Abstract
Scientists continually relate information from the published literature to their current research. The challenge of this essential and time-consuming activity increases as the body of scientific literature continues to grow. In an attempt to lessen the challenge, we have developed an Electronic Laboratory Notebook (ELN) application. Our ELN functions as a component of another application we have developed, an open-source knowledge management system for the neuroscientific literature called NeuroScholar (http://www.neuroscholar.org/). Scanned notebook pages, images, and data files are entered into the ELN, where they can be annotated, organized, and linked to similarly annotated excerpts from the published literature within Neuroscholar. Associations between these knowledge constructs are created within a dynamic node-and-edge user interface. To produce an interactive, adaptable knowledge base. We demonstrate the ELN’s utility by using it to organize data and literature related to our studies of the neuroendocrine hypothalamic paraventricular nucleus (PVH). We also discuss how the ELN could be applied to model other neuroendocrine systems; as an example we look at the role of PVH stressor-responsive neurons in the context of their involvement in the suppression of reproductive function. We present this application to the community as open-source software and invite contributions to its development.
Index Entries: Paraventricular hypothalamus, neuroendocrine, knowledge engineering
Introduction
The Electronic Laboratory Notebook (ELN) has emerged as an important application in commercial research and development and Laboratory Informatics Management Systems (LIMS). Several mature technologies competing within this growing market are commercially available, including applications targeting academic audiences (e.g., Axiope’s “Catalyzer”; Goddard et al., 2003) or the open-source community (e.g., http://www.amphora.com/). These and similar systems developed within academia (such as “NeuroSys,” Pittendrigh and Jacobs, 2003), facilitate the construction of computational laboratory records for semistructured text, numerical, and image data. Also noteworthy is the Senselab EAV/CR (Entity, Attribute, Value with Classes, and Relationships) data storage approach (Shepherd et al., 1998; download: http://ycmi.med.yale.edu/trialdb/) that supports multiple neuroinformatics applications to store and serve data, including those located at Neuroscience Database Gateway of the Society for Neuroscience (http://big.sfn.org/NDG).
ELN technology promises advantages over traditional methods of recording laboratory-based information by allowing scientists to organize, retrieve, and share their data more coherently, thereby promoting a culture of consistency, accountability, and accessibility regarding acquired knowledge. Currently, however, ELNs are rarely used within academic institutions, underscoring the enduring practicality of using paper laboratory notebooks (PLNs). This tendency may result from academic culture, in which researchers tend to work more autonomously than their counter-parts in industry. Importantly, demands for ELN technology may now increase because of the recently announced data sharing policy of the U.S. National Institutes of Health (NIH), which specifies that all grants requesting federal funding exceeding $500K in any single year must include a plan for how the project’s data will be made publicly available (http://grants.nih.gov/grants/policy/data_sharing/). This is very likely to develop into more stringent data-sharing requirements in the future. Policies advocating open-access publishing and data sharing are also promoted in other funding agencies such as the Wellcome Trust in the United Kingdom.
In this report, we present an ELN extension for “NeuroScholar” (Burns et al., 2003), a knowledge management system that helps neuroscientists construct, manage, analyze, and publish knowledge models (useful representations of known information linked to supporting primary data). Originally, the NeuroScholar system was designed to help users superimpose structured ontologies over information “fragments” derived from the primary scientific literature, thereby allowing them to construct knowledge representations (KRs) of published work relevant to their research (Burns, 2001; Burns et al., 2003). However, as the neuroscientists use published literature in conjunction with their own laboratory notes and raw data, we have now extended NeuroScholar’s functionality to also support information fragments derived from scanned laboratory notebook pages and local raw data files.
With this extended ELN functionality, NeuroScholar provides a unique alternative to existing ELNs by allowing users to directly relate their own raw, unpublished data with data published by other laboratories. The application has many features
it supports the use of PLNs so that traditional lab practices are not replaced;
it safeguards data by requiring researchers to create “digital backups” of their PLNs;
it permits text/voice annotations for excerpts from PLNs and research papers;
it reproduces the indexing and cross-referencing capabilities of other ELNs;
it allows users to search readily for key words and data; and
it places raw and published data within a single KR.
Although such technology does not guarantee good scientific content, it does promote good scientific practice by requiring clearly delineated data structures and rigorously managed data instances.
In this report, we first explain the philosophy underlying our neuroinformatics approach to experimental neuroscience. This is followed by a description of how the ELN extension for NeuroScholar was designed and implemented. We use two examples from our current work to highlight how NeuroScholar’s ELN component is useful for the experimental neuroscientist. In the first example, we used the ELN to manage raw data and primary literature relevant to our own experiments, which concern the activation of brain areas during glucoprivation, a specific form of metabolic stress (Khan and Watts, 2003, 2004). In our second example, we discuss potential uses of NeuroScholar to link this information to a related line of inquiry concerning how glucoprivic stress could suppress reproductive function (Hahn et al., 2003). These specialized examples serve to illustrate the use of the system in a specific context, and the reader is invited to apply the general design of the system for other scientific questions. Finally, we compare our approach with two existing ELN applications (Axiope’s Catalyzer and the NeuroSys hierarchical database).
Design Rationale: Experimental Science is a Process of Inscription and Synthesis
We invite the reader to consider that the key action performed by scientists in the laboratory is the process of inscription (Latour and Woolgar, 1979). This refers to the act of taking notes, recording data, labeling experimental samples, writing papers, writing proposals, or even making sketches on a whiteboard. A process of synthesis typically generates new inscriptions, in which sets of existing inscriptions are brought together (e.g., in the process of data analysis, publication preparation, grant writing, laboratory discussions, and so on) to produce new inscriptions that may themselves contribute to the next cycle of synthesis. Importantly, different types of inscription are involved in this process; key among these are descriptions, interpretations, and citations (Burns, 2001). This philosophy forms the underlying rationale for NeuroScholar as a computational knowledge management system in which the processes of inscription and synthesis are formalized within the heuristic framework of a software application.
A laboratory’s scientific integrity depends not only on the accuracy of its inscriptions but also on the correctness and predictive power of its theoretical approach. Within a traditional laboratory’s workflow, the structures that support this process are noncomputational and highly specialized. The objective of building computational support structures is to empower researchers to
track and share their inscriptions easily;
perform synthesis from increasingly larger sets of inscriptions; and
be able to share and communicate inscriptions easily.
The development of NeuroScholar is a natural instantiation of this philosophical approach.
Knowledge Engineering Concepts Within NeuroScholar
Like most knowledge management systems, NeuroScholar operates within an artificially defined universe of discourse (or “ontology,” Gruber, 1993). This is defined programmatically within the framework used by the NeuroScholar system (Burns et al., 2003). Neuroscientists without mathematical or computational training may have difficulty in using knowledge engineering concepts, such as ontologies, classes, constraints and instances. We have, however, incorporated them in our design as a network of nodes and edges that users may interact with to query, display, and edit their knowledge. By providing documentation tailored to noncomputational users, and by designing a system that is easily navigable and usable, we hope to train neuroscientists in the use of our knowledge engineering tools so that they adopt our system in their work (see online documentation available at http://www.neuroscholar.org/).
