/cc12885. Available online at http://ccforum.com/content/17/5/R191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes MR, Oliveira MA, Pereira VO, et al. Goal-directed fluid management based on pulse pressure variation monitoring during high-risk surgery: a pilot randomized controlled trial. Crit Care. 2007;11:R100. doi: 10.1186/cc6117. Available online at http://ccforum.com/content/11/5/R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Brienza N, Giglio MT, Marucci M, et al. Does perioperative hemodynamic optimization protect function in surgical patients? A meta-analytic study. Crit Care Med. 2009;37:2079–2090. doi: 10.1097/CCM.0b013e3181a00a43. [DOI] [PubMed] [Google Scholar]
- 37*.Dalfino L, Giglio MT, Puntillo F, et al. Haemodynamic goal-directed therapy and postoperative infections: earlier is better. A systematic review and meta-analysis. Crit Care. 2011;15:R154. doi: 10.1186/cc10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Qiao H, He Z, et al. Intraoperative fluid management in open gastrointestinal surgery: goal-directed versus restrictive. Clinics. 2012;67:1149–1155. doi: 10.6061/clinics/2012(10)06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng H, Guo H, Ye J, et al. Goal-directed fluid therapy in gastrointestinal surgery in older coronary heart disease patients: randomized trial. World J Surg. 2013;37:2820–2829. doi: 10.1007/s00268-013-2203-6. [DOI] [PubMed] [Google Scholar]
- 40.Barochia AV, Chi X, Vitberg D, et al. Bundled care for septic shock: an analysis of clinical trials. Crit Care Med. 2010;38:668–678. doi: 10.1097/CCM.0b013e3181cb0ddf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Chamberlain DJ, Willis EW, Bersten AB. The severe sepsis bundles as processes of care: a meta-analysis. Aust Crit Care. 2011;24:229–243. doi: 10.1016/j.aucc.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Shim HJ, Jang JY, Lee SH, et al. The effects of positive balance on the outcomes of critically ill noncardiac postsurgical patients: a retrospective cohort study. J Crit Care. 2014;29:43–8. doi: 10.1016/j.jcrc.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Boyd JH, Forbes J, Nakada T, et al. Fluid resuscitation in septic shock: a positive fluid balance and elevated central venous pressure are associated with increased mortality. Crit Care Med. 39:259–265. doi: 10.1097/CCM.0b013e3181feeb15. [DOI] [PubMed] [Google Scholar]
- 44.Futier E, Constantin J, Petit A, et al. Conservative vs. restrictive individualized goal-directed fluid replacement strategy in major abdominal surgery. Arch Surg. 2010;145:1193–1200. doi: 10.1001/archsurg.2010.275. [DOI] [PubMed] [Google Scholar]
- 45.Lobo SM, Ronchi LS, Oliveira NE, et al. Restrictive strategy of intraoperative fluid maintenance during optimization of oxygen delivery decreases major complications after high-risk surgery. Crit Care. 2011;15:R226. doi: 10.1186/cc10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayes MA, Timmins AC, Yau EHS, et al. Elevation of systemic oxygen delivery in the treatment of critically ill patients. N Engl J Med. 1994;330:1717–1722. doi: 10.1056/NEJM199406163302404. [DOI] [PubMed] [Google Scholar]
- 47.Pearse R, Dawson D, Fawcett J, et al. Early goal-directed therapy after major surgery reduces complications and duration of hospital stay. A randomised, controlled trial. Crit Care. 2005;9:R687–R693. doi: 10.1186/cc3887. Available online at http://ccforum.com/content/9/6/R687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairchild KD, O’Shea TM. Heart rate characteristics: physiomarkers for detection of late-onset neonatal sepsis. Clin Perinatal. 2010;37:581–598. doi: 10.1016/j.clp.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godin PJ, Buchman TG. Uncoupling of biological oscillators: a complementary hypothesis concerning the pathogenesis of multiple organ dysfunction syndrome. Crit Care Med. 1996;24:1107–1116. doi: 10.1097/00003246-199607000-00008. [DOI] [PubMed] [Google Scholar]
