Abstract
Purpose
To examine the extent to which commonly ordered laboratory values obtained from large health care databases are representative of the distribution of laboratory values from the general population as reflected in the National Health and Nutrition Examination Survey.
Methods
Means of test values from commercial insurance laboratory data and National Health and Nutrition Examination Survey data were compared. Inverse probability of selection weighting was used to account for possible selection bias and to create comparability between the two data sources. Results: The average values of most of the laboratory results from routine care were very close to their population means as estimated from NHANES. Tests that were more selectively ordered tended to differ. The inverse probability of selection weighting approach generally had a small effect on the estimated means but did improve estimation of some of the more selected tests.
Conclusions
Commonly ordered laboratory tests appear to be representative of values from the underlying population. This suggests that trends and other patterns in biomarker levels in the population may be reasonably studied using data collected during the routine delivery of medical care.
Keywords: National Health and Nutrition Examination, Survey, Laboratories, Selection bias, Epidemiologic methods
Introduction
Laboratory test results are increasingly available in large health care databases and may be helpful for controlling confounding, identifying subgroups of interest, and characterizing populations. However, laboratory tests are often ordered to diagnose disease or monitor disease progress; therefore, patients with laboratory tests ordered may be a highly selected sample of the overall population. For example, patients with chronic kidney disease may be more likely to have frequent measures of serum creatinine to track renal function decline [1]. Similarly, patients with hyperlipidemia may be more likely to have lipid panels ordered so that physicians can make decisions about statin treatment [2]. Furthermore, even if a physician orders a laboratory test, it is often necessary for a patient to return later to have blood drawn. This introduces an additional selection process as many patients may fail to return for follow-up appointments. Finally, in some settings, laboratory tests are only available through certain laboratory testing companies. All factors governing the availability of laboratory values in data from routine care suggest that patients with a specific test result available may not be representative of the general population.
The selection bias created by this problem could be addressed theoretically by inverse probability of selection weighting (IPSW) [3], a semiparametric approach for missing data problems. The approach would first require a model for estimating the probability that a patient would have a laboratory test result available. The fitted model would then be used to generate IPSW that would be applied to individual patients in an analysis restricted to patients with the test result available. The approach would downweight patients who are likely to have the test result available, as they are overrepresented in the data. Similarly, patients unlikely to have a test result available would be upweighted to account for the many patients like them who are not present in the sample. The validity of the method requires that the analyst have measurements of the variables that influence both selection and the laboratory value. It is unknown how well this approach would work using typical administrative health care databases.
Using nationally representative data from the National Health and Nutrition Examination Survey (NHANES) and routine care data from a large population of patients with commercial insurance, we examined the extent to which commonly ordered laboratory values were representative of the distribution of laboratory values from the general population as reflected in the NHANES data. We hypothesized that the values collected in routine care would be substantially biased, reflecting a population of patients with more disease and abnormal test results. We then examined the distribution of laboratory values within the IPSW sample. We hypothesized that the IPSW sample would result in distributions of laboratory results that were more comparable with those from NHANES.
Methods
Data and cohort identification
We created a cohort of patients receiving routine medical care using the Truven Health Analytics MarketScan Commercial Claims and Encounters and Laboratory databases. These databases contain individual-level information on outpatient services, procedures, diagnoses, and medication information from pharmacy records for a very large population of patients in the United States with employer-provided commercial insurance, and their dependents. This enables researchers to have longitudinal views of pharmacy and health care utilization [4]. Laboratory values are available on patients who have a test ordered and submitted to a specific national testing company. Truven links the laboratory testing data to the claims data. Using these data, we identified a retrospective cohort of patients aged 40 to 64 years who had an office visit during the calendar years 2009 to 2010 using Current Procedural Terminology codes (99211-99215, 99201-99205) to identify office visits. Next, we examined a 2-week period after the office visit to see if a patient received various laboratory tests of interest. We dropped observations in which a patient had fewer than 6 months of eligibility before the index date (date of the office visit) and less than two weeks of eligibility after the index date. To ensure that patients with multiple visits were not over-represented in the analysis data set, we selected one physician visit at random per year to include in the analysis data set. For the selected visits, we then looked in the 2-week period after the index date to see if a patient had a specific test performed and an available result. To minimize the size of the data set, we sampled 1% of the visits with no available laboratory value.
