Abstract
Understanding biological diversity elevational pattern and the driver factors are indispensable to develop the ecological theories. Elevational gradient may minimize the impact of environmental factors and is the ideal places to study soil microbial elevational patterns. In this study, we selected four typical vegetation types from 1000 to 2800 m above the sea level on the northern slope of Shennongjia Mountain in central China, and analysed the soil bacterial community composition, elevational patterns and the relationship between soil bacterial diversity and environmental factors by using the 16S rRNA Illumina sequencing and multivariate statistical analysis. The results revealed that the dominant bacterial phyla were Acidobacteria, Actinobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Verrucomicrobia, which accounted for over 75% of the bacterial sequences obtained from tested samples, and the soil bacterial operational taxonomic unit (OTU) richness was a significant monotonous decreasing (P < 0.01) trend with the elevational increasing. The similarity of soil bacterial population composition decreased significantly (P < 0.01) with elevational distance increased as measured by the Jaccard and Bray–Curtis index. Canonical correspondence analysis and Mantel test analysis indicated that plant diversity and soil pH were significantly correlated (P < 0.01) with the soil bacterial community. Therefore, the soil bacterial diversity on Shennongjia Mountain had a significant and different elevational pattern, and plant diversity and soil pH may be the key factors in shaping the soil bacterial spatial pattern.
Introduction
Understanding patterns of biological diversity along elevational gradients and the factors driving such patterns are indispensable to gaining a comprehensive understanding of the response of ecosystems to global climate change, aid in determining the broad scale distributions of species and in the development ecological theories (Lomolino, 2001; Rahbek, 2005; Malhi et al., 2010; Wang et al., 2011). In the past, most studies have focused on plants and animals (Lomolino, 2001; McCain, 2005; Renaud et al., 2009), although soil microbes are abundant and play important roles in ecosystems as decomposers, primary producers and drive many important biogeochemical cycles. Because traditional methods fail to allow researchers to adequately classify microbial communities, little information is available related to the patterns of diversity in soil microbes along elevational gradients.
Recently, the development of techniques related to molecular biology which use the extraction of DNA from environmental samples has allowed researchers to classify soil microbes in different environments and promoted an understanding of how soil microbial diversity changes along elevational gradients (Bryant et al., 2008; Fierer et al., 2011; Wang et al., 2012a; Shen et al., 2013). These studies and reports yielded different results related to elevational patterns of microbial communities showing either no trend (Fierer et al., 2011; Wang et al., 2012a; Shen et al., 2013), a monotonous decrease (Bryant et al., 2008), an increase (Wang et al., 2011) or a humpbacked trend (Singh et al., 2012a,b) for soil microbial communities. However, some studies suffered from certain basic limitations. For example, some studies assessed only one or several particular phyla (Bryant et al., 2008; Singh et al., 2012b) while other studies mixed different latitudes, climatic zones or geological substrates (Lauber et al., 2009). Therefore, the primary question of how microbial diversity changes along an elevational gradient and whether such a pattern resembles the patterns observed for macroorganisms remains unanswered (Shen et al., 2013). To gain a better understanding, additional studies related to the changes in microbial patterns along elevational gradients are needed.
Shennongjia Mountain (31°15′–31°57'N and 109°59′–110°58'E) stands in northwestern Hubei Province as the highest peak (3105.4 m above sea level) in central China (Ma et al., 2008). The unique geographical location and complex terrain of this region makes it one of three centres having the highest biodiversity in China. The region includes the most well-preserved subtropical virgin and mature forest in a mid-latitudinal area on earth. The vertical distribution of vegetation on Shennongjia Mountain transitions very distinctly from evergreen broadleaved forest at low elevations to subalpine shrub at high elevations (Zhao et al., 2005). Therefore, Shennongjia Mountain presents an ideal location to study elevational patterns of animal, plant and soil microbial diversity. At present, some studies have shown that plant diversity exhibited a single peak distributional pattern with increasing elevation (Shen et al., 2004; Zhao et al., 2005; Cong et al., 2013).