For these reasons, the ontology structure defined within NeuroScholar and its appearance to the user are both important design concerns. When users open a “knowledge base” within NeuroScholar, they are presented with a dynamic, interactive graph (Fig. 1, generated by TouchGraph, an open-source java library, http://touchgraph.sourceforge.net/) in which each node represents a “view.” This is simply a rich encapsulation of data pertaining to a specific item of interest. In this report, names of views appear in bold italics, in which each of these names is described in the following pages (please consult Fig. 1 to understand its relationship with other nodes in the view graph).
Fig. 1.
An expanded black and white screen shot of the complete view definition graph of NeuroScholar’s systems involving fragments. This may be separated into two main “branches” of KnowledgeStatement view: BibliographicFragment views and NotebookFragment views. The actual view in the working software is in color. (A) View definitions directly involved in ELN-related functions of the system, (B) parent- and related-view definitions that enable ELN-based views in the system.
A small graphical icon is used to identify nodes representing instances of a particular view definition. Each view may require a complex query involving multiple underlying database tables. We insulate the user from this complexity through a transactional mechanism that operates on the view as a whole. The mechanism of this interaction is based on the “View-Primitive-Data Model framework” (VPDMf), which has been described in detail elsewhere (Burns, 2001; Burns et al., 2003).
For the purposes of orienting readers to Fig. 1, we describe some characteristics of the graph. Views have object-oriented properties, forming “parent—child” inheritance hierarchies with other views. We use visual notation from the Universal Modeling Language (UML) to denote this inheritance within the view graph: a triangular arrow pointing from the child to the parent (Rumbaugh et al., 1999). A view may be dependent on other views (represented by a dashed line with an open arrow). Relations may exist between views, appearing graphically as open nodes connected by gray edges. At present, relations are directional and non-commutative. Given our definition of “knowledge” as “information placed into the context of other information” (Blum, 1986), relations are useful for the construction and delineation of knowledge within NeuroScholar. When views require additional functionality we use specialized “form controls” (a small component included in the standard data form) and “plug-ins” (a specialized panel that replaces the whole form). In theory, this plug-in mechanism may be used to incorporate any Java application into NeuroScholar, as long as an adapter class can be written to convert the application’s data into the VPDMf schema used by the system.
Knowledge Statements, Fragments, and Knowledge Acquisition in NeuroScholar
KnowledgeStatement view definitions are central in NeuroScholar. Instances of this view are simply statements of scientific relevance, forming the system’s central currency as a computational distillation of scientific knowledge. Importantly, all KnowledgeStatement instances (and by extension, all instances of children of this view definition) may be inter-related by contradicts or supports relations, permitting the construction of a scientific argument within the knowledge management system. Each KnowledgeStatement view may contain attributes that are relevant to a specific data point (values and error values). These are constrained by the definition of the variable being used (definition name, units, whether this is nominal, ordinal, interval, ratio data, etc.). This provides users with the capability of entering specific values for data points within the ELN.
The Fragment view definition (Fig. 1) inherits from KnowledgeStatement and is a key data entity for this article. Instances of this view provide the entry point for knowledge into NeuroScholar system by capturing statements originally made external to the system (Burns et al., 2003). Essentially, this is simply that users of the system may delineate some external digital source of information and then make an assertion based on that external information. Fragment views therefore may be derived from any digital source. Within this article, we provide a mechanism to define Fragment views based on local laboratory data. This includes image data captured from microscopes, scanned pages from paper laboratory notebooks, data files, and the tools that act on full-text journal articles.
This functionality is expressed by inheritance hierarchies of views within the knowledge engineering framework of NeuroScholar. There are two children of the Fragment view: NotebookFragment views are based on excerpts taken from scanned laboratory notebook pages or from binary data files and BibliographicFragment views are based on excerpts from full text articles distributed as PDF files. All of the other views in the design shown above provide organization and navigation tools for fragments. These include the documents from which the fragments are derived (the Resource, Notebook, and Publication view definitions); the scientist who wrote the originating documents (the Scientist view definition); and a collection view that contains multiple instances of the documents (the ResourceCollection view definition).
We briefly describe the subsystem that acts as a framework for BibliographicFragment views (Fig. 1) because this implementation has not appeared in press before now. These views are constructed from the various types of Publication view that are available within the system. These are Articles, Books, Book Chapters, ConferencePresentations, or Theses views. Some of these publications require additional citation information: an Article view is published in a Journal; a ConferencePresentation is published in a ConferenceProceedings; and a BookChapter is published in a Book. PDF files may be rendered using functions from the Multivalent system (http://multivalent.sourceforge.net/, Phelps and Wilensky, 2001) and has the appearance shown in Fig. 2. We mention this here for two reasons: the ELN application is built from this framework and relating fragments from publications to fragments derived from the ELN is an important functionality offered by the system.
Fig. 2.
An unembellished screen shot from the NeuroScholar system, showing the system displaying a BibliographicFragment from a journal article (Itoi et al., 1999). The fragment is found in the right panel as a shaded box labeled 1. The image in the right hand panel shows the relevant figure that substantiates the knowledge statement present in the system; this statement is included as a node in the view panel on the left.
Technical Description of NeuroScholar’s Electronic Laboratory Notebook (ELN)
In this section, we describe the terminology and computational usage of the ELN. This subsystem provides a framework for the generation and management of NotebookFragment views (Fig. 1), which have all the characteristics of a KnowledgeStatement with controlled vocabulary representations of the type of knowledge and the statement’s format. Data within this view are concerned with the location of excerpts and the annotations placed over them (combined with a reference to the notebook that contains them). Central to this definition is the Notebook view, which contains essential indexing and descriptive information and which keeps track of all pages and authors. DataFile view instances actually contain the contents of the files themselves within the system, along with the name of the file and a user-entered description of the file’s contents. Note that the MySQL database’s default configuration limits the size of stored files to 1 or 2 MB, which system administrators can increase by adjusting the maximum size of allowable packets (see MySQL’s online documentation pertaining to the error “Packet too large,” http://www.mysql.com/). See Appendix A for full UML class diagrams describing the organization of this subsystem.
System Implementation Status
NeuroScholar version 1.3 runs reliably on systems running the Windows 32 bit operating system. At present, the system is not supported on machines running Mac OS X or Unix. The system was coded using Borland’s JBuilder X programming interface and on IBM’s Eclipse interface. An extensive test framework for the NeuroScholar application ensures that the system functions reliably when changes are made to the data model of a specific knowledge base. This framework is distributed with the source distribution of NeuroScholar from SourceForge (http://www.sourceforge.net/projects/neuroscholar).
Using the ELN Component of NeuroScholar to Manage Raw and Published Data Concerning Brain Regions Sensitive to Stress
In this section, we describe how the ELN helps relate raw and published data relating to experiments in our laboratory. These experiments concern the effects of stress on the activity of neuroendocrine neurons (hormone-releasing neurons) within the hypothalamus. We first provide a brief overview of this neural system, including the experimental methods for testing our hypotheses. We then describe (1) how we have used NeuroScholar, with its ELN extension, to manage raw and published data concerning these studies, and (2) how we have used the ELN to link these data with other published work about this neuron population in the context of reproductive function. Because NeuroScholar has been designed to be as universally applicable as possible, we invite neuroscientists to extrapolate from these two examples to work performed in their own field of interest.