We created a large number of covariates based on claims occurring in the 6-month period before the index date. To ensure broad capture of potentially relevant covariates, we identified all generic medications and three-digit International Classification of Diseases, Ninth Revision diagnostic codes that occurred with a prevalence of greater than 0.5% and procedure codes that occurred with a prevalence of greater than 1%. We created indicator variables to represent the presence of these codes. This approach is similar to other data-driven approaches to variable creation [5]. We also identified various demographic variables, such as age, sex, and region of residence.
To obtain estimates of the distribution of laboratory values from the underlying population, we used data from the 2009 to 2010 NHANES. NHANES uses probability sampling to characterize the health and nutritional status of the United States civilian population. The survey examines a sample of approximately 5000 persons each year. These persons are located in counties across the country, 15 of which are visited each year. The NHANES interview includes demographic, socioeconomic, dietary, and health-related questions. The examination component consists of medical, dental, and physiological measurements, and laboratory tests administered by trained medical personnel. To make the NHANES population directly comparable with our routine care population, we restricted the NHANES cohort to people aged 40 to 64 years with private insurance. People with private insurance were identified from those with a positive response to the question, “Are you/Is SP (survey participant) covered by private insurance?” as part of the household questionnaire. Appropriate examination sample weights from the mobile examination center data were used for national summary statistics of laboratory test value distributions.
Laboratory tests
The laboratory tests selected for study were those commonly ordered and available in NHANES. To account for data entry errors, we worked with a clinician (A.K.) to create trimming rules for the laboratory values (Appendix Table 1). In the instance of laboratory results reported in different units, when possible, we converted those units. We examined most of the laboratories that make up the comprehensive metabolic panel including the following: electrolytes (potassium, sodium, and chloride); proteins (total protein and albumin); measures of kidney function (blood urea nitrogen and creatinine); measures of liver function (alkaline phosphatase, alanine aminotransferase, aspartate aminotransferase, and bilirubin); glucose, and calcium. We also examined components of the standard lipid panel (low-density lipoprotein, high-density lipo-protein, triglycerides, and total cholesterol), and hemoglobin concentration, white blood cell count, glycohemoglobin, C-reactive protein, and gamma-glutamyl transferase. Laboratories were identified in the routine care data using Logical Observation Identifiers Names and Codes (LOINC) codes.
Statistical analysis
Using the supplied survey sampling weights, we computed summary statistics (means, standard deviations, and medians) for all selected laboratories from the NHANES cohort. These statistics were also computed for the trimmed laboratory values from routine care cohort. These statistics were then computed within strata of sex and age group. We attempted to reduce possible selection bias in the routine care cohort through IPSW. For each patient, we used a logistic model to determine the probability of having each particular laboratory value available in the 2-week period after the index visit. These models included all available covariates. Using the estimated probability of selection from the logistic regression model, we then created for each patient an inverse probability weight that was the inverse of a patient’s predicted probability of having a laboratory result available given his or her observed covariate vector. Using these weights, we then computed the summary statistics of the laboratory value distribution in the reweighted sample. To diagnose possible problems with the estimated weights, we also computed the C-statistic from the fitted logistic regression model and summary statistics for the distribution of the IPSW. This process was repeated for each laboratory value.
We considered a laboratory result from routine care to be meaningfully different from the underlying population distribution if its mean was more than half a standard deviation from the NHANES mean (using the standard deviation of the laboratory result from NHANES).
All statistical analyses were performed using SAS version 9.2 software (SAS Institute, Cary, NC). The University of North Carolina Institutional Review Board approved this research.