This study addressed three questions: (i) What is the composition and structure of soil bacterial communities on Shennongjia Mountain? (ii) How do bacterial diversity and richness vary along the mountainside elevational gradient? and (iii) What environmental factors drive the elevational patterns observed in the soil bacterial community? To answer these questions, we selected four typical plant community types along the elevation gradient from 1000 to 2800 m, including evergreen broadleaved forest (EBF1050), deciduous broadleaved forest (DBF1750) and coniferous forest (CF2550) as well as a subalpine shrub (SAS2750) (Table 1), and analysed the soil bacterial diversity using a 16S rRNA Illumina sequencing technique.
Table 1.
Site information and soil biogeochemical characteristic in this study
| Study site | EBF1050 | DBF1750 | CF2550 | SAS2750 |
|---|---|---|---|---|
| Elevation | 1009–1057 m | 1725–1844 m | 2530–2590 m | 2720–2776 m |
| Vegetation types | Evergreen broadleaved forest | Deciduous broadleaved forest | Coniferous forest | Subalpine shrub |
| Soil organic carbon (g kg−1) | 52.58 ± 10.21b | 28.22 ± 1.01a | 59.51 ± 5.25b | 60.95 ± 4.98b |
| Total nitrogen (g kg−1) | 4.14 ± 0.63b | 1.83 ± 0.12a | 4.22 ± 0.30b | 4.50 ± 0.36b |
| Available nitrogen (g kg−1) | 0.29 ± 0.03b | 0.18 ± 0.01a | 0.33 ± 0.03bc | 0.42 ± 0.04c |
| The ratio of SOC to TN | 12.25 ± 0.53a | 15.74 ± 0.73c | 14.00 ± 0.50b | 13.52 ± 0.13ab |
| Soil pH | 6.58 ± 0.32c | 5.36 ± 0.19b | 4.99 ± 0.05b | 4.38 ± 0.03a |
| Soil moisture (%) | 37.47 ± 2.30b | 49.13 ± 1.69c | 44.52 ± 1.60c | 28.86 ± 1.10a |
| Soil temperature at a 10 cm depth (°C) | 20.02 ± 1.78d | 16.71 ± 0.12c | 10.83 ± 0.18a | 12.07 ± 0.35b |
| Shannon index of plant | 2.73 ± 0.16c | 2.41 ± 0.14bc | 2.10 ± 0.09b | 0.84 ± 0.10a |
Data present the mean value and standard error (n = 8). The same lowercase letters within the same row mean the difference was not significant, whereas the difference was significant P < 0.05.
Results
Overall soil bacterial community composition and structure
A total of 1 122 910 quality 16S rRNA sequences were detected in all study sites, with 21 098–49 741 (mean 35 090) sequences per sample. The complete linkage clustering method was used to define operational taxonomic units (OTUs), using 97% identity as a cut-off and resulting in 79 494 OTUs being detected. Rarefaction analysis and Chao1 estimator showed that the diversity in these soil samples was within the same range (Fig. S1, Table 2).
Table 2.
Summary of OTU number, Chao 1, Shannon and Simpson indices based on relative abundance of bacterial sequences at four different sites
| Sample | No. of OTUs (0.03) | Chao1 | Shannon index | Simpson index |
|---|---|---|---|---|
| EBF1050 | 8208.38 ± 269.82c | 18284.82 ± 1382.19ab | 8.14 ± 0.09c | 894.37 ± 163.85b |
| DBF1750 | 7490.50 ± 154.72b | 20801.44 ± 1531.48b | 7.75 ± 0.04b | 330.32 ± 65.76a |
| CF2550 | 6544.00 ± 272.93a | 18494.40 ± 1361.45ab | 6.99 ± 0.19a | 82.96 ± 37.67a |
| SAS2750 | 6250.50 ± 187.08a | 15972.55 ± 547.60a | 7.43 ± 0.09b | 253.66 ± 70.68a |
Data present the mean value and standard error (n = 8). The same lowercase letters within the same row mean the difference was not significant, whereas the difference was significant P < 0.05.
The classified phylotypes were counted at different taxonomical levels and 36 bacterial phyla were presented in these forest sites (Table S1), and all phylotypes were detected at all sample sites. Ninety class and 153 order were found in the tested samples (Table S1). The relatively abundant dominant phyla were Acidobacteria, Actinobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Verrucomicrobia, which accounted for more than 75% of the bacterial sequences from each of the soils (Table S2). In addition, Bacteroidetes, Planctomycetes, Firmicutes and Deltaprotecobacteria were presented in most soils but at low relatively abundance, and the other phyla were rarely found in these soil samples (Table S2). The OTUs number at different taxonomical level were counted and showed the different dominant phylotypes were existed (Tables S3 and S4). For example, the Alphaproteobacteria is the most dominant phylotype at the class level, and the Rhizobiales is the most dominant phylotype at the order level (Table S4).
