SUMMARY
Fundamental to cellular processes are directional movements driven by molecular motors. A common theme for these and other molecular machines driven by ATP is that controlled release of hydrolysis products is essential to use the chemical energy efficiently. Mechanochemical transduction by myosin motors on actin is coupled to unknown structural changes that result in the sequential release of inorganic phosphate (Pi) and MgADP. We present here a myosin structure possessing an actin-binding interface and a tunnel (back door) that creates an escape route for Pi with a minimal rotation of the myosin lever arm that drives movements. We propose that this state represents the beginning of the powerstroke on actin, and that Pi translocation from the nucleotide pocket triggered by actin binding initiates myosin force generation. This elucidates how actin initiates force generation and movement, and may represent a strategy common to many molecular machines.
Keywords: Molecular motor, chemo-mechanical transduction, force generation, allosteric transition, Pi release
INTRODUCTION
Force production and force sensing in cells is of fundamental importance since mechanotransduction and directed movement are the basis of numerous cellular processes. Cytoskeleton motors interacting with cellular tracks play essential roles in such cellular functions as cell division, cell migration, intra-cellular trafficking and proper formation and maintenance of the cell’s specialized compartments (Hartman et al. 2011; Hirokawa et al. 2009 ; Roberts et al., 2013 ; Franker & Hoogenraad, 2013). Despite extensive investigations, how force and movement is produced by the sequential structural rearrangements of cytoskeleton motors triggered by interactions with their track remains unknown for microtubule-based motors (kinesins and dyneins) and actin-based motors that belong to the myosin superfamily. Currently, data is lacking to reveal how the sequential binding events of a molecular motor to its track can trigger force production. The critical initial track-binding event is linked to the release of inorganic phosphate (Pi) in the case of myosin and dynein molecular motors. In the case of kinesin motors, Pi release controls the end of the force producing state. Thus, how the track controls phosphate release from these molecular motors is directly linked to the force production mechanism.
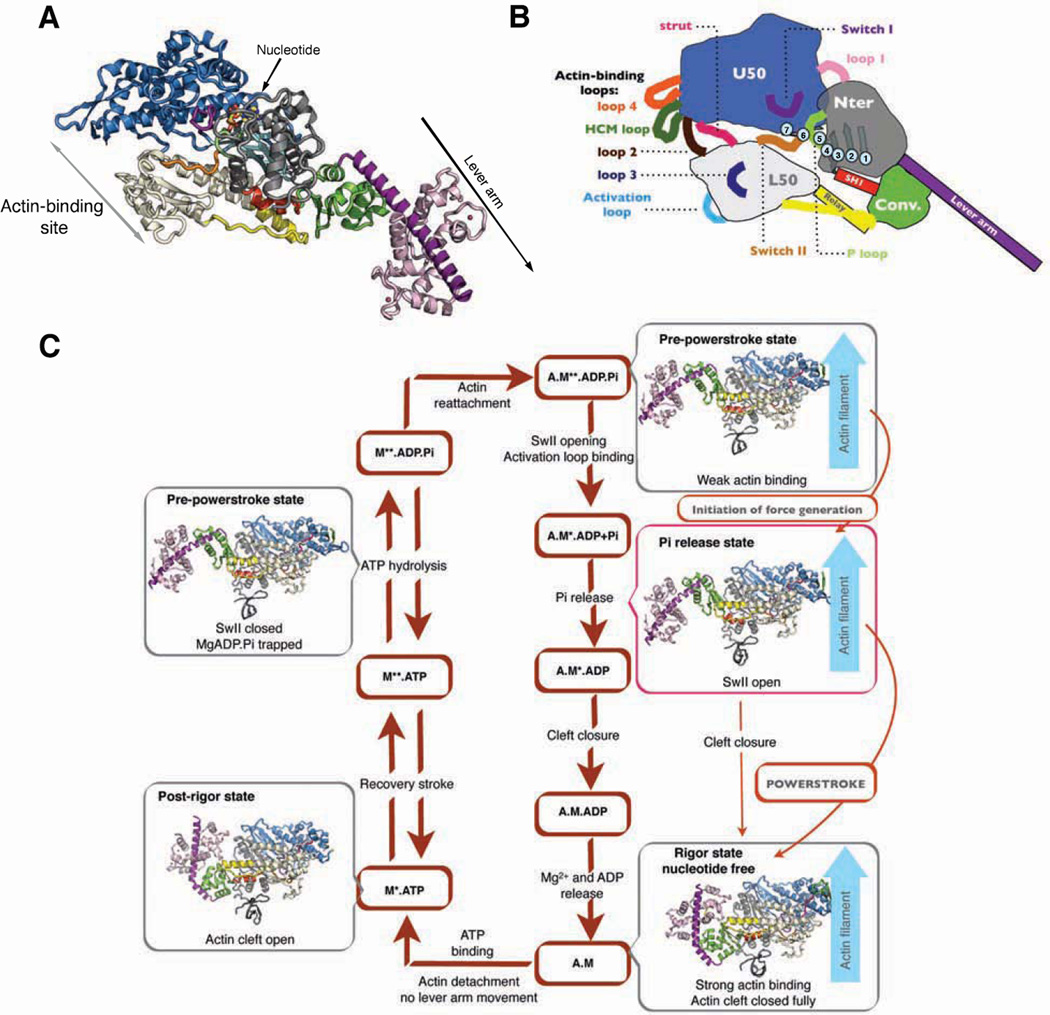
The myosin motor proteins power muscle contraction, as well as movement or force on actin filaments in all eukaryotic cells, via the cyclic interactions between myosin motors and actin filaments. Much progress has been made in understanding the changes in the myosin motor that lead to dissociation from actin by ATP, and the subsequent conformational changes, known as the recovery stroke, that allow hydrolysis of ATP. Following ATP hydrolysis, the myosin motor is in the Pre-powerstroke (PPS) state, and is primed for force production and movement on actin with MgADP and inorganic phosphate (Pi) trapped within the motor. Insights into the subsequent structural changes that actin promotes to generate force and movement are lacking (Sweeney and Houdusse, 2010), although there has been considerable speculation as to how this may occur (Geeves and Holmes, 2005 ; Cecchini et al., 2010 ; Kull and Endow, 2013 ; Preller and Holmes, 2013). There is general agreement that the motor activity of myosin on actin is driven by actin’s ability to catalyze the sequential releases of Pi and MgADP, which are coupled to conformational changes in the myosin motor that allow movement and force generation. What is commonly known as the myosin “head” contains all of the elements necessary for force generation and movement. Figure 1 describes the structural elements of the myosin motor. As also shown in Figure 1, the myosin head can be subdivided into a motor domain, which is the site of both actin-binding and ATPase activity, and the “lever arm”, which consists of the C-terminal subdomain of the motor (converter) followed by an extended helix of variable length containing a number of consensus calmodulin or calmodulin-like light chain binding sites. The myosin lever arm is followed by a coiled coil for two-headed myosin classes. Both single- and two-headed myosin classes contain C-terminal sequences that allow binding of the myosin to its cellular target(s)/cargo(s).
Figure 1. Actin-myosin force-generating cycle and allosteric rearrangements in myosin motors.
(A) Ribbon diagram of the Myosin VI motor domain in the Pi release state depicting the four subdomains: Upper 50 (blue), Lower 50 (white), N-terminal (gray), Converter (green). The Insert 2 (violet) with a bound calmodulin (light pink) is an insertion that repositions the lever arm, acting as a reverse gear.