Results
Patient characteristics
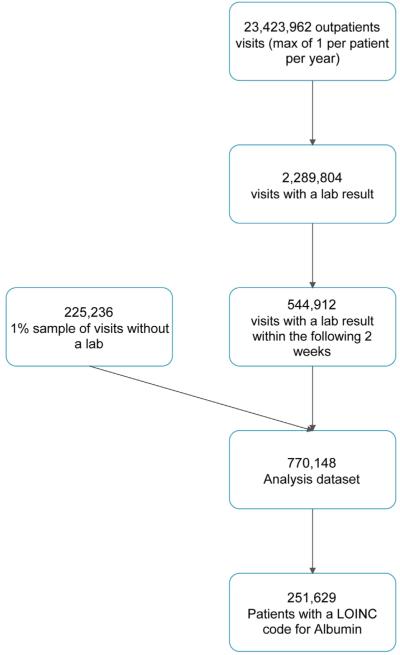
From the source population, we identified more than 23 million eligible office visits. Of these, 544,912 had a laboratory value of interest that was available in the following 2 weeks. The mean age of patients with a laboratory value was 52 years, and 58% of these patients were female. These patients came from the southern (56.2%), northeastern (11.1%), north central (15.9%), and western (16.7%) regions of the United States. For our analysis population, we selected all of the visits that were followed by an available laboratory result and a 1% sample of the remaining visits. This resulted in a final analysis data set of 770,148 patients. We depict the cohort construction process in Figure 1.
Fig. 1.
Cohort construction flow diagram illustrating inclusion criteria using the example of albumin laboratory results.
Summary statistics
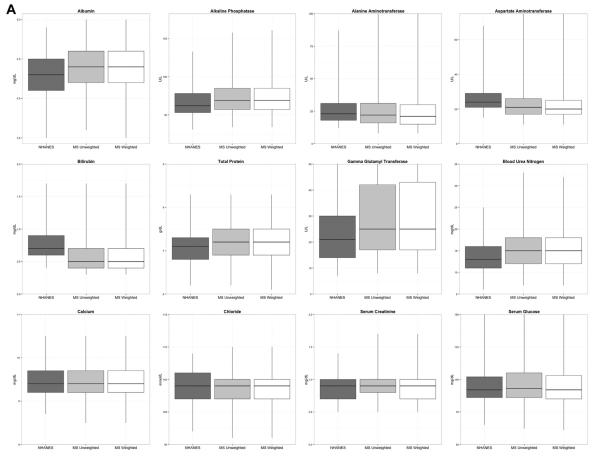
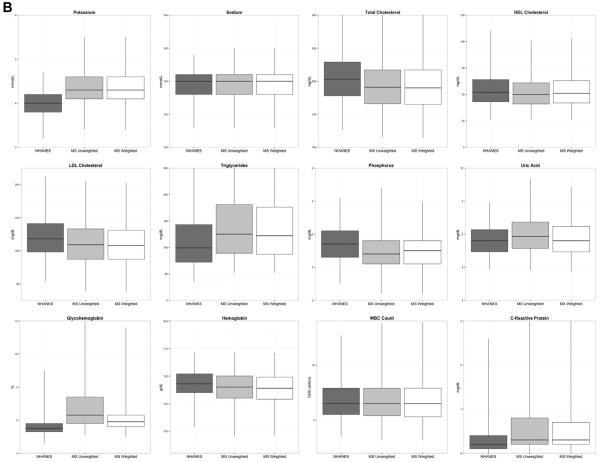
In Table 1 and Figure 2, we report and depict the basic summary statistics of the laboratory values in NHANES, the routine care population, and the inverse probabilityeweighted sample. This table reveals that the means and medians of most of the laboratory values in the routine care population were similar to the corresponding statistics from NHANES. However, several tests differed meaningfully. For example, the means for C-reactive protein and potassium in routine care were each nearly a standard deviation above the means from NHANES. Blood urea nitrogen, glycohemoglobin, alkaline phosphatase, uric acid, and gamma-glutamyl transferase levels were slightly higher (around 0.5 standard deviation) in routine care. We observed similar patterns when we looked within subgroups defined by age and sex (data not presented).
Table 1.
