Abstract
To establish a productive infection, HIV-1 must counteract cellular innate immune mechanisms and redirect cellular process towards viral replication. Recent studies have discovered that HIV-1 and other primate immunodeficiency viruses subvert cell cycle regulatory mechanisms to achieve these ends. The viral Vpr and Vpx proteins target cell cycle controls to counter innate immunity. The cell cycle-related protein Cyclin L2 is also utilized to counter innate immunity. The viral Tat protein utilizes Cyclin T1 to activated proviral transcription, and regulation of Cyclin T1 levels in CD4+ T cells has important consequences for viral replication and latency. This review will summarize this emerging evidence that primate immunodeficiency viruses subvert cell cycle regulatory mechanisms to enhance replication.
CD4+ T lymphocytes and myeloid cells -- macrophages and dendritic cells -- are the two major cell types infected in vivo by HIV-1, HIV-2, and other primate immunodeficiency viruses. To successfully replicate in these cells, the viruses must overcome the antiviral action of multiple proteins of the innate immune system. The importance of escaping innate immunity is clear, as despite having relatively small genomes of ~10,000 nucleotides, primate immunodeficiency viruses encode several proteins that act to counter innate immunity -- Nef, Vpr, Vpu, and Vpx.
While all lineages of primate lentiviruses encode Vpr, the HIV-2/SIV sooty mangabey (SIVsmm) and SIV/red-capped mangabey/mandrill (rcm/mnd2) lineages also encode the paralog Vpx. Early genetic studies demonstrated that both Vpr and Vpx are dispensable for viral replication in vitro, but the proteins play important roles in vivo in experimental infection of macaques. Deletion of either the vpr or vpx gene attenuated SIVmac replication in rhesus macaques, although the infected animals eventually progressed to AIDS (Gibbs et al., 1995). Interestingly, animals infected with the vpx mutant had lower viral burdens and slower CD4+ T cell decline than animals inoculated with the vpr mutant. Deletion of both vpr and vpx severely attenuated the virus. Furthermore, studies of SIVsmm and SIV/macaca nemestrina (mne) in pigtailed macaques demonstrated that deletion of vpx compromised mucosal transmission and disease (Belshan et al., 2012; Hirsch et al., 1998). Although studies have consistently demonstrated an effect of Vpx on macrophage tropism of SIV in vitro, deletion of vpx also appears to severely attenuate the spread of virus through the CD4+ T-cell population in vivo (Belshan et al., 2012) Thus, for SIVs that encode both vpr and vpx (SIVsmm and SIVrcm/mnd2), Vpx may be more critical than Vpr for replication in Old World Monkeys and it likely makes important contributions to viral replication in both macrophages and CD4+ T cells.
Cell cycle-regulated CDKs overcome SAMHD1
The importance of Vpx was determined in quiescent CD4+T cells, monocytes, and dendritic cells. These are non-dividing cells that are generally not permissive for HIV-1 infection, but infection of these cells could be enhanced by incorporation of the HIV-2 Vpx protein into HIV-1 virions (Goujon et al., 2008). Early work indicated that Vpx functions in this experimental system to overcome a restriction factor in non-dividing cells that acts early at a post-entry stage to inhibit reverse transcription (Fletcher, III et al., 1996). The analysis of mutant Vpx proteins established a correlation between the ability of Vpx to enhance reverse transcription and to associate with an ubiquitin E3 ligase complex composed of DCAF1-DDB1-CUL4A-RBX1 (Le Rouzic et al., 2007; Srivastava et al., 2008). Thus, it was believed that Vpx enhances reverse transcription through proteasome-mediated proteolysis of a restriction factor. Using mass spectrometry technology to identify cellular proteins that co-immunoprecipitated with wild type but not a mutant Vpx unable to associate with DCAF1, SAMHD1 was identified as this key restriction factor in non-dividing cells (Hrecka et al., 2011; Laguette et al., 2011).