(B) Schematic representation of Myosin VI showing important nucleotide-binding loops [switch I, switch II, P-loop] and connectors: [SH1 helix, Relay helix, strut] as well as the five actin-binding loops [Loop2, HCM loop, Loop3, Loop4 and Activation loop]. The transducer (light blue) is a β-sheet of seven strands belonging to the Nter and U50 sub-domains. There is an important distortion in the transducer conformation between the Rigor state and the subsequent ATP-bound Post-Rigor and Pre-powerstroke (PPS) states. Between the U50 and L50 subdomains, an internal cleft (so-called 50-kDa cleft) can form either near actin (outer cleft) or near the nucleotide (inner cleft defining the backdoor).
(C) Actomyosin ATPase cycle showing the known structural states of Myosin VI in the force-generating cycle. The motor domain of Myosin VI is depicted in four structural states: Rigor (nucleotide free, on F-actin); Post-Rigor (detached from F-actin, bound to an ATP analog); PPS (bound to ADP.Pi, representing post hydrolysis with ADP.Pi trapped in the active site) and the Pi release state presented in this paper (bound to ADP+Pi or ADP, representing the state in which the Pi can be released). Note that the priming of the lever arm occurs upon the recovery stroke, prior to hydrolysis, when myosin is detached from actin. Quite different structural transitions within the myosin head trigger the powerstroke when myosin is bound on actin, and we propose that these are triggered after Pi release to produce directional movement on F-actin. See also Figure S1.
While the force generation mechanism is conserved, each motor has evolved to perform specific actions and to participate in multiple cellular processes. The rates of the force production transitions on the track and their force sensitivity differ among cytoskeleton motors to tune them for different cellular actions. Structural and functional information on the motor mechanism is thus essential to investigate these differences and to understand the cellular processes in which multiple motors often work in synergy. From the standpoint of delineating the fundamental basis of chemo-mechanical transduction by myosin motors, the actin-induced structural changes represent the most important area to investigate.
Myosin rapidly hydrolyzes ATP in the absence of actin, but rapid product release requires interaction with actin. Once phosphate and ADP have been released, ATP rapidly rebinds to the actin-bound myosin, causing rapid dissociation from actin. All forms of myosin have the same basic kinetic cycle (shown in Figure 1). In the absence of actin, Pi release is quite slow. Soon after the publication of the initial myosin structure (Rayment et al, 1993), it was noted that in order for actin-activated Pi release to precede ADP release, actin would likely create an escape route for phosphate that was an alternative to the normal exit to the nucleotide binding pocket (Yount et al, 1995). There has been much speculation as to the nature of the creation of the back door (Geeves and Holmes, 2005 ; Sweeney and Houdusse, 2010). What is clear is that in order for the phosphate to dissociate, actin binding must cause some rearrangement in either switch I or switch II (Figures 1B and S1). These elements, along with MgADP, block any possible dissociation of Pi. It is not obvious how either element can be induced to move by actin binding. The majority of investigators in the field favor a mechanism in which switch I is pulled away in order to create a phosphate escape route, which is denoted as the side door in Figure S1 (Geeves and Holmes, 2005 ; Cecchini et al., 2010 ; Kull and Endow, 2013).
The appeal of switch I movement creating the exit site is largely based on the bias that switch II rearrangements must be coupled to lever arm movements, as in the recovery stroke transition (Geeves and Holmes, 2005). But either no or minimal lever arm movement should occur until after myosin has bound strongly to actin, in order to allow the maximal possible movement and force generation on actin to occur. Thus models have been proposed in which cleft closure would allow most interactions seen in Rigor to form in the beginning of the powerstroke state, so that myosin with a lever arm in the pre-stroke position would bind strongly prior to either Pi release or lever arm movement (Preller and Holmes, 2013). However, no data exists that suggests that cleft closure can promote an opening of the active site to allow Pi release. A major movement of switch I would result in loss of MgADP as well as Pi, leading to a myosin state that MgATP can rapidly detach from actin. Since myosin with MgADP bound is the primary force bearing/generating state on actin, this would greatly limit force generation.
Herein, we present evidence that there is not formation of the rigor-like actin-interface prior to Pi release, in sharp contrast to prevailing models (Geeves and Holmes, 2005 ; Cecchini et al., 2010 ; Kull and Endow, 2013 ; Preller and Holmes, 2013). We show that the major cleft closure at the actin interface follows Pi release. We present a structure that we propose represents the state that actin stabilizes for the initial release of Pi. In this Pi release state, the lever arm is in its “primed” or pre-stroke position and the escape route for Pi release is created by a movement of switch II, with no movement of switch I. We assess whether this state indeed represents the Pi release state for myosin classes in general by creating a number of mutations and performing kinetic experiments in parallel on class VI, V and II myosins.
This data demonstrates that the initial binding to the track (actin in the case of myosin) induces a tunnel that allows phosphate to translocate away from the active site and exit the protein by first promoting a different type of structural change at the interface with the track, as compared to the conformational change that must follow in order to drive the lever arm swing (powerstroke). The importance of gating of the force producing states by the track and the role of controlled sequential release of ATPase products to couple ATP usage to force production, as we report for myosin, elucidates what may be a general strategy for ATP-powered cellular machines. Namely, that the effector protein induces a conformation that displaces the trapped phosphate from the active site, allowing the mechanical transitions to proceed.
RESULTS
A structural state of myosin with an open phosphate escape route
To create a back door for Pi release, there are only two options. Either switch I can rearrange in such a way as to maintain coordination of MgADP and yet create an escape route for phosphate or switch II must rearrange to create the opening without a major change in the position of the lever arm (Figure S1). A series of crystallization experiments with fragments of myosin VI bound to MgADP yielded a previously unseen structural state that has unexpected attributes and could possibly be the missing Pi release structure (Figure 1A and 2). The structure was determined to 1.75Å resolution (PiR, Table S1). It was also obtained in two different crystal forms (Table S1 and Figure S2A), thus demonstrating that it did not arise from crystal packing. The characteristics of this structural state include the lever arm remaining in a primed position and a new actin interface. The transducer is similar to that found in the PPS state, but movement of the lower 50 (L50) subdomain has opened the inner cleft near where the Pi is trapped in the active site in the PPS state. While the inner cleft has opened, the outer cleft near actin is more closed than in the PPS state.
Figure 2. Features of the Pi release structure.
(A) Hallmarks of the Pi release structure are highlighted in this figure. To open the phosphate release route, switch II adopts a position that allows the converter to remain in its PPS conformation with a primed lever arm. This is in part made possible by a kink in the SH1 helix (red). The SH1 helix links the Nter subdomain (not shown for clarity) and the converter (green) and interacts with the relay helix and loop (yellow), thus playing a central role in the coordination of the movements between these subdomains.
(B) Comparison between the Pre-powerstroke (PPS, grey) and the Pi release (PiR, colour code as in Figure 1) Myosin VI structures, upon superimposition on their U50 and Nter subdomains. The black arrows (1a and 1b) indicate the movement of the L50 subdomain (white) and switch II (orange), necessary for the backdoor opening, giving rise to the PiR structure. In the PiR structure there are no conformational changes in switch I (violet) or the P-loop (lime), and limited change for the converter (green). A 90° rotation shows also these movements in more detail. See also Figure S2 and Table S1.
An important feature of the structure is that switch II has moved by 4Å compared to its position in the PPS structure (Figure 2), opening a possible escape route for Pi, while the switch I and the P-loop positions are unchanged, as is the MgADP coordination (Movie S1). What this structure demonstrates is that a large switch II opening compared to the PPS state, linked to the formation of this actin interface, does not trigger a large change in the lever arm position. As described in Supplemental Information, we obtained a closely related Dictyostelium myosin II structure that also shows that a large switch II movement in myosins can occur without a significant lever arm swing (Figure S2B).