Distribution of laboratory results from NHANES and weighted and unweighted samples
| Variable | Cohort | N | Mean | SD | Min | Max | Med |
|---|---|---|---|---|---|---|---|
| Albumin (g/dL) | NHANES | 4.3 | 0.3 | 3.1 | 5.2 | 4.3 | |
| Routine care | 251,629 | 4.4 | 0.3 | 1.5 | 5.3 | 4.4 | |
| Routine care, weighted | 251,629 | 4.4 | 0.3 | 1.5 | 5.3 | 4.4 | |
| Alkaline phosphatase (U/L) | NHANES | 66.5 | 19.8 | 22.0 | 194.0 | 62.0 | |
| Routine care | 205,979 | 73.8 | 30.7 | 15.0 | 1917.0 | 69.0 | |
| Routine care, weighted | 205,979 | 73.9 | 30.5 | 15.0 | 1917.0 | 69.0 | |
| Alanine aminotransferase (U/L) | NHANES | 27.3 | 15.1 | 8.0 | 218.0 | 23.0 | |
| Routine care | 259,427 | 27.3 | 29.1 | 7.0 | 3650.0 | 22.0 | |
| Routine care, weighted | 259,427 | 26.6 | 30.8 | 7.0 | 3650.0 | 21.0 | |
| Aspartate aminotransferase (U/L) | NHANES | 26.8 | 12.3 | 12.0 | 296.0 | 24.0 | |
| Routine care | 258,407 | 24.0 | 21.2 | 8.0 | 2555.0 | 21.0 | |
| Routine care, weighted | 258,407 | 23.8 | 23.3 | 8.0 | 2555.0 | 20.0 | |
| Total bilirubin (mg/dL) | NHANES | 0.8 | 0.3 | 0.2 | 2.8 | 0.7 | |
| Routine care | 237,058 | 0.6 | 0.4 | 0.3 | 45.2 | 0.5 | |
| Routine care, weighted | 237,058 | 0.6 | 0.4 | 0.3 | 45.2 | 0.5 | |
| Total protein (g/dL) | NHANES | 7.1 | 0.4 | 5.8 | 9.0 | 7.1 | |
| Routine care | 252,016 | 7.2 | 0.5 | 3.9 | 10.5 | 7.2 | |
| Routine care, weighted | 252,016 | 7.2 | 0.5 | 3.9 | 10.5 | 7.2 | |
| Gamma-glutamyl transferase (U/L) | NHANES | 27.2 | 31.9 | 4.0 | 1116.0 | 21.0 | |
| Routine care | 13,824 | 44.1 | 94.7 | 5.0 | 3889.0 | 25.0 | |
| Routine care, weighted | 13,824 | 47.9 | 117.1 | 5.0 | 3889.0 | 25.0 | |
| BUN (mg/dL) | NHANES | 13.6 | 4.0 | 3.0 | 33.0 | 13.0 | |
| Routine care | 281,574 | 15.5 | 5.6 | 2.0 | 158.0 | 15.0 | |
| Routine care, weighted | 281,574 | 15.3 | 5.4 | 2.0 | 158.0 | 15.0 | |
| Calcium (mg/dL) | NHANES | 9.5 | 0.4 | 7.5 | 11.0 | 9.4 | |
| Routine care | 223,396 | 9.5 | 0.4 | 5.0 | 20.0 | 9.4 | |
| Routine care, weighted | 223,396 | 9.5 | 0.4 | 5.0 | 20.0 | 9.4 | |
| Chloride (mmol/L) | NHANES | 104.0 | 2.6 | 90.0 | 114.0 | 104.0 | |
| Routine care | 229,423 | 103.5 | 2.9 | 62.0 | 130.0 | 104.0 | |
| Routine care, weighted | 229,423 | 103.6 | 2.9 | 62.0 | 130.0 | 104.0 | |
| Creatinine (mg/dL) | NHANES | 0.9 | 0.2 | 0.4 | 2.5 | 0.9 | |
| Routine care | 286,811 | 0.9 | 0.3 | 0.3 | 19.3 | 0.9 | |
| Routine care, weighted | 286,811 | 0.9 | 0.3 | 0.3 | 19.3 | 0.9 | |
| Serum Glucose (mg/dL) | NHANES | 99.7 | 33.1 | 52.0 | 777.0 | 92.0 | |
| Routine care | 249,463 | 103.6 | 39.9 | 20.0 | 848.0 | 93.0 | |
| Routine care, weighted | 249,463 | 101.3 | 36.9 | 20.0 | 848.0 | 92.0 | |
| Potassium (mmol/L) | NHANES | 4.0 | 0.3 | 2.8 | 5.6 | 4.0 | |
| Routine care | 282,852 | 4.3 | 0.4 | 2.1 | 8.1 | 4.3 | |
| Routine care, weighted | 282,852 | 4.3 | 0.4 | 2.1 | 8.1 | 4.3 | |
| Sodium (mmol/L) | NHANES | 139.4 | 2.1 | 124.0 | 146.0 | 140.0 | |