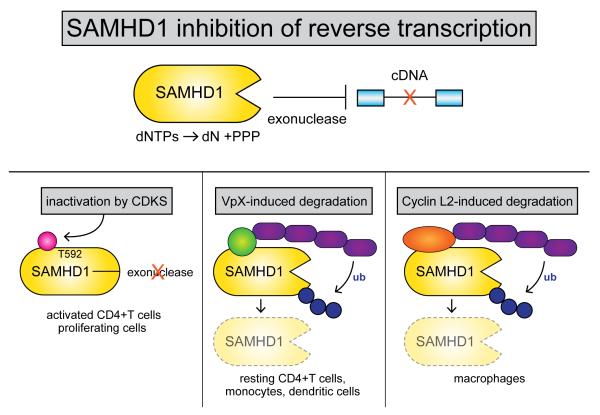
Prior to its discovery as a target of Vpx, SAMHD1 was linked to the immune system, as mutations in SAMHD1 were known to cause Aicardi-Goutieres syndrome, a condition involving chronic inflammation and reminiscent of persistent viral infections (Rice et al., 2009). SAMHD1 is a phosphohydrolase that cleaves dNTPs into deoxynucleotides and inorganic triphosphates. In quiescent CD4+ T cells, monocytes, and dendritic cells, SAMHD1 activity depletes the dNTP pool required for efficient HIV-1 reverse transcription. SAMHD1 also possesses a 3′-to-5′ exonuclease activity that degrades single-stranded RNA and single strand DNA overhangs, and this activity has been associated with inhibition of HIV-1 reverse transcription. Vpx overcomes the antiviral activity of SAMHD1 by loading it onto the DCAF1-DDB1-CUL4A-RBX1 E3 ubiquitin ligase complex, resulting in efficient proteasome-mediated degradation of SAMHD1 (Figure 1).
Figure 1. Inhibition of SAMHD1 antiviral activity.
SAMHD1 possesses phosphohydrolase and exonuclease activities that have been shown to inhibit retroviral cDNA synthesis (top panel). In activated CD4+ T cells and proliferating cell lines, CDKs inactivate SAMHD1 through phosphorylation of T592 (left bottom panel). In resting CD4+ T cells, monocytes, and dendritic cells, the viral Vpx protein functions to degrade SAMHD1 through loading it onto the DCAF1/DDB1/CUL4A/RBX1 E3 ubiquitin complex (bottom middle panel). In macrophages, the cellular Cyclin L2 protein functions to degrade SAMHD1 through loading it onto the E3 ubiquitin complex (bottom right panel).
With the discovery of SAMHD1 as a potent HIV-1 restriction factor in quiescent CD4+ T cells, monocytes, and dendritic cells, a conundrum arose as SAMHD1 is expressed at high levels in activated CD4+ T cells and monocytic cell lines – proliferating cells that support robust HIV-1 replication. Clearly, the expression level of SAMHD1 is not sufficient to inhibit HIV-1 replication. This conundrum was resolved when it was discovered that cyclin-dependent kinases (CDKs) activated during the cell cycle of proliferating cells can inactivate SAMHD1 through phosphorylation of threonine 592 (T592) (White et al., 2013; Cribier et al., 2013). The mechanism whereby this phosphorylation inactivates SAMDH1 is presently unclear, as there are conflicting reports on the effects on phosphohydrolase activity (White et al., 2013; Yan et al., 2015; Ruiz et al., 2015). Additional studies are required to clarify to what extent T592 phosphorylation inhibits SAMHD1 phosphohydrolase and nuclease activity, and the relative importance of each activity for anti-HIV-1 activity.
To date, three cyclin-dependent kinases -- CDK1, CDK2, and CDK6 -- have been reported to phosphorylate SAMHD1 T592 in proliferating cells (Pauls et al., 2014; White et al., 2013; Cribier et al., 2013; St-Gelais et al., 2014). Because these CDKs are generally not active in non-proliferating cells, SAMHD1 lacks T592 phosphorylation and is able to function as a restriction factor to inhibit HIV-1 reverse transcription in these cells. In proliferating cells, cell cycle-regulated CDKs are activated and HIV-1 can take advantage of this regulation to evade SAMHD1 and achieve robust replication (Figure 1).