Overview: An Experiment for Evaluating Chemical Changes in the Brain During Glucoprivic Challenge
We are interested in understanding the effects of stress on brain chemistry (e.g., Watts, 1996). One of the stress models used in our lab involves monitoring how nerve cells respond to sudden impairments in the body’s ability to utilize glucose, a condition known as glucoprivic stress or glucoprivation. Such responses to glucoprivation manifest themselves as chemical changes within nerve cells, which we can detect using various anatomical techniques. In our laboratory, we focus on glucoprivation-induced chemical changes taking place within a cluster of small neuroendocrine neurons located within the paraventricular nucleus of the hypothalamus (PVH; Fig. 3). These neurons together constitute the principal center that coordinates the brain’s response to stress. Much evidence over the past few decades, including recent studies conducted by our laboratory and by our collaborators (Ritter et al., 1998, 2001, 2003; Hudson and Ritter, 2004; Khan and Watts, 2004) suggests that these PVH neuroendocrine neurons respond to glucoprivation in two ways
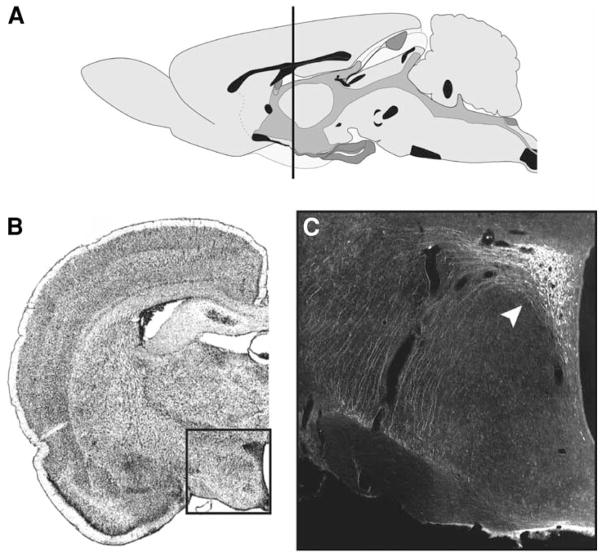
Fig. 3.
Location of the paraventricular nucleus of the hypothalamus (PVH) in the rat brain. (A) Sagittal section (side view) of the rat brain. The black line delineates the rostrocaudal position of plane of the tissue shown in B. (B) Cross-section showing one hemisphere of the rat brain at the level of the PVH. (C) Chemically processed tissue of the rat, displaying antibody staining for the hormone CRH in the PVH. The region shown in C corresponds approximately with the boxed outline overlaid on B.
They release the hormone CRH into the blood vessels that supply the pituitary gland, ultimately leading to the release of the hormone corticosterone, which helps the body adapt to stress by mobilizing energy stores.
They begin synthesizing more CRH, by initiating a series of chemical reactions that use the crh gene (DNA) blueprint to create a CRH message (mRNA), which is finally translated into CRH peptide. Newly synthesized CRH is then packaged and stored for later release.
In our experiments, we track both of these physiological events in vivo using laboratory rats, shortly after they receive a glucoprivic stressor. Here we focus on point (2) above, i.e., tracking the glucoprivation-induced chemical reactions that trigger CRH synthesis.
Hypotheses About Glucoprivation-Induced CRH Synthesis and the Techniques Used to Test Them
As noted earlier, much evidence supports the idea that glucoprivation increases CRH synthesis within PVH neuroendocrine neurons through a series of intracellular chemical reactions. We are currently testing the hypothesis that two intracellular molecules, in particular, may participate in these reactions to control CRH synthesis. These molecules are (1) extracellularly-regulated kinase (ERK) and (2) cyclic AMP- and/or calcium-sensitive response element binding protein (CREB). We hypothesize that during glucoprivation, neural signals activate ERK and CREB inside PVH neuroendocrine neurons, which increase the amount of CRH mRNA, the product synthesized from the crh gene. To test this hypothesis, we inject rats with a drug to induce glucoprivation, and then apply chemical probes to their brain tissue to determine whether ERK and CREB are activated in PVH neurons.
Tracking ERK and CREB Activation
To track ERK and CREB following presentation of the glucoprivic stressor, we use immunocytochemistry (ICC) to detect ERK and CREB activation. This technique involves the use of specific antibodies that can recognize the activated forms of ERK and CREB. When brain tissue sections containing PVH neurons are exposed to these antibodies, the antibodies will preferentially bind to all ERK and CREB in the tissue that is activated in response to glucoprivation. All ERK and CREB thus bound by the antibodies will be stained a dark brown-black color (see Fig. 4A), or, if fluorescently colored antibodies are used, with bright fluorescent colors. Stained neurons can then be visualized using a microscope, photographed using digital cameras attached to the microscope, and counted either manually or using image analysis software (Khan and Watts, 2004). Counts of positively stained neurons from injected rats vs control rats are then compared using commercially available statistics software. Appropriate steps are taken to control for nonspecific reactions of the antibodies.
Fig. 4.

(A) Example of immunostaining for phosphorylated ERK in PVH tissue from a rat subjected to stress. The darkened cells are displaying a reaction product brought about as a result of antibodies binding to phosphorylated ERK molecules within PVH neurons. The presence of such staining suggests that ERK is chemically activated only in these neurons. Unstressed rats do not show such immunostaining, suggesting that ERK activation tracks stressful events. (B) Example of in situ hybridization (ISH). This image depicts hybridized CRH mRNA within the PVH. Note the robust presence of this transcript throughout much of the PVHmp, which is displayed as the bright punctate patterns within the neurons of this nucleus. The patterns of bright labeling are formed as a result of silver grain photographic emulsion deposits forming over the hybridized tissue.
Tracking CRH Transcription
To monitor the activation of the crh gene and its transcription to CRH mRNA following glucoprivic stress, we use in situ hybridization (ISH). ISH involves the specific binding of a probe to the target mRNA sequence. This probe is an oligonucleotide probe (cRNA) is made up of a nucleotide sequence that is complementary to the target and may therefore bind to it. In our laboratory, we employ a radiolabeled cRNA probe that recognizes various forms of the CRH transcript. The reactions can be visualized using X-ray film, as well as on microscope slides in the form of a photographic emulsion of silver grains localized over the hybridized tissue (Fig. 4B).