| Routine care | 269,218 | 139.8 | 2.4 | 111.0 | 162.0 | 140.0 | |
| Routine care, weighted | 269,218 | 139.8 | 2.4 | 111.0 | 162.0 | 140.0 | |
| Total cholesterol (mg/dL) | NHANES | 205.6 | 41.1 | 93.0 | 528.0 | 203.0 | |
| Routine care | 249,808 | 193.4 | 40.3 | 100.0 | 771.0 | 191.0 | |
| Routine Care, Weighted | 249,808 | 192.9 | 40.7 | 100.0 | 771.0 | 190.0 | |
| Direct HDL (mg/dL) | NHANES | 55.0 | 17.6 | 21.0 | 133.0 | 52.0 | |
| Routine care | 244,912 | 52.4 | 15.7 | 10.0 | 197.0 | 50.0 | |
| Routine care, weighted | 244,912 | 53.8 | 16.7 | 10.0 | 197.0 | 51.0 | |
| LDL (mg/dL) | NHANES | 121.7 | 33.4 | 33.0 | 266.0 | 118.0 | |
| Routine care | 238,061 | 111.2 | 34.7 | 1.0 | 441.0 | 109.0 | |
| Routine care, weighted | 238,061 | 110.2 | 35.1 | 1.0 | 441.0 | 108.0 | |
| Triglycerides (mg/dL) | NHANES | 128.2 | 121.7 | 28.0 | 2742.0 | 99.0 | |
| Routine care | 237,943 | 153.9 | 137.0 | 50.0 | 9523.0 | 125.0 | |
| Routine care, weighted | 237,943 | 149.5 | 129.4 | 50.0 | 9523.0 | 122.0 | |
| Phosphorus (mg/dL) | NHANES | 3.7 | 0.6 | 2.0 | 5.7 | 3.7 | |
| Routine care | 12,354 | 3.5 | 0.7 | 1.1 | 15.4 | 3.4 | |
| Routine care, weighted | 12,354 | 3.5 | 0.6 | 1.1 | 15.4 | 3.5 | |
| Uric acid (mg/dL) | NHANES | 5.4 | 1.4 | 1.5 | 10.9 | 5.4 | |
| Routine care | 22,091 | 6.0 | 1.8 | 1.0 | 16.3 | 5.8 | |
| Routine care, weighted | 22,091 | 5.6 | 1.7 | 1.0 | 16.3 | 5.4 | |
| Glycohemoglobin (%) | NHANES | 5.6 | 0.8 | 4.1 | 14.2 | 5.5 | |
| Routine care | 79,859 | 6.9 | 1.7 | 3.4 | 19.0 | 6.3 | |
| Routine care, weighted | 79,859 | 6.2 | 1.2 | 3.4 | 19.0 | 5.9 | |
| Hemoglobin (g/dL) | NHANES | 14.3 | 1.4 | 7.5 | 19.5 | 14.3 | |
| Routine care | 190,980 | 13.9 | 1.6 | 4.0 | 20.6 | 14.0 | |
| Routine care, weighted | 190,980 | 13.9 | 1.5 | 4.0 | 20.6 | 13.9 | |
| WBC count (1000 cells/uL) | NHANES | 6.9 | 2.3 | 2.8 | 99.9 | 6.5 | |
| Routine care | 189,223 | 6.8 | 2.3 | 0.5 | 91.0 | 6.5 | |
| Routine care, weighted | 189,223 | 6.8 | 2.3 | 0.5 | 91.0 | 6.5 | |
| C-reactive protein (mg/dL) | NHANES | 0.3 | 0.7 | 0.0 | 18.0 | 0.2 | |
| Routine care | 11,959 | 1.0 | 2.9 | 0.0 | 90.7 | 0.3 | |
| Routine care, weighted | 11,959 | 0.9 | 2.8 | 0.0 | 90.7 | 0.3 |
BUN = blood urea nitrogen; HDL = high-density lipoprotein; LDL = low-density lipoprotein; WBC = white blood cell.
Our empirical approach to covariate identification created 218 indicators for diagnosis codes, 79 indicators for generic medication names, and 207 indicators for procedures. Therefore, the models of laboratory availability that we constructed included slightly more than 500 variables. All models converged. The C-statistics are provided in Table 2. Most of the models were moderately discriminating, with C-statistics around 0.7. The models of urine creatinine and glycohemoglobin availability resulted in C-statistics more than 0.8. For glycohemoglobin, the model was driven by diabetes-related diagnoses and medications.