- 50.Pinsky MR. Complexity modeling: identify instability early. Crit Care Med. 2010;38(Suppl):S649–S655. doi: 10.1097/CCM.0b013e3181f24484. [DOI] [PubMed] [Google Scholar]
- 51.Seely AJE, Christou NV. Multiple organ dysfunction syndrome: Exploring the paradigm of complex nonlinear systems. Crit Care Med. 2000;28:2193–2200. doi: 10.1097/00003246-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 52.Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Heart rate variability: standards of measurement, physiological interpretation, and clinical use. Circulation. 1996;93:1043–1065. [PubMed] [Google Scholar]
- 53.Fathizadeh P, Shoemaker WC, Wo CCJ, et al. Autonomic activity in trauma patients based on variability of heart rate and respiratory rate. Crit Care Med. 2004;32:1300–1305. doi: 10.1097/01.ccm.0000127776.78490.e4. [DOI] [PubMed] [Google Scholar]
- 54.Omboni S, Parati G, Di Rienzo M, et al. Blood pressure and heart rate in autonomic disorders: a critical review. Clin Auton Res. 1996;6:171–182. doi: 10.1007/BF02281905. [DOI] [PubMed] [Google Scholar]
- 55.Kamath MV, Fallen EL. Power spectral analysis of heart rate variability: a noninvasive signature of cardiac autonomic function. Crit Rev Biomed End. 1993;21:245–311. [PubMed] [Google Scholar]
- 56.Kasaoka S, Nakahara T, Kawamura Y, et al. Real-time monitoring of heart rate variability in critically ill patients. J Crit Care. 2010;25:313–316. doi: 10.1016/j.jcrc.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 57.Kleiger RE, Miller JP, Bigger JT, Jr, et al. Decreased heart rate variability and its association with increased mortality after acute myocardial infarction. Am J Cardiol. 1987;59:256–262. doi: 10.1016/0002-9149(87)90795-8. [DOI] [PubMed] [Google Scholar]
- 58.Buchan CA, Bravi A, Seely AJ. Variability analysis and the diagnosis, management, and treatment of sepsis. Curr Infect Dis Rep. 2012;14:512–521. doi: 10.1007/s11908-012-0282-4. [DOI] [PubMed] [Google Scholar]
- 59.Norris PR, Morris JA, Ozdas A, et al. Heart rate variability predicts trauma patient outcomes as early as 12 hours: Implications for military and civilian triage. J Surg Res. 2005;129:122–128. doi: 10.1016/j.jss.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 60.Chen W, Kuo C. Characteristics of heart rate variability can predict impending shock in emergency department patients with sepsis. Acad Emerg Med. 2007;14:392–397. doi: 10.1197/j.aem.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 61.Barnaby D, Ferrick K, Kaplan DT, et al. Heart rate variability in emergency department patients with sepsis. Acad Emerg Med. 2002;9:661–670. doi: 10.1111/j.1553-2712.2002.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 62.Ahmad S, Ramsay T, Huebsch L, et al. Continuous multi-parameter heart rate variability analysis heralds onset of sepsis in adults. PLoS One. 2009;4:e6642. doi: 10.1371/journal.pone.0006642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batchinsky AI, Cooke WH, Kuusela T, et al. Loss of complexity characteristics the heart rate response to experimental hemorrhagic shock in swine. Crit Care Med. 2007;35:519–525. doi: 10.1097/01.CCM.0000254065.44990.77. [DOI] [PubMed] [Google Scholar]
- 64.Cooke WH, Salinas J, Convertino VA, et al. Heart rate variability and its association with mortality in prehospital trauma patients. J Trauma. 2006;60:363–370. doi: 10.1097/01.ta.0000196623.48952.0e. [DOI] [PubMed] [Google Scholar]
- 65.Bradley B, Green GC, Batkin I, et al. Feasibility of continuous multiorgan variability in the intensive care unit. J Crit Care. 2012;27:218.e9–218.e20. doi: 10.1016/j.jcrc.2011.09.009. Epub 2011. [DOI] [PubMed] [Google Scholar]
- 66.Cannesson M. Arterial pressure variation and goal-directed fluid therapy. J Cardiothorac Vasc Anesth. 2010;24:487–497. doi: 10.1053/j.jvca.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Barbeito A, Mark JB. Arterial and central venous pressure monitoring. Anesthesiology Clin. 2006;24:717–735. doi: 10.1016/j.atc.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 68*.Marik PE, Baram M, Vahid B. Does central venous pressure predict volume responsiveness? A systematic review of the literature and the tale of seven mares. Chest. 2008;134:172–178. doi: 10.1378/chest.07-2331. [DOI] [PubMed] [Google Scholar]