Detrended correspondence analysis showed that four distinct clusters were formed and were well separated from among samples (Fig. 1). The results of the Multi-Response Permutation Procedure, Adonis and Anosim also showed significant differences (P < 0.01) existed among different study sites (Table S5). Therefore, the soil bacterial communities were sharply different among these study sites.
Fig 1.
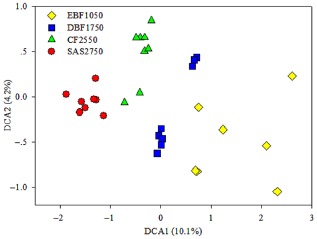
Detrended correspondence analysis of soil bacteria community structure based on the relative abundance of detected bacterial sequence number.
Bacterial community distribution pattern along an elevational gradient
The soil bacterial community was analysed by calculating the Shannon and Simpson indices, and OTUs numbers (richness). The soil bacterial Shannon index ranged from 6.99 ± 0.19 to 8.14 ± 0.09, Simpson index ranged from 82.96 ± 37.67 to 894.37 ± 163.85, and the number of OTUs ranged from 6250.50 ± 187.08 to 8208.38 ± 269.82 in these four sites respectively (Table 2). The results showed that the soil bacterial Shannon index decreased significantly (P < 0.05) with increasing elevation from EBF1050 to CF2550 (Table 2). However, the richness monotonously decreased significantly (R2 = 0.614, P < 0.01) with increasing elevation (Table 2, Fig. 2).
Fig 2.
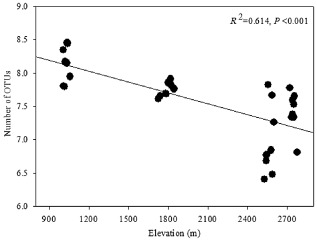
The regression relationship between soil microbial OTUs richness and elevation.
The relative abundance of bacteria at the phylum level varied among the four sites. We documented distributional patterns of relative abundance for the six dominant phyla (Fig. 3). One of three trends were observed for each phylum: (i) the relative abundance decreased from EBF1050 to CF2550 and increased in the SAS2750 sites, such as Acidobacteria, Actinobacteria, Alphaproteobacteria; (ii) the relative abundance increased from EBF1050 to CF2550 and decreased SAS2750, such as Verrucomicrobia and Betaproteobacteria and (iii) no significant difference was observed, such as Gammaproteobacteria. Therefore, the relative abundance of the dominant phyla showed different elevational patterns (Fig. 3). In addition, the richness also exhibited similar trends in different dominant phyla (Table S3). The different phyla were dominant at different study sites (Fig. 3, Table S2). For example, the most dominant phyla were Acidobacteria (18.85%) and Alphaproteobacteria (17.78%) in EBF1050, Alphaproteobacteria (17.77%) and Verrucomicrobia (17.63%) in DBF1750, Betaproteobacteria (36.41%) in CF2550 and Acidobacteria (21.34%) in SAS2750.
Fig 3.
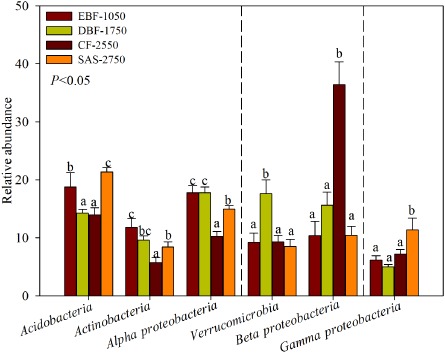
Relative abundance of the dominant bacterial phyla in soil separated according to study sites. All data are presented as mean± SE. Different letters indicated statistical differences at a P value of < 0.05 among sampling sites by ONE ANOVA.