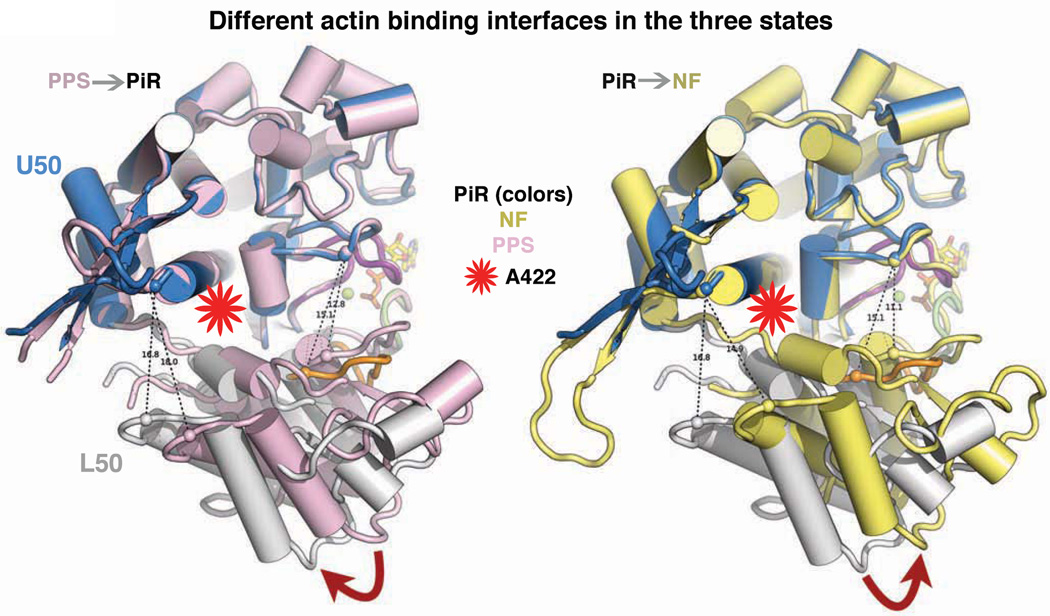
Interestingly, the outer cleft near F-actin is closed to a greater extent than in the PPS state, due to a movement of the L50 subdomain (Figure 3). However, this involves a different relative rotation of subdomains than what is necessary to form the Rigor state (Figure 3). This PiR state thus presents a different actin interface as compared to the PPS state (Figure 3), which may be necessary to allow stereo-specific binding. While it has been proposed that actin binding would close this cleft, it has generally been assumed that cleft closure would occur as seen in Rigor-like structures (Preller and Holmes, 2013) and would be coupled to lever arm movement, thus preceding Pi release (Geeves and Holmes, 2005; Muretta et al., 2013). As demonstrated below, the cleft closure sensed by pyrene-actin quenching occurs after Pi has dissociated from the protein and likely represents a much larger cleft closure than is seen in our putative Pi release state. A recent attempt to use molecular dynamics simulations to explore a state in which the cleft was forced to close in a Rigor-like manner, while not allowing the lever arm to move, resulted in a structure that did not open the inner cleft leaving phosphate trapped (Preller and Holmes, 2013). Thus this simulation did not capture the key features of the PiR structure.
Figure 3. Actin-binding elements and subdomains of myosin in different states.
The actin-binding interface of myosin, which includes elements of the U50 (blue) and L50 (white) subdomains, are presented as viewed from F-actin. Comparison of the PiR state (U50, blue and L50, grey), PPS (pink) state, and Rigor (NF, yellow) state show that these three states have three very different actin interfaces. The outer cleft near F-actin is closed in a greater extent in the PiR state than in the PPS state (left). However the closure of the cleft involves a different rotation of the subdomains than those needed to form the Rigor interface (yellow, right). The position of the A422L mutation, which slows cleft closure, is shown with a red star. See also Figure S3 to visualize how the lever arm stays primed despite these subdomain movements in the motor domain.
In this putative Pi release state, a kink of the SH1 helix occurs and is linked to several changes that allow the N-terminal (Nter) subdomain, the relay, and the SH1 helix to interact in such a way to keep the converter/lever arm primed (Figure S3. The interface between the relay and both the SH1 helix and the converter remain largely the same in the PPS and Pi release states, but a kink of the myosin VI SH1 helix between Val697 and Leu698 allows the formation of new interactions with the Nter subdomain.
Introduction of Pi into the putative Pi release structure
To demonstrate that this structure allows access of phosphate to the active site (i.e. reverse transit of the release), crystals were soaked with 25–100 mM Pi. A series of quick freezing experiments generated three distinct structures. The most rapid freezing (see Methods) gave rise to one of two crystal structures that were unchanged except for the inclusion of Pi. In the first type of crystal, the phosphate was at the exit of the putative Pi release tunnel (referred to as PiR1 in Table S1), coordinated by S153, T197, S203, R205, and E461 (Figure 4A, 4C and S4A left panel). In the second type of crystal (PiR2 in Table S1), the Pi is near ADP (Figure 4A and S4A middle panel). With delays before freezing, the PPS state was reformed with Pi and MgADP trapped. These structures provide strong evidence for the observed tunnel being able to allow phosphate to transit from solution to the active site and vice versa. Thus this tunnel can serve as the Pi escape route (Movie S2). It further demonstrates that Pi reentry to the tunnel promotes closure of the back door and isomerization to the PPS state. This has important implications for the interpretation of a number of experiments in muscle fibers and with myosin Va, as discussed below.
Figure 4. Pi release tunnel.
Opening of switch II and formation of a tunnel allowing the release of the inorganic phosphate (Pi).
(A) Superposition of the Pre-powerstroke (PPS) and the Pi release (PiR) states showing the opening of switch II (orange) while the conformation of switch I (violet) and the P-loop (lime) stay unchanged. The position of the Pi in the structures obtained after Pi soaking is indicated: the Pi release1 (PiR1, red) at the end of the tunnel, the Pi release2 (PiR2, blue) close to ADP and the PPS (cyan, site in which the Pi is generated upon ATP hydrolysis).
(B) Shown are the side chains of residues in the active site (stick representation) and in particular S203, R205, E461 define the Pi release pathway in the PiR state.
(C) Interactions that stabilize the presence of Pi at the end of the putative phosphate release tunnel (PiR1 state). The Pi interacts directly with S153, T197, S203, R205, E461 and via a water molecule with E242.
(D) The residues mutated to test the Pi release tunnel are coloured in red. The S203A was designed to impede Pi entry into the phosphate tunnel seen in PiR; the A458E to add a bulky and charged residue in the tunnel (the red star shows how this mutation will perturb the Pi pathway coloured in cyan) and E152A was designed to remove a bulky and charged residue from the alternative Pi release pathway (Cecchini et al., 2010). See also Figure S4 and Table 1.
As an important control, Post-Rigor crystals (containing MgADP) were also soaked with Pi. No matter how long the crystals were soaked, the Pi was only detected at the end of the tunnel, coordinated by residues S153, T197, R199, S203 and R205 (Figure S4A right panel).
Assessing the putative phosphate release route
Assays of the kinetic cycle of myosin motors on actin (outlined in Figure 1) rely on the existence of probes that report the structural changes. For the release of phosphate, a phosphate binding protein PiBP is used that changes its fluorescence when phosphate is released into the solution and binds the PiBP. Thus its limitation for the current study is that it cannot report upon the translocation of Pi from its position in the PiR2 state described above, to the PiR1 state, but only once the Pi is released from PiR1 binding and moves into the solvent.
To begin to test whether the structure does in fact represent the Pi release state induced by actin binding, we made parallel sets of mutations in our putative Pi release tunnel in myosin VI and in myosin V, as well as in Dictyostelium myosin II (DdII). Myosin VI and myosin V have rapid Pi release, while DdII generally has been thought to have slow, rate-limiting Pi release, as do all studied myosin II isoforms. However, at ultra-low ionic strength, fast Pi release has been reported for DdII as well as for skeletal muscle myosin II (Muretta et al., 2013; White et al., 1997). While these low ionic strength conditions are not of physiological relevance, the observation does suggest that it is the formation of the initial interactions with actin that are weak and rate limiting, rather than Pi release per se. It is particularly important to examine DdII, as its structures have been used to generate proposals of the Pi release mechanism involving a switch I movement (Geeves and Holmes, 2005 ; Cecchini et al., 2010). Using the low ionic strength conditions that allow rapid Pi release also provided the opportunity to observe that the major cleft closure (the so-called weak to strong transition) follows Pi release in myosin II, as it does for Myosin V and VI (Table 1).