Example 1: Using ELN Technology to Manage Data From These Experiments
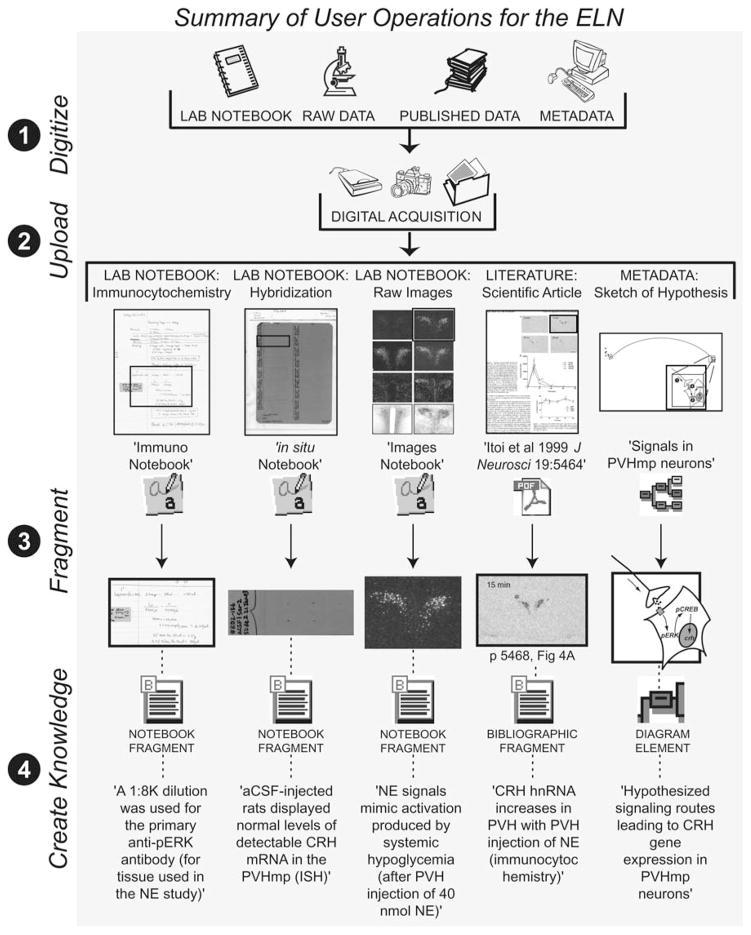
Over the past 2 years, our laboratory has conducted numerous experiments similar to the one outlined in the preceding section. Specifically, we have used ICC and ISH techniques to track the activation of intracellular molecules within PVHmp neuroendocrine neurons during stress, including ERK, CREB, and CRH mRNA. This has resulted in hundreds of raw data images, digitally captured using a microscope, similar to those shown in Fig. 4. In the process, we have generated hundreds of pages of laboratory notes and protocols related to these raw data, and have also catalogued large numbers of scientific papers within the primary literature that contain published data related to these experiments. This situation is typical within a neuroscience laboratory and provides us with a realistic example to illustrate the utility and functionality of the ELN. We have, therefore, recently begun assimilating all of these pieces of information into the ELN component of NeuroScholar, and have presented demonstrations of the populated, functional system at the 2004 Society for Neuroscience Annual Meeting (Khan et al., 2004). To date, seven laboratory notebooks have been scanned into the ELN along with the raw data produced from experiments outlined within them. From these, 690 notebook pages and data images, and 15 notebook fragments have been uploaded into the ELN extension of NeuroScholar. Additionally, 136 primary research articles related to these data, with 28 bibliographic fragments, have also been uploaded. A summary of this effort is presented in Fig. 5, which also highlights the four major operations we performed using the ELN to manage these data: digitizing, uploading, fragmenting, and knowledge creation. Each of these steps will now be discussed in more detail, in relation to the data classes depicted in Fig. 5.
Fig. 5.
User operations within the ELN. Laboratory records and raw data from our ongoing experiments are shown in relation to the published literature related to these experiments. Four major operations are highlighted: digitizing records, uploading them into the ELN, fragmenting portions that are of interest, and associating these fragments with knowledge statements to create knowledge.
ELN Operations
Digitization
To use the ELN, a laboratory user must first create digital versions of the laboratory records, raw data, published literature, and associated metadata files relevant to a given experiment (Fig. 5, Step 1). The digitized files can be produced, in principle, from any digital source. Thus, scanned notebook pages, digitally captured microscope images, as well as portable document format (PDF) files can all be utilized by the ELN. It takes relatively little time to digitize lab notebooks, a procedure which is likely to be the most rate-limiting within this class of operation. For example, a 241 page loose-leaf notebook of immunocytochemical notes was scanned in a little under 3 h. However, this notebook represented experiments performed over a 2-yr time period; “scan as you go” operations after each experiment could take as little as a minute per page using a dedicated workstation with attached scanner. With this we emphasize two points; first, the more organized a user is about setting up dedicated lab space for scanning records, the more likely will throughput be markedly improved; second, that “laboratory culture” may have to shift, i.e., users would have to be willing to accommodate ELN technology by developing some sound laboratory practices that would be required on a daily basis. By diligently capturing the lab’s daily inscriptions in a digital format, users would also help ensure that there is a second location where their records are kept secure. As for other resources, published data within PDFs are already in digital format on the user’s local hard drive, and metadata such as drawings created in programs like Adobe Illustrator are compatible so long as they are saved as an SVG file (Burns and Khan, 2003).
Uploading
Once local copies of all digitized files are on the user’s hard disk, they can be uploaded into NeuroScholar, (see documentation at http://www.neuroscholar.org/ for instructions on how to do this) as described in the “Technical Descriptions” sections earlier. Users are allowed to fragment and annotate the data contained within each uploaded file (Fig. 5, Step 2).
Fragmenting
This feature is the heart of the ELN’s functionality, and involves highlighting portions of any uploaded digital object within the ELN, and saving it as a “fragment,” complete with associated annotations and notes, which can be either text strings or digital voice recordings (Fig. 5, Step 3). Users can create many different types of fragment, depending on the file being examined. Thus, Bibliographic Fragments are fragments from portable document format (PDF) files of the scientific literature, and NotebookFragments can be from scanned notebook pages or microscope image collections. Fragments can then be stored within the ELN and called up again for future evaluation and updating. This allows a user to archive their annotations concerning specific, delineated parts of an image or a scientific paper. Multiple fragments can be made of a single digital object within the ELN, allowing for newer observations and rationale to be recorded as needed without losing the old annotations. Fragmenting will now be discussed in more detail in relation to the creation of knowledge statements.
Creating Knowledge
The previous three operations (digitizing, uploading, and fragmenting) constitute the mechanisms by which NeuroScholar allows one to manage and archive information concerning a particular experiment or project within a laboratory (Fig. 5, Step 4). The final operation in this sequence enables users to go beyond simple data management, and move toward data integration. Specifically, the design of the system encourages users to treat each fragment as a KnowledgeStatement, so that users may not only manage fragments, but also relate them to one another in a way that allows for conclusions to be drawn and for evidence to serve as support for a user’s interpretations. What is a KnowledgeStatement? Currently, it consists of a simple English sentence (e.g., “CRH is synthesized in the PVHmp”). Such statements can be associated with particular Fragments, and these KnowledgeStatements, in turn, can be related to one another. Consider this example from our populated data set concerning glucoprivic stress. As shown in Fig. 6, the left panel (a graph-like representation of the knowledge within the system) contains the following KnowledgeStatement: “NE signals mimic activation produced by systemic hypoglycemia.”
Fig. 6.
A screen shot from the NeuroScholar system, showing the system displaying a fragment from a laboratory notebook. The image in the right hand panel shows activation of ERK in the PVHmp under conditions of hypoglycemic stress.
This KnowledgeStatement is, in fact, a Fragment view supported by data shown in the right-hand panel of the same figure, which depict pCREB immunostaining. In Fig. 2, a similar Fragment is generated for a published article (Itoi et al., 1999). That Fragment is the following KnowledgeStatement: “CRH hnRNA increases with PVH injection of NE” (see Fig. 2).
Notice now, however, how this statement is set in relation to the previous Knowledge Statement of Fig. 6, with the relation “supports” (see Fig. 6). In other words, we assigned the relation between the two KnowledgeStatements such that it is clearly delineated in the system that the first statement supports the second.