Bacterial elevational distance-decay patterns
To detect and compare the level of beta diversity among different sites along an elevational gradient, we analysed the beta diversity using Jaccard and Bray–Curtis indices (Fig. 4). The Jaccard and Bray–Curtis indices ranged from 0.52 to 0.63 and 0.42 to 0.50 within the study sites and 0.78 to 0.91 and 0.64 to 0.84 among the sites respectively (Table S6). The pairwise bacterial compositional dissimilarities across the elevational distance increased significantly (P < 0.01) with increasing elevation (Fig. 4). Thus, the soil bacterial diversity on Shennongjia Mountain showed a significant elevational distance-decay.
Fig 4.
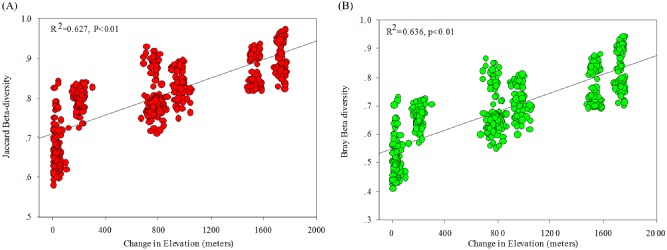
The regression relationship between the beta diversity of Jaccard (A) and Bray–Curtis (B) index and change in elevation distance.
Linking bacterial community structure and environmental factors
Mantel tests and canonical correspondence analysis (CCA) were performed to analyse the major environmental factors that shaped the bacterial community and structure. The results of a partial Mantel test showed that the bacterial community and structure were significantly linked (P < 0.01) to the soil and plant factors (Table 3). For example, the Acidobacteria, Actinobacteria, Firmicutes, Alphaproteobacteria, Betaproteobacteria and Planctomycetes exhibited a significant positive relationship (P < 0.05) with soil and plant factors, while Bacteroidetes, Verrucomicrobia and Deltaproteobacteria were significant positively linked (P < 0.05) with soil factors. Therefore, the soil bacterial community and structure at the phylum level was controlled by different environmental factors.
Table 3.
Partial Mantel analysis on the relationship between the relative abundance of dominant phyla and soil characteristics or plant properties
| Phylum | Soil,a partial plant |
Plant,b partial soil |
||
|---|---|---|---|---|
| r | P | r | P | |
| All phylotypes | 0.634 | 0.001 | 0.330 | 0.001 |
| Acidobacteria | 0.578 | 0.001 | 0.196 | 0.023 |
| Actinobacteria | 0.536 | 0.001 | 0.191 | 0.045 |
| Bacteroidetes | 0.343 | 0.002 | 0.125 | 0.133 |
| Chloroflexi | 0.101 | 0.163 | 0.444 | 0.001 |
| Firmicutes | 0.481 | 0.001 | 0.281 | 0.001 |
| Alphaproteobacteria | 0.572 | 0.001 | 0.319 | 0.002 |
| Betaproteobacteria | 0.196 | 0.015 | 0.306 | 0.001 |
| Deltaproteobacteria | 0.555 | 0.001 | 0.105 | 0.219 |
| Gammaproteobacteria | 0.389 | 0.001 | 0.250 | 0.004 |
| Gemmatimonadetes | 0.424 | 0.001 | 0.309 | 0.002 |
| Planctomycetes | 0.505 | 0.001 | 0.159 | 0.046 |
| Verrucomicrobia | 0.551 | 0.001 | −0.033 | 0.635 |
Selected soil variables: soil organic carbon, total nitrogen, available nitrogen, soil moisture, soil pH and soil temperature at the depth of 0–10 cm.
Selected plant variables: the Shannon index of tree and shrub.
CCA was used to identify the major environmental variables that significantly controlled the soil bacterial community structure (P = 0.005). The Shannon index for plants and soil pH appeared to be the most important environmental factors in controlling the bacterial community structure since they had long-term projections, which represented major variations among microbial communities (Fig. 5A). The regression analysis also showed that the number of soil bacterial OTUs was significant linked to plant Shannon index (R2 = 0.472, P < 0.01) and soil pH (R2 = 0.485, P < 0.01) (Figs S2 and S3). Therefore, soil temperature may directly affect the relative abundance of soil microbes, and soil pH and plant diversity may directly control the bacterial species present or absent at any particular site.
Fig 5.