Table 2.
C-statistics for weight models
| Laboratory model | C-statistic |
|---|---|
| Albumin (g/dL) | 0.702 |
| Alkaline phosphatase (U/L) | 0.702 |
| Alanine aminotransferase (U/L) | 0.699 |
| Aspartate aminotransferase (U/L) | 0.699 |
| Total bilirubin (mg/dL) | 0.702 |
| Total protein (g/dL) | 0.702 |
| Gamma-glutamyl transferase (U/L) | 0.791 |
| BUN (mg/dL) | 0.703 |
| Calcium (mg/dL) | 0.700 |
| Chloride (mmol/L) | 0.701 |
| Creatinine (mg/dL) | 0.701 |
| Serum glucose (mg/dL) | 0.701 |
| Potassium (mmol/L) | 0.702 |
| Sodium (mmol/L) | 0.705 |
| Total cholesterol (mg/dL) | 0.756 |
| Direct HDL (mg/dL) | 0.756 |
| LDL (mg/dL) | 0.753 |
| Triglycerides (mg/dL) | 0.753 |
| Phosphorus (mg/dL) | 0.786 |
| Uric acid (mg/dL) | 0.781 |
| Hemoglobin (g/dL) | 0.684 |
| Glycohemoglobin (%) | 0.825 |
| WBC count (1000 cells/uL) | 0.682 |
| C-reactive protein (mg/dL) | 0.761 |
BUN = blood urea nitrogen; HDL = high-density lipoprotein; LDL = low-density lipoprotein; WBC = white blood cell.
Inverse probability of selection weighting
In Table 3, we report summary statistics of the IPSW. Note the weights were multiplied by the marginal probability that a given test would be ordered for a patient in our sample. The means of these weights were mostly close to 1, as expected, indicating no obvious pathologies in the models. The lipid panels resulted in the largest weights, with maximum weights around 250. In most cases, the weights had little effect on the estimated means. In some instances, the weighting moved the estimates substantially closer to the mean from NHANES (e.g., glycohemoglobin and uric acid).
Table 3.
Distribution of the Inverse-Probability of Selection Weights
| Laboratory model | Mean | Min | Max | P25* | Median | P75* |
|---|---|---|---|---|---|---|
| Albumin (g/dL) | 1.01 | 0.34 | 82.95 | 0.65 | 0.84 | 1.12 |
| Alkaline phosphatase (U/L) | 1.01 | 0.33 | 96.04 | 0.65 | 0.84 | 1.12 |
| Alanine aminotransferase (U/L) | 1.01 | 0.35 | 61.78 | 0.66 | 0.85 | 1.12 |
| Aspartate aminotransferase (U/L) | 1.01 | 0.35 | 58.38 | 0.66 | 0.85 | 1.12 |
| Total bilirubin (mg/dL) | 1.00 | 0.33 | 95.86 | 0.65 | 0.83 | 1.11 |
| Total protein (g/dL) | 1.01 | 0.34 | 89.85 | 0.65 | 0.84 | 1.12 |
| Gamma-glutamyl transferase (U/L) | 1.08 | 0.02 | 54.71 | 0.18 | 0.57 | 1.24 |
| BUN (mg/dL) | 1.01 | 0.37 | 46.62 | 0.67 | 0.85 | 1.12 |
| Calcium (mg/dL) | 1.00 | 0.30 | 54.76 | 0.64 | 0.83 | 1.13 |
| Chloride (mmol/L) | 1.00 | 0.31 | 47.84 | 0.64 | 0.83 | 1.13 |
| Creatinine (mg/dL) | 1.01 | 0.38 | 47.08 | 0.68 | 0.85 | 1.12 |
| Serum glucose (mg/dL) | 1.00 | 0.33 | 69.63 | 0.65 | 0.84 | 1.13 |
| Potassium (mmol/L) | 1.01 | 0.38 | 49.33 | 0.67 | 0.85 | 1.12 |
| Sodium (mmol/L) | 1.01 | 0.36 | 74.56 | 0.66 | 0.84 | 1.12 |
| Total cholesterol (mg/dL) | 1.05 | 0.34 | 247.24 | 0.55 | 0.75 | 1.10 |
| Direct HDL (mg/dL) | 1.04 | 0.33 | 257.82 | 0.54 | 0.74 | 1.11 |
| LDL (mg/dL) | 1.04 | 0.33 | 254.16 | 0.55 | 0.75 | 1.11 |
| Triglycerides (mg/dL) | 1.04 | 0.33 | 254.65 | 0.55 | 0.75 | 1.10 |
| Phosphorus (mg/dL) | 1.03 | 0.02 | 64.37 | 0.20 | 0.55 | 1.33 |
| Uric acid (mg/dL) | 1.03 | 0.03 | 143.51 | 0.25 | 0.63 | 1.20 |
| Glycohemoglobin (%) | 1.01 | 0.26 | 66.86 | 0.66 | 0.83 | 1.11 |
| Hemoglobin (g/dL) | 1.00 | 0.11 | 130.35 | 0.21 | 0.38 | 1.33 |
| WBC count (1000 cells/uL) | 1.01 | 0.26 | 52.64 | 0.67 | 0.83 | 1.11 |
| C-reactive protein (mg/dL) | 0.93 | 0.02 | 14.71 | 0.27 | 0.80 | 1.31 |
BUN = blood urea nitrogen; HDL = high-density lipoprotein; LDL = low-density lipoprotein; WBC = white blood cell.
P25, 25th Percentile; P75, 75th Percentile.
Discussion