Evolutionary analyses have shown that SAMHD1 antagonism is a highly conserved activity of primate lentiviruses infecting some Old World Monkeys but not hominoids (Lim et al., 2012). Vpx and some Vpr proteins from SIVs are able to degrade the SAMHD1 protein of their natural hosts, and the ability of SIVs to degrade SAMHD1 preceded the evolution of the vpx gene, which presumably occurred through recombination/duplication of the vpr gene and subsequent divergence of one allele into vpx. Moreover, Vpr/Vpx-mediated antagonism resulted in strong positive selection of SAMHD1 in the primate subfamily Cercopithecinae, but not hominoids or New World Monkeys, indicating the importance of targeting SAMHD1 for infection of many Old World Monkeys. Vpx, and the ability to inhibit SAMHD1, was deleted during the evolution of SIVcpz from two SIV lineages of Old World Monkeys and the absence of SAMHD1 degrading activity was maintained with adaptation of HIV-1 to humans (Etienne et al., 2013). However, this phenotype may not be a prerequisite for infection of a species, as in contrast to HIV-1, HIV-2 does degrade SAMHD1 in human cells. Furthermore, mandrills are also infected by SIVs that encode either only Vpr (SIVmnd1) or both Vpr and Vpx (SIVmnd2). Thus, the lack of SAMHD1 antagonism by certain primate lentiviruses may be the consequence of the evolution of unique replication mechanisms that obviate a requirement to inhibit SAMHD1.
Cyclins exploited by HIV-1
The term “Cyclin” was first given to a protein detected in fertilized sea urchin eggs that was degraded at certain points in the cell cycle (Evans et al., 1983). This degradation was subsequently shown to occur via proteasome-mediated proteolysis and approximately 30 Cyclin proteins are now known to be encoded in the human genome. Some Cyclins are key regulators of the cell cycle through their binding to and activation of CDKs, followed by the subsequent proteolysis of the Cyclin at define points in the cell cycle. Other Cyclin-CDK pairs regulate cellular processes that are not directly related to cell cycle control, such as stimulation of RNA Polymerase II (RNAP II) transcriptional elongation by Cyclin T1/CDK9 (discussed below).
Cyclin L2 degrades SAMHD1 in macrophages
Efficient replication of HIV-2 in macrophages requires the interaction of Vpx with DCAF1, and a yeast two-hybrid system screen was therefore carried out to identify DCAF1-associated proteins (Kyei et al., 2015). Cyclin L2 was identified in this screen and this Cyclin is involved in cell cycle regulation and pre-mRNA splicing, although the mechanisms whereby it is involved in these processes are poorly understood. Cyclin L2 forms a complex with DCAF1 and SAMHD1, resulting in the proteasome-mediated degradation of SAMHD1. siRNA knock-down of cyclin L2 reduces HIV-1 replication in macrophages but not proliferating cells (where SAMHD1 is inactivated by CDKs). In macrophages, therefore, Cyclin L2 inactivates SAMHD1 through proteolysis, thereby enhancing HIV-1 replication in this cell type (Figure 1).
Cyclin T1 affects HIV-1 latency in CD4+ T cells
Cyclin T1 has long been known to be targeted by HIV-1. Cyclin T1 is the regulatory subunit of CDK9 and the Cyclin T1/CDK9 heterodimer is the catalytic core of a general RNAP II elongation factor termed P-TEFb (Mbonye and Karn, 2014). In the absence of the retroviral trans-activator protein Tat, RNAP II initiates transcription from the viral 5′ LTR sequences, but elongation is restricted by the action of two cellular factors termed DSIF and NELF. To stimulate elongation, Tat binds directly to Cyclin T1 and CDK9 in P-TEFb, and this complex binds to the viral TAR RNA element that forms at the 5′ end of viral transcripts. CDK9 is then positioned to phosphorylate both the carboxyl terminal domain of RNA Polymerase II and the negative factors that restrict elongation, leading to a potent stimulation of elongation.