TABLE 1.
Kinetics of myosin constructs: rates of Pi release, cleft closure (weak to strong transition), and actin-activated ATPase activity for myosin constructs
| Construct Design |
Myosin (Mutation) |
Phosphate Release (s−1) |
Cleft Closure (s−1) |
Steady State ATPase | |
|---|---|---|---|---|---|
| Vmax (head−1 s−1) |
KATPase (µM) |
||||
| Wild Type motor | Myosin VI | 90±6 | 30±3 | 6±0.1 | 3±0.5 |
| Myosin Va | 143±7 | 38±4 | 17±0.5 | 2±0.4 | |
| Myosin II | 103±31 | 28±4 | - | - | |
| Impede Pi entry into putative phosphate tunnel seen in PiR structure | Myosin VI (S203A) | 43±5 | ND | 2.4±0.1 | 9±1.3 |
| Myosin Va (S217A) | 41±5 | ND | 5.0±0.3 | 11±1.6 | |
| Myosin II (S236A) | 0.2±0.51 | ND | - | - | |
| Bulk and charge in putative phosphate tunnel seen in PiR structure | Myosin VI (A458E) | 29±3 | ND | 1.3±0.1 | 2±0.3 |
| Myosin Va (Y439E) | 117±5 | ND | 4.0±0.4 | 1±0.2 | |
| Myosin II (S456E) | 0.4±0.21 | ND | - | - | |
| Removal of bulk and charge from alternative Pi release pathway (Cecchini et al. 2010) | Myosin VI (E152A) | 94±5 | 20±2 | 1.0±0.2 | 4±0.6 |
| Myosin Va (E164A) | 154±6 | 23±3 | 2.7±0.4 | 3±0.5 | |
| Myosin II (E180A) | 101±51 | 16±3 | - | - | |
| Impede closure of actin-binding cleft to slow “weak to strong” transition | Myosin VI (A422L) | 98±8 | 6±3 | 0.15±0.05 | 15±4 |
| Myosin Va (A402L) | 140±9 | 4±1 | 0.26±0.09 | 10±2 | |
| Myosin II (A420L) | 98±61 | 9±4 | - | - | |
| Reversal of charge in activation loop to slow Pi release state formation | Myosin VI (R521E) | 36±5 | 17±32 | 4.4±0.3 | 25±4 |
| Myosin Va (K502E) | 10±1 | 8±32 | 5.1±0.8 | 15±3 | |
| Myosin II (R520E) | 28 ± 61 | 15±0.51,2 | - | - | |
| Activation loop mutation + Bulk and charge in phosphate tunnel | Myosin Va (K502E + Y439E) | 1.1±0.3 | ND | 0.36±0.1 | 11±3 |
Mean values (±S.D.) of 3–5 independent protein preparations are shown for each construct and condition
ND = Not Determined. Since Pi release precedes cleft closure, for mutations that result in a marked slowing of phosphate release, an apparent slowing of the subsequent rate (cleft closure) is observed even if the true rate is unchanged. Thus we only attempted to measure the cleft closure rate in constructs that had a Pi release rate of ~100/sec or greater. The one exception was for the mutations in the activation loop. The apparent slowing of cleft closure in those cases was consistent with an unchanged actual rate of cleft closure.
Low ionic strength buffer (0.4mM MgCl2; 1mM MOPS pH=7.0). Note that the steady state assay used in this study cannot be performed at this low ionic strength (noted by “−”).
Apparent rate (follows Pi release), consistent with actual rate being ~30sec−1
The first mutation, S203A in myosin VI, was designed to either impede entry of the Pi into the tunnel (serine 203 in myosin VI plays the role of guiding the Pi away from the active site). We also attempted to impede the exit of Pi from the tunnel via introducing a combination of bulk and repulsive charge (A458E in myosin VI). These two myosin VI mutations correspond to S217A and Y439E in myosin Va, and S236A and S456E in DdII. A summary of the kinetic results from the mutations, as well as the rationale for each mutation, are given in Table 1.
A structure of the myosin VI Pi release state (PiR) with the A458E mutation was also obtained. Figure 4 illustrates the Pi tunnel (with Pi in two positions) with these altered residues. The structure of the A458E mutant also reveals that there is sufficient room for the E side chain to assume an alternative, non-blocking position. Thus we would expect this mutation to slow, rather than prevent Pi release, and that the magnitude of the effect may be context (i.e. isoform) dependent. This is consistent with the data shown in Table 1.
The results in Table 1 are all consistent with the Pi release route revealed in the PiR structure being the Pi release route used by myosins II, V and VI when they interact with actin. Previous studies on the S217A mutation in myosin V revealed that it slows hydrolysis as well as Pi release, thus slowing the overall cycling rate (Forgacs et al., 2009). The most convincing mutation is the one that was created to slow Pi moving into the putative channel by a combination of bulk and repulsive charge (A458E, Y439E and S456E for myosin VI, V and DdII), which is depicted in Figure 4D). The largest effect was in myosin II at low ionic strength, where Pi release was slowed more than 100-fold, followed by myosin VI with a 3-fold slowing, while myosin Va showed only a modest slowing of about 20%. We noted that the mutations tended to slow the steady state actin-activated ATPase (Table 1) of myosin V and VI, which appears to be the result of slowing ADP release from myosin bound to actin in addition to Pi release (myosin VI WT=6.0±0.1/sec, A458E=1.3±0.1/sec; myosin V WT=17.0±0.5/sec, Y439E=4.0±0.4/sec).
The modest effect of the Y439E mutation on Pi release for myosin V may relate to the relatively high affinity of myosin V in its “weak” binding states (Yengo et al., 2002), which could allow it to dwell on actin long enough in the Pi release conformation for a rearrangement of the glutamate side chain position to unblock the Pi release tunnel. To further assess this conjecture, we weakened the binding of myosin V by introducing a mutation in the activation loop (K502E, discussed below), and then introduced the Y439E mutation. In this context the glutamate slowed Pi release approximately 10-fold (Table 1). Note that results with this switch II mutation are not consistent with a switch I / P-loop movement occurring to allow Pi release, as previously proposed (Geeves and Holmes, 2005; Cecchini et al., 2010). In this model, Pi release should not be affected by this switch II mutation since this residue does not impact switch I / P-loop movements and it is positioned far from the proposed alternative Pi exit route.
To further test this alternative Pi release route, we made a mutation (E152A in myosin VI, E164A in myosin V, and E180A in DdII) that had been suggested by molecular dynamics simulations (Cecchini et al., 2010). As discussed above, rearrangement of switch I that would maintain MgADP coordination would be necessary if this were the case. This mutation had no affect on Pi release (Table 1), consistent with there being no major rearrangement of switch I prior to MgADP release. Interestingly, removal of this glutamate residue slows ADP release (Table 1), which could indicate a role for promoting inner cleft closure necessary for ADP release.