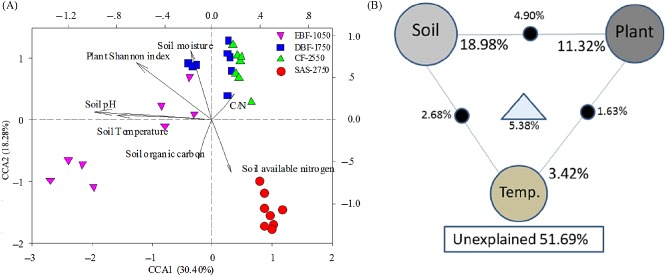
The relationship analysis between soil microbial functional gene diversity and environmental variables.A. Canonical correspondence analysis (CCA) of relative abundance data and soil environmental variables that were significantly related to microbial variations, C/N: the ratio of soil organic carbon to soil total nitrogen.B. Variation partitioning analysis (VPA) of microbial diversity explained by soil chemical factors (soil, including soil organic carbon, available nitrogen, the ratio of soil organic carbon to total nitrogen, soil moisture and pH), soil temperature (temp) and plant diversity (the Shannon index of trees and shrubs), and their relationships. Each diagram represents the biological variation partitioned into the relative effects of each factor or a combination of factors, in which geometric areas were proportional to the percentages of explained variation.
Variance partitioning analysis was used to quantify the contributions of soil chemical factors (soil), soil temperature (temp) and plant diversity (plant) to variation in the bacterial community. A total of 48.31% of the variation was explained by these three environmental variables (Fig. 5B). Soil, temp and plant independently explained 18.98%, 3.42% and 11.32% of the variation in the bacterial community structure, respectively, verifying that they were major factors shaping bacterial community structure.
Discussion
Microorganisms are the most abundant and diverse group of species on our planet and play important roles in the biogeochemical cycling of various materials and elements; however, the spatial pattern of microbial diversity at the community level remains poorly studied (Martiny et al., 2011). The development and increasing availability of high-throughput molecular techniques has helped improve our understanding of the composition and spatial distribution of microbial communities at different scales. This is especially true for the use of direct sequencing of metagenomic DNA that has become the most accurate approach for the assessment of the taxonomic composition of microbial communities (Lin et al., 2010; Martiny et al., 2011). Fierer and Jackson (2006) indicated that Acidobacteria, Actinobacteria, Proteobacteria and Bacteroidetes dominated all biomes, and the bacterial community composition does not vary significantly across different biomes. In recently years, more and more reports have shown that the dominant bacterial community compositions in soils are similar in various ecosystems (Fierer et al., 2011; Wang et al., 2011; Shen et al., 2013). For example, Acidobacteria, Alphaproteobacteria, Actinobacteria, Betaproteobacteria and Gammaproteobacteria accounted for over 75% of the bacteria in Changbai Mountain, in northeastern China (Shen et al., 2013). Acidobacteria and Proteobacteria were dominant in a mountainous forest of eastern Peru (Fierer et al., 2011). Proteobacteria accounted for more than 50% of the soil bacterial community in pristine perhumid forests in the Yuanyang Lake ecosystem in Taiwan (Myers et al., 2001). Proteobacteria, Acidobacteria, Actinobacteria and Bacteroidetes are the dominant phyla on Mount Fuji in Japan (Singh et al., 2012b). In our study, the Proteobacteria, Acidobacteria, Actinobacteria and Verrucomicrobia dominated and accounted for over 75% of all the classified bacterial sequences on Shennongjia Mountain. Simultaneously, the similar forest types may have the same dominant microbial phylum; for example, the Betaproteobacteria is the most dominant phylum in the dark-coniferous spruce–fir forest in Changbai Mountain (Shen et al., 2013), and in Shennongjia Mountain, the Acidobacteria is the most dominant phylum in the broadleaved forest in Peru (Fierer et al., 2011) and in Shennongjia Mountain. Therefore, the dominant phyla of soil microbial communities may be similar in various ecosystems; forest types may be one of the key factors shaping the dominant phyla.