Regulation of Cyclin T1 in CD4+ T lymphocytes is of considerable significance for HIV-1 replication and latency. HIV-infected individuals treated with suppressive antiviral therapy contain a population of quiescent memory CD4+ T cells with transcriptionally silent or latent HIV-1 proviruses. Upon cessation of anti-viral drugs, viruses in this latent reservoir reactivate and rekindle infection, thereby precluding a cure of infection by current antiviral drugs. Tat activity is limiting for productive transcription of latent proviruses in CD4+ T cells from these infected individuals (Lassen et al., 2004) and this is due in part to limiting levels of Cyclin T1. Cyclin T1 protein levels are low in resting CD4+ T cells due to repression by several miRNAs: miR-27b, miR-29b, miR-150, and miR-223 (Chiang et al., 2012). T cell activation reduces the level of these miRNAs relative to Cyclin T1 mRNA, leading to a strong increase in Cyclin T1 protein expression. Unlike Cyclin T1, CDK9 protein levels are high in resting CD4+ T cells. However, CDK9 catalytic activity is quite low in resting cells due to the lack of phosphorylation of threonine 186 in the CDK9 T-loop -- phosphorylation of the T-loop allows access of substrates to the enzyme’s catalytic core. The phosphatase PPM1A is involved in dephosphorylation of the T-loop in resting CD4+ T cells (Budhiraja et al., 2012). Upon T cell activation, there is a rapid phosphorylation of the CDK9 T-loop leading to active P-TEFb (Ramakrishnan et al., 2009) and CDK7 has been identified as the kinase that phosphorylates the CDK9 T-loop (Larochelle et al., 2012).
Although Cyclin T1 has not been shown to be directly involved in cell cycle regulation, it is degraded by proteasome-mediated proteolysis when activated CD4+ T cells returns to quiescence and this appears to be important in the establishment of latent infection (Budhiraja et al., 2013). A PEST sequence, which is thought to initiate degradation, at the carboxyl terminus of Cyclin T1 is likely to be a signal for proteolysis in cells returning to quiescence. Up-regulation of Cyclin T1 following activation of quiescent CD4+ cells is required for reactivation of latent HIV-1, further indicating that Cyclin T1 is a limiting factor for HIV-1 replication in quiescent CD4+ T cells (Tyagi et al., 2010).
Vpr cell cycle arrest and SLX4complex
In 1995, several groups reported that the HIV-1 Vpr protein induces cell cycle arrest at the G2/M transition [reviewed in (Andersen and Planelles, 2005)]. Subsequent studies found that the cell cycle arrest involves the DNA damage checkpoint network and activation of the ATR and Chk1 kinases (Andersen and Planelles, 2005). However, the importance of cell cycle arrest to HIV-1 replication proved remarkably difficult to elucidate. Critical data were obtained when Vpr was found to associate with a DCAF1-DDB1-Cul4 E3 ligase complex and analyses of mutant Vpr proteins indicated that this association is required for cell cycle arrest (Belzile et al., 2007; Dehart et al., 2007; Hrecka et al., 2007; Le Rouzic et al., 2007; Schrofelbauer et al., 2007). These findings let to the expectation that Vpr cell cycle arrest is the result of degradation of a key cellular protein whose down-regulation induced cell cycle arrest.