Cleft closure must follow phosphate release
The major cleft closure that leads to what has been characterized as strong actin binding is detected in kinetic experiments as a quenching of a pyrene label on actin (De La Cruz et al., 1999). The transition that is sensed is known as the weak to strong transition on actin. Since the labeled residue on actin (Cys374) is thought to be outside of the actin-myosin interface, this pyrene signal may indicate that as the cleft in myosin fully closes and allow a wider interaction surface with the track, the F-actin filament structure is also perturbed. This pyrene-quenching cleft closure precedes the release of MgADP (Sweeney and Houdusse, 2004), and thus the major cleft closure on actin occurs prior to the formation of the Rigor state. To provide strong evidence that this major cleft closure occurs after Pi has been released, we designed mutants that should slow cleft closure by introducing a bulky side chain (leucine) into the myosin outer cleft near the strut and the actin-binding site (A422L, A402L and A420L for myosin VI, V and DdII) (Table 1). This mutation should interfere with formation of Rigor-like cleft closure, but not the rather different cleft closure seen in the PiR structure (Figures 3 and 5). Importantly, these mutations did not affect the Pi release rate, but greatly slowed the rate of pyrene-actin quenching (weak to strong transition), consistent with the major closure at the outer cleft not being necessary for Pi release and occurring subsequent to Pi release, during the transition that may be coupled to lever arm movement (as will be investigated in experiments described below). Interestingly, the steady state actin-activated ATPase was slowed to a much greater extent than was the rate of pyrene-actin quenching, suggesting that multiple actin-associated transitions are affected by this cleft mutation
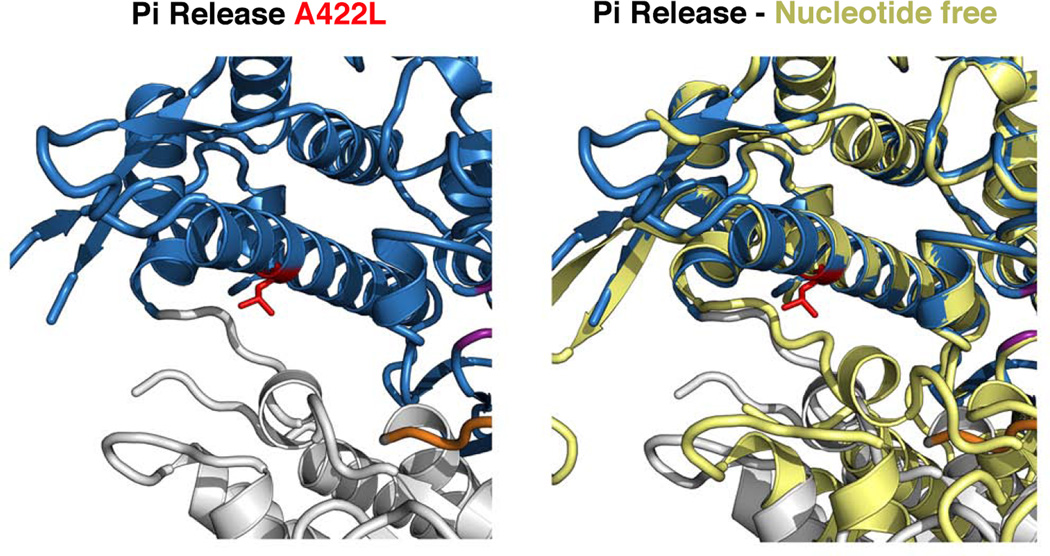
Figure 5. Impairment of cleft closure in the Myosin VI A422L mutant.
Note that the mutant side chain of L422 (red) which belongs to the HO helix of the U50 subdomain doesn’t impair the movement required to populate the Pi release state, in which the closure of the outer cleft is drastically different from that found in Rigor. In contrast, the position of the strut and the L50 subdomain found in the Rigor state (nucleotide-free) drastically differ upon full closure of the cleft. A clash occurs between the L422 side chain and the new position of the strut and slows the transition required to close the cleft.
Surface loops allow formation of the phosphate release state
Binding to actin is necessary to stabilize this Pi release state since myosin does not normally release Pi at a high rate in the absence of actin. The docking on actin of the myosin PPS state is initiated by non-stereo-specific, electrostatic interactions. Exploration of the actin surface guided by electrostatic interactions must catalyze the formation of stereospecific actin binding of the PiR state, promoting release of Pi. From a variety of experimental data, a number of the loops on the myosin surface could create these initial electrostatic interactions with actin (Sasaki et al., 1999; Wang et al., 2000; Joel et al., 2001; Onishi et al., 2006). For this study, we focused on one of the actin-binding loops that has previously been implicated in triggering the initiation of force generation, and has been termed the activation loop (Várkuti et al., 2012). This loop is in the L50 subdomain, interacts with the N-terminus of actin, and is in position to help create an interface for our Pi release structure with actin, possibly promoting the L50 subdomain rotation away from its PPS position. We created a mutation that was previously shown (Várkuti et al., 2012) to interfere with force generation in myosin II and Myosin V (R520E in DdII, K502E and R521E in myosin V and VI). In all of the myosin classes, this mutation greatly slowed the rate of Pi release (Table 1), consistent with the activation loop playing a role in stabilization of the PiR state on actin and thus in the initiation of force generation. The mutations in this loop did not appear to impact either the actual rate of cleft closure (although the apparent rate was slowed; Table 1), or the rate of ADP release from myosin bound to actin for myosin VI (WT=6.0±0.1/sec vs. R521E=6.3±1.1/sec) or myosin V (WT=17.0±0.5/sec vs. K502E=15.9±1.8/sec).
DISCUSSION
The Pi release (PiR) state that we present has all of the hallmarks that are necessary for initiation and optimization of myosin force generation, including creating a different actin binding interface than in the PPS state, that allows Pi release to occur before the bulk of the lever arm movement, while maintaining high affinity for MgADP. What is revealed by the structure is that while there is some degree of cleft closure at the outer cleft near actin, this occurs differently than is necessary for the major cleft closure seen in Rigorlike structures (Figure 3). The cleft position has similarities with that found in the Post-Rigor state, but is formed by a different switch II movement. The changes at the actin interface in the transition from the PPS to the Pi release state are achieved by a coordinated movement of switch II and the L50 subdomain from their PPS positions. A large shift in switch II position creates an opening for Pi release while having a minimal impact on the lever arm position, which remains in a “primed” position. Note that different switch II positions in the Rigor and Post-Rigor states allow the lever arm to remain in the post-stroke position upon detachment from actin, even though the motor is undergoing major internal rearrangements (Coureux et al., 2003 and 2004). Thus switch II controls lever arm position, but not in a simple two-state manner (“in” and “out”), as is often described in the literature (Reubold et al., 2003).
While our structural data is based primarily on myosin VI, which is a reverse-direction myosin (Wells et al., 1999), the reversal of directionality is due to the unusual myosin VI lever arm and not to changes within the motor. When the unusual lever arm is removed or replaced, the core motor of myosin VI is revealed to be a plus-ended motor (Bryant et al., 2007; Park et al., 2007), and undoubtedly shares the common motor mechanism found in all myosin classes. Further support for the generality of this Pi release state comes from our demonstrating that Dictyostelium myosin II can also form this state (Figure S2B) and by the fact that mutations in class VI, V and II myosins have similar impact on Pi release.
The emerging view is that myosin is initially bound to actin via electrostatic interactions with loop 2, and likely other flexible surface loops, such as the HCM loop (Onishi et al., 2006; Sasaki et al., 1999; Wang et al., 2000; Liu et al., 2005). Once these loops orient the myosin PPS state on actin, presumably mostly via electrostatic interactions (given the acceleration of Pi release from myosin II at extremely low ionic strength), actin can trigger Pi release by inducing the transition to the more strongly and stereo-specifically bound Pi release state. Formation of the stereo-specific binding interface involves hydrophobic residues, including the actin-binding region (helix-loop-helix) in the L50 subdomain (Kojima et al., 2001; Sasaki et al., 2002), which may initially be positioned by the activation loop. This loop indeed plays an important role for promoting the transition that populates the Pi release state on actin and thus accelerates release of Pi (Table 1; Várkuti et al., 2012). What the Pi release structure reveals is that the sequential myosin-actin interactions during the powerstroke promote different states of the motor that are initiated and gated by the control of Pi release. Pi release must occur only upon stereo-specific binding to actin, and must trigger subsequent rearrangements that control force production and lever arm movement.