We examined the extent to which commonly ordered laboratory values collected in routine care were representative of those in the underlying population. We further explored whether the use of an inverse probability weighting approach would improve the comparability of the routine care laboratory results with their corresponding distribution in the general population.
Contrary to our expectation, we found that the average values of most of the laboratory results from routine care were very close to their population means as estimated from NHANES. Only a few of the many tests that we considered differed meaningfully. The most extreme divergences were seen with C-reactive protein, potassium, gamma-glutamyl transferase, and glycohemoglobin. Except for potassium, these tests tend to be ordered more selectively for patients with known or strongly suspected disease. C-reactive protein is an inflammatory biomarker used to monitor the progress of autoimmune conditions and detect systemic inflammation or infection. Glycohemoglobin can be used to diagnose diabetes and measure long-term blood glucose control in people with diabetes. Gamma-glutamyl transferase is usually ordered in conjunction with other liver tests, such as alkaline phosphatase, and used to detect liver disease and bile duct obstructions. With some of the other tests, we did observe slightly increased (or decreased) levels in routine care, but we had expected to observe more divergence between the two samples. In some cases, the means appeared to be moderately skewed by a small number of outliers. In these cases, the median values were more comparable. Previous research has found reasonably good comparability between various lipid measures collected in routine care and their corresponding population levels in NHANES [6,7].
The relative comparability between the laboratory results from routine care and those from NHANES could partly be attributed to our study design that restricted the sample to one laboratory result per patient during the 2-year period. This design reduced the influence of patients receiving frequent testing. Furthermore, because many of these laboratory values are commonly ordered in relatively healthy adults, it may be that our sample of patients receiving a given laboratory value is not too different from a random sample of the underlying population. For example, the National Cholesterol Education Program’s Adult Treatment Panel III, sponsored by the National Institutes of Health and endorsed by the American Heart Association, recommends that all adults aged older than 20 years receive a fasting lipoprotein profile every 5 years [8]. Metabolic panels are often ordered on relatively healthy patients taking prescription medications to monitor for renal or hepatic side effects of medications or underlying conditions being treated. For example, patients receiving antihypertensives are recommended to have serum creatinine and potassium levels checked one to two times per year [9].
The IPSW approach had a relatively small effect on the means of most routine laboratory tests. With uric acid and glycohemoglobin, two of the more selected laboratories, IPSW moved the estimated mean substantially closer to toward the NHANES mean. However, the bias was not fully eliminated. A look-back period longer than 6 months may be needed to more fully correct this bias. It is also possible that to fully control selection effects, variables not available in health care claims are required, such as better measures of co-morbidity. Alternative methods for handling missing data, such multiple imputation, that are based on models for the missing laboratory values rather than the selection mechanism may also more fully correct bias [10].