In effect, we have made a stable annotation concerning our raw data about pCREB immunostaining, and have set this in relation to data published by Itoi et al. (1999). This example illustrates how we can determine which data support and contradict our own unpublished findings. For our experiments, associated lab records and relevant published literature, the bottom of Fig. 5 shows five examples of KnowledgeStatements as Fragments.
Example 2: Using ELN Technology to Interrelate Two Datasets Concerning the Same Population of Neurons
Importantly, the same population of neurons described in the example above could serve a critical role within overlapping neuroanatomic circuits underlying two distinct physiological processes. This poses an interesting challenge for the neuroinformatician taking a knowledge engineering approach to neurobiological systems, particularly if the neurons in question were examined within these overlapping circuits using both functional and neuroanatomic studies. To illustrate this challenge and how the ELN component of NeuroScholar can help users address it, we provide the following example.
To briefly restate Example 1 above, we described our functional experiments concerning CRH neurons in the PVH region of the brain that are responsive to stress (in this case, impaired glucose utilization or glucoprivation). In contrast to the KnowledgeStatements that were Fragments from the published studies described in Example 1, one could also use the following KnowledgeStatement to summarize the key finding from our experiments: “CRH neurons in the PVH respond rapidly to glucoprivation stress.”
In a related neuroanatomic study performed by a member of our group (Hahn et al., 2003), CRH neurons in the PVH were also examined in the context of their potential involvement in the suppression of reproductive function. Specifically, CRH has been implicated as a mediator for the inhibitory effects of stress (including glucoprivic stress) on reproductive function (Rivier and Vale, 1984; Tsukahara et al., 1999). A group of neurons expressing the peptide luteinizing hormone releasing hormone (LHRH) control the release of luteinizing hormone (LH) and are essential for reproductive function. Hahn et al. used a tract-tracing technique to determine whether any of the major CRH neuron populations in the brain project directly to the vicinity of the LHRH neuron cell bodies. In addition, because estrogen potentiates stress-induced suppression of reproductive function, Hahn et al. used immunocytochemistry to probe for nuclear estrogen receptors (ERs) in CRH-expressing neurons. The results of the study revealed that tracer molecule was not detected in CRH neurons at any of the sites examined (including the PVH), but that some CRH neurons do express nuclear ERs. With respect to our focus in example 1 of CRH neurons within the PVH, these results are summarized with the following KnowledgeStatements:
“CRH neurons in the PVH do not project directly to LHRH neurons”
“Some CRH neurons within the caudal PVH express the β sub-type of the nuclear estrogen receptor (ERβ)”
The related findings of these two sets of experiments are rendered concisely and explicitly by the KnowledgeStatements associated with each of them. Within the ELN, the relationship between these two studies, both of which are concerned with the same population of CRH neurons (those found in the PVH) but in the context of two distinct behaviors, can now be simply represented by formally relating the KnowledgeStatements. In this way, overlapping characteristics of two sets of neural circuits underlying distinct physiological processes can be interrelated. Moreover, by comparing these studies and the Knowledge Statements contained within, the scientist can construct more informative Knowledge Statements, for example: “CRH neurons in the PVH respond rapidly to glucoprivation, but do not directly influence LHRH neurons during glucoprivation-induced suppression of reproductive function.” Moreover, the scientist can even develop new testable hypotheses by interrelating KnowledgeStatements in this way, for example: “Do CRH neurons in the PVH respond differently to glucoprivic stress depending on whether or not they express nuclear ERβ?.” This ability of the system to store large numbers of these stable, open-ended annotations that capture the elements of a scientific argument in a human-readable format lies at the heart of this approach. Future directions for our work will involve placing computational KRs within the same framework to enable computational inference to be performed over these statements.
Comparison With Other Systems, Pros and Cons
Three interrelated fields inform the discipline of KR: “logic” provides formal structure and inference rules, “ontology” defines the “universe of discourse,” and “computation” distinguishes KR from pure philosophy (Sowa, 2000). We have designed NeuroScholar to strongly emphasize computation as its primary focus. Thus, our principal challenge is to provide neuroscientists with a knowledge engineering system that not only streamlines their workflow, but also generates knowledge that would otherwise be impossible to obtain because of the information overload. To this end, we help users construct computational representations of their scientific knowledge from both the primary literature and from laboratory notes. This distinguishes NeuroScholar from other ELN and LIMS developers who seek primarily to provide a data management system tailored for a laboratory setting. In this section, we explain how NeuroScholar differs from two representative ELN systems (Axiope’s Catalyzer, and NeuroSys).
The most obvious difference concerns data structure definition. In Catalyzer and NeuroSys, users may define the structure of their laboratory records within the system itself. This provides them with a powerful, searchable tabulated representation of numbers, text, and images. In contrast, NeuroScholar users may add attribute/value pairs to KnowledgeStatement instances, meaning that the system is primarily organized around the text of scanned notes. Thus, existing ELNs require that users construct data structures ahead of time and then process the archived data, whereas NeuroScholar’s ELN extension requires users to “postprocess” their laboratory notes, so that the data they enter into the system is defined after, rather than during, the execution of the experiment. Importantly, all approaches are robust enough to permit users to redefine their attribute types without the risk of breaking and losing access to previously archived data.
The View-Primitive Data Model framework (VPDMf, Burns et al., 2003) provides the infrastructure for the NeuroScholar system and determines the system’s characteristics. At present, this operates as scripts that load an object-oriented schema expressed in the UML with some additional specialized semantics, which then builds a set of MySQL and Java source code files. These files are used by NeuroScholar to build and populate the MySQL database and then define the ontology used by the system (see Fig. 7 in Appendix A). Source code for the VPDMf is available from the NeuroScholar project page at Sourceforge (http://www.sourceforge.net/projects/neuroscholar/). NeuroScholar does not provide end-users with the same functionality as existing ELN systems (i.e., the opportunity to define their own ontological representation of their data). Rather, this task requires knowledge of the UML and an understanding of the additional semantics used within view specification files. This is straightforward for programmers with computational expertise, requiring familiarization in the scripts that generate the code and a certain amount of debugging when preparing the view specification files (based on the authors’ experience of using the VPDMf). However, this is far beyond what would be expected for users of the NeuroScholar system.
Fig. 7.
A UML class diagram of the data model of the NeuroScholar system that underlies the views shown in Fig. 1. This is presented so that other neuroinformatics developers might better grasp the detailed informatics design of the system.
One feature of Catalyzer not currently implemented within NeuroScholar’s ELN is the definition of meta-data file templates for specific data files, so that the standard attributes used in specific file types are automatically defined within relevant records (e.g., different data file types from different confocal microscope manufacturers can be parsed to provide appropriate imaging parameters). In addition to this, Catalyzer has a rich functionality, providing spreadsheet-like interfaces, drag-and-drop import functions, and specialized control-building capabilities. It is supported as required by its development as a commercial product. As such, it is suited to providing computational data support to whole laboratories with a suitable budget and a commitment to alter their working habits in order to benefit from the use of the ELN software. As with all sophisticated software tools, it does require training in setting it up effectively and although its license is not pejoratively expensive, it would require the commitment of the head of a laboratory to use it.