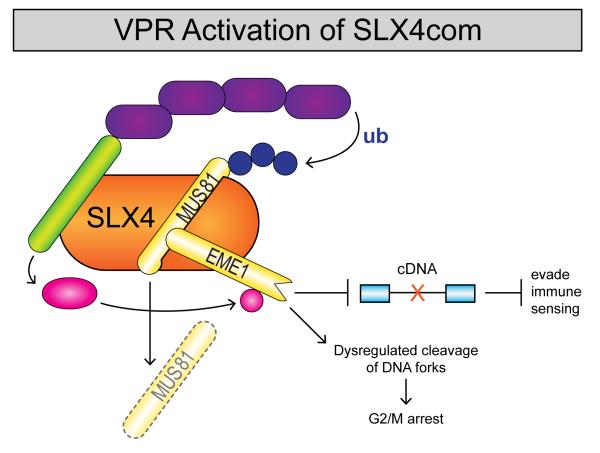
In a recent comprehensive study, Vpr was found to interact with the SLX4 scaffold protein and the structure-specific endonucleases (SSEs) MUS81-EME1 and ERCC1-ERCC4 (Laguette et al., 2014). The complexes formed between SLX4 and these SSEs, termed SLX4coms, resolve secondary DNA structures generated during DNA repair and recombination such as Holliday junctions. SLX4coms are under strict cell cycle regulation as their inappropriate activation can result in chromosome instability. Biochemical experiments by Laguette and colleagues showed that Vpr interacts directly with the carboxyl terminus of SLX4, and the viral protein is present in a distinct SLX4com consisting of DCAF1, MUS81-EME1 and SLX4. The interaction between SLX4com and Vpr is quite specific, as the related HIV-2 Vpx protein does not associate with the SLX4com. The endonuclease activity of the MUS8-EME1 SSE is stimulated by phosphorylation of EME1 by Polo-Like Kinase 1 (PLK1), and Laguette and colleagues found that Vpr and DCAF1 recruit active PLK1 into the SLX4com (Figure 2). Additional biochemical experiments showed that association of Vpr and DCAF1 with the SLX4com stimulates endonuclease activity in vitro, and the association in vivo results in the accumulation of punctate structures termed FANCD2 foci, which are markers of damaged DNA due to deficient cleavage of replication intermediates.
Figure 2. Vpr activation of SLX4com and cell cycle arrest.
The viral Vpr protein binds to the SLX4complex that contains the MUS81/EME1 structure-specific endonuclease. Vpr also recruits the kinase PLK1 which phosphorylates EME1 to stimulate endonuclease activity. The nuclease activity of the SLX4complex degrades viral cDNA, thereby preventing detection of infection by the innate immune system. Inappropriate activation of MUS81/EME1 nuclease activity by Vpr results in G2/M cell cycle arrest. Vpr also recruits the DCAF1/DDB1/CUL4A/RBX1 E3 ubiquitin complex to the SLX4complex, leading to degradation of MUS81.
Substantial data indicate that inappropriate activation of the SLX4com by Vpr induces G2/M arrest. Formation of the Vpr/SLX4com precedes cell cycle arrest, and siRNA depletion of SLX4, EME1, or MUS81 reduces Vpr-induced G2/M arrest (Laguette et al., 2014). Mutant Vpr proteins unable to associate with the SLX4com do not induce cell cycle arrest. An analysis of Vpr proteins from a number of SIVs identified a correlation in human cells between the ability of SIV Vpr proteins to associate with SLX4 and induce the G2/M arrest (Berger et al., 2015). Formation of the Vpr-SLX4com leads to ubiquitination and degradation of the MUS81 subunit of the complex (Laguette et al., 2014; DePaula-Silva et al., 2015). However, degradation of MUS81 can be rather modest, and as stated above, MUS81 siRNA depletions reduce rather than promote Vpr-mediated cell cycle arrest.
Vpr activation of SLX4com prevents Interferon induction
How does Vpr activation of the SLX4com benefit HIV-1 replication? A major benefit is likely to be impairment of the host cell’s ability to sense infection and induce IFN. SLX4 belongs to the Fanconi Anemia (FA) family of proteins in which mutations result in increased susceptibility to cancer and bone marrow failure, as well as abnormal production of IFN and proinflammatory cytokines. Vpr activation of the SLX4com appears to allow the endonuclease to degrade HIV-1 reverse transcripts and help the virus to evade detection by the innate immune system (Figure 2). SiRNA depletion of MUS81, DCAF1, or SLX4 leads to induction of IFN following HIV-1 infection, while infection with a Vpr-deleted virus induces IFN (Laguette et al., 2014). Furthermore, SLX4 co-immunoprecipitated with HIV-1 reverse transcription products and this was largely dependent upon Vpr, presumably by promoting the interaction of the SLX4com with RT products and the subsequent degradation of cDNA.