One important limitation of our research was that we considered only distributions of laboratory results in a population of privately insured patients between 35 and 64 who were receiving outpatient care. Our results may not generalize to younger populations, or patients with Medicaid or Medicare. Our results may also not generalize to selected subpopulations. This is particularly likely when membership in the subpopulation is partly determined by a laboratory value, such as users of a medication or a group of patients with a certain diagnosis. For example, initiation of statin therapy is due to either elevated lipid levels or the presence of other cardiovascular risk factors such as diabetes. Therefore, patients with lipid levels indicating hyperlipidemia may be more likely to be new users of statins and have higher lipid levels compared with the general population of new users. This may have implications for the use of laboratory values to characterize certain subgroups of patients. There are other limitations of our study to consider. Our results will not necessarily generalize to the many other types of laboratory tests that we did not consider. Selection effects and availability of results may also be different in an integrated health care delivery system. Also, we have only considered single laboratory values. In many studies, researchers may want to control for multiple laboratory values. Patients with multiple laboratory values available may represent a more selected subgroup. Finally, we used an arbitrary criterion for determining whether laboratory values from the two databases were substantively different. The actual meaningfulness of any difference will depend on the specific research context.
Biomarkers levels from NHANES have been widely used to describe the health and nutritional status of the US population. Laboratory values from health care databases are not a replacement for rigorously collected samples from broadly representative studies such as NHANES. However, NHANES data are geographically and temporally sparse and may not contain certain laboratory values of interest. Our study has found that many of the most commonly ordered laboratory tests are fairly representative of the distribution of laboratories from NHANES. This is an important observation and suggests that trends and other patterns in biomarker levels in the population may be reasonably studied using data collected during the routine delivery of medical care.
Supplementary Material
Fig. 2.
(A) and (B): Distribution of laboratory values: NHANES, MarketScan unweighted, and MarketScan weighted results.
Acknowledgments
This project was funded through a subcontract to University of North Carolina Chapel Hill from the Brigham and Women’s Hospital and the Agency for Healthcare Research and Quality (AHRQ), US Department of Health and Human Services (DHHS) as part of the Developing Evidence to Inform Decisions about Effectiveness (DECIDE) program, HHSA29020050016I task order #10. Dr. Brookhart also receives investigator-initiated research funding from the National Institutes of Health (R01 AG042845, R21 HD080214, and R01 AG023178).
References
- [1].Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012;379(9811):165–80. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- [2].Drozda J, Jr, Messer JV, Spertus J, Abramowitz B, Alexander K, Beam CT, et al. ACCF/AHA/AMA-PCPI 2011 performance measures for adults with coronary artery disease and hypertension: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Performance Measures and the American Medical Association-Physician Consortium for Performance Improvement. Circulation. 2011;124(2):248–70. doi: 10.1161/CIR.0b013e31821d9ef2. [DOI] [PubMed] [Google Scholar]
- [3].Bang H, Robins JM. Doubly robust estimation in missing data and causal inference models. Biometrics. 2005;61(4):962–73. doi: 10.1111/j.1541-0420.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- [4].Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58(4):323–37. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- [5].Schneeweiss S, Rassen JA, Glynn RJ, Avom J, Mogun H, Brookhart MA. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2009;20(4):512–22. doi: 10.1097/EDE.0b013e3181a663cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martin SS, Blaha MJ, Elshazly MB, Brinton EA, Toth PP, McEvoy JW, et al. Friedewald-estimated versus directly measured low-density lipoprotein cholesterol and treatment implications. J Am Coll Cardiol. 2013;62(8):732–9. doi: 10.1016/j.jacc.2013.01.079. [DOI] [PubMed] [Google Scholar]
- [7].Martin SS, Blaha MJ, Toth PP, Joshi PH, McEvoy JW, Ahmed HM, et al. Very large database of lipids: rationale and design. Clin Cardiol. 2013;36(11):641–8. doi: 10.1002/clc.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults Executive summary of the third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285(19):2486–97. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- [9].Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, et al. The seventh report of the Joint National Committee on prevention, detection, evaluation, and treatment of sssshigh blood pressure: the JNC 7 report. JAMA. 2003;289(19):2560–72. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- [10].Rubin DB, Schenker N. Multiple imputation in health-care databases: an overview and some applications. Stat Med. 1991;10(4):585–98. doi: 10.1002/sim.4780100410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.