In comparing myosin Va, which releases Pi rapidly on actin, to myosin II, which releases Pi slowly except at extremely low ionic strength, it has been shown that the weak interactions with actin in the PPS state are stronger in the case of myosin Va (Yengo et al., 2002). This is consistent with the assertion that the loop interactions are critical for stabilizing binding of the PPS state promoting the subsequent transition to the PiR state, triggering Pi release. While the general features of the PiR state are likely conserved within the family, how much of the tuning of the rates occurs via the actin binding loops or the internal sequence of the motor remains to be studied and will elucidate how the cellular motors are perfectly suited to their precise action. Interestingly, Pi release is the step affected for the deafness R156W mutation in Myo1c (Lin et al., 2011). Structural insights can now explain how this mutation affects the duty ratio, since the tryptophan side chain slows down the opening of the backdoor.
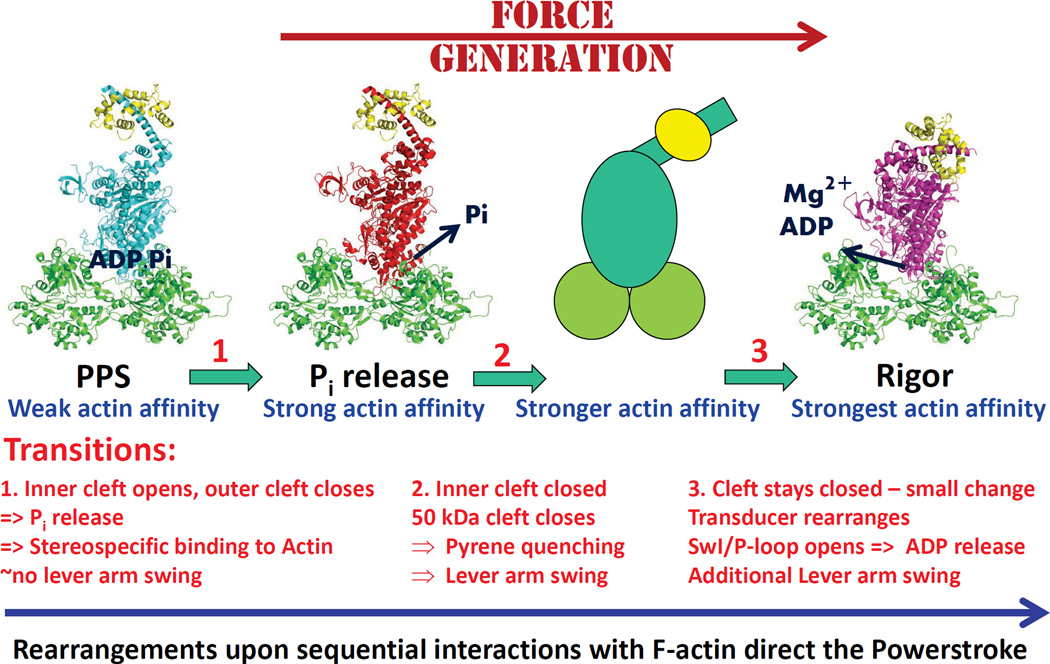
As illustrated in (Figure 6), force production would begin when a myosin head attached to actin in the PPS state via ionic interactions transitions to the PiR state, which then binds in the first stereo-specific, force-bearing state on actin. The PiR state is likely equivalent to the state first described by Sleep and Hutton (1980), in which force production is initiated and Pi rebinding is possible (Caremani et al., 2008). In the absence of strain, Pi release is virtually irreversible due to the next structural transition involving cleft closure coupled to lever arm movement. In this scheme, the difference between a myosin that releases Pi slowly on actin, such as Dictyostelium myosin II, and a myosin that releases Pi rapidly, such as myosin Va or myosin VI, is based on the affinity that these myosin loops mediate for F-actin as well as their influence on the equilibrium between the PPS state and PiR state when the myosin is bound to actin. Thus variability in the nature of the myosin surface loops strongly influences the rate of entry into the force generating states.
Figure 6. Model for myosin force generation.
Shown are the structural transitions that underlie chemo-mechanical transduction by myosin. With the description of the PiR state, herein, only the structural changes associated with the strong ADP-binding, actin bound state remains to be elucidated.
Creation of a strong binding actin interface
While the pyrene probe at Cys374 on actin has been used to monitor the so-called “weak to strong” transition on actin, we propose it reports on the major cleft closure, but not the initial formation of a strong actin binding state, which would be the beginning of force generation. We propose that the initial closure of the outer cleft and strong binding to actin occurs upon formation of the PiR state. This initial strong binding state forms with minimal movement of the lever arm. The major movement of the lever arm (powerstroke) is coupled to the further cleft closure following Pi release, and presumably cannot occur until Pi moves away from the active site. In this regard, Pi at the active site may act as a wedge to prevent Rigor-like cleft closure. This mechanism would also prevent the post-Rigor, myosin.MgATP state from reforming a strong actin interface due to the presence of the gamma-phosphate in the active site. Note that the myosin.MgADP Post-Rigor state can readily undergo the weak to strong transition on actin.
It is also clear that once Pi is released and the weak to strong transition occurs, the resulting myosin.MgADP complex affinity for actin is not as high as is the Rigor binding affinity (De La Cruz et al., 1999) suggesting that until the MgADP coordination is broken and the active site is completely open (Coureux et al., 2004), the cleft cannot completely close. Thus a model emerges of at least three distinct types of cleft closure associated with strong binding to actin, the one seen in the Pi release state, the one yet to be seen at high resolution in the strong MgADP-binding state, and the one that has been shown for Rigor (Coureux et al., 2003). In order to drive the cycle forward, the affinity of each of these states for actin should increase, beginning with Pi release and ending with Rigor.
The three distinct cleft conformations described above are likely coupled to distinct positions of the myosin lever arm. To optimize force generation, only a small lever arm movement should occur prior to strong binding of actin in the state necessary to initiate Pi release. The remainder of the power stroke would then occur in at least two discrete movements in the absence of load. A large movement would be expected to accompany the major cleft closure at the actin interface (i.e. the pyrene-actin quenching transition), and there is a well-documented second swing upon the release of MgADP (Whittaker et al., 1995; Veigel et al., 2002). Our Pi release structure has a slight repositioning of the myosin lever arm as compared to the PPS structure. The magnitude of this movement may vary among myosin isoforms, given that there is variability in the positioning of the lever arm in the PPS state (Houdusse et al., 2000; Kollmar et al., 2002; Münnich et al., 2014), but in all cases the bulk of the powerstroke should follow formation of the PiR state.
The general concept of the lever arm swing following Pi release fits well with single molecule studies of either skeletal muscle myosin II (Takagi et al., 2006) or myosin Va (Sellers and Veigel, 2010) undergoing rapid feedback to counter movement. These experiments suggest that the Pi release step is only reversible when the lever arm swing is prevented. Furthermore, kinetic studies of dimeric myosin V demonstrated that Pi is released from the lead head of a dimer at essentially the unstrained rate, while MgADP release from the lead head is greatly slowed (Rosenfeld and Sweeney, 2004). Lastly, muscle fiber studies suggest that the Pi release step is only reversible if the myosin lever arms are at a position near the beginning of the powerstroke (Dantzig et al., 1992; Caremani et al., 2008). All of these results support the concept that the major component of the power stroke follows Pi release.