Patterns of change in microbial diversity along elevational gradients make an extremely interesting topic among ecologists because of the existence of fundamental biogeographic patterns underlying microbial biogeography (Martiny et al., 2011; Wang et al., 2012b). Elevation is well known to nearly always be a complex and direct driving factor for different climatic gradients, and climatic factors have been shown to have the strongest positive associations with patterns of elevational diversity for animals and plants (Storch et al., 2012b; Forister et al., 2010). Elevational patterns on mountainsides may minimize the impact of environmental factors (Singh et al., 2012a). At present, studies have produced different results related to elevation patterns of microbial communities; this indicates some other factors may be driving the elevational distribution of microbial communities (Bryant et al., 2008; Fierer et al., 2011). In our study, four typical forest types were formed along an elevational gradient, and plant diversity had a monotonic decrease with elevation. Both the CCA and relationship analysis showed that plant diversity and soil microbial diversity are significantly (P < 0.01) correlated. Therefore, plant diversity may be one of the key impact factors that shapes the microbial community and its diversity along elevational gradients. Plant communities can influence the associated soil microbial communities through the types of communities present, the amounts of carbon and nutrient inputs, and by influencing the temperature and water content of the soil (Myers et al., 2001; Waldrop and Firestone, 2004). Different plant species can be associated with different communities as evidenced by fatty acid (Lin et al., 2010), physiological (Myers et al., 2001) and DNA techniques (Kuske et al., 2002). Therefore, the different plant community types that have formed along elevational gradients serve as an important factor in shaping the elevational patterns of the microbial community.
Soil pH is usually one of the best predictors of variation in microbial diversity in the various horizons of soils (Shen et al., 2013), including at continental scales (Fierer and Jackson, 2006; Chu et al., 2010), in the presence of land use changes (Jenkins et al., 2009) and at small scales (Baker et al., 2009). Some specific phyla exhibit unique patterns of response to variations in pH. For example, the relative abundance of Acidobacteria increased with decreasing pH (Jones et al., 2009), while the relative abundance of Alphaproteobacteria decreased with increasing pH (Chu et al., 2010). Recent reports also indicated that pH has a strong correlation with microbial diversity and composition along an elevational gradient in soils or in other environments. For example, Shen et al. (2013) observed soil pH is the primary factor in controlling bacteria diversity and community composition across an elevational gradient on Changbai Mountain (Singh et al., 2012b). Xiong and colleagues (2012) revealed pH is a better predictor of bacterial community structure in alkaline sediment than in soils with other pH levels. In our study, CCA and the Mantel test all showed that pH (P < 0.01) is an important factor that influenced the elevational patterns at the community level of some microbial phyla. The strong correlation between soil pH and microbial diversity could be a result of soil pH interacting with a number of other individual soil and site variables (Fierer and Jackson, 2006). Some authors have expressed the idea that pH maybe an independent driver of soil microbial diversity because the microorganisms themselves have specific pH values and a significant deviation in pH causes environmental stresses (Madigan et al., 1997). Therefore, pH is a universally good predictor of bacterial distribution patterns (Singh et al., 2012b).
Beta diversity is central to many ecological and evolutionary topics and is a universal biogeographic pattern observed in communities from all domains of life (Green et al., 2004); however, patterns of beta diversity for microorganisms have been left understudied (Wang et al., 2012b). At present, several studies have examined the beta diversity of bacterial communities on mountainsides (Bryant et al., 2008) and in aquatic environments (Wang et al., 2012b). Bryant and colleagues (2008) showed that bacterial lineages of Acidobacteria were not randomly distributed but exhibited significant (P < 0.05) spatial structure across elevational gradients, and suggested that bacterial lineages harbour increasingly disparate ecological features (or functions) at increased elevational distances as a probable consequence of abiotic filtering. Wang and colleagues (2012b) examined the aquatic microbial beta diversity in Laojun Mountain, China, and indicated that species turnover was mostly related to environmental heterogeneity and spatial gradients including horizontal distance and elevation. In this study, we first observed that bacterial compositional similarity at the whole community level significantly decreased (P < 0.01) with elevational distance based on the Jaccard and Bray–Curtis indices. Therefore, these results indicated that bacteria may have a strong ability to disperse to a wide variety of environmental conditions (Wang et al., 2012b).