- 69.Michard F, Boussat S, Chemla D, et al. Relation between respiratory changes in arterial pulse pressure and fluid responsiveness in septic patients with acute circulatory failure. Am J Respir Crit Care Med. 2000;162:143–138. doi: 10.1164/ajrccm.162.1.9903035. [DOI] [PubMed] [Google Scholar]
- 70.Huang C, Fu J, Hu H, et al. Prediction of fluid responsiveness in acute respiratory distress syndrome patients ventilated with low tidal volume and high positive end-expiratory pressure. Crit Care Med. 2008;36:2810–2816. doi: 10.1097/CCM.0b013e318186b74e. [DOI] [PubMed] [Google Scholar]
- 71*.Zhang Z, Lu B, Sheng X, et al. Accuracy of stroke volume variation in predicting fluid responsiveness: a systematic review and meta-analysis. J Anesth. 2011;25:904–916. doi: 10.1007/s00540-011-1217-1. [DOI] [PubMed] [Google Scholar]
- 72.Schereen TW, Wiesenack C, Gerlach H, et al. Goal-directed intraoperative fluid therapy guided by stroke volume and its variation in high-risk surgical patients: a prospective randomized multicentre study. J Clin Monit Comput. 2013;27:225–233. doi: 10.1007/s10877-013-9461-6. [DOI] [PubMed] [Google Scholar]
- 73.Belloni L, Pisano A, Natale A, et al. Assessment of fluid-responsiveness parameters for off-pump coronary artery bypass surgery: a comparison among LiDCO, transesophageal echocardiography, and pulmonary artery catheter. J Cardiothorac Vasc Anesth. 2008;22:243–248. doi: 10.1053/j.jvca.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Kramer A, Zygan D, Hawes H, et al. Pulse pressure variation predicts fluid responsiveness following coronary artery bypass surgery. Chest. 2004;126:1563–1568. doi: 10.1378/chest.126.5.1563. [DOI] [PubMed] [Google Scholar]
- 75.Natalini G, Rosano A, Taranto M, et al. Arterial versus plethysmographic dynamic indices to test responsiveness for testing fluid administration in hypotensive patients: a clinical trial. Anesth Anal. 2006;103:1478–1484. doi: 10.1213/01.ane.0000246811.88524.75. [DOI] [PubMed] [Google Scholar]
- 76.Reuter DA, Felbinger TW, Kilger E, et al. Optimizing fluid therapy in mechanically ventilated patients after cardiac surgery by on-line monitoring of left ventricular stroke volume variations. Comparison with aortic systolic pressure variations. BrJ Anaesth. 2002;88:124–126. doi: 10.1093/bja/88.1.124. [DOI] [PubMed] [Google Scholar]
- 77.Tavernier B, Makhotine O, Lebuffe G, et al. Systolic pressure variation as a guide to fluid therapy in patients with sepsis-induced hypotension. Anesthesiology. 1998;89:1313–1321. doi: 10.1097/00000542-199812000-00007. [DOI] [PubMed] [Google Scholar]
- 78.Jeleazcov C, Krajinovic L, Munster T, et al. Precision and accuracy of a new device (CNAP) for continuous non-invasive arterial pressure monitoring: assessment during general anesthesia. Br J Anaesth. 2010;105:264–272. doi: 10.1093/bja/aeq143. [DOI] [PubMed] [Google Scholar]
- 79.Kako H, Corridore M, Rice J, et al. Accuracy of the CNAP monitor, a noninvasive continuous blood pressure device, in providing beat-to-beat blood pressure readings in pediatric patients weighing 20–40 kilograms. Ped Anesth. 2013;23:989–993. doi: 10.1111/pan.12173. [DOI] [PubMed] [Google Scholar]
- 80.Martina JR, Westerhof BE, van Goudoever J, et al. Noninvasive continuous arterial blood pressure monitoring with Nexfin. Anesthesiology. 2012;116:1092–1103. doi: 10.1097/ALN.0b013e31824f94ed. [DOI] [PubMed] [Google Scholar]
- 81.Cannesson M, Besnard C, Durand PG, et al. Relation between respiratory variations in pulse oximetry plethysmographic waveform amplitude and arterial pulse pressure in ventilated patients. Crit Care. 2005;9:R562–R568. doi: 10.1186/cc3799. Available online at http://ccforum.com/content/9/5/R562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forget P, Lois F, de Kock M. Goal-directed fluid management based on the pulse oximeter-derived pleth variability index reduces lactate levels and improves fluid management. Anesth Analg. 2010;111:910–914. doi: 10.1213/ANE.0b013e3181eb624f. [DOI] [PubMed] [Google Scholar]