Initiation of Lever Arm movement
We are postulating that Pi occupancy of the nucleotide pocket near the MgADP (PiR2 in Table S1, Figures 4A and S4A) prevents full cleft closure and the lever arm movement associated with the powerstroke. We further postulate that once the PiR2 state forms on actin, the Pi can rapidly translocate through the phosphate tunnel and occupy the position at the mouth of the Pi release tunnel, as seen in the structure referred to as PiR1 in Table S1 and Figure 4A. Once in this position, it is likely that the motor can begin to close the cleft to form a stronger interface with actin with a concomitant movement of the lever arm (i.e. the beginning of the powerstroke). This would make the Pi release essentially irreversible in the absence of load. As discussed above, if the lever arm is prevented from moving, then the Pi release tunnel would remain open allowing Pi to return to the active site.
Note that our Pi release assay only detects the Pi once it is in solution, having left its position at the mouth of the release tunnel. If the structural rearrangements that begin to close the actin cleft and move the lever arm are more rapid than the movement of Pi from its position in the PiR1 state, then it could appear that lever arm movement and cleft closure precede Pi release, but this would simply reflect the limitations of our assays. There is a report (Muretta et al. 2013) that suggests that Pi is released after the major lever arm movement. While they interpret this as evidence for a lever arm swing gating Pi release, we argue that it may simply reflect that it is the movement of Pi from its PiR2 to its PiR1 positions in the PiR state that gates lever arm movement, but that this is not directly detected by the phosphate release protein assay which only senses the Pi once it is in solution. Since the lever arm swing they detect likely is accompanied by cleft closure, but is much faster that the weak to strong transition on actin, it is unclear if the pyrene actin detects a further closure of the cleft accompanied by additional movement of the lever arm, or it could represent a rearrangement of the actin that follows myosin cleft closure that allows an even stronger actin-myosin interface to form. Clarification of these points will require additional kinetic probes of myosin structural rearrangements.
Note that our model based on the structural and kinetic data in this paper differs significantly from a recent molecular dynamics simulation of the beginning of the powerstroke (Preller and Holmes, 2013). In this simulation, the cleft near actin was forced into a Rigor-like interface, the lever arm remained close to the primed position seen in PPS and both Pi and MgADP remained trapped at the active site. We demonstrate in fact that an actin interface much different from the Rigor interface must first form to allow departure of Pi from the active site, and that subsequent to this event, further cleft closure coupled to lever arm movement occur promoting formation of the Rigor actin-binding interface. Thus in our model, Pi translocation away from the active site gates the cleft closure that is coupled to the major movement of the lever arm, which is known as the powerstroke.
Conclusion
Our study highlights a mechanism for opening the escape route for Pi that might also be used for other cellular machines powered by ATP and GTP. It differs from the mechanism supported by studies of motors such as F1ATPase (Menz et al., Cell 2001), in which a rearrangement of the central beta sheet must occur. Lessons from myosin could shed light on models for dynamins or ATPases, such as FlaI (Reindl et al., 2013), in which binding to partners gates and precedes force production.
Given all of the data, we assert that the structural state of myosin that we present in this paper is in fact the Pi release state, the first actin bound state in force generation. This study provides insights into how force production is controlled and tuned in the myosin superfamily. Visualizing this structure is necessary for understanding how myosin motors choose their cellular track, and how they perform so different cellular actions. It provides essential information to account for the effect of numerous mutations that have been reported in human genetic disorders in more than 12 myosin classes, including cardiomyopathy (cardiac myosin) and microvillus disease (myosin 5b). This structure also provides a framework to rationally design drugs that can slow, block or accelerate force production in myosins that have been mis-tuned by disease-causing mutation.
EXPERIMENTAL PROCEDURES
Expression Constructs, Production and Purification
Recombinant DNA of porcine myosin VI, chicken myosin Va or Dictyostelium myosin II were generated to express various truncated myosin constructs containing the motor domain of these myosins using the baculovirus expression system. For the Pi release myosin VI crystal structures, a C-terminal truncation was made corresponding to I789 (MD). This truncation is at the end of the first (proximal) helix of insert 2, and precedes the CaM-binding site of insert 2. All the kinetic studies described with myosin VI were performed from WT or point-mutation introduced in the construct (MDins2) truncated at the end of insert 2, after residue A816, prior to the CaM-binding IQ motif. For the soaking of Post-rigor crystals, the construct (MDins2-delta.ins1) was used since it crystallizes in this state. In this MDins2 construct, insert 1 (residues C278– A303) was removed, as previously described (Ménétrey et al, 2005; Ménétrey et al, 2008). Each of these three constructs (MD, MDins2, MDins2-delta.ins1) had a Flag tag (encoding DYKDDDDK) appended via a glycine to the N-terminus to facilitate purification. For the Dictyostelium MD, the DNA corresponds to the construct truncated after the codon corresponding to R761 with a C-terminal flag. All kinetic studies described for myosin Va were performed from WT or point-mutation introduced in the chicken Myosin Va construct (MD1IQ) truncated after the codon corresponding to R792 with a C-terminal flag, as previously studied (De la Cruz et al. 1999). This construct encompassed the motor domain and the first light chain/calmodulin-binding site of myosin Va. The myosin Va-expressing virus was co-infected with a virus encoding a truncated human essential light chain (LC-1sa) (De la Cruz et al. 2000) as previously described (Coureux et al 2003). These constructs were used to create recombinant baculoviruses for expression in SF9 cells, as previously described (De La Cruz et al, 1999). All of the expressed myosin molecules were purified as previously described (Sweeney et al, 1998).
Crystallisation and data collection
Crystals of myosin VI in the Pi release state (PiR) were obtained with the MD (1–789) construct using the hanging-drop vapor-diffusion method. Spontaneous nucleation occurred at 277K with equal amounts of reservoir solution containing 6.25% PEG8k, 50 mM TRIS, pH 8.5, 1mM TCEP, 3% Glycerol and stock solution of the protein (10 mg.ml-1 in 10 mM HEPES pH 7.5, 50 mM NaCl, 1 mM TCEP, 1 mM NaN3 with 2 mM Mg2+ADP). The best crystals were obtained using seeding approaches. Further details of the experiments performed for this study are indicated in the extended experimental procedures. Crystals of proteins were obtained and were cryo-cooled prior to data collection at either the ESRF or SOLEIL Synchrotron beamlines. The data sets were processed with XDS (Kabsch, 2010). Statistics on the data collection for five new Pi release myosin VI structures, one Post-Rigor structure frozen after long Pi soaking, one PPS myosin VI structure obtained after long Pi soaking of a Pi release myosin VI crystal, as well as for a Dictyostelium R238E, E459R myosin II structure, are indicated in Table S1. Statistics of the final models are also summarized in Table S1. Details of structure determination, model building and refinement are indicated in extended experimental procedures.
Kinetic experiments
For the transient kinetic experiments, which are essentially single turnover experiments, the general strategy can be summarized as follows. For the measurement of Pi release, Pi that is released from myosin is detected by binding to a phosphate binding protein that is labeled so that a change in fluorescence is observed upon Pi binding (White et al., 1997). Turnover is inhibited after the initial transient by including MgADP in the final mix. Cleft closure is detected by the quenching of a pyrene label on actin. However, this rate can only be accurately measured if phosphate release, which as demonstrated in this study, precedes cleft closure, is much faster than cleft closure as it is in wild type myosin V and myosin VI. Again, turnover is inhibited after the initial transient by including MgADP in the final mix. In general, we only attempted this measurement for constructs that displayed a Pi release rate of 100/sec or greater. The exception was for mutations in the activation loop (Table 1), which greatly slowed Pi release and the apparent rate of cleft closure, but the calculated rate of cleft closure (assuming the observed rate is limited by both Pi release and cleft closure) is similar to the wild type rate. The rate of ADP release from the actin-myosin complex was measured for a subset of constructs by binding mantADP to the myosin, then competing it off with unlabeled ADP and measuring the rate of fluorescence decrease as the mant signal was quenched by exposure to solvent.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiaoyan Liu for the expression and purification of the protein for structural studies. We thank Pierre Legrand and Andy Thompson as well as beamline scientists of PX1 (SOLEIL synchrotron), ID23-2 and ID29 (ESRF synchrotron) for excellent support during data collection. A.H. was supported by grants from the CNRS, FRM équipes, ANR blanche BLAN07 and BLAN10, Ligue contre le cancer and ARC subvention fixe. HLS was supported by NIH grants DC009100 and HL110869. The AH team is part of Labex CelTisPhyBio 11-LBX-0038.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The atomic coordinates and structure factors of eight structures have been deposited in the Protein Data Bank, www.pdb.org, with accession numbers 4PFO (PiR), 4PFP (PiR-P21), 4PJN (PiR1), 4PJM (PiR2), 4PK4 (PPS-0), 4PJJ (PR-Pi), 4PJL (PiR-A458E), 4PJK (Dy-R238E.E459R–PiR), see Table S1.