Conclusions
The soil bacterial diversity and richness patterns along an elevational gradient on Shennongjia Mountain were analysed using the 16S rRNA Illumina sequencing. The dominant phyla were Acidobacteria, Actinobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Verrucomicrobia, which accounted for more than 75% of the bacterial sequences. Bacterial diversity decreased significantly (P < 0.01) with increasing elevation from 1050 to 2550 m and increased at 2750 m; however, OTUs richness tended to decrease monotonously with increasing elevation. We also found that the soil bacterial compositional similarity decreased significantly (P < 0.01) with elevational distance increased based on the Jaccard and Bray–Curtis indices. Partial Mantel tests and CCA analysis showed soil pH and plant diversity may be the key factors in shaping soil bacterial community structure and diversity on Shennongjia Mountain.
Acknowledgments
We also gratefully acknowledge Jingyuan Yang, Kun Liu and Min Wang for helping survey the plant plots.
Conflict of interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Rarefaction curves for OTUs were calculated with sequences normalized to 20 000 for each sample using 0.03 distance OTUs.
Fig. S2. The regression relationship between soil microbial OTUs richness and plant diversity.
Fig. S3. The regression relationship between soil microbial OTUs richness and soil pH.
Table S1. The classified phylotypes detected at different taxonomical levels.
Table S2. Relative abundances of detected phylum in four forest sites at different elevation.
Table S3. Numbers of detected OTUs at phylum level in four forest sites.
Table S4. The OTU number of the top 10 dominant phylotypes detected at different taxonomical levels.
Table S5. Statistical analysis of differences in the microbial community composition and structure between different sites.
Table S6. Microbial beta diversity of Jaccard and Bray–Curtis index along elevational distance on Shennongjia Mountain.
Appendix S1. Materials and methods.
References
- Baker KL, Langenheder D, Nicol GW, Ricketts D, Killham K. Gampbell CD. Environmental and spatial characterization of bacterial community composition in soil to inform sampling strategies. Soil Biol Biochem. 2009;41:2292–2298. [Google Scholar]
- Bryant JA, Lammanna C, Morlon H, Keroff AJ, Enquist BJ. Green JL. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. PNAS. 2008;105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HY, Fierer N, Lauber CL, Caporaso JG, Knight R. Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- Cong J, Yin H, Lu H, Su X, Yang J, Li D. Zhang Y. Species diversity and environmental interpretation of typical vegetation types in the Shennongjia natural reserve. Sci Silvae Sinicae. 2013;49:30–35. [Google Scholar]
- Fierer N. Jackson RB. The diversity and biogeography of soil bacterial communities. PNAS. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM. Silaman MR. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011;92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- Forister ML, McCall AC, Sanders NJ, Fordyce JA, Thorne JH. O'Brien J. Compounded effects of climate change and habitat alteration shift patterns of butterfly diversity. PNAS. 2010;107:2088–2092. doi: 10.1073/pnas.0909686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D. Dangerfield M. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Jenkins SN, Waite IS, Blackburn A, Husband R, Rushton SP. Manning DC. Actinobacterial community dynamics in long term managed grasslands. Antonie Van Leeuwenhoek. 2009;95:319–334. doi: 10.1007/s10482-009-9317-8. [DOI] [PubMed] [Google Scholar]
- Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R. Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Int Soc Microb Ecol J. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuske CR, Ticknor LO, Miller ME, Dunbar JM, Davis JA. Barns SM. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl Environ Microbiol. 2002;68:1854–1863. doi: 10.1128/AEM.68.4.1854-1863.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R. Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Huang YJ, Tang SL, Whitman WB, Coleman DC. Chiu CY. Bacterial community diversity in undisturbed perhumid montane forest soils in Taiwan. Microb Ecol. 2010;59:369–378. doi: 10.1007/s00248-009-9574-0. [DOI] [PubMed] [Google Scholar]
- Lomolino MV. Elevational gradients of species-density: historical and prospective views. Glob Ecol Biogeogr. 2001;10:3–13. [Google Scholar]