- 83*.Yin JY, Ho KM. Use of plethysmographic variability index derived from the Massimo pulse oximeter to predict fluid or preload responsiveness: a systematic review and meta-analysis. Anaesthesia. 2012;67:777–783. doi: 10.1111/j.1365-2044.2012.07117.x. [DOI] [PubMed] [Google Scholar]
- 84.O’Rourke MF. Time domain analysis of the arterial pulse in clinical medicine. Med Biol Eng Comput. 2009;47:119–129. doi: 10.1007/s11517-008-0370-7. [DOI] [PubMed] [Google Scholar]
- 85.Fujita Y, Hayashi D, Wada S, et al. Central venous pulse pressure analysis using an R-synchronized pressure measurement system. J Clin Monit Comput. 2006;20:385–389. doi: 10.1007/s10877-006-9035-y. [DOI] [PubMed] [Google Scholar]
- 86.Roy S, Couture P, Qizilbash B, et al. Hemodynamic pressure waveform analysis in predicting volume responsiveness. J Cardiothorac Vasc Anesth. 2013;27:676–680. doi: 10.1053/j.jvca.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 87.Scheuer ML, Wilson SB. Data analysis for continuous EEG monitoring in the ICU. seeing the forest and the trees. J Clin Neurophysiol. 2004;21:353–378. [PubMed] [Google Scholar]
- 88.De Melis M, Morbiducci U, Rietzschel ER, et al. Blood pressure waveform analysis by means of wavelet transform. Med Biol Eng Comput. 2009;47:165–173. doi: 10.1007/s11517-008-0397-9. [DOI] [PubMed] [Google Scholar]
- 89.Cohen MJ. Use of models in identification and prediction of physiology in critically ill surgical patients. Br J Surg. 2012;99:487–493. doi: 10.1002/bjs.7798. [DOI] [PubMed] [Google Scholar]
- 90.Dubrawski A. Detection of events in multiple streams of surveillance data: Multivariate, multi-stream and multi-dimensional approaches. In: Zeng D, Chen H, Castillo-Chavez C, Lober WB, Thurmond M, editors. Infectious disease informatics and biosurveillance. New York: Springer; 2011. pp. 145–171. [Google Scholar]
- 91.Rasouli K, Hsieh WW, Cannon AJ. Daily streamflow forecasting by machine learning methods with weather and climate inputs. J Hydrology. 2012;414–415:284–293. [Google Scholar]
- 92.Buckeridge DL. Outbreak detection through automated surveillance: a review of determinants of detection. J Biomed Inform. 2007;40:370–379. doi: 10.1016/j.jbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 93.Chang Y, Yeh M, Li Y, et al. Predicting hospital-acquired infections by scoring system with simple parameters. PLoS One. 2011;6:e23137. doi: 10.1371/journal.pone.0023137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dumont TM, Rughani AI, Tranmer BI. Prediction of symptomatic cerebral vasospasm after aneurysmal subarachnoid hemorrhage with an artificial neural network: feasibility and comparison with logistic regression models. World Neurosurg. 2011;75:57–63. doi: 10.1016/j.wneu.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 95.Paul M, Nielsen AD, Goldberg E, et al. Prediction of specific pathogens in patients with sepsis: evaluation of TREAT, a computerized decision support system. J Antimicrob Chemother. 2007;59:1204–1207. doi: 10.1093/jac/dkm107. [DOI] [PubMed] [Google Scholar]
- 96.Purwento, Eswaran C, Logeswaran R, et al. Prediction models for early risk detection of cardiovascular event. J Med Syst. 2012;36:521–531. doi: 10.1007/s10916-010-9497-9. [DOI] [PubMed] [Google Scholar]
- 97.Ong ME, Ng CH, Goh K, et al. Prediction of cardiac arrest in critically ill patients presenting to the emergency department using a machine learning score incorporating heart rate variability compared with the modified early warning score. Crit Care. 2012;16:R108. doi: 10.1186/cc11396. Available online at http://ccforum.com/content/16/3/R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clermont G, Angus DC, DiRusso SM, et al. Predicting hospital mortality for patients in the intensive care unit: a comparison of artificial neural networks with logistic regression models. Crit Care Med. 2001;29:291–296. doi: 10.1097/00003246-200102000-00012. [DOI] [PubMed] [Google Scholar]