AUTHOR CONTRIBUTIONS
P.L., T.I. and L.S. contributed equally to this work. P.L. and T.I. determined the structures and analyzed them with A.H. Biochemical experiments were performed by V.R., H.B., S.S. and C.K.; C.M., L.S. and B.Z performed kinetic studies. A.H. and H.L.S. conceived the project, oversaw the experiments, analyzed them, and wrote the manuscript.
REFERENCES
- Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci U S A. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caremani M, Dantzig J, Goldman YE, Lombardi V, Linari M. Effect of inorganic phosphate on the force and number of myosin cross-bridges during the isometric contraction of permeabilized muscle fibers from rabbit psoas. Biophys. J. 2008;95:5798–5808. doi: 10.1529/biophysj.108.130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini M, Alexeev Y, Karplus M. Pi release from myosin: a simulation analysis of possible pathways. Structure. 2010;18:458–470. doi: 10.1016/j.str.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux P-D, Wells A, Ménétrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A. A structural state of the myosin V motor without bound nucleotide. Nature (London) 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- Coureux P-D, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzig JA, Goldman YE, Millar NC, Lacktis J, Homsher E. Reversal of the cross-bridge force-generating transition by photogeneration of phosphate in rabbit psoas muscle fibres. J. Physiol. 1992;451:247–278. doi: 10.1113/jphysiol.1992.sp019163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- Forgacs E, Sakamoto T, Cartwright S, Belknap B, Kovács M, Tóth J, Webb MR, Sellers JR, White HD. Switch 1 mutation S217A converts myosin V into a low duty ratio motor. J. Biol. Chem. 2009;284:2138–2149. doi: 10.1074/jbc.M805530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker MA, Hoogenraad CC. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–155. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel PB, Trybus KM, Sweeney HL. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J. Biol. Chem. 2001;276:2998–3003. doi: 10.1074/jbc.M006930200. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintses B, Gyimesi M, Pearson DS, Geeves MA, Zeng W, Bagshaw CR, Málnási-Csizmadia A. Reversible movement of switch 1 loop of myosin determines actin interaction. EMBO J. 2007;26:265–274. doi: 10.1038/sj.emboj.7601482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Konishi K, Katoh K, Fujiwara K, Martinez HM, Morales MF, Onishi H. Functional roles of ionic and hydrophobic surface loops in smooth muscle myosin: their interactions with actin. Biochemistry. 2001;40:657–664. doi: 10.1021/bi0011328. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Dürrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. EMBO J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J. Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd. New York: Springer; 2006. pp. 443–468. [Google Scholar]
- Liu X, Shu S, Kovács M, Korn ED. Biological, Biochemical and Kinetic effects of mutations of the Cardiomyopathy loop of Dictyostelium Myosin II. J. Biol. Chem. 2005;280:26974–26983. doi: 10.1074/jbc.M504453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétrey J, Llinas P, Cicolari J, Squires G, Liu X, Li A, Sweeney HL, Houdusse A. The Post-rigor structure of myosin VI and implications for the recovery stroke. EMBO J. 2008;27:244–252. doi: 10.1038/sj.emboj.7601937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- Münnich S, Taft MH, Manstein DJ. Crystal structure of human myosin 1c--the motor in GLUT4 exocytosis: implications for Ca2+ regulation and 14-3-3 binding. J Mol Biol. 2014;426:2070–2081. doi: 10.1016/j.jmb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Muretta JM, Petersen KJ, Thomas DD. Direct real-time detection of the actin-activated power stroke within the myosin catalytic domain. Proc. Natl. Acad. Sci. USA. 2013;110:7211–7216. doi: 10.1073/pnas.1222257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi H, Mikhailenko SV, Morales MF. Toward understanding actin activation of myosin ATPase: the role of myosin surface loops. Proc. Natl. Acad. Sci. USA. 2006;103:6136–6141. doi: 10.1073/pnas.0601595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Li A, Chen LQ, Houdusse A, Selvin PR, Sweeney HL. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc Natl Acad Sci U S A. 2007;104:778–783. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preller M, Holmes KC. The myosin start-of-power stroke state and how actin binding drives the power stroke. Cytoskeleton. 2013;70:651–660. doi: 10.1002/cm.21125. [DOI] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Bäde K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, Manstein DJ. A structural model for actin-induced nucleotide release in myosin. Nat. Struct. Biol. 2003;10:826–830. doi: 10.1038/nsb987. [DOI] [PubMed] [Google Scholar]
- Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche AL, Albers SV, Tainer JA. Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol. Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Asukagawa H, Yasuda R, Hiratsuka T, Sutoh K. Deletion of the myopathy loop of Dictyostelium myosin II and its impact on motor functions. J. Biol. Chem. 1999;274:37840–37844. doi: 10.1074/jbc.274.53.37840. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Ohkura R, Sutoh K. Dictyostelium myosin II as a model to study the actin-myosin interactions during force generation. J. Muscle Res. Cell Motil. 2002;23:697–702. doi: 10.1023/a:1024415409406. [DOI] [PubMed] [Google Scholar]
- Sellers JR, Veigel C. Direct observation of the myosin-Va power stroke and its reversal. Nat. Struct. Mol. Biol. 2010;17:590–595. doi: 10.1038/nsmb.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleep JA, Hutton RL. Exchange between inorganic phosphate and adenosine 5’-triphosphate in the medium by actomyosin subfragment 1. Biochemistry. 1980;19:1276–1283. doi: 10.1021/bi00548a002. [DOI] [PubMed] [Google Scholar]
- Sun M, Oakes JL, Ananthanarayanan SK, Hawley KH, Tsien RY, Adams SR, Yengo CM. Dynamics of the upper 50-kDa domain of myosin V examined with fluorescence resonance energy transfer. J. Biol. Chem. 2006;281:5711–5717. doi: 10.1074/jbc.M508103200. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Rosenfeld S, Brown F, Faust L, Smith J, Stein L, Sellers J. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J. Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. The motor mechanism of myosin V: insights for muscle contraction. Phil. Trans. R. Soc. Lond. B. 2004;359:1829–1842. doi: 10.1098/rstb.2004.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. Structural and Functional Insights into the Myosin Motor Mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várkuti BH, Yang Z, Kintses B, Erdélyi P, Bárdos-Nagy I, Kovács AL, Hári P, Kellermayer M, Vellai T, Málnási-Csizmadia A. A novel actin binding site of myosin required for effective muscle contraction. Nat. Struct. Mol. Biol. 2012;19:299–306. doi: 10.1038/nsmb.2216. [DOI] [PubMed] [Google Scholar]
- Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol. Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Wang F, Harvey EV, Conti MA, Wei D, Sellers JR. A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA, Sweeney HL. A 35Å movement of smooth muscle myosin on ADP release. Nature. 1995;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- Yount RG, Lawson D, Rayment I. Is myosin a “back door” enzyme? Biophys. J. 1995;68:44S–47S. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.