- Ma C, Zhu C, Zheng C, Wu C, Guan Y. Zhao Z. High-resolution geochemistry records of climate changes since late-glacial from Dajiuhu peat in Shennongjia Mountains, Central China. Chin Phys Lett. 2008;53(Suppl. 1):28–41. [Google Scholar]
- McCain CM. Elevational gradients in diversity of small mammals. Ecology. 2005;86:366–372. [Google Scholar]
- Madigan M, Martinko J. Parker J. Brock Biology of Microorganisms. Upper Saddle River, NJ, USA: Prentice Hall; 1997. [Google Scholar]
- Malhi Y, Silman M, Salinas N, Bush M, Meir P. Saatchi S. Introduction, elevation gradients in the tropics, laboratories for ecosystem ecology and global change research. Glob Chang Biol. 2010;16:3171–3175. [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD. Horner-Devine MC. Drivers of bacterial beta-diversity depend on spatial scale. PNAS. 2011;108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RT, Zak DR, White DC. Peacock A. Landscape-level patterns of microbial community composition and substrate in upland forest ecosystems. Soil Sci Soc Am J. 2001;65:359–367. [Google Scholar]
- Rahbek C. The role of spatial scale and the perception of large-scale species richness patterns. Ecol Lett. 2005;8:224–239. [Google Scholar]
- Renaud PE, Webb T, Bjorgesaeter A, Karakassis I, Kedra M. Kendall M. Continental-scale patterns in benthic invertebrate diversity: insights from the MarBEF database. Mar Ecol Prog Ser. 2009;382:239–252. [Google Scholar]
- Shen C, Xiong J, Zhang H, Feng Y, Lin X. Li X. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai mountain. Soil Biol Biochem. 2013;57:204–211. [Google Scholar]
- Shen Z, Hu H, Zhou Y. Fang J. Altitudinal patterns of plant species diversity on the southern slope of Mt. Shennongjia Hubei, China. Biodivers Sci. 2004;12:99–107. [Google Scholar]
- Singh D, Takahashi K. Adams JM. Elevational patterns in Archaeal diversity on Mt. Fuji. PLoS ONE. 2012a;7:e44494. doi: 10.1371/journal.pone.0044494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Takahashi K, Kim M, Chun J. Adams JM. A hump-backed trend in bacterial diversity with elevation on mount Fuji, Japan. Microb Ecol. 63:429–437. doi: 10.1007/s00248-011-9900-1. [DOI] [PubMed] [Google Scholar]
- Storch HV, Zorita E, Jones JM, Dimitriev Y, Gonzalez-Rouco F. Tett SFB. Reconstructing past climate from noisy data. Science. 2012b;306:679–682. doi: 10.1126/science.1096109. 2006. [DOI] [PubMed] [Google Scholar]
- Waldrop MP. Firestone MK. Microbial community utilization of recalcitrant and simple carbon compounds: impact of oak-woodland plant communities. Oecologia. 2004;138:275–284. doi: 10.1007/s00442-003-1419-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Soininen J, Zhang Y, Wang B, Yang X. Shen J. Contrasting patterns in elevational diversity between microorganisms and macroorganisms. J Biogeogr. 2011;38:595–603. [Google Scholar]
- Wang J, Soininen J, He J. Shen J. Phylogenetic clustering increases with elevation for microbes. Environ Microbiol Rep. 2012a;4:217–226. doi: 10.1111/j.1758-2229.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Soininen J, Zhang Y, Beixin W, Yang X. Shen J. Patterns of elevational beta diversity in micro- and macroorganisms. Glob Ecol Biogeogr. 2012b;21:743–750. [Google Scholar]
- Xiong JB, Liu YQ, Lin XG, Zhang HY, Zeng J. Hou JZ. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol. 2012;14:2457–2466. doi: 10.1111/j.1462-2920.2012.02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Chen W, Tian Z. Xie Z. Altitudinal pattern of plant species diversity in Shennongjia Mountains, Central China. J Integr Plant Biol. 2005;47:1431–1449. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Rarefaction curves for OTUs were calculated with sequences normalized to 20 000 for each sample using 0.03 distance OTUs.
Fig. S2. The regression relationship between soil microbial OTUs richness and plant diversity.
Fig. S3. The regression relationship between soil microbial OTUs richness and soil pH.
Table S1. The classified phylotypes detected at different taxonomical levels.
Table S2. Relative abundances of detected phylum in four forest sites at different elevation.
Table S3. Numbers of detected OTUs at phylum level in four forest sites.
Table S4. The OTU number of the top 10 dominant phylotypes detected at different taxonomical levels.
Table S5. Statistical analysis of differences in the microbial community composition and structure between different sites.
Table S6. Microbial beta diversity of Jaccard and Bray–Curtis index along elevational distance on Shennongjia Mountain.
Appendix S1. Materials and methods.