- 99.Silva Á, Cortez P, Santos MF, et al. Rating organ failure via adverse events using data mining in the intensive care unit. Artif Intell Med. 2008;43:179–193. doi: 10.1016/j.artmed.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 100.Ribas VJ, López JC, Ruiz-Sanmartin A, et al. Severe sepsis mortality prediction with relevance vector machines. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:100–3. doi: 10.1109/IEMBS.2011.6089906. [DOI] [PubMed] [Google Scholar]
- 101.Jaimes F, Farbiarz J, Alvarez D, et al. Comparison between logistic regression and neural networks to predict death in patients with suspected sepsis in the emergency room. Crit Care. 2005;9:R150. doi: 10.1186/cc3054. Available online at http://ccforum.com/content/9/2/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gultepe E, Green JP, Nguyen H, et al. From vital signs to clinical outcomes for patients with sepsis: a machine learning basis for a clinical decision support system. J Am Med Inform Assoc. 2014;21:315–25. doi: 10.1136/amiajnl-2013-001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schurink CAM, Lucas PJF, Hoepelman IM, et al. Computer-assisted decision support for the diagnosis and treatment of infectious diseases in intensive care units. Lancet Infect Dis. 2005;5:302–312. doi: 10.1016/S1473-3099(05)70115-8. [DOI] [PubMed] [Google Scholar]
- 104.Andersson B, Andersson R, Ohlsson M, et al. Prediction of severe acute pancreatitis at admission to hospital using artificial neural networks. Pancreatology. 2011;11:328–335. doi: 10.1159/000327903. [DOI] [PubMed] [Google Scholar]
- 105.Tang CHH, Middleton PM, Savkin AV, et al. Non-invasive classification of severe sepsis and systemic inflammatory response syndrome using a nonlinear support vector machine: a preliminary study. Physiol Meas. 2010;31:775–793. doi: 10.1088/0967-3334/31/6/004. [DOI] [PubMed] [Google Scholar]
- 106.Xiao Y, Griffin P, Lake DE, et al. Nearest-neighbor and logistic regression analyses of clinical and heart rate characteristics in the early diagnosis of neonatal sepsis. Med Decis Making. 2010;30:258–266. doi: 10.1177/0272989X09337791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Glass TF, Knapp J, Ambrun P, et al. Use of artificial intelligence to identify cardiovascular collapse in a model of hemorrhagic shock. Crit Care Med. 2004;32:450–456. doi: 10.1097/01.CCM.0000109444.02324.AD. [DOI] [PubMed] [Google Scholar]
- 108.Convertino VA, Ryan KL, Rickards CA, et al. Physiological and medical monitoring for en route care of combat casualties. J Trauma. 2008;64:S342–S353. doi: 10.1097/TA.0b013e31816c82f4. [DOI] [PubMed] [Google Scholar]
- 109.Tarrasenko L, Hann A, Young D. Integrated monitoring and analysis for early warning of patient deterioration. Br J Anaesth. 2006;97:64–68. doi: 10.1093/bja/ael113. [DOI] [PubMed] [Google Scholar]
- 110.Watkinson PJ, Barber VS, Price JD, et al. A randomized controlled trial of the effect of continuous electronic physiological monitoring on the adverse event rate in high risk medical and surgical patients. Anaesthesia. 2006;61:1031–1039. doi: 10.1111/j.1365-2044.2006.04818.x. [DOI] [PubMed] [Google Scholar]
- 111.Hravnak M, DeVita MA, Clontz A, et al. Cardiorespiratory instability before and after implementing an integrated monitoring system. Crit Care Med. 2011;39:65–72. doi: 10.1097/CCM.0b013e3181fb7b1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shoemaker WC, Bayard DS, Wo CCJ, et al. Outcome prediction in chest injury by a mathematical search and display program. Chest. 2005;128:2739–2748. doi: 10.1378/chest.128.4.2739. [DOI] [PubMed] [Google Scholar]
- 113.Shoemaker WC, Bayard DS, Botnen A, et al. Mathematical program for outcome prediction and therapeutic support for trauma beginning within 1 hr of admission: a preliminary report. Crit Care Med. 2005;33:1499–1506. doi: 10.1097/01.ccm.0000162641.92400.aa. [DOI] [PubMed] [Google Scholar]