By contrast, NeuroSys is open-source, freely downloadable and undergoing development. Users may build a front-end for their ELN from a set of “widgets” (i.e., text fields, text areas, pull-down boxes, and so on) and then populate the system with data from their experimental work. The system then allows the data to be queried and retrieved. Both NeuroSys and Catalyzer contain the database components themselves, requiring no installation of an underling database system. The ELN extension of NeuroScholar is quite different from these (and other) existing ELN applications. Rather than provide a reprogrammable database interface with web-publishing capabilities, NeuroScholar includes laboratory notes in a wider representation of scientific knowledge. This is advantageous because the system can be used without needing to predefine data structures within it. Rather than being a data management system, NeuroScholar is geared to represent knowledge (i.e., structured information within the context of other information) so that experimental data are related explicitly to information from the literature. The definition of typed data values is represented within the system and can support tabulated data forms, although the user presently has much less control on the final output of these tables than with other ELN systems.
Beyond the scope of ELN applications, the NeuroScholar system itself is designed to work with other software to provide a useful knowledge engineering framework. For example, the system can read citations from bibliographic software, browse and download the relevant full-text article with a web-browser, and render the PDF file to be fragmented by the user. The software is not designed to replicate functionality that is better performed by other tools. Instead the system simply provides a way to bring knowledge together from different modalities so that scientific knowledge may be extracted and used.
Finally, NeuroScholar is still under development, as are all “live” ELN applications. Thus, it may be possible (and desirable) to seek ways to develop direct computational interaction between other ELN systems and NeuroScholar. This could consolidate the strengths of both applications. Interestingly, the two examples mentioned (Catalyzer and NeuroSys) are both programs written in the Java programming language. NeuroScholar’s “plug-in” framework that permits separate Java applications to be incorporated directly into NeuroScholar (Burns et al., 2003). The process is straightforward if developers have access to the source code of the target application being incorporated. As an open-source, academic ELN Java application, NeuroSys is an excellent candidate for such incorporation.
Discussion
In this article, we have presented a new ELN software application, which compliments and extends the functionality of our NeuroScholar knowledge management/engineering application. We have illustrated how the ELN is amenable to working with experimental data, providing a basis for its utility. We aim to continue to develop this software as a component of our NeuroScholar system. The current implementation of our ELN application provides users with the ability to create, store and retrieve structured annotations on data of interest, and within Neuroscholar allows users to view, navigate and interact with experimental data directly in relation to information from the primary literature.
Although retooling ELNs so that they contain bibliographic information is simple to do, we are not aware of another system that can extract, display and manipulate the printed contents of primary research articles and allow neuroscientists to link their original research data to that information. The apparent absence of such systems prompts us to address the reasons for their absence and in so doing we consider these questions: do academic neuroscientists desire these systems and what is the evidence that they will find them useful? The initial idea for an ELN extension to NeuroScholar arose from a need to link raw data to the primary literature, a need expressed by research scientists within our group. The general response of colleagues to discussion of the ELN has been enthusiastic, with most concerns focused on three issues: (a) ease-of-use, (b) reliability, and (c) privacy and security measures. Within the development process, these concerns must be addressed by building a reliable, well-engineered, well-supported system with good documentation. When these conditions are met, it is reasonable to speculate that such a system would be desired by scientists and would be of use to them.
Tools and approaches such as this are attractive to high-level policy makers within the field: a prime impetus for the creation of a neuroinformatics field was the development of computational infrastructure to standardize and share data (Bloom 1978, 1995). Also, development of this technology is stated as a clear objective of the Society for Neuroscience (http://www.sfn.org/ndgwhitepaper). Policy-makers have also emphasized the benefits that would be available to the community if neuroscientists share their data more actively by, for example, submitting it to a public database upon publication (Koslow 2000; Alper 2003). One strategy for building the necessary computational infrastructure is for individual neuroscientists to develop small-scale databases in collaboration with neuroinformaticians. This has produced many working systems (see http://big.sfn.org/NDG/site/). ELN technology could provide a complimentary strategy that would allow scientists to digitize their data at an early stage in the scientific process, thus making it far easier to submit data to databases, at a later stage.
Regarding whether neuroscientists would find ELN technology useful, we refer to how ELN technology is being adopted by industry. In fact, commercial ELN software defines a large, rapidly expanding market and there are many existing industrial-strength products that are in widespread use (Atrium Research, 2005). An indication of the high value of ELN technology as a commodity is given by the price of a recent 2005 market research study of the technology: a single of copy of the study report costs almost $3000 (Atrium Research, 2005). The primary benefits of these systems are increased efficiency and data quality, creating an institutional memory and protecting intellectual property (Elliot, 2004).
Many areas of the biotechnology industry require ELN technology, but it has not yet had a noticeable impact within research in an academic setting; we offer three explanations for this. First, a private research company weighs the cost of implementing a new technology against the financial benefits it offers; whereas for academic scientists monetary gain is not the prime objective. Second, within academia, concerns about intellectual property are more focused on publishing data and less focused on keeping it internal to the organization. Third, funding within academia tends to be rather limited in comparison to industry, and this reduces the ability of those working in academia to divert resources to the purchase and development of specialized software. Finally, much of the research performed in academic research laboratories is hypothesis-driven and complex. This type of research relies heavily on individual human expertise and does not lend itself to high-throughput, high-volume processing.
The issues of dealing with complex, hypothesis-driven research questions is being addressed by the construction of KR structures within NeuroScholar that are fabricated from KnowledgeStatement constructs. In this article, we have introduced the definition of Fragment views as our initial representation of knowledge derived from sources outside the system. The most important long-term contribution of this approach is to encourage the definition of more structured KR objects to describe individual experiments and their interpretations. For example, a structured knowledge statement may be paraphrased in English as “when an animal of species A is exposed to a stressor B, neurons located in regions (C1, C2, … Cn) stain positive under a histological labeling protocol D.” This statement could be represented computationally under any one of several formats including the “VPDMf” as described here, the Ontology Web Language (OWL) or the Resource Description Framework (RDF), or any one of many KR formats. If a large number of these statements can be collated, they may then be aggregated into summaries and analyzed to provide authoritative ontologies, vocabularies, and models supported by direct links to the primary data from the literature and from the laboratory. The development of mathematically based KRs for systems-level neuroscience has been preempted by their emerging importance within molecular biology. As the body of molecular knowledge increases in size and mathematical sophistication, it is vital that our representations for the larger-scale systems in which molecules are acting are also defined in a compatible framework with the same mathematical rigor.
Our system design is informed by the philosophy that science involves the inscription and synthesis of statements that have varying degrees of reliability. The goal of scientific work is to transform these statements so that they are more reliable or “fact-like” (Latour and Woolgar, 1979). Knowledge management systems make these inscriptions explicit and provide tools that can help manage and transform them. Given that the reliability of any given statement depends on the fidelity of its inscription (and subsequent transcription) and the reliability of the statements that support or refute it, a system capable of linking these statements provides a means to represent and visualize reliability explicitly as well as methods for constructing scientific arguments (Uren et al., 2003). The current implementation of NeuroScholar permits users to add their opinions to definitions and instances. In our future work, this will provide a means of running detailed analyses to substantiate or discredit a statement based on consensus among several users of the same system.