It has been estimated that up to 90% of HIV-1 reverse transcripts do not integrate (Suspene and Meyerhans, 2012) and these viral products are potential signals of infection that can be recognized by the innate immune system. The exonucleases TREX1 and RNASEH2 have been implicated in degrading excessive HIV-1 reverse transcripts, thereby preventing IFN induction (Yan et al., 2010; Genovesio et al., 2011). Given the recent findings by Laguette and colleagues, Vpr activation of the endonuclease activity of SLX4com also contributes to the degradation of viral transcripts. The finding that multiple cellular nucleases degrade HIV-transcripts suggests that reverse transcription produces a large excess of transcripts over that needed for replication. Reverse transcription of the HIV-1 genome is clearly an effective but apparently inelegant step in the viral replication strategy -- the virus synthesizes far too much viral cDNA and must utilize cellular nucleases to degrade the excess to avoid detection by the innate immune system.
Future Perspectives
The recent findings described above have led to the appreciation that HIV-1 and other primate immunodeficiency viruses subvert cell cycle regulatory mechanisms as part of their replication strategy, especially to counter innate immune mechanisms. A number of intriguing questions have been raised by these findings and there will surely be rapid progress in this area. Does SAMHD1 function as a restriction factor for other viruses, especially other families of retroviruses and DNA viruses? If so, what counter measures do these viruses utilize to overcome SAMHD1? The Vpx protein is involved in spread of SIV through the population of CD4+ T cells in vivo (Belshan et al., 2012). Does Vpx enhance viral replication in CD4+ T cells in vivo through mechanisms that do not involve SAMHD1? Furthermore, previous studies found that in the early stages of human infection, high levels of HIV-1 replication could be observed in resting CD4+ T cells (Zhang et al., 1999). As SAMHD1 activity should be high in these resting cells, what mechanisms allow HIV-1 replication in resting CD4+ T cells? What are the molecular events by which activation of the SLX4com by Vpr leads to G2/M arrest, and will elucidation of these events provide general insight into the cell cycle? In addition to associating with the MUS81/EME1 endonuclease in the SLX4com, Vpr also associates with the structure-specific ERCC1/ERCC4 endonuclease (Laguette et al., 2014). Does this endonuclease also degrade viral cDNA, or might ERCC1/ERCC4 and MUS91/EME1 have separate functions that contribute to HIV-1 replication? Finally, mutations in SAMHD1 are associated with Aicardi-Goutieres syndrome, a chronic inflammatory disease, and mutations in SLX4 are associated with Fanconi Anemia, a disease involved in susceptibility to cancer and abnormal production of proinflammatory cytokines. Future studies of the dysregulation of these proteins by the primate immunodeficiency viruses are likely to provide insight into these and related human genetic diseases.