- 114.Convertino VA, Grudic G, Mulligan J, et al. Estimation of individual-specific progression to impending cardiovascular instability using arterial waveforms. J Appl Physiol. 2013;115:1196–1202. doi: 10.1152/japplphysiol.00668.2013. [DOI] [PubMed] [Google Scholar]
- 115.Rothman MJ, Rothman SI, Beals J., IV Development and validation of a continuous measure of patient condition using the electronic medical record. J Biomed Inform. 2013;46:837–848. doi: 10.1016/j.jbi.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 116.Bradley EH, Yakusheva O, Horwitz LI, et al. Identifying patients at risk for unplanned readmission. Med Care. 2013;51:761–766. doi: 10.1097/MLR.0b013e3182a0f492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Finlay GD, Rothman MJ, Smith RA. Measuring the modified early warning score and the Rothman index: advantages of utilizing the electronic medical record in an early warning system. J Hosp Med. 2014;9:116–119. doi: 10.1002/jhm.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tepas JJ, III, Rimar JM, Hsiao AL, et al. Automated analysis of electronic medical record data reflects the physiology of operative complications. Surgery. 2013;154:918–926. doi: 10.1016/j.surg.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 119.Fairchild KD. Predictive monitoring for early detection of sepsis in neonatal ICU patients. Curr Opin Pediatr. 2013;25:172–9. doi: 10.1097/MOP.0b013e32835e8fe6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fairchild KD, Aschner JL. HeRO monitoring to reduce mortality in NICU patients. Res Rep Neonatol. 2012;2:65–76. [Google Scholar]
- 121.Moorman JR, Delos JB, Flower AA, et al. Cardiovascular oscillations at the bedside: early diagnosis of neonatal sepsis using heart rate characteristics monitoring. Physiol Meas. 2011;32:1821–1832. doi: 10.1088/0967-3334/32/11/S08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Griffin MP, Moorman JR. Toward the early diagnosis of neonatal sepsis and sepsis-like illness using novel heart rate analysis. Pediatrics. 2001;107:97–104. doi: 10.1542/peds.107.1.97. [DOI] [PubMed] [Google Scholar]
- 123.Griffin MP, O’Shea TM, Bissonette EA, et al. Abnormal heart rate characteristics preceding neonatal sepsis and sepsis-like illness. Pediatr Res. 2003;53:920–926. doi: 10.1203/01.PDR.0000064904.05313.D2. [DOI] [PubMed] [Google Scholar]
- 124.Moorman JR, Carlo WA, Kattwinkel J, et al. Mortality reduction by heart rate variability characteristic monitoring in very low birth weight neonates: A randomized controlled trial. J Pediatr. 2011;159:900–906. doi: 10.1016/j.jpeds.2011.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Griffin MP, Lake DE, O’Shea TM, et al. Heart rate characteristics and clinical signs in late-onset neonatal sepsis. Pediatr Res. 2007;61:222–227. doi: 10.1203/01.pdr.0000252438.65759.af. [DOI] [PubMed] [Google Scholar]
- 126.Seely AJE, Green GC, Bravi A. Continuous multiorgan variability monitoring in critically ill patients- complexity science at the bedside. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:5503–6. doi: 10.1109/IEMBS.2011.6091404. [DOI] [PubMed] [Google Scholar]
- 127.Green GC, Bradley B, Bravi A, et al. Continuous multiorgan variability analysis to track severity of organ failure in critically ill patients. J Crit Care. 2013;28:879.e1–11. doi: 10.1016/j.jcrc.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 128.Bradley B, Green GC, Batkin I, et al. Feasibility of continuous multiorgan variability analysis in the intensive care unit. J Crit Care. 2012;27:218.e9–20. doi: 10.1016/j.jcrc.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 129.Das A, Wong RCK. Prediction of outcome in acute lower gastrointestinal hemorrhage: role of artificial neural network. Eur J Gastroenterol Hepatol. 2007;19:1064–9. doi: 10.1097/MEG.0b013e3282f198f7. [DOI] [PubMed] [Google Scholar]
- 130.Vallée F, Fourcade O, Marty P, et al. The hemodynamic “target”: a visual tool of goal-directed therapy for septic patients. Clinics. 2007;62:447–454. doi: 10.1590/s1807-59322007000400012. [DOI] [PubMed] [Google Scholar]