The ELN extension to NeuroScholar was a product of the generality of the definition of the Fragment construct as “knowledge external to the system.” It is worth noting that the NeuroScholar system’s current source of fragments, namely the primary literature and laboratory-based data may be complemented by other sources. These could include data from online databases, seminars and multimedia webcasts, which may provide further valuable sources of knowledge to be used in conjunction with the contents of the ELN. This approach may permit other web-accessible ELN technology (such as Axiope or NeuroSys) to be linked to NeuroScholar. It is interesting to note that NeuroScholar’s plug-in framework provides a way of incorporating third party, open-source software into the inscription process if the software is written in the Java programming language. Fortunately, this is true for a number of neuroinformatics tools (Axiope and NeuroSys are both Java applications).
Ultimately, the goal of the whole approach is to study the neural basis of specified behaviors on a large scale (Camhi, 1984; Burns, 2001). The construction of formal, computational representations for raw data generated in neuroscience experiments is the first stage of this process and the success of our approach is critically tied to the usefulness of the tools we develop for neuroscientists in their everyday work. This directly leads us to the development of an ELN product that is maximally compatible with current PLN-based methodology. Additionally, this technology is geared towards supporting the development of a culture of communication and explicit argumentation in neuroscience that can interoperate seamlessly with computational tools available to biologists in other fields.
NeuroScholar, with its ELN extension, is a functional open-source system that may be downloaded, installed and run by neuroscientific researchers. The full functionality of the system can be utilized with additional training from online resources. Given a large enough user-base, community-based open-source software has the potential to provide zero cost solutions to the community that are as well-designed and reliable as top-flight commercial software. It is hoped that the promise of providing powerful knowledge engineering tools to the community will encourage open-source developers to participate in NeuroScholar’s future development.
Concluding Remarks
We have described a new open-source ELN application that may be used to organize laboratory-based information. This application extends the functionality of an existing bibliographic knowledge management system, enabling raw and published data to be incorporated directly into a unified structure. The system operates by serving up knowledge statements that are each supported by links to either the primary literature or to raw data; together, these statements and their supporting links constitute a knowledge model. Our aim in this endeavor is to formalize knowledge model construction in order to accelerate the sharing and distribution of scientific knowledge.
Acknowledgments
This work was made possible by funding from the National Library of Medicine (LM 07061 to Burns), the National Institute of Mental Health (MH 071108 to Khan), and the National Institute of Neurological Disorders and Stroke (NS 29728 to Watts). We thank all members of the Watts and Swanson laboratories for their encouragement and critical suggestions.
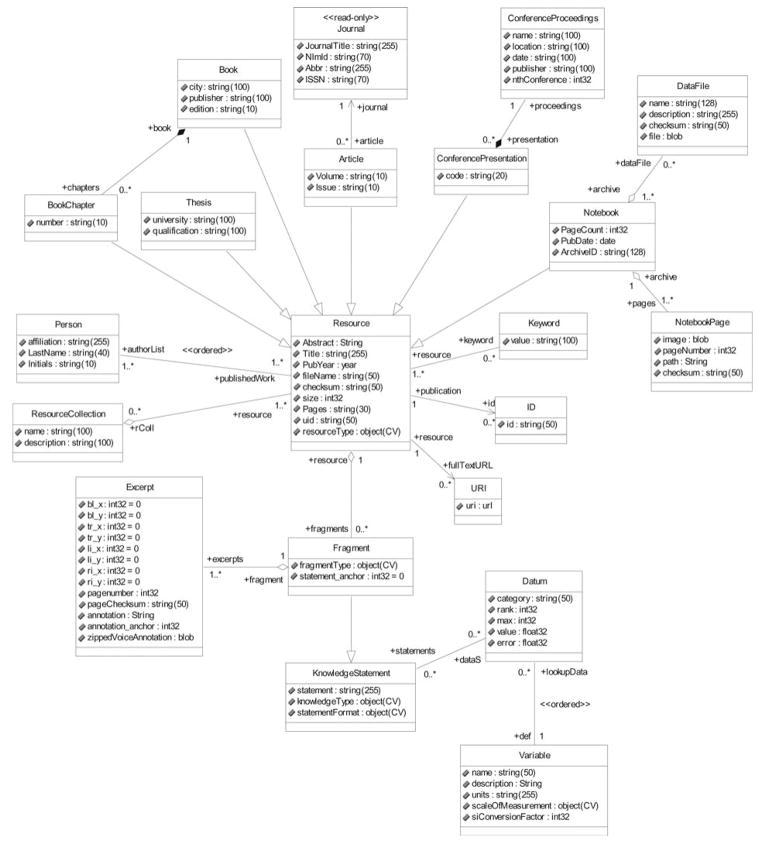
Appendix A: Data Schemata for Laboratory Notebook and Bibliographic Packages
Here, we present the object-oriented design underlying the knowledge model for the parts of the NeuroScholar system presented in this paper as a UML schema. The process of generating the functional system using the VPDMf operates on this model directly and generates the necessary relational database tables and the java interface required to interface with the model. This is provided here as a reference for programmers and as an illustration of the methodology.
The logical structure of the data model serving the Laboratory Notebook is shown below in Fig. 7. This shows the precise representation within the design of the core knowledge base of version 1.3. Note that this schema describes the organization of classes before the VPDMf transformation is applied and four separate actions are performed on the schema with the VPDMf system-builder: (a) primary- and foreign-key attributes are added, (b) set-backing tables are added to govern n-to-n relationships, (c) unidirectional associations are added to represent attribute types referring to “object” types, and (d) additional system classes are added into the model to manage generic data required for NeuroScholar to function. Attribute types conform to the specifications of the VPDMf, and are largely self-explanatory. The class composition of each view is shown in Table 1, breaking the structure of the view into those classes forming the primary primitive of the view and listing all others involved in the class definition. The reader is referred to online documentation available for download from the NeuroScholar website for more information (http://www.neuroscholar.org/).
Table 1.
| View | Primary primitive classes | All other classes |
|---|---|---|
| Class composition of shared views in fragment packages | ||
| Scientist | Person | |
| ResourceCollection | ResourceCollection | Notebook, Publication |
| KnowledgeStatement | KnowledgeStatement | CV, Variable |
| Fragment | KnowledgeStatement, Fragment | CV, Variable |
| Class composition of views in the Electronic Laboratory Notebook package | ||
| Notebook | Resource, Notebook | Scientist, NotebookPage |
| NotebookFragment | KnowledgeStatement, Fragment, NotebookFragment | CV, Variable, Excerpt, Notebook |
| DataFile | DataFile | Notebook |
| Class composition of views in the Bibliography package | ||
| Publication | Resource, Publication | ID, Person |
| Article | Resource, Publication, Article | URI, Journal, ID, Person |
| Journal | Journal | – |
| Book | Resource, Publication, Book | ID, Person |
| BookChapter | Resource, Publication, BookChapter | Book, ID, Person |
| ConferencePresentation | Resource, Publication, Conference Presentation | ConferenceProceedings, ID, Person |
| ConferenceProceedings | ConferenceProceedings | – |
| BibliographicFragment | KnowledgeStatement, Fragment, NotebookFragment | CV, Variable, Excerpt, Publication |
References
- Alper J. Neuroscientists Have Better Tools On the Brain. Bio-IT World; 2003. http://www.bioitworld.com/news/041503_report2345.html. [Google Scholar]
- Atrium Research. 2005 Electronic Laboratory Notebook Survey. Atrium Research; Wilton, CT: 2005. [Google Scholar]
- Bloom FE. New solutions for science communication problems needed now. Trends Neurosci. 1978;1(1):1. [Google Scholar]
- Bloom FE. Neuroscience-knowledge management: slow change so far. Trends Neurosci. 1995;18(2):48, 49. doi: 10.1016/0166-2236(95)93864-t. [DOI] [PubMed] [Google Scholar]
- Blum B. Clinical Information Systems. Springer; New York: 1986. [Google Scholar]
- Burns GA. Knowledge management of the neuroscientific literature: the data model and underlying strategy of the NeuroScholar system. Philos Trans R Soc Lond B Biol Sci. 2001;356:1187–1208. doi: 10.1098/rstb.2001.0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns GAPC, Khan AM. An example of a neuroinformatics knowledge model derived from the primary literature: catecholaminergic regulation of hypothalamic paraventricular neuroendocrine neurons. Ann. Meeting for Soc. Neuroscience; New Orleans. 2003. p. Abstract #758.2. [Google Scholar]
- Burns GAPC, Khan AM, Ghandeharizadeh S, O’Neill MA, Chen Y-S. Tools and approaches for the construction of knowledge models from the neuroscientific literature. Neuroinformatics. 2003;1:81–109. doi: 10.1385/NI:1:1:081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camhi J. Neuroethology: Nerve Cells and the Natural Behavior of Animals. Sinauer; Sunderland, MA: 1984. [Google Scholar]
- Elliot MH. It’s Not About the Paper. Scientific Computing & Instrumentation September 2004: Feature. 2004 http://www.scimag.com/
- Goddard NH, Cannon RC, Howell FW. Axiope tools for data management and data sharing. Neuroinformatics. 2003;1:271–284. doi: 10.1385/NI:1:3:271. [DOI] [PubMed] [Google Scholar]
- Gruber TR. Towards Principles for the Design of Ontologies Used for Knowledge Sharing. International Workshop on Formal Ontology; Padova, Italy: 1993. [Google Scholar]
- Hahn JD, Kalamatianos T, Coen CW. Studies on the neuroanatomical basis for stress-induced oestrogen-potentiated suppression of reproductive function: evidence against direct corticotropin-releasing hormone projections to the vicinity of luteinizing hormone-releasing hormone cell bodies in female rats. J Neuroendocrinol. 2003;15:732–742. doi: 10.1046/j.1365-2826.2003.01056.x. [DOI] [PubMed] [Google Scholar]
- Hudson B, Ritter S. Hindbrain catecholamine neurons mediate consummatory responses to glucoprivation. Physiol Behav. 2004;82:241–250. doi: 10.1016/j.physbeh.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Itoi K, Helmreich DL, Lopez-Figueroa MO, Watson SJ. Differential regulation of corticotropin-releasing hormone and vasopressin gene transcription in the hypothalamus by norepinephrine. J Neurosci. 1999;19:5464–5472. doi: 10.1523/JNEUROSCI.19-13-05464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AM, Watts AG. Norepinephrine rapidly elevates crh hnRNA, c-fos mRNA and levels of phosphorylated MAP kinases (ERK 1 and 2) in hypothalamic parvocellular paraventricular neurons in vivo. Soc Neurosci Abstr (program #39718) 2003 [Google Scholar]
- Khan AM, Watts AG. Intravenous 2-deoxy-D-glucose injection rapidly elevates levels of the phosphorylated forms of p44/42 mitogen-activated protein kinases (Extracellularly regulated kinases 1/2) in rat hypothalamic parvicellular paraventricular neurons. Endocrinol. 2004;145:351–359. doi: 10.1210/en.2003-0539. [DOI] [PubMed] [Google Scholar]
- Khan AM, Cheng W-C, Watts AG, Burns GAPC. Histochemical studies of stress-activated paraventricular hypothalamic neuroendocrine neurons: A neuroinformatics-based digital lab notebook to relate the primary literature to raw, unpublished data. Soc Neurosci Abstr (program # 92229) 2004 [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data? Nat Neurosci. 2000;3(9):863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Latour B, Woolgar S. Laboratory Life. Princeton University Press; Princeton, NJ: 1979. [Google Scholar]
- Phelps TA, Wilensky R. Document Engineering 2001. Atlanta, Georgia: 2001. The Multivalent Browser: A Platform for New Ideas. [Google Scholar]
- Pittendrigh S, Jacobs G. NeuroSys: a semistructured laboratory database. Neuroinformatics. 2003;1:167–176. doi: 10.1007/s12021-003-0003-0. [DOI] [PubMed] [Google Scholar]
- Ritter S, Bugarith K, Dinh TT. Immunotoxic destruction of distinct catecholamine subgroups produces selective impairment of glucoregulatory responses and neuronal activation. J Comp Neurol. 2001;432:197–216. doi: 10.1002/cne.1097. [DOI] [PubMed] [Google Scholar]
- Ritter S, Llewellyn-Smith I, Dinh TT. Subgroups of hindbrain catecholamine neurons are selectively activated by 2-deoxy-D-glucose induced metabolic challenge. Brain Res. 1998;805:41–54. doi: 10.1016/s0006-8993(98)00655-6. [DOI] [PubMed] [Google Scholar]
- Ritter S, Watts AG, Dinh TT, Sanchez-Watts G, Pedrow C. Immunotoxin lesion of hypothalamically projecting norepinephrine and epinephrine neurons differentially affects circadian and stressor-stimulated corticosterone secretion. Endocrinol. 2003;144:1357–1367. doi: 10.1210/en.2002-221076. [DOI] [PubMed] [Google Scholar]
- Rivier C, Vale W. Influence of corticotropin-releasing factor on reproductive functions in the rat. Endocrinology. 1984;114:914–921. doi: 10.1210/endo-114-3-914. [DOI] [PubMed] [Google Scholar]
- Rumbaugh J, Jacobson I, Booch G. The Unified Modeling Language Reference Manual. Reading, MA: Addison-Wesley; 1999. [Google Scholar]
- Shepherd GM, Mirsky JS, Healy MD, et al. The Human Brain Project: neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends Neurosci. 1998;21:460–468. doi: 10.1016/s0166-2236(98)01300-9. [DOI] [PubMed] [Google Scholar]
- Sowa J. Knowledge Representation, Logical, Philosophical and Computational Foundations. Brooks/Cole; Pacific Grove: 2000. [Google Scholar]
- Tsukahara S, Tsukamura H, Foster DL, Maeda KI. Effect of corticotropin-releasing hormone antagonist on oestrogen-dependent glucoprivic suppression of luteinizing hormone secretion in female rats. J Neuroendocrinol. 1999;11:101–105. doi: 10.1046/j.1365-2826.1999.00312.x. [DOI] [PubMed] [Google Scholar]
- Uren V, Buckingham Shum SJ, Li G, Domingue J, Motta E. WWW 2003. Budapest, Hungary: ACM Press; 2003. Scholarly publishing and argument in hyperspace. [Google Scholar]
- Watts AG. The impact of physiological stimuli on the expression of corticotropin-releasing hormone (CRH) and other neuropeptide genes. Front Neuroendocrinol. 1996;17:281–326. doi: 10.1006/frne.1996.0008. [DOI] [PubMed] [Google Scholar]