Acknowledgements
Research in the Authors’ laboratories is supported by the National Institutes of Health (APR: R21 AI110263, R21 AI114335, R21 AI116173; JTK: R56 AI108467; Baylor-UTHouston CFAR: P30 AI036211). We apologize to colleagues for not citing relevant publications due to the limit on number of references).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen JL, Planelles V. The role of Vpr in HIV-1 pathogenesis. Curr. HIV Res. 2005;3:43–51. doi: 10.2174/1570162052772988. [DOI] [PubMed] [Google Scholar]
- Belshan M, Kimata JT, Brown C, Cheng X, McCulley A, Larsen A, Thippeshappa R, Hodara V, Giavedoni L, Hirsch V, Ratner L. Vpx is critical for SIVmne infection of pigtail macaques. Retrovirology. 2012;9:32. doi: 10.1186/1742-4690-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 2007;3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Lawrence M, Hue S, Neil SJ. G2/M cell cycle arrest correlates with primate lentiviral Vpr interaction with the SLX4 complex. J Virol. 2015;89:230–240. doi: 10.1128/JVI.02307-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja S, Famiglietti M, Bosque A, Planelles V, Rice AP. Cyclin T1 and CDK9 T-Loop Phosphorylation Are Downregulated during Establishment of HIV-1 Latency in Primary Resting Memory CD4+ T Cells. J Virol. 2013;87:1211–1220. doi: 10.1128/JVI.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja S, Ramakrishnan R, Rice AP. Phosphatase PPM1A negatively regulates P-TEFb function in resting CD4T+ T cells and inhibits HIV-1 gene expression. Retrovirology. 2012;9:52. doi: 10.1186/1742-4690-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K, Sung TL, Rice AP. Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4+ T Lymphocytes. J Virol. 2012;86:3244–3252. doi: 10.1128/JVI.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Dehart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Arganaraz ER, Planelles V. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol J. 2007;4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaula-Silva AB, Cassiday PA, Chumley J, Bosque A, Monteiro-Filho CM, Mahon CS, Cone KR, Krogan N, Elde NC, Planelles V. Determinants for degradation of SAMHD1, Mus81 and induction of G2 arrest in HIV-1 Vpr and SIVagm Vpr. Virology. 2015;477:10–17. doi: 10.1016/j.virol.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Hahn BH, Sharp PM, Matsen FA, Emerman M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe. 2013;14:85–92. doi: 10.1016/j.chom.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, III, Brichacek B, Sharova N, Newman MA, Stivahtis G, Sharp PM, Emerman M, Hahn BH, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- Genovesio A, Kwon YJ, Windisch MP, Kim NY, Choi SY, Kim HC, Jung S, Mammano F, Perrin V, Boese AS, Casartelli N, Schwartz O, Nehrbass U, Emans N. Automated genome-wide visual profiling of cellular proteins involved in HIV infection. J Biomol. Screen. 2011;16:945–958. doi: 10.1177/1087057111415521. [DOI] [PubMed] [Google Scholar]
- Gibbs JS, Lackner AA, Lang SM, Simon MA, Sehgal PK, Daniel MD, Desrosiers RC. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Arfi V, Pertel T, Luban J, Lienard J, Rigal D, Darlix JL, Cimarelli A. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol. 2008;82:12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Sharkey ME, Brown CR, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins WR, Hahn BH, Lifson JD, Stevenson M. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei GB, Cheng X, Ramani R, Ratner L. Cyclin L2 Is a Critical HIV Dependency Factor in Macrophages that Controls SAMHD1 Abundance. Cell Host Microbe. 2015;17:98–106. doi: 10.1016/j.chom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Bregnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell. 2014;156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to elongation switch of RNA polymerase II. Nat Struct. Mol Biol. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K, Han YF, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends in Molecular Medicine. 2004;10:525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454-455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R, Dow EC, Rice AP. Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4(+) T lymphocytes. J Leukoc Biol. 2009;86:1345–1350. doi: 10.1189/jlb.0509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Pauls E, Badia R, Torres-Torronteras J, Riveira-Munoz E, Clotet B, Marti R, Ballana E, Este JA. Cyclin D3-dependent control of the dNTP pool and HIV-1 replication in human macrophages. Cell Cycle. 2015 doi: 10.1080/15384101.2015.1030558. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, Skowronski J. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 2008;4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Gelais C, de SS, Hach JC, White TE, Diaz-Griffero F, Yount JS, Wu L. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol. 2014;88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R, Meyerhans A. Quantification of unintegrated HIV-1 DNA at the single cell level in vivo. PLoS One. 2012;7:e36246. doi: 10.1371/journal.pone.0036246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M, Pearson RJ, Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol. 2010;84:6425–6437. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J. Cyclin A2 - CDK regulates SAMHD1 phosphohydrolase domain. J Biol Chem. 2015 doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, Veazey RS, Notermans D, Little S, Danner SA, Richman DD, Havlir D, Wong J, Jordan HL, Schacker TW, Racz P, Tenner-Racz K, Letvin NL, Wolinsky S, Haase AT. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science JID - 0404